Abstract
Summary
Cyanobacteria, among Earth's oldest organisms, have evolved sophisticated biosynthetic pathways to produce a rich arsenal of bioactive natural products. In consequence, cyanobacterial secondary metabolites have been an incredibly fruitful source of lead compounds in drug discovery efforts. Investigations into the biochemistry responsible for the creation of these compounds, complemented by genome sequencing efforts, are revealing unique enzymatic mechanisms not described or rarely described elsewhere in the natural world. Herein, we discuss recent advances in understanding the biosynthesis of three cyanobacterial classes of natural product: mixed polyketide synthase/non ribosomal peptide synthetase (PKS/NRPS) metabolites, aromatic amino acid-derived alkaloids, and ribosomally encoded cyclic peptides. The unique biosynthetic mechanisms employed by cyanobacteria are inspiring new developments in heterologous gene expression and biotechnology.
B. Introduction
Unarguably, natural products have played an enormous role in the development of modern medicines, especially in the areas of cancer and infection [1]. The last 30 years has seen a focused and purposeful exploration of the marine environment for drug leads from natural products, and recently it has emerged that marine microbes are an especially rich source of structurally novel and bioactive compounds [2••,3]. One particularly noteworthy group, the cyanobacteria, produces a surprisingly diverse array of metabolites that derive largely from the integration of non-ribosomal peptide synthetases with polyketide synthases [4-6]. These nitrogen-rich frameworks are often decorated with unusual modifications such as halogenations, methylations and oxidations. Many have potent cellular toxicity due to inhibition of tubulin or actin mediated processes, although a growing number have other more novel sites of action [7,8]. Hence, there is great interest to better understand how these unusual structures are created, with special focus on mechanistic biochemistry, gene evolution, transcriptional regulation and biosynthetic logic [9,10]. It is hoped that these studies will improve access to the full richness of cyanobacterial natural products as well as an ability to employ these gene clusters and biosynthetic gene motifs in heterologous expression and combinatorial biosynthesis.
C. Curacin A biosynthesis
Curacin A, a metabolite isolated from a Curaçao strain of the marine cyanobacterium Lyngbya majuscula [11], exhibits potent anti-proliferative and cytotoxic activity against colon, renal, and breast-cancer derived cell lines [12]. Its biosynthetic pathway was identified and partially characterized by genetic and precursor labeling studies [13] as a mixed polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS) (Figure 1). Based on this initial analysis, it was predicted that a number of genes encoding unusual catalytic domains and enzymes were present in this pathway, including a GCN5-related N-acetyltransferase (GNAT)-like domain in the chain initiation module, a HMG enzyme cassette, an α-KG dependent non-heme halogenase, and a sulfotransferase (ST) domain in the chain termination module (Figure 1). These intriguing catalytic elements embedded within the Cur biosynthetic system illustrate the combinatory flexibility of modular polyketide assembly lines as well as provide valuable objectives for mechanistic enzymology and protein evolution studies.
Figure 1.
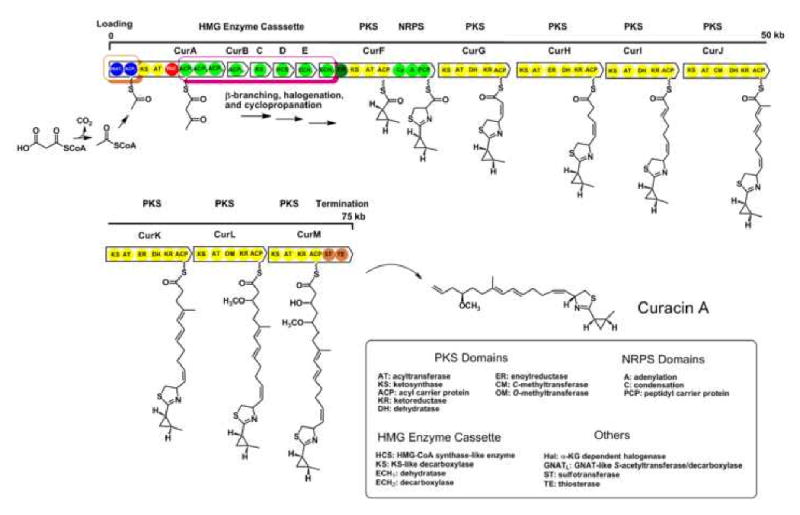
The curacin A biosynthetic pathway. The chain initiation module and HMG enzyme cassette are highlighted in orange and magenta, respectively.
The GNAT-like domain (GNATL) in the CurA chain initiation module is an ideal example of functional diversification and gain-of-function in an enzyme scaffold. Previously, the GNAT superfamily of enzymes was only reported to catalyze N-acetyl transfer from acetyl-CoA or ACP to primary amino groups on diverse acceptor substrates [14]. In curacin A as well as a series of other biosynthetic pathways, GNAT-like domains embedded in chain initiation modules were predicted to transfer acetyl groups to the terminal thiol of their adjacent ACP phosphopantetheine arm (ACPLs) [13,15••,16••, 17-18]. Unexpectedly, malonyl-CoA was found to be loaded onto CurA ACPL, but only an acetyl product was detected on the ACP phosphopantetheine (PPant) arm [19••]. Thus GNATL was demonstrated to catalyze decarboxylation of malonyl-CoA to form acetyl-CoA followed by S-acetyltransfer to the adjacent ACPL (Figure 1). Both of these steps represent new functional activities for the GNAT superfamily.
The insertion of an HMG enzyme cassette in the curacin pathway reveals an interesting convergence of two distinct biosynthetic systems, type I polyketide synthases and enzymes involved in isoprenoid assembly. In the curacin A pathway, this cassette catalyzes β-branching modification during the biosynthetic assembly line process at β-carbonyls of the polyketide chain elongation intermediate (dehydration and decarboxylation) [20•]. These modifications are combined with halogenation and cyclopropane ring formation to generate the unusual β-branched cyclopropyl group in the final product [21]. A most interesting aspect of this cassette is how it is structured and incorporated into PKS modules. It comprises tandem triple ACPs (ACPI, ACPII, ACPIII), a discrete ACP (ACPIV), ketosynthase-like decarboxylase (KS), HMG-CoA synthase (HCS), and a dehydratase (ECH1)/decarboxylase (ECH2) pair related to the crotonase superfamily of enzymes (Figure 1) [13, 20•]. The polyketide intermediate tethered to the tandem triple ACPs undergoes multiple modifications to convert a β-carbonyl group to β-branched methyl group. The HCS catalyzes a key Claisen condensation using C-2 of acetate as a nucleophile, the PhyH halogenase chlorinates at the γ position [21], and then ECH1 and ECH2 catalyze the consecutive dehydration and decarboxylation of the chlorinated HMG-like intermediate to form the β-branched product [22•]. Related β-branching modifications have been observed in several biosynthetic pathways, and provide chemical diversity by variation in the number and type of HMG cassette enzymes [23, 24] or by using alternative starter units [25] to create additional structural variation in natural products. The recently solved curacin ECH2 structure suggests that specific amino acid residues in a hypervariable region play a role in regiochemical control of ECH2 decarboxylation, which might provide a facile strategy for metabolic diversification [26].
D. Jamaicamide
Jamaicamide A is a neurotoxic metabolite of a Jamaican collection of Lyngyba majuscula that possesses several highly intriguing structural features, including an interdigitated polyketide and NRPS overall construction with acetylenic bromide, pendant vinyl chloride, and terminal pyrrolidone ring functionalities [27••]. The original isolation work also mapped out the biosynthetic components and characterized the gene cluster coding for jamaicamide assembly and tailoring reactions. One key insight from the gene cluster analysis was realization that a six-carbon carboxylic acid serves as the starter unit for the pathway. An innovative mass spectrometric method was subsequently used to gain insight into the relative timing of bromination to form the bromoalkyne functionality [28•]. Recombinant proteins JamA [hexanoyl acyl carrier protein (ACP) synthetase] and JamC (ACP) were provided with ATP and a choice of substrates, hexenoic and bromo-hexynoic acids (Figure 2). The results of incubation were queried by Fourier transform ion cyclotron resonance mass spectrometry (FT ICR MS) and only a hexenoyl chain was shown to be tethered to JamC via a thioester bond. The enzyme system was completely unreactive to bromohexynoic acid under any circumstances, showing conclusively that bromination occurs subsequent to hexanoic, hexenoic or hexynoic acid activation and covalent tethering.
Figure 2.

A) Mass Spectrometric method for determining the identity of preferred substrate initiating the jamaicamide A biosynthetic pathway [28•]. B) Reductive off-loading of the NRPS tethered dipeptide intermediate in lyngbyatoxin biosynthesis shown to be a 4e- reduction involving two equivalents of NADPH [31].
E. Lyngbyatoxin
Lyngbyatoxin, a prenylated cyclic dipeptide, was originally isolated from a Hawaiian strain of the marine cyanobacterium Lyngbya majuscula and shown to be responsible for a condition known as “Swimmer's Itch” due to its potent activation of protein kinase C [29]. Subsequently, the lyngbyatoxin gene cluster was cloned from this strain, and found to be composed of only four genes that code for several novel biochemical features [30]. The first gene encodes LtxA, a didomain NRPS required for assembly of the dipeptide that terminates with a reductase domain for release of a presumed primary alcohol. LtxB is a cytochrome P450 believed to oxidize the indole ring and possibly act in the cyclization of the molecule. LtxC is a reverse prenyltransferase, and LtxD functions as a short chain acyl dehydrogenase involved in formation of the saturated geranyl chain in lyngbyatoxins B and C. Recently, mechanistic aspects of the reductive off-loading of the dipeptide were studied using alternative substrates and the PCP/reductase components of the NRPS (Figure 2) [31]. Using the N-acetylcysteamine thioester (S-NAC) analog of the proposed natural dipeptide thioester substrate, the four electron reduction of the thioester to a primary alcohol was conclusively demonstrated. Furthermore, performing the reaction in H218O allowed deduction of an aldehyde intermediate in this reaction sequence, and use of chiral forms of 2H-labeled NADPH showed only the pro-S hydrogen was transferred in both reductive steps.
F. Scytonemin
Scytonemin is a dimeric indolic-phenolic alkaloid that acts as a passive sunscreen in the protection of cyanobacteria against ultraviolet light in marine and freshwater environments [32, 33]. Studies of its unique structure, gene cluster, and enzymatic mechanisms have given new insights on the biosynthesis of this compound class in cyanobacteria [33-35]. In 2007, a scytonemin deficient Nostoc punctiforme ATCC 29133 mutant was created using random transposon mutagenesis [34]. Analysis of this mutant revealed the transposon was embedded within NpR1273, an open reading frame (ORF) that encodes a putative protein with a signal peptide. NpR1273 was one of 18 contiguous ORFs, likely all co-transcribed [34], and shown to be upregulated by UV light based on semi-quantitative reverse transcriptase-PCR analysis (CM Sorrels et al., unpublished). These ORFs appear to be involved in tryptophan and tyrosine biosynthesis and have sequence similarity to corresponding operons in five other cyanobacterial species (CM Sorrels et al., unpublished). Bioinformatics analyses showed that NpR1271-NpR1274 had little sequence similarity to other characterized proteins and led to the prediction that they were involved in scytonemin biosynthesis [34]. Further analysis of the cluster prompted a recent investigation into the mechanistic biochemistry required for scytonemin assembly through study of NpR1275 and NpR1276 (Figure 3) [35]. NpR1275, which resembles a leucine dehydrogenase, was shown to catalyze the oxidative deamination of tryptophan to form indole-3-pyruvic acid (IPA). IPA and p-hydroxyphenylpyruvic acid then act as the substrates for an acyloin reaction catalyzed by NpR1276, a homolog to a thiamine diphosphate (ThDP) dependent acetolactate synthase. The acyloin product arises from a single β-ketoacid regioisomer, indicative of a highly selective reaction by the ThDP-dependent NpR1276. The mechanism of this early reaction in scytonemin biosynthesis has rarely been described in any other natural system [35].
Figure 3.
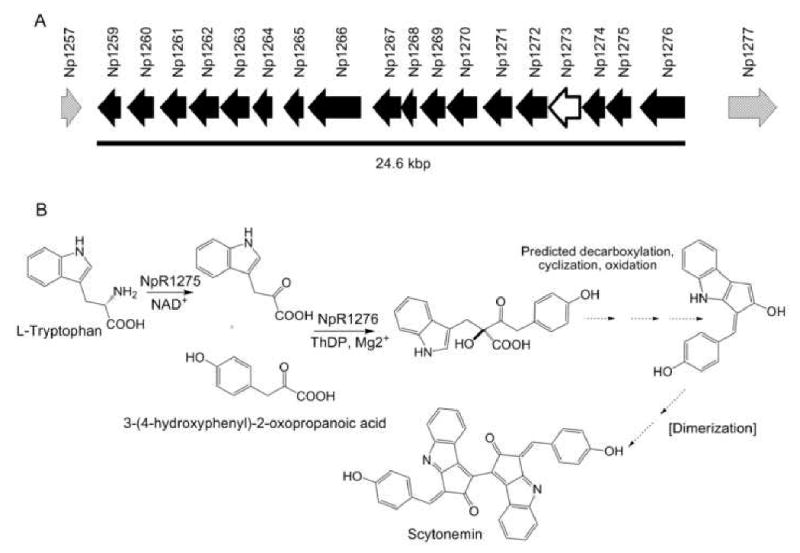
A) Proposed scytonemin gene cluster shown by solid black arrows with transposon insertion site indicated with a white arrow outlined in black. Patterned arrows represent boundary genes of the proposed cluster [34]. B) Summary of results from initial studies in the biochemistry of scytonemin biosynthesis including functions of NpR1275 and NpR1276 [35]. Predicted gene functions of the scytonemin gene cluster are: NpF1257 - Short-chain dehydrogenase; NpR1259 - Hypothetical protein; NpR1260 - 3-deoxy-D-arabino-heptolosonate-7-phosphate (DAHP) synthase; NpR1261 - Anthranilate phosphoribosyltransferase; NpR1262 -Tryptophan synthase (β subunit); NpR1263 - Putative tyrosinase; NpR1264 - Tryptophan synthase (α subunit); NpR1265 - Indole-3-glycerol phosphate synthase; NpR1266 - Anthranilate synthase; NpR1267 - 3-Dehydroquinate synthase; NpR1268 - Dithiol-disulfide isomerase; NpR1269 - Prephenate dehydrogenase; NpR1270 - Putative glycosyltransferase; NpR1271 -Hypothetical protein; NpR1272 - Hypothetical protein; NpR1273 - Hypothetical protein; NpR1274 - Hypothetical protein; NpR1275 - Leucine dehydrogenase; NpR1276 - Thiamine diphosphate requiring enzyme; NpR1277 - PAS/PAC sensor signal transduction histidine kinase.
G. Prochloron biosynthesis
In 1976, a new subclass of algae, the Prochlorophyta, was proposed to describe a unique symbiont living in association with didemnid ascidians from tropical Pacific locations [36]. This symbiont, given the generic name “Prochloron” [37], is now recognized as a cyanobacterium despite its plant-like use of chlorophylls a and b and lack of phycobilins. Although Prochloron has evaded culture attempts, this cyanobacterium is now known to be the biosynthetic source of the patellamides, cyclic peptides first isolated from one of the host ascidians, Lissoclinum patella [38••]. Unlike other peptides from cyanobacteria, the patellamides are remarkable because they are generated from a ribosomally encoded gene cluster rather than from a NRPS mediated process. The pat gene cluster is composed of seven ORFs (patA-patG) in which the precursor peptide patE directly encodes the amino acids found in patellamides A and C. While none of the other genes in the pat cluster appear to function as epimerases, nonenzymatic epimerization of amino acids to D-isomers appears possible (e.g., see the lissoclinamides below) [39]. The involvement of this gene cluster in patellamide biosynthesis was confirmed through successful heterologous expression of patellamide A in E. coli [3,38,40].
In a PCR based screening for the patE gene from Prochloron spp. obtained from 46 tropical Pacific ascidian samples [41], over 20 patE variants were detected. The majority of these gene clusters were virtually identical, except for the patellamide amino acid encoding region. The variable regions in the clusters were predicted to correspond to three cyclic peptide classes: the patellamides, ulithiacyclamides, and lissoclinamides. Engineered mutation of the region encoding the ulithiacyclamides led to heterologous expression of a novel compound, eptidemnamide. Using quantitative PCR, it was found that each Prochloron strain likely contains a single pathway, and thus, ascidians can store an entire chemical library by harboring multiple strains of the cyanobacterium.
Continuing exploration of other cyanobacteria, both free living as well as obligate symbionts, for biosynthetic gene clusters homologous to the patellamide pathway [42] has revealed that this ribosomally encoded process is widespread (more than 100 compounds have been identified or implied from gene analysis), and given rise to a new term to describe these cyclic peptides as the ‘cyanobactins’ (Figure 4) [43•]. Recent advances with the cyanobactins include structural diversification through the biosynthetic addition of side chains such as prenyl groups (patellins, trunkamide), and improved recombinant expression of symbiont natural products [44•].
Figure 4.
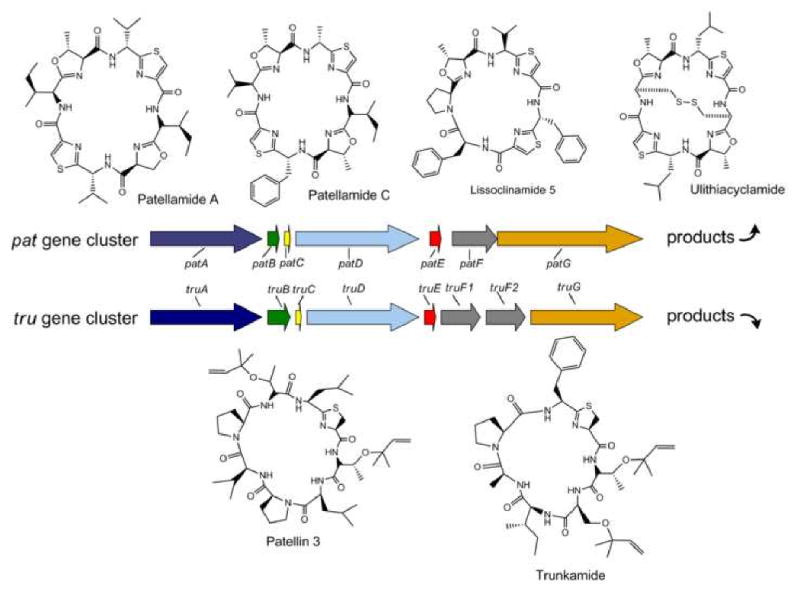
Two examples of cyanobactin gene clusters from Prochloron spp. isolated from the ascidian Lissoclinum patella (represented by arrows) [43•]. Conserved genes between the patellamide (pat, top) and trunkamide (tru, bottom) clusters are shown in matching colors. Subsamples of compounds encoded by variations in patE and truE are provided above and below the gene clusters, respectively. The absolute stereoconfiguration of lissoclinamide 5 is based on recent synthetic efforts [39].
H. Visualizing biosynthesis
Most organisms exist in Nature as complex assemblages. For example, marine cyanobacteria are commonly found living in association with various invertebrates, such as sponges and tunicates (e.g. see above), and are richly populated themselves by diverse heterotrophic bacteria. Thus, it can be quite difficult to rigorously determine the actual biosynthetic source of a given metabolite. Nevertheless, a variety of gene-based [44•, 45••] as well as mass spectrometric imaging techniques are emerging to provide powerful insights to these long standing questions. A recent illustration involved MALDI imaging to demonstrate that mixtures of cyanobacterial filaments could be easily distinguished by their metabolite profile, and that chlorinated peptides could be localized to regions of tissue from the sponge Dysidea herbacea populated by the cyanobacterium Oscillatoria spongeliae [2••, 46]. Other soft ionization techniques such as Desorption Electrospray Ionization (DESI) and Direct Analysis in Real Time (DART) will certainly find innovative application in answering questions concerning the biosynthesis of natural products from complex assemblages, including those containing cyanobacteria [47, 48].
I. Conclusions
We are at a particularly interesting juncture in our understanding of how the remarkable natural product structures of marine cyanobacteria are created, mainly as a result of modular NRPS and PKS pathways with a variety of novel tailoring steps. Functional groups of unprecedented structure, such as a variety of pendant one carbon units at C-1 positions of polyketides (terpene-like β branches) and halogen atoms located at traditionally unreactive sites (radical halogenases) [49, 50], are now comprehensible in terms of their biosynthetic origin. While the functions of these enzyme systems are now better appreciated, the mechanistic features are only slowly being revealed and much exciting biochemical investigation remains. Moreover, new pathways of assembly with only partially defined biosynthetic processes are being described in some cyanobacteria, such as the Prochloron symbionts of ascidians. While these studies are making rapid progress, realization of the ultimate goal of pathway engineering and heterologous expression will require significant advances in underlying gene manipulation technologies as well as understanding of the regulation, storage, and excretion of these metabolites. Indeed, these ancient prokaryotic algae have much new biochemistry to teach us!
Acknowledgments
On behalf of the work of the various authors described above, we thank the countries of Curaçao, Jamaica, Papua New Guinea and Palau, as well as the State of Hawaii, for permission to collect research specimens. Research in the author's laboratories has been supported by NIH CA108874 and Sea Grant 100-TECH-N.
Footnotes
Dedicated to Ralph A. Lewin, longtime inspirational professor of phycology at Scripps Institution of Oceanography and discoverer of Prochloron – 1921-2008.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Adam C. Jones, Email: acjones@ucsd.edu.
Liangcai Gu, Email: gulc@umich.edu.
Carla M. Sorrels, Email: csorrels@ucsd.edu.
David H. Sherman, Email: davidhs@umich.edu.
William H. Gerwick, Email: wgerwick@ucsd.edu.
References
- 1.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Last 25 Years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2••.Simmons TL, Coates RC, Clark BR, Engene N, Gonzalez D, Esquenazi E, Dorrestein PC, Gerwick WH. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc Nat Acad Sci USA. 2008;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reviews the literature on defining the biosynthetic source of natural products, and describes a new imaging mass spectrometric approach for visualizing these relationships.
- 3.Dunlap WC, Battershill CN, Liptrot CH, Cobb RE, Bourne DG, Jaspars M, Long PF, Newman DJ. Biomedicinals from the phytosymbionts of marine invertebrates: A molecular approach. Methods. 2007;42:358–376. doi: 10.1016/j.ymeth.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Gademann K, Portmann C. Secondary metabolites from cyanobacteria: Complex structures and powerful bioactivities. Curr Org Chem. 2008;12:326–341. [Google Scholar]
- 5.Van Wagoner RM, Drummond AK, Wright JLC. Biogenetic diversity of cyanobacterial metabolites. Adv Appl Microbiol. 2007;61:89–217. doi: 10.1016/S0065-2164(06)61004-6. [DOI] [PubMed] [Google Scholar]
- 6.Tidgewell K, Clark BR, Gerwick WH. Comprehensive Natural Products Chemistry. Vol. 8. Pergamon Press; 2009. The natural products chemistry of cyanobacteria. In press. [Google Scholar]
- 7.Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Izpisua Belmonte JC, Schultz PG. A functional genomics approach to the mode of action of apratoxin A. Nat Chem Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 8.Wrasidlo W, Mielgo A, Torres VA, Barbero S, Stolenov K, Suyama TL, Klemke RL, Gerwick WH, Carson DA, Stupack DG. The marine lipopeptide somocystinamide A triggers apoptosis via caspase 8. Proc Nat Acad Sci USA. 2008;105:2313–2318. doi: 10.1073/pnas.0712198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswamy AV, Flatt PM, Edwards DJ, Simmons TL, Han B, Gerwick WH. The secondary metabolites and biosynthetic gene clusters of marine cyanobacteria Applications in biotechnology. In: Proksch P, Mueller WEG, editors. Frontiers in Marine Biotechnology. Horizon Bioscience; 2006. pp. 175–224. [Google Scholar]
- 10.Smith JL, Sherman DH. An enzyme assembly line. Science. 2008;321:1304–1305. doi: 10.1126/science.1163785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerwick WH, Proteau PJ, Nagle DG, Hamel E, Blokhin A, Slate D. Structure of curacin A, a novel antimitotic, antiproliferative, and brine shrimp toxic natural product from the marine cyanobacterium Lyngbya majuscula. J Org Chem. 1994;59:1243–1245. [Google Scholar]
- 12.Verdier-Pinard P, Lai JY, Yoo HD, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Structure-activity analysis of the interaction of curacin A, the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells. Mol Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 13.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 14.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Nat Acad Sci USA. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated the origin of the biosynthetic gene cluster for pederin by isolation and characterization of gut-derived bacterial genomic DNA from the Paederus beetle.
- 16••.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Nat Acad Sci USA. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work revealed the origin of the natural product onnamide based on analysis of metagenomic DNA derived from a marine sponge.
- 17.Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Müller R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthetases, and trans-acting acyltransferases. Chembiochem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- 18.Partida-Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- 19••.Gu L, Geders TW, Wang B, Gerwick WH, Hakansson K, Smith JL, Sherman DH. GNAT-like strategy for polyketide chain extension. Science. 2007;318:970–974. doi: 10.1126/science.1148790. [DOI] [PubMed] [Google Scholar]; This analysis revealed a new mechanism for polyketide chain initiation mediated by the well-known GNAT enzyme family that has evolved two new catalytic functions.
- 20•.Gu L, Jia J, Liu H, Hakansson K, Gerwick WH, Sherman DH. Metabolic coupling of dehydration and decarboxylation in the curacin A pathway: Functional identification of a mechanistically diverse enzyme pair. J Amer Chem Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]; This work demonstrated the role of two important enzymes in the curacin biosynthetic pathway that ultimately leads to introduction of the cyclopropane ring system from a β-branch early in the chain elongation process.
- 21.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Håkansson K, Wipf P, Smith JL, Gerwick WH, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009 doi: 10.1038/nature07870. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: Isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc Nat Acad Sci USA. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revealed the biochemical process for introduction of a β-branch methyl group in polyketide structures now known to be the bacillaene biosynthetic pathway.
- 23.Tang GL, Cheng YQ, Shen B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 25.Calderone CT, Iwig DF, Dorrestein PC, Kelleher NL, Walsh CT. Incorporation of nonmethyl branches by isoprenoid-like logic: Multiple β-alkylation events in the biosynthesis of myxovirescin A1. Chem Biol. 2007;14:835–846. doi: 10.1016/j.chembiol.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geders TW, Gu L, Mowers JC, Liu H, Gerwick WH, Hakansson K, Sherman DH, Smith JL. Crystal structure of the ECH2 catalytic domain of CurF from Lyngbya majuscula: Insights into a decarboxylase involved in polyketide chain β-branching. J Biol Chem. 2007;282:35954–35963. doi: 10.1074/jbc.M703921200. [DOI] [PubMed] [Google Scholar]
- 27••.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]; This study reports the isolation, structure, biosynthesis and gene cluster analysis for the highly unusual natural product jamaicamide A.
- 28•.Dorrestein PC, Blackhall J, Straight PD, Fischbach MA, Garneau-Tsodikova S, Edwards DJ, McLaughlin S, Lin M, Gerwick WH, Kolter R, et al. Activity screening of carrier domains within nonribosomal peptide synthetases using complex substrate mixtures and large molecule mass spectrometry. Biochem. 2006;45:1537–1546. doi: 10.1021/bi052333k. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes an efficient mass spectrometric approach for defining the nature of substrates naturally loaded onto acyl carrier proteins involved in assembly line type biosyntheses.
- 29.Cardellina JH, II, Marner FJ, Moore RE. Seaweed dermatitis: Structure of lyngbyatoxin A. Science. 1979;204:193–195. doi: 10.1126/science.107586. [DOI] [PubMed] [Google Scholar]
- 30.Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Amer Chem Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 31.Read JA, Walsh CT. The lyngbyatoxin biosynthetic assembly line: Chain release by four-electron reduction of a dipeptidyl thioester to the corresponding alcohol. J Amer Chem Soc. 2007;129:15762–15763. doi: 10.1021/ja077374d. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Pichel F, Sherry ND, Castenholz RW. Evidence for an ultraviolet sunscreen role of the extracellular pigment scytonemin in the terrestrial cyanobacterium Chlorogloeopsis sp. Photochem Photobiol. 1992;56:17–23. doi: 10.1111/j.1751-1097.1992.tb09596.x. [DOI] [PubMed] [Google Scholar]
- 33.Proteau PJ, Gerwick WH, Garcia-Pichel F, Castenholz R. The structure of scytonemin, an ultraviolet sunscreen pigment from the sheaths of cyanobacteria. Experientia. 1993;49:825–829. doi: 10.1007/BF01923559. [DOI] [PubMed] [Google Scholar]
- 34.Soule T, Stout V, Swingley WD, Meeks JC, Garcia-Pichel F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J Bacteriol. 2007;189:4465–4472. doi: 10.1128/JB.01816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balskus EP, Walsh CT. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J Amer Chem Soc. 2008;130:15260–15261. doi: 10.1021/ja807192u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewin RA. Prochlorophyta as a proposed new division of algae. Nature. 1976;261:697–698. doi: 10.1038/261697b0. [DOI] [PubMed] [Google Scholar]
- 37.Lewin RA. Prochloron and the theory of symbiogenesis. Annal NY Acad Sci. 1981;361:325–329. doi: 10.1111/j.1749-6632.1981.tb46528.x. [DOI] [PubMed] [Google Scholar]
- 38••.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Nat Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper was the first to conclusively demonstrate that Prochloron didemni is the biosynthetic source of the patellamide class of cyclic peptides found in its ascidian host, and is one of the few examples of cyanobacterial natural product heterologous expression in E. coli.
- 39.Boden CDJ, Pattenden G. Total syntheses and re-assignment of configurations of the cyclopeptides lissoclinamide 4 and lissoclinamide 5 from Lissoclinum patella. J Chem Soc Perkin Trans 1. 2000;6:875–882. [Google Scholar]
- 40.Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 41.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 42.Sudek S, Haygood MG, Youssef DTA, Schmidt EW. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl Env Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses several examples of ribosomally encoded peptides Prochloron and other bacteria to establish the “cyanobactin assembly line” as an important biosynthetic source of natural products in marine cyanobacteria. Using genetic engineering, the authors successfully expressed the antitumor compound trunkamide in E. coli.
- 44•.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Evidence for the biosynthesis of bryostatins by the bacterial symbiont Candidatus Endobugula sertula of the bryozoan Bugula neritina. Appl Env Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper revealed the source of the important anticancer lead bryostatin from a symbiotic bacterium that resides within the marine invertebrate host Bugula neritina
- 45••.Flatt PM, Gautschi JT, Thacker RW, Musafija-Girt M, Crews P, Gerwick WH. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar Biol. 2005;147:761–774. [Google Scholar]; This paper describes the use of a fluorescently labeled gene probe, developed to a unique halogenase involved in chlorinated peptide biosynthesis, to demonstrate biosynthesis occurs in a symbiotic cyanobacterium and not in the host sponge.
- 46.Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Molec Biosyst. 2008;4:562–570. doi: 10.1039/b720018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisemen JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc Nat Acad Sci USA. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu S, Crawford E, Tice J, Musselman B, Wu JT. Bioanalysis without sample cleanup or chromatography: The evaluation and initial implementation of direct analysis in real time ionization mass spectrometry for the quantification of drugs in biological matricies. Anal Chem. 2008;81:193–202. doi: 10.1021/ac801734t. [DOI] [PubMed] [Google Scholar]
- 49.Flatt PM, O'Connell SJ, McPhail KL, Zeller G, Willis CL, Sherman DH, Gerwick WH. Characterization of the initial enzymatic steps of barbamide biosynthesis. J Nat Prod. 2006;69:938–944. doi: 10.1021/np050523q. [DOI] [PubMed] [Google Scholar]
- 50.Galonic DP, Vaillancourt FH, Walsh CT. Halogenation of unactivated carbon centers in natural product biosynthesis: Trichlorination of leucine during barbamide biosynthesis. J Amer Chem Soc. 2006;128:3900–3901. doi: 10.1021/ja060151n. [DOI] [PubMed] [Google Scholar]


