Abstract
The Drosophila system has proven a powerful tool to help unlock the regulatory processes that occur during specification and differentiation of the embryonic heart. In this review, we focus upon a temporal analysis of the molecular events that result in heart formation in Drosophila, with a particular emphasis upon how genomic and other cuttingedge approaches are being brought to bear upon the subject. We anticipate that systemslevel approaches will contribute greatly to our comprehension of heart development and disease in the animal kingdom.
Introduction
A central goal of developmental biology is to identify and characterize the genes which control the formation of specific cells, tissues, or organs within the body. Much recent progress has been driven by the increasing realization that genes controlling organ formation are implicated in congenital birth defects, as well as diseases in later life. Thus, genes whose normal function is to create cell types in the embryo, can also destroy those structures when mutated. Of equal importance, it is now apparent that genes showing evolutionary conservation in sequence frequently show evolutionary conservation in function. Thus, the analysis of development in model organisms has provided important insight into developmental mechanisms (reviewed in [1]).
It is also apparent that the complex tissues and organs of higher animals arise from the concerted actions of very large numbers of genes influencing cell behavior and function. These genes, many of them regulatory in nature, are known to function as components of regulatory networks; and there is increasing evidence that the genetic networks have also been conserved through evolution. Defining the parameters of such regulatory networks, and how the networks have evolved, has become a central challenge in the field (reviewed in [2]).
One of the earliest organs to form in the mammalian body is the heart. Significant progress has been made in defining the genes which contribute to heart development, in a diverse array of animal models (reviewed in [3-5]). Many of these critical genes play regulatory roles, and the roles of individual genes in contributing to the overall genomic regulatory network for cardiogenesis is becoming clear.
We recently presented a developmental regulatory network for cardiogenesis in Drosophila and mammals, drawing attention to the striking conservation in function for several named factors [3]. While this network provides insight into the cadre of genes controlling cardiac development, it does not provide a sense for how regulatory interactions change over time. In addition, it does not take into account the plethora of new information and new technologies available to the developmental cardiologist. As more data become available, it is incumbent upon us to find ways to analyze the data and to use it to generate predictive models regarding both normal cardiac development and cardiac disease mechanisms.
In this review, we have updated the gene regulatory interactions that are known to take place during Drosophila cardiac development. In doing this, we have parsed the embryonic period to a series of critical stages, and we have focused upon the known regulatory events taking place at each stage. This allows us to visualize cardiac development as a series of interdependent processes, starting prior to gastrulation, and culminating in the formation of a functioning heart tube only 20 hours later.
The embryology of Drosophila cardiac development
Cardiac tissue in Drosophila, as in other animals, arises from the mesoderm. The cellular details of cardiac cell specification and morphogenesis are described in Figure 1. Mesodermal cells are specified early in fly development as a ventral group of cells, which invaginates to form a layer within the embryo (Figure 1A-A”). Mesodermal cells then spread laterally and dorsally, and those that migrate the furthest become restricted to dorsal mesoderm fate via the action of the dorsally-derived TGFß signal, Decapentaplegic (Dpp). This process results in the formation of two bilateral lines of precardiac cells (Figure 1B-B”).
Figure 1. Morphological aspects of cardiogenesis in the Drosophila embryo.
A. The lateral side of a stage 6 embryo immunohistochemically stained for expression of a Mef2-lacZ transgene, to outline the newly forming mesoderm (arrow). A'. Schematic representation of the lateral view of an early embryo (stages 4-6) outlining the position of the mesodermal layer (dark brown) and the gradient of nuclear Dorsal (brown). The arrow shows the direction of movement of the extending germ band. A". The same as in A' but transverse section is shown. Arrows indicate directions of spreading for invaginating mesodermal cells. B. The lateral view of a stage 9 embryo immunostained for Mef2-lacZ expression to reveal the dorsal mesoderm (arrow). B'. Representation of the embryo shown in B, highlighting in yellow the dorsal ectodermal region that produces Dpp. B". Transverse section of the same embryo, depicting zones of Dpp-producing dorsal ectoderm (yellow) and underlying Dpp-receiving dorsal mesoderm (orange). C. The lateral view of a middle-stage embryo immunostained for Mef2-lacZ expression to reveal clusters of cardiac mesoderm (arrow). C'. The cartoon of the embryo shown in C demarcating the ectodermal zones secreting Dpp (yellow) and Wg (blue), at the intersection of which the cardiac mesoderm is formed. C". Simplified flow chart of events leading to cardiac progenitor specification: cardiac mesoderm (pink) is specified under the intersection of Dpp (yellow stripe) and Wg (blue stripe) ectodermal signals; within cardiac mesoderm local clusters of potency groups (red) appear due to ectopic local activation of the RTK pathway; within each potency group a progenitor (dark red) is singled out as a result of the Notch signal activity. D. The lateral view of a late-stage transgenic embryo expressing lacZ under control of the cardioblast-specific Sur enhancer; immunostaining for beta-galactosidase reveals a row of cardioblasts. D'. The cartoon of a similar-stage embryo as shown in D depicting a row of cardioblasts migrating dorsally (arrows) to merge with cardioblasts from the opposite side to form the dorsal vessel. D". Predicted models of cell division events leading to formation of cardioblasts. The symmetrical division model (bracket s; which gives rise to Tin cardial cells) is where progenitors (P) give rise to identical founders (F) and, subsequently, cardioblasts (cb). In the asymmetrical model (bracket a; which specifies Svp cells) founder cells give rise to two cardiac cells subtypes: cardioblasts (cb) and adjacent pericardial cells (pc, grey color) due to Notch-mediated asymmetrical divisions. E. The dorsal view of a stage 16 embryo showing the mature embryonic dorsal vessel (arrow) visualized by immunostaining for beta-galactosidase expressed under the control of the cardioblast-specific Sur enhancer. E'. Organization of the embryonic dorsal vessel. The morphologically distinctive thinner aorta lies anteriorly; the thicker heart proper, containing inflow tracts (termed ostia; yellow), are indicated. Hemolymph flow is shown by dashed arrows. Note, that the number of grey pericardial cells (pc) is more than shown and additional cells are located both above and below of the cardioblasts (cb). Anterior is to the left and dorsal is uppermost, in all panels unless indicated.
Cells within the dorsal mesoderm become directed to either cardiac fate or trunk visceral mesoderm fate. This diversification occurs in each segment of the embryo and is controlled by the actions of the ectodermally-derived signaling molecules Wingless (Wg), which promotes cardiac fate [6]; and Hedgehog (Hh), which promotes visceral mesoderm fate ([7]; Figure 1C-C”). Whereas visceral mesoderm precursors subsequently migrate internally, the cardiac cells remain close to the ectoderm [8]. Here, a series of intrinsic and extrinsic signals diversify the cardiac cell population to generate distinct subsets of cardiac cell types (Figure 1D). As this process elaborates, the bilateral cardiac precursors migrate dorsally and medially during the embryonic process of dorsal closure. Ultimately, the precursors meet at the dorsal midline to generate an antero-posteriorly oriented tube (Figure 1E, E').
The mature cardiac tube thus comprises a series of distinct cell types: cardial cells comprise the muscular portion of the vessel, and express either the homeodomain protein Tinman (Tin) or the orphan nuclear receptor Seven-up (Svp). In addition cardial cells are patterned along the antero-posterior (AP) axis, such that inflow tracts in the embryo form from the three most posterior sets of Svp cells [9-12]). Surrounding the cardial cells is a group of pericardial cells, which are characterized by diverse patterns of gene expression [13, 14]. Pericardial cells are thought to function in detoxification of the hemolymph [15], although given their great diversity as defined by distinct patterns of gene expression, they probably have a number of distinct functions. Indeed, ablation of even-skipped (eve) -expressing pericardial cells affects cardiac physiology [16]. More recently, a subset of anterior eve-expressing pericardial cells was shown to contribute to the adult wing hearts [17]. As will be discussed, the embryonic origin of the Eve pericardial cells is a paradigm to understand how ectodermal and mesodermal cells interact in order to program the formation of a specific cell type within the cardiac mesoderm.
Viewed in this manner, the process of cardiogenesis can be considered as a series of distinct developmental decisions: dorso-ventral patterning of the embryo and mesoderm specification; specification of the dorsal mesoderm and of cardiac fate within the dorsal mesoderm; diversification of the cardioblast cell pool in each segment; AP diversification of cardial cells; and initiation of cardiac differentiation. In the following sections, we shall describe what is known concerning the regulatory circuits for each of these developmental events.
The dorsoventral axis: several domains contribute to cardiac specification
The first critical decision made during the formation of cardiac tissue is to pattern the embryo along the dorsoventral axis. The dorsoventral patterning mechanism promotes the formation of a gradient of nuclear Dorsal protein from low in the dorsal region, through moderate in lateral regions, to high in ventral regions (Figure 2). On a superficial level, this decision is important since it results in the specification of the mesoderm as the ventral-most group of cells in response to the highest levels of nuclear Dorsal. However, as will be discussed, each dorsoventral domain is critical for cardiac development.
Figure 2. Gene regulatory networks controlling specification of the dorsoventral axis.
The fertilized embryo can be functionally subdivided into three main regions. Ventrally, the mesoderm is specified by the highest levels of nuclear Dorsal protein, which results in the activation of genes characteristic of the mesoderm, and by active repression of neuroectodermal genes by the sna gene product. Laterally, the neuroectoderm is specified by moderate levels of nuclear Dorsal, which are sufficient to activate genes such as rho and ths, but which are at insufficient levels to activate the sna repressor gene. Dorsally, the relative lack of nuclear Dorsal protein permits expression of genes such as zen and Dpp which specify dorsal ectoderm and amnioserosa fate.
In the most ventral region, the mesoderm is specified. This occurs via the action of Dorsal upon the twist gene promoter, for which Dorsal is a direct transcriptional activator [18-20]. twist transcription is subsequently maintained by positive autoregulation [21]. Dorsal and Twist, in turn, activate expression of the zinc finger transcription factor snail [22] and the FGF receptor heartless (htl; [23]), among other targets. Snail is a transcriptional repressor of genes whose expression is not required in the ventral region, including short gastulation (sog) and the FGF ligand thisbe (ths; [23]). Direct transcriptional targets of Twist in the ventral mesoderm include the cardiogenic gene tinman and the muscle regulatory factor Myocyte enhancer factor-2 [24-26]. Thus, the highest levels of nuclear Dorsal create a mesodermal cell fate, which is robustly maintained by positive autoregulation, and repression of non-mesodermal genes.
In lateral regions of the embryo, the neuroectoderm is specified. Here, low to moderate nuclear concentrations of Dorsal activate expression of at least two sets of genes defining dorsal and ventral subcomponents of the neuroectoderm. These genes include sog and ths [23, 27], each of which encode secreted proteins critical to embryonic development. In this lateral region Twist and Snail are not activated, since their respective Dorsal binding sites are of lower affinity than those of the enhancers for markers of the neuroectoderm. By contrast, sog and ths are activated by Dorsal in lateral regions since the Dorsal binding sites in their enhancers are of higher affinity and they can thus be activated at lower nuclear concentrations of the Dorsal transcriptional activator.
The most dorsal regions of the embryo are specified by the relative lack of Dorsal protein in their nuclei. Cells in this region are fated to become dorsal ectoderm, and will express such genes as dpp.
What is the relationship between specification of these three broad embryonic regions, and cardiac development? Following invagination of the mesoderm, Twist-expressing cells spread laterally over the inside surface of the embryo, and many cells migrate dorsally. Cells that migrate the furthest become restricted to dorsal mesoderm fate, since they come into the vicinity of the dorsally-derived Dpp signal, which is essential for the next step in cardiac cell specification ([28]; see next section).
The importance of the dorsal migration of the mesoderm has been elegantly demonstrated by genetic analysis: mutant embryos which fail to exhibit mesoderm migration have been isolated and characterized, and most of these mutants show defects in the formation of cardiac tissue, since mesodermal cells in such embryos do not receive the Dpp signal. Analysis of the genes exhibiting such phenotypes has revealed that most are members of the fibroblast growth factor receptor (FGF-R) signaling pathway, implicating this signaling pathway in mesoderm migration and (indirectly) in cardiac specification [29-31].
While the FGF-R pathway is activated within the mesoderm, the ligand for the receptor is encoded by a pair of FGF-like genes named pyramus and ths. As indicated above, pyr and ths are expressed in the neuroectoderm, and their combined mutation also results in a failure of dorsal mesoderm specification [32]. These findings suggest that the release of ligand from the lateral ectoderm provides a source towards which the mesodermal cells might move. As development proceeds, expression of pyr and ths refines to more dorsal regions, potentially drawing mesodermal cells with them ultimately to contact the Dpp signal emanating from the dorsal ectoderm. The cellular details of this migration have yet to be fully established.
Figure 2 summarizes the regulatory network of events leading to the specification of the dorsoventral domains in the early embryo. A more complete treatment of this process can be found in [33]; here, we have concentrated upon events of direct relevance to cardiac specification.
What is the cadre of genes that is activated during dorsoventral axis specification? This question has been addressed by recent genome-scale studies, which have identified numerous new genes contributing to the realization of dorsoventral fate. The first study [34] used a microarray approach to identify genes expressed in response to either high, moderate, or low levels of nuclear Dorsal protein. This was achieved by comparing transcript levels for mutants which showed either fully ventralized phenotypes (high levels of nuclear Dorsal throughout the embryo in Toll10B mutants), fully dorsalized phenotypes (low levels of nuclear dorsal in pipe mutants), or mutants in which there was a uniform moderate level of nuclear Dorsal (Tollrm9/Tollrm10 mutants).
This study identified roughly 100 genes enriched in each region, several of which had been shown previously to be markers of the respective domains. Moreover, several new genes were identified, which were shown by in situ hybridization analysis to be expressed in the location predicted by their calculated levels of expression in the different mutants. Some of the newly-identified genes also have predicted regulatory regions with binding sites consistent with their patterns of activation.
A complementary approach carried out by Zeitlinger et al. [35] utilized ChIP-on-chip to identify targets of Dorsal, Twist and Snail. Here, antibodies against the regulatory factors were used to immunoprecipitate genomic fragments from young embryos. Immunoprecipitated DNAs were then labeled and used in a microarray analysis to identify at the genomic level the many targets of these factors. In addition to identifying enhancer regions predicted based upon existing information or bioinformatics, these studies indicated that there were significantly more target enhancers for each of the tested factors than were previously expected.
Specification of the cardiac field: integration of intrinsic and extrinsic events
The dorsal mesoderm
Specification of the dorsal mesoderm begins when the underlying mesodermal layer comes into the vicinity of the dorsal ectoderm (Figure 1B-B”). In response to the ectodermally produced Dpp signal, the dorsal ridge of the mesodermal layer is specified, as expression of a number of molecular markers becomes restricted to this region. Specifically, expression of the cardiogenic factor tinman (tin) narrows down from the broad expression in trunk mesoderm to a slimmer band corresponding to the dorsal mesoderm, as ventral mesoderm rapidly loses both tin RNA and protein [36-38]. Genetic evidence in support of this model is very strong. Dpp is both required and sufficient for the maintenance of the dorsal mesoderm in stage 9 embryos [28]. Moreover, the function of Dpp receptors is also required for dorsal mesoderm specification [39].
The change in tin expression from pan-mesodermal to dorsal mesoderm is a result of switching in its enhancer activities, and has been studied in some detail. Originally, expression of tin at the early stages of development is under control of twist (Figure 3A). Twist protein directly binds to the enhancer region termed TinB and located in the first intron of tin, and activates its broad expression in trunk mesoderm, similar to the Twist distribution pattern itself [26]. During formation of the dorsal mesoderm, however, transcription of tin comes under the control of a separate enhancer, TinD.
Figure 3. Gene regulatory interactions in the formation of the dorsal mesoderm and cardiogenic mesoderm.
Direct and indirect interactions are shown in solid and dashed lines, respectively. Protein factors are shown as circles; genes are represented as boxes with their transcription status indicated in color. A. In early mesoderm Twi initiates expression of tin. B. In the dorsal mesoderm the tin expression is maintained through the Dpp signaling and self-activation. Dpp also probably stimulates a number of genes important for early cardiac specification (shaded in grey circle). Additionally, Dpp along with Tin activate expression of the pro-visceral mesodermal gene bap. C. At the moment when the dorsal mesoderm receives Wg signal it becomes cardiac mesoderm (left panel) in which cardiac specification genes (shaded in grey circle) are activated by products of tin and slp genes, controlled by Dpp and Wg, respectively. At the same time Slp effectors suppress expression of bap. Genes expressed in cardial and pericardial cells, like Hand, also depend on tin activation and additionally require a product of one of the cardiac specification genes (e.g. tup). Meanwhile, in the absence of inhibitory Wg signal in the visceral mesoderm (right panel), the transcriptional activity of bap goes on, fed by stimulatory inputs from Tin and the genetically downstream gene bin. Bap suppresses the expression of pro-cardiac genes.
The dorsal mesoderm enhancer of tin is directly responsive to Dpp (Figure 3B). Dpp affects the transcriptional program of cells via binding to transmembrane receptors and activating intracellular messenger Smads (for general references see [40] and [41]). In Drosophila Dpp binds to the receptors encoded by Thickvein (tkv) and punt (put) (Dpp receptors type I and II, respectively [42, 43]), which leads to phosphorylation within the dorsal mesodermal cells of a Smad protein termed MAD (Mothers Against Dpp), heterooligomerization of the latter with yet another Smad protein, Medea, and translocation of this complex into the nucleus. In the nucleus of dorsal mesodermal cells the Smad complex binds to the TinD enhancer, located in the 3' region of the tin gene [44]. Since Smads are active only in the region where the mesoderm receives Dpp signals from the dorsal ectoderm, this makes the tin expression pattern narrower, confining it to the dorsal part of the mesoderm.
An interesting observation from this work was that in order for the Smads to activate the TinD enhancer, Tin protein must also be present [44]. Such autoregulation of tin is an important requirement for confining the cardiac field to the mesodermal cells, and not other germ layers which also receive the Dpp signal. Thus, pre-existing Tin protein, accumulated as a result of tin transcription and activated initially by Twist, provides a mesodermal context for reception of the Dpp signal (Figure 3B).
Several other genes with critical cardiogenic roles also become expressed at this stage of cardiac specification. These genes encode the GATA factor Pannier (Pnr; [45, 46]); the three T-box genes Dorsocross 1-3 (Doc1-3, [47]); and the LIM homeodomain transcription factor Tailup/Islet 1 (Tup; [48]). Other than the observation that Tin is a direct activation of the pnr gene [49], the direct regulators of these genes have yet to be defined in detail. Nevertheless, it is highly likely that both Tin and the Dpp pathways play major roles in their activation in the dorsal mesoderm.
The cardiogenic mesoderm
As development proceeds, the regions of the dorsal mesoderm that give rise to cardiac tissue are further refined. This confining of cardiac mesoderm is due to an interplay of activating and inhibitory stimuli received on top of the Dpp/Smad pathway, mediated by the segment polarity gene wingless (wg), encoding a member of the Wnt family [50, 51]. Wg is produced in the ectoderm in transverse stripes that intersect perpendicularly with the Dpp secreting regions to generate ten clusters of cardiogenic cells (Figure 1C'). The Wg signaling is essential for cardiogenesis because in wg mutants, and in mutants for members of the Wg signaling pathway, no cardiac mesoderm is formed [6, 52]. Meanwhile, dorsal mesoderm patches that do not receive the Wg signal but that still express Tin, differentiate into visceral mesoderm (Figure 3C; [7, 28, 53]).
Genetical analyses have identified several important intracellular transducers of the Wg signal in cardiac specification, including the scaffold protein Dishevelled (Dsh) and the genetically more distal transducer Armadillo [52, 54, 55]. Detailed mechanistic studies in several cell types have revealed that Dsh works through aggregating Wg-bound receptors (Arrow [56] and Frizzled-2 [57]) into aggresomes on the plasma membrane, followed by phosphorylation of intracellular receptor domains and thereby transduction of the Wg signal intracellularly [58]. Based upon the data of Park et al. [52] this pathway also functions in cardiac specification.
One of the terminal acceptors of the Wg signal is the beta-catenin transcriptional regulator Armadillo (Arm). In the absence of signaling, Arm is phosphorylated and rapidly degraded. However, upon stimulation (probably by recruiting the effector kinase, GSK3/zw3/shaggy, to the Wg receptor aggresomes), hypophosphorylated Arm translocates into the nucleus, where it performs multiple regulatory functions. One of the well-know partners of Arm is Pangolin (Pan), the only Drosophila member of the TCF family of transcriptional regulators [50]. Arm binds to Pan at its N-terminus and the resulting complex gains transcriptional activity. Interestingly without Arm, Pan mediates a quite opposite function of inhibition of the same set of target genes, because in its default state it is bound to the transcriptional repressor Groucho [59]. Hence, Armadillo is a classical example of a transactivational switch. There have been proposed additional partners of Armadillo (of both a positive and negative nature) including Pygopus, [60], CtBP [61], and others [50, 51].
To date, direct transcriptional impact of the Wg signaling pathway in cardiogenesis has been demonstrated in only a few cases. The most striking, as well as the most studied, example is activation of the even-skipped (eve) gene. eve expression is commonly used as a genetical marker for progenitors that give rise to subpopulations of pericardial cells and dorsal muscles [62-64]. These progenitors appear first at places within the early cardiac mesoderm, where the Dpp and Wg signals intersect [38], and will be discussed in greater detail in the next section.
The role of the Wg signaling pathways in the activation of other cardiogenic genes has yet to be fully established. Indeed, given the critical role for Wg signaling in cardiac specification, surprisingly few direct targets of Wg in the cardiac cells have been identified. A possible explanation for this fact is that Wg acts, at least partially, through intermediary factors. One such pair of factors are the forkhead domain proteins encoded by the sloppy paired genes (slp-1, slp-2). Slp1 and Slp2 are critical to cardiac specification [65], and their genes are induced by the Wg signal in transverse mesodermal stripes corresponding to the overlying patterns of ectodermal Wg expression [53]. In the case of slp-1, this results from direct transcriptional activation by Pan [66].
A quite opposite situation is found in the Wg-dependent regulation of another dorsal mesodermal gene, bagpipe (bap; Figure 3C). bap codes for a NK-homeobox transcription factor participating in visceral mesoderm development [37]. The bap product is inhibitory to cardiogenesis and thereby must be suppressed in the cardiac mesoderm. Still, the early enhancer of bap bears a remarkable similarity to that of eve: there are Tin and Smad binding sites functioning in a similar manner to that of the TinD enhancer. However, unlike TinD, the bap enhancer additionally contains a binding site for Slp proteins. This high affinity Slp-binding site overlaps with a weaker binding site for the transcription factor Biniou and seems to have inhibitory function, repressing bap expression in cardiac mesoderm, where Slp proteins are present [53]. Thus, in this example, the Wg signal acts strictly as a suppressor, extending its action remotely via induction of secondary effectors, Slp.
Specification of distinct progenitors within the cardiac mesoderm: the Eve pericardial cell
By the end of stage 10 of Drosophila embryogenesis, the newly formed cardiac mesoderm comprises spaced clusters of cells situated at the dorsal ridge of the mesoderm, under the intersection points between Dpp and Wg ectodermal signals (Lockwood and Bodmer, 2002). From this presumptive cardiac field, three elements emerge: a subset of dorsal body wall muscles; cardial cells (cardiomyocytes); and pericardial cells. There is still much to learn about the mechanisms that specify cardial cell progenitors at this stage. By contrast, we have some appreciation of how combinatorial inputs of signaling networks specify the segregation and formation of a specific pair of pericardial cells: those which express the homeobox gene even-skipped (eve). This knowledge serves as a paradigm in our understanding of the complexity of cross-talk between different regulatory signals in the dorsal mesoderm, and several of the principles here probably also apply to cardiogenesis.
At late stage 10, mesodermal patches receiving Dpp and Wg signals start expressing the lethal of scute (l'sc) gene. Although originally l'sc has been reported to act in neurogenesis [67], its expression also can be found in the dorsal, lateral and ventral somatic mesoderm: in the mesoderm, expression of l'sc appears first in larger clusters of cells, which then condense into smaller groups, each comprising a few expressing cells - these groups of cells will ultimately be further restricted to single cells showing maximum expression of l'sc [64]. This temporo-spatial pattern of l'sc expression in the mesoderm is thought to mark and outline the processes of selection and specification of muscle and cardiac progenitors [64, 68].
Cardiac expression of eve appears at early stage 11 in each hemisegment, in a small cell cluster shortly after initiation of l'sc expression [62]. eve expression is subsequently restricted, similarly to l'sc, to a single progenitor cell, named P2 [64]. The P2 cell then divides to produce two founder cells (F2) that give rise to the dorsal muscle DO2 and pericardial cells [69]. The spatial nature of eve expression that rapidly subsides to a single cell suggests a sequential programming of cell fate, that firstly establishes the eve-expressing cell clusters, and secondly restricts eve expression to a single progenitor.
It was found that in addition to the regulatory mechanisms that specify the cardiac field (Tin, Dpp signaling, and Wg signaling), expression of eve also depends on a locally activated receptor tyrosine kinase (RTK) transduction pathway, including the FGF receptor Htl [64, 69], the G-protein Ras, and MAP kinase signaling [63]. Thereby, the ectoderm continues to be actively involved in specification of the mesoderm. Moreover, it is clear that proper transduction and transcriptional interpretation of the signal from activated Htl can only be achieved in cells that have been made competent by Wg and Dpp signals [63]. The terminal effector of the RTK/Ras signal, Pointed, has been proposed to participate in full activation of the eve enhancer. Nevertheless, forced misexpression of activated Ras did not induce ectopic eve expression outside of Dpp-Wg intercross regions [64].
Consistent with the genetic evidence suggesting a plethora of regulatory factors that converge upon the eve gene, the enhancer responsible for cardiac expression of eve is particularly complex. A series of papers describing the identification and analysis of this enhancer have been published [63, 70-72]. These several regulatory interactions are summarized in Figure 4.
Figure 4. Interaction events governing expression of the eve gene.
Direct and indirect interactions are shown in solid and dashed lines, respectively. Protein factors are shown as circles; genes are represented as boxes. A. The specification of future eve expression is mediated by the Dpp signaling that activates Smad effectors (Mad and Medea) that oligomerize and positively control tin expression. In parallel, Wg signaling stabilizes the transcription activator Arm and promotes its formation of a complex with the DNAbinding factor Pan and releasing it from a transcription repressor (not shown). Dpp and Wg specify the location of future eve expression as their contribution is equally important and thereby eve activation can be achieved only at Dpp and Wg signals intersection zones (see Fig.1 C'); the tin gene is active only in the mesoderm and thereby provides the mesodermal context to eve activation. However, the activity of the transcriptional repressor Anterior open (Aop) puts eve expression on hold (yellow shading). B. Within the cardiac mesoderm, a local activation of the receptor Htl by Pyr or Ths ligands subsequently leads to activation of the MAPK pathway, attendant activation of the transcription factor Pnt, and inactivation of the repressor Aop. The cooperative stimulatory inputs activate eve transcription (green) in small clusters of cardiac mesoderm. C. The final process of singling out a progenitor cell (left side) from adjacent neighboring cells (right side) within a cluster of eve-expressing cells. In the progenitor, the active RTK signaling pathway activates expression of the eve marker gene along with components of the Notch signal: the receptor Notch (N) and its ligand Delta (Dl). Here, the components of the RTK signaling, Htl and Hbr, are also self-maintained, while the Notch receptors are not active (shown in subdued colors) in the absence of Delta received from the neighboring cells. In contrast, neighboring cells receive the Delta signal produced by the progenitor cell, which leads to activation of N and suppression of the RTK signaling (subdued) and most of its targets, including the eve and Delta genes. Meanwhile, the Notch receptor stimulates its own expression to maintain and amplify the N signal. The neighboring cell cannot activate N signaling in the progenitor cell, as its own Delta production is suppressed by active Notch.
The refining of eve expression from a cluster (potency group) to a single cell (progenitor) arises from the lineage specifying signaling of Notch (N), and its inhibitory effects upon the RTK/Ras signaling. Genetical studies have revealed that activated Ras induces expression of the membrane-bound Notch ligand Delta [69]. Delta binds to Notch receptors on the surface of neighboring cells and activates N signal (for review see [73]). Activated Notch receptors start suppressing the activating stimuli of the RTK/Ras pathway, while self-stimulating their own activity. According to the current view of the lateral inhibition model, the progenitor cell that becomes singled out of a potency group produces a fraction more Delta than its neighboring cells, thereby shifting the balance between activating (RTK/Ras) and inhibitory (Notch) signals in its neighbors in favor of the latter [74]. These interactions are detailed in Figure 4C.
Recently some progress has been achieved in our understanding of how different signaling pathways which participate in progeny cell specification are integrated. Besides the Notch ligand Delta that communicates between the Ras and N signal pathways [69], another factor Argos [75] inhibits RTK activities responsible for Ras activation [69]. In addition, Canoe has been shown recently to mediate cross-talks between Ras, N, and Wg pathways. Canoe is an intracellular protein that, via its multiple domains, is capable of physical binding to and modulating activities of important signal transducers: Notch, Ras, and Dsh (the most proximal component of the Wg signal) [76].
Another important contribution of the N signal in lineage establishment occurs at the first (asymmetrical) division, when individual progenitors (P) produce two founder cells (F) with unequal fates. In the case of the eve-positive P2 cells discussed above, one of the resulting F2 cells becomes a muscle founder and gives rise to the DO2 dorsal muscle; while the other daughter cells establishes a subpopulation of pericardial cells [69]. Besides Notch itself [77], genetical studies revealed two genes, Sanpodo and Numb, participating in regulation of asymmetrical division of pericardial progenitors [13, 78].
In summary, although a comprehensive picture of how different types of cardiac progenitors emerge is far from being complete, we may assume that the process of progenitor specification follows this general scenario. Wg and Dpp (and probably Twist) signals create potency groups in the dorsal mesoderm, that further undergoes clustering through local activation of RTK signaling and l'sc expression. Within each cluster, or equivalence group, a progenitor is singled out as a result of lateral inhibition via the N signal. This concept, originally applicable to formation of a subset of pericardial and dorsal muscles progenitors, may be extended to cardioblasts. The lack of a reliable molecular marker for early cardioblast progenitors complicates studying mechanisms of cardial lineage specification. Nevertheless, recent studies of the origins of Ladybirdexpressing cardioblasts [79], and the identification of the NK homeobox gene C15 as being expressed in the cardiac mesoderm [72] substantially improve these prospects.
Recent bioinformatic studies have also begun to shed light upon the identification of enhancers which integrate multiple intrinsic and extrinsic signals in the Drosophila embryo. These approaches depend upon the identification of cis-regulatory regions which show enrichment of binding sites for candidate transcriptional regulators (see for example [80]). This work was highly effective in predicting the locations of tissue-specific enhancers, in this instance for genes known to be expressed in skeletal muscle founder cells. A similar methodology should be effective for defining enhancers which function within the cardiac mesoderm.
Diversifying the cardiac tube: the anteroposterior axis
As the linear cardiac tube is being formed via the convergence of two rows of cardial cells, cells along the anteroposterior (AP) axis are programmed to assume distinct cell fates. This programming is most apparent in the patterning of Svp cells in the developing dorsal vessel: whereas ten trunk segments contribute cells to the cardiac tube, only in the most posterior seven segments do Svp cells form. There are additionally a number of genes, both regulatory and structural, whose patterns of expression show distinct AP diversification (Figure 5).
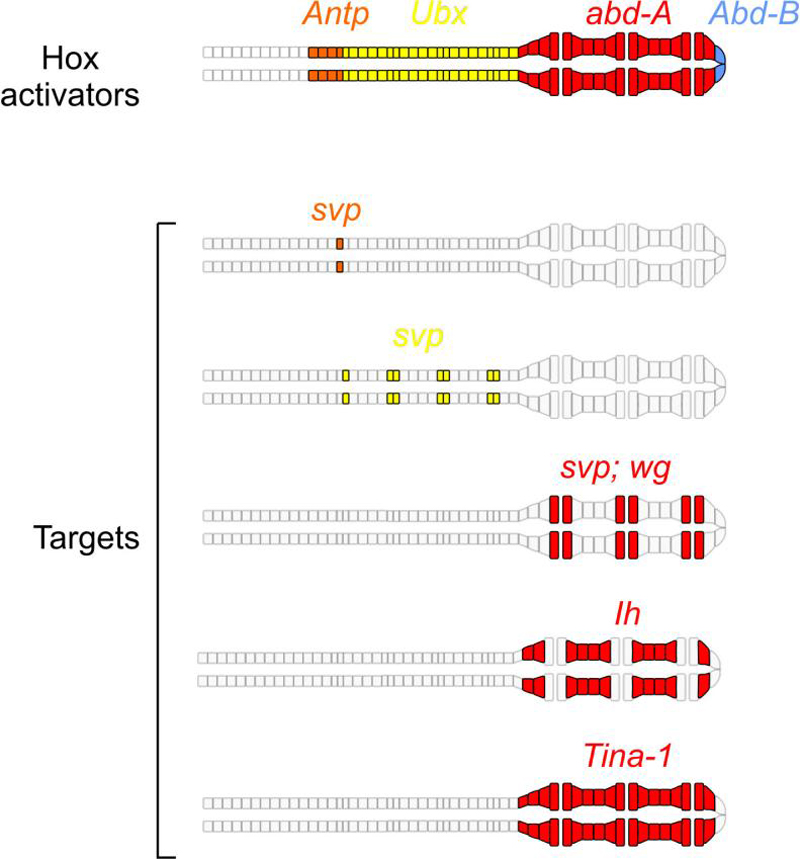
Figure 5. Schematic representation of the spatial expression of various Hox regulators and their putative targets in the Drosophila dorsal vessel.
Top panel: a cartoon of dorsal vessel showing the color-coded expression patterns of indicated Hox genes: Antennapedia (Antp, orange), Ultrabithorax (Ubx, yellow), abdominal-A (abd-A, red), and Abdominal-B (Abd-B, blue). Note the sequential expression of Antp and Ubx in the aorta and mostly exclusive expression of abd-A in the heart proper. No Hox genes are expressed in the most anterior region of the aorta. Anterior is to the left. Lower panel: expression of putative Hox target genes, color-coded to match their potential Hox activators (from top to bottom): Antp-dependent seven-up (svp) expression in the anterior-most pair of Seven-up expressing cells (future adult ostia cells); Ubx-dependent svp expression in the rest of the aorta Seven-up cells; svp and wg expression in ostia cells in the heart proper depend on abd-A; expression of Ih and Tina-1 are also abd-A-dependent although these genes are expressed in different patterns. There is no Abd-Bdependent cardiac target found up to date.
The specification of Svp cell fate is important, since Svp cells form the inflow tracts (also called ostia) of the cardiac tube: in the embryo and larva, the most posterior three sets of Svp cells express the Wg signaling molecule and develop into ostia [9, 10, 12]. Svp cells in the remaining four segments do not form ostia until pupal development, whereupon the Svp cells in these four segments are remodeled into ostia for the adult heart [9, 81, 82]. There are now a number of genes whose patterns of expression reflect AP fate in the cardiac tube [82, 83], and a current challenge is to define how this pattern of gene expression is encoded at the genomic regulatory level.
Important insight into the identity of the factors which must control the AP-restricted expression of cardiac genes has come from analyses of homeotic selector genes of the Bithorax- and Antennapedia Complexes (BX-C and ANT-C, respectively). Studies from several groups have demonstrated that Hox genes are responsible for the AP patterning of the cardiac tube (Figure 5). Firstly, abdominal-A (abd-A) expression in the posterior heart region is required and sufficient for heart fate. This was demonstrated by both loss- and gain- of functions experiments using abd-A, and the analysis of such treatments upon patterns of gene expression and cell function [10-12]. Secondly, Ultrabithorax (Ubx) expressed in four segments of the aorta is required and sufficient for Svp cell fate in those segments. This was proven by removal of Ubx expression, which resulted in a loss of most of the Svp cells in the aorta; and by ectopic expression of Ubx, which resulted in supernumerary Svp cells in the anterior-most three segments [84, 85].
A reasonable conclusion from these studies is that Hox gene products directly impact the enhancers of AP-restricted cardiac genes. However, the direct influence of a Hox protein upon a cardiac enhancer has yet to be demonstrated, perhaps because several of the target genes are large and complex, thus complicating traditional approaches for enhancer analysis.
Some progress on this front has come from the recent identification of the svp cardiac enhancer (SCE). Ryan et al. [86] used a simple bioinformatics approach to identify a candidate regulatory region for svp: locate a 1-kb sequence in the vicinity of svp that contains at least two consensus binding sites for the cardiac regulator Tinman, and determine if this sequence is conserved in other Drosophila species. The analysis identified a single location which might correspond to the svp enhancer, from a genomic region in excess of 50kb. Testing of the candidate enhancer region confirmed that it was active in the developing Svp cells of the dorsal vessel. Ryan et al. [86] also demonstrated that the SCE was a direct transcriptional target of Tinman during Svp cell specification, however they did not yet determine if this enhancer is a direct target of Hox gene products [86].
Based upon the observation by Ponzielli et al. [10] that svp expression is activated within each segment under the positive influence of Hedgehog signaling from the ectoderm, we can hypothesize that the svp cardiac enhancer must integrate at least three distinct types of signals: a cardiac-restricted signal, presumably Tinman; an axial signal such as Ubx and abd-A; and a segment-polarity signal from the Hedgehog pathway.
Once activated in the cardiac tissue, svp expression must very rapidly become independent of Tin, since studies have indicated that over-expression of svp in the cardiac tube functions to inhibit tin transcription [87, 88]. How svp expression is maintained in the cardiac tissue remains to be determined.
Gene expression studies have also identified several additional cardiac structural genes which show AP polarity in their expression patterns ([83]; Figure 5). Based upon the in situ hybridization data, and upon the known expression patters of regulatory genes expressed in the cardiac mesoderm, it is possible to predict the gene regulatory interactions that are acting. Further, given the recent publication of the genome sequences of several Drosophila species, the identification and characterization of enhancers for Hox targets in the dorsal vessel should proceed rapidly.
Cardiac structural genes: a common code for a common pattern of expression?
Expression of a set of genes that directly determine cardiac function is the ultimate goal of the differentiation process, yet as described below, significant diversity is apparent both in patterns of structural gene expression and mechanisms of structural gene regulation. Cardiac structural genes can be tentatively grouped into sub-categories of (i) general muscle-specific genes and (ii) cardiac-specific genes, although there are also several genes, such as svp and brokenhearted, which show expression in rather disparate tissues including the cardiac mesoderm and the central nervous system, among others. In the first category are genes that encode proteins forming and regulating the contractile apparatus: muscle-specific actin (Act57B), myosin heavy-chain (Mhc), and several others. The second category contains genes comprising diverse functions, including those whose products code for ion transporters (Sur, Ih; [83, 89, 90]), structural proteins (Pericardine; [91]), receptors (Toll; [92]), and enzymes (Transglutaminase; [93]).
The number of genes that are expressed in the dorsal vessel is under continuous growth, as more in situ hybridization data become available via public databases, mainly the Berkeley Drosophila Genome Project [94]. A recent query via the FlyExpress search engine (url: http://www.flyexpress.net) for genes associated with the ontology term `embryonic dorsal vessel' returned over 60 hits. More candidate cardiac genes may be obtained using a computational prediction approach: recently a prediction for the identification of novel genes expressed in muscle was made, based on microarray expression data and employing machine learning algorithms [95]. A similar methodology might be used for the identification of genes expressed in cardiac tissue.
For the transcriptional activation of pan-muscular structural genes, the crucial input most likely comes from the activity of Myocyte enhancer factor-2 (MEF2). In Mef2 mutants all types of muscles (including cardioblasts) are specified normally, but the mutants entirely lack expression of muscle structural genes [96-98]. In several cases MEF2-binding sites within an enhancer region of the target structural gene contribute much of the transcriptional activity [99-101] [102]. However, despite a strong requirement for MEF2 function in terminal muscle differentiation, ectopic expression of Mef2 was unable to induce the myogenic program [103]. A possible explanation for such a failure may be a requirement for other co-factors that could synergize with MEF2 upon its target genes. Thus, for early stages of development a role for Twist as a co-activator acting along with MEF2 was deduced, based upon ChIP-on-chip assay results [104]. For the late stages of muscle development, another MEF2 co-regulator - zinc-finger domain factor Chorion Factor-2 - was recently reported [105].
Consistent with the single-gene identification of MEF2 targets, ChIP-on-chip analysis revealed active MEF2 binding sites in the intronic and/or flanking genomic sequences of many known and putative structural muscle genes [98], and it is anticipated that a large number of these target genes are expressed in cardiac tissue. In addition, over 600 total genomic regions were shown to interact with MEF2 during embryogenesis, confirming that MEF2 plays a major role in gene activation.
In contrast to our understanding of the regulation of structural muscle genes, there is much less known about the regulation of cardiac-specific or cardiac-enriched structural genes. The expression of these genes is MEF2-independent in many cases, and more likely depends upon cardiac factors such as Tinman. Indeed, it has been shown that Tinman directly regulates expression of a number of such genes. These include: Sulfonylurea receptor (Sur; [89, 90]); the trans-membrane receptor gene Toll, which predominantly requires the function of a single Tinman-binding site to drive expression in cardioblasts, [92]; the ß3tubulin gene [106]; and the basic, helix-loop-helix factor Hand [71]. These examples suggest a significant role of Tinman in the late stages of heart development.
To test this notion, Zaffran et al [107] generated a genomic rescue fragment of tin, comprising the entire transcribed region, and all of the tin enhancers with the exception of TinC, which is active in the mature cardiobasts. tin null mutants rescued with this construct showed normal cardiac specification and tube formation, however significant aspects of cardiac differentiation were abrogated. This study confirmed a critical and sustained role for tin in cardiac development.
Since tin is only expressed in a subset of muscular cells in the dorsal vessel, one must also consider how genes expressed in the Svp cells are regulated. To date, relatively few genes whose expression is restricted to these cells have been characterized as to their regulators. One of these is svp itself, for which the SCE is activated at stage 12 by Tin [86]. Activity of this enhancer does not persist at stage 16, however, when tin and svp expression are mutually exclusive [87]. A Svp cell marker active in the mature dorsal vessel is the crosslinking factor encoded by Transglutaminase (Tg), which is expressed predominantly in Svp cells from stage 14 onwards. Tg is activated in the cardiac tube via a MEF2-dependent enhancer, yet how the Svp cell specificity of this enhancer is determined has yet to be clarified [93]. A summary of cardiac structural gene regulatory mechanisms is presented in Figure 6.
Figure 6. Transcriptional regulatory pathways in cardiac differentiation.
The functioning of the cardiac tube is characterized by the expression of some genes throughout the cardiac tube and other genes detected only is specific subsets of the cardiac musculature, such as the Tin and Svp cells. Muscle structural genes, which in flies are expressed in all muscles types, depend centrally upon: MEF2, which is essential for muscle structural gene expression; and CF2, which collaborates with MEF2 in activation of structural genes. Of the genes whose expression is restricted to the Tin cells, all of those currently characterized are direct transcriptional targets of Tin. Tin can collaborate with the GATA factor Pnr (which is also activated by Tin) in activation of target genes. For the relatively few genes whose expression is predominant in the Svp cells Tg is a MEF2 target, whereas svp expression is initially activated by Tin but quickly becomes independent of it.
Concluding Comments
Significant progress has been made over the last few years in identifying how cardiac cells are specified in the embryo and how their differentiation is regulated. It is now apparent that the process of cardiogenesis, in organisms from flies to man, arises from the action of a conserved network of regulatory interactions. New technological approaches are further identifying genes which are expressed in the cardiac tissue, defining their functions in that tissue, and demonstrating how their expression might be regulated during development.
We can anticipate a time when the entire set of genes that are expressed in the cardiac tissue during specification and differentiation has been identified. The Berkeley Drosophila Genome Project is in the middle of a sustained effort to define the expression patterns of all predicted genes in the genome [94]. This information will be critical in identifying what genes contribute to the cardiac phenotype.
Coupled with this, the identification of transcriptional targets for all of the critical cardiogenic factors (Tinman, Pannier, T-box factors (H15, Midline, Doc1-3), Tailup/Islet-1 and Hand) should be uncovered. Data in this area is likely to arise from analysis of single-gene enhancers according to traditional methodologies. It is also likely to be overtaken by array-based studies of changes in transcription factor levels in mutant backgrounds, and ChIP-on-chip studies to define the cadre of genomic regions bound by candidate transcription factors. This has already been achieved for critical mesodermal factors such as Twist and MEF2; parallel studies for the cardiogenic factors discussed above cannot be far behind. Also, computational approaches are rapidly developing to a stage where the enhancers for cardiac-expressed genes can be identified by searching for clusters of bindings sites for cardiac regulatory factors.
These innovations will significantly move us towards solving the problems of cardiac development and disease discussed at the start of this communication. Nevertheless, while each new discovery brings us closer to a more systemic understanding of cardiac biology, we anticipate that there is still much more to be learned. The transcriptional techniques and studies that are currently predominant in the cardiac development field do not take into account the modifications that both primary transcripts and polypeptides undergoes within the cell, nor do they integrate epigenetic effects upon gene expression and regulation. How this problem will be solved at the systems level remains to be determined.
Furthermore, only recently have we become aware of the importance of non-coding RNAs in biological systems. While micro-RNAs are already being associated with gene regulation in the mesoderm in Drosophila [108], and in cardiac development in mammals [109, 110], a concerted effort to identify and characterize micro-RNAs expressed in the cardiac tissue has yet to be carried out, and such studies could have a profound effect upon our appreciation of the cardiac developmental regulatory network. These considerations remind us that, the more we discover in our search for understanding a biological system, the more complex and elegant that system becomes.
Acknowledgements
Research in the Cripps Laboratory is supported by grants from the NIH (GM61738, HL080545) American Heart Association, Pacific-Mountain Affiliate, and the March of Dimes Birth Defects Foundation.
References
- [1].Carrol SB, Klaiss JP, Olds LM. Endless forms most beautiful: the new science of evo devo and the making of the animal kingdom. Norton; New York: 2005. [Google Scholar]
- [2].Davidson D, Baldock R. Bioinformatics beyond sequence: mapping gene function in the embryo. Nat Rev Genet. 2001;2:409–417. doi: 10.1038/35076500. [DOI] [PubMed] [Google Scholar]
- [3].Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- [4].Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- [5].Tao Y, Schulz RA. Heart development in Drosophila. Semin Cell Dev Biol. 2007;18:3–15. doi: 10.1016/j.semcdb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- [6].Wu X, Golden K, Bodmer R. Heart development in Drosophila requires the segment polarity gene wingless. Dev Biol. 1995;169:619–628. doi: 10.1006/dbio.1995.1174. [DOI] [PubMed] [Google Scholar]
- [7].Azpiazu N, Lawrence PA, Vincent JP, Frasch M. Segmentation and specification of the Drosophila mesoderm. Genes Dev. 1996;10:3183–3194. doi: 10.1101/gad.10.24.3183. [DOI] [PubMed] [Google Scholar]
- [8].Dunin-Borkowski OM, Brown NH. Mammalian CD2 is an effective heterologous marker of the cell surface in Drosophila. Dev Biol. 1995;168:689–693. doi: 10.1006/dbio.1995.1115. [DOI] [PubMed] [Google Scholar]
- [9].Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–59. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- [10].Ponzielli R, Astier M, Chartier A, Gallet A, Therond P, Semeriva M. Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development. 2002;129:4509–4521. doi: 10.1242/dev.129.19.4509. [DOI] [PubMed] [Google Scholar]
- [11].Lovato TL, Nguyen TP, Molina MR, Cripps RM. The Hox gene abdominal-A specifies heart cell fate in the Drosophila dorsal vessel. Development. 2002;129:5019–5027. doi: 10.1242/dev.129.21.5019. [DOI] [PubMed] [Google Scholar]
- [12].Lo PC, Skeath JB, Gajewski K, Schulz RA, Frasch M. Homeotic genes autonomously specify the anteroposterior subdivision of the Drosophila dorsal vessel into aorta and heart. Dev Biol. 2002;251:307–319. doi: 10.1006/dbio.2002.0839. [DOI] [PubMed] [Google Scholar]
- [13].Ward EJ, Skeath JB. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development. 2000;127:4959–4969. doi: 10.1242/dev.127.22.4959. [DOI] [PubMed] [Google Scholar]
- [14].Alvarez AD, Shi W, Wilson BA, Skeath JB. pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development. 2003;130:3015–3026. doi: 10.1242/dev.00488. [DOI] [PubMed] [Google Scholar]
- [15].Poulsen DF. Histogenesis, organogenesis, and differentiation in the embryo of Drosophila melanogaster meigen. In: Demerec M, editor. Biology of Drosophila. Hafner Publishing Company; London: 1950. [Google Scholar]
- [16].Fujioka M, Wessells RJ, Han Z, Liu J, Fitzgerald K, Yusibova GL, Zamora M, Ruiz-Lozano P, Bodmer R, Jaynes JB. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ Res. 2005;97:1108–1114. doi: 10.1161/01.RES.0000191546.08532.B2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Togel M, Pass G, Paululat A. The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev Biol. 2008;318:29–37. doi: 10.1016/j.ydbio.2008.02.043. [DOI] [PubMed] [Google Scholar]
- [18].Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- [19].Pan DJ, Huang JD, Courey AJ. Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev. 1991;5:1892–1901. doi: 10.1101/gad.5.10.1892. [DOI] [PubMed] [Google Scholar]
- [20].Jiang J, Kosman D, Ip YT, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- [21].Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- [22].Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- [23].Stathopoulos A, Levine M. Whole-genome analysis of Drosophila gastrulation. Curr Opin Genet Dev. 2004;14:477–484. doi: 10.1016/j.gde.2004.07.004. [DOI] [PubMed] [Google Scholar]
- [24].Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nguyen HT, Xu X. Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev Biol. 1998;204:550–566. doi: 10.1006/dbio.1998.9081. [DOI] [PubMed] [Google Scholar]
- [26].Yin Z, Xu XL, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]
- [27].Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Frasch M. Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- [29].Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM. heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 1996;10:3003–3017. doi: 10.1101/gad.10.23.3003. [DOI] [PubMed] [Google Scholar]
- [30].Beiman M, Shilo BZ, Volk T. Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev. 1996;10:2993–3002. doi: 10.1101/gad.10.23.2993. [DOI] [PubMed] [Google Scholar]
- [31].Vincent S, Wilson R, Coelho C, Affolter M, Leptin M. The Drosophila protein Dof is specifically required for FGF signaling. Mol Cell. 1998;2:515–525. doi: 10.1016/s1097-2765(00)80151-3. [DOI] [PubMed] [Google Scholar]
- [32].Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- [34].Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- [35].Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- [37].Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- [38].Lockwood WK, Bodmer R. The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech Dev. 2002;114:13–26. doi: 10.1016/s0925-4773(02)00044-8. [DOI] [PubMed] [Google Scholar]
- [39].Yin Z, Frasch M. Regulation and function of tinman during dorsal mesoderm induction and heart specification in Drosophila. Dev Genet. 1998;22:187–200. doi: 10.1002/(SICI)1520-6408(1998)22:3<187::AID-DVG2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [40].Raftery LA, Sutherland DJ. TGF-beta family signal transduction in Drosophila development: from Mad to Smads. Dev Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- [41].Affolter M, Marty T, Vigano MA, Jazwinska A. Nuclear interpretation of Dpp signaling in Drosophila. Embo J. 2001;20:3298–3305. doi: 10.1093/emboj/20.13.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shimmi O, O'Connor MB. Physical properties of Tld, Sog, Tsg and Dpp protein interactions are predicted to help create a sharp boundary in Bmp signals during dorsoventral patterning of the Drosophila embryo. Development. 2003;130:4673–4682. doi: 10.1242/dev.00684. [DOI] [PubMed] [Google Scholar]
- [43].Letsou A, Arora K, Wrana JL, Simin K, Twombly V, Jamal J, Staehling-Hampton K, Hoffmann FM, Gelbart WM, Massague J, et al. Drosophila Dpp signaling is mediated by the punt gene product: a dual ligand-binding type II receptor of the TGF beta receptor family. Cell. 1995;80:899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- [44].Xu X, Yin Z, Hudson JB, Ferguson EL, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gajewski K, Fossett N, Molkentin JD, Schulz RA. The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development. 1999;126:5679–5688. doi: 10.1242/dev.126.24.5679. [DOI] [PubMed] [Google Scholar]
- [46].Klinedinst SL, Bodmer R. Gata factor Pannier is required to establish competence for heart progenitor formation. Development. 2003;130:3027–3038. doi: 10.1242/dev.00517. [DOI] [PubMed] [Google Scholar]
- [47].Reim I, Frasch M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development. 2005;132:4911–4925. doi: 10.1242/dev.02077. [DOI] [PubMed] [Google Scholar]
- [48].Tao Y, Wang J, Tokusumi T, Gajewski K, Schulz RA. Requirement of the LIM homeodomain transcription factor tailup for normal heart and hematopoietic organ formation in Drosophila melanogaster. Mol Cell Biol. 2007;27:3962–3969. doi: 10.1128/MCB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gajewski K, Zhang Q, Choi CY, Fossett N, Dang A, Kim YH, Kim Y, Schulz RA. Pannier is a transcriptional target and partner of Tinman during Drosophila cardiogenesis. Dev Biol. 2001;233:425–436. doi: 10.1006/dbio.2001.0220. [DOI] [PubMed] [Google Scholar]
- [50].Roose J, Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424:M23–37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- [51].Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- [52].Park M, Wu X, Golden K, Axelrod JD, Bodmer R. The wingless signaling pathway is directly involved in Drosophila heart development. Dev Biol. 1996;177:104–116. doi: 10.1006/dbio.1996.0149. [DOI] [PubMed] [Google Scholar]
- [53].Lee HH, Frasch M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;132:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- [54].Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- [55].Noordermeer J, Klingensmith J, Perrimon N, Nusse R. dishevelled and armadillo act in the wingless signalling pathway in Drosophila. Nature. 1994;367:80–83. doi: 10.1038/367080a0. [DOI] [PubMed] [Google Scholar]
- [56].Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- [57].Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- [58].Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- [59].Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- [60].Hoffmans R, Stadeli R, Basler K. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol. 2005;15:1207–1211. doi: 10.1016/j.cub.2005.05.054. [DOI] [PubMed] [Google Scholar]
- [61].Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. Embo J. 2006;25:2735–2745. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. Embo J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- [64].Carmena A, Gisselbrecht S, Harrison J, Jimenez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Riechmann V, Irion U, Wilson R, Grosskortenhaus R, Leptin M. Control of cell fates and segmentation in the Drosophila mesoderm. Development. 1997;124:2915–2922. doi: 10.1242/dev.124.15.2915. [DOI] [PubMed] [Google Scholar]
- [66].Lee HH, Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- [67].Martin-Bermudo MD, Gonzalez F, Dominguez M, Rodriguez I, Ruiz-Gomez M, Romani S, Modolell J, Jimenez F. Molecular characterization of the lethal of scute genetic function. Development. 1993;118:1003–1012. doi: 10.1242/dev.118.3.1003. [DOI] [PubMed] [Google Scholar]
- [68].Carmena A, Bate M, Jimenez F. Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes Dev. 1995;9:2373–2383. doi: 10.1101/gad.9.19.2373. [DOI] [PubMed] [Google Scholar]
- [69].Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, Michelson AM. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- [70].Knirr S, Frasch M. Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol. 2001;238:13–26. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- [71].Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development. 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- [72].Liu J, Qian L, Han Z, Wu X, Bodmer R. Spatial specificity of mesodermal even-skipped expression relies on multiple repressor sites. Dev Biol. 2008;313:876–886. doi: 10.1016/j.ydbio.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–138. [PubMed] [Google Scholar]
- [74].Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- [75].Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- [76].Carmena A, Speicher S, Baylies M. The PDZ protein Canoe/AF-6 links Ras-MAPK, Notch and Wingless/Wnt signaling pathways by directly interacting with Ras, Notch and Dishevelled. PLoS ONE. 2006;1:e66. doi: 10.1371/journal.pone.0000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hartenstein AY, Rugendorff A, Tepass U, Hartenstein V. The function of the neurogenic genes during epithelial development in the Drosophila embryo. Development. 1992;116:1203–1220. doi: 10.1242/dev.116.4.1203. [DOI] [PubMed] [Google Scholar]
- [78].Park M, Yaich LE, Bodmer R. Mesodermal cell fate decisions in Drosophila are under the control of the lineage genes numb, Notch, and sanpodo. Mech Dev. 1998;75:117–126. doi: 10.1016/s0925-4773(98)00098-7. [DOI] [PubMed] [Google Scholar]
- [79].Liu J, Qian L, Wessells RJ, Bidet Y, Jagla K, Bodmer R. Hedgehog and RAS pathways cooperate in the anterior-posterior specification and positioning of cardiac progenitor cells. Dev Biol. 2006;290:373–385. doi: 10.1016/j.ydbio.2005.11.033. [DOI] [PubMed] [Google Scholar]
- [80].Philippakis AA, Busser BW, Gisselbrecht SS, He FS, Estrada B, Michelson AM, Bulyk ML. Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Comput Biol. 2006;2:e53. doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sellin J, Albrecht S, Kolsch V, Paululat A. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr Patterns. 2006;6:360–375. doi: 10.1016/j.modgep.2005.09.012. [DOI] [PubMed] [Google Scholar]
- [82].Monier B, Astier M, Semeriva M, Perrin L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;132:5283–5293. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- [83].Monier B, Tevy MF, Perrin L, Capovilla M, Semeriva M. Downstream of homeotic genes. Fly. 2007;1:59–67. doi: 10.4161/fly.3993. [DOI] [PubMed] [Google Scholar]
- [84].Perrin L, Monier B, Ponzielli R, Astier M, Semeriva M. Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev Biol. 2004;272:419–431. doi: 10.1016/j.ydbio.2004.04.036. [DOI] [PubMed] [Google Scholar]
- [85].Ryan KM, Hoshizaki DK, Cripps RM. Homeotic selector genes control the patterning of seven-up expressing cells in the Drosophila dorsal vessel. Mech Dev. 2005;122:1023–1033. doi: 10.1016/j.mod.2005.04.007. [DOI] [PubMed] [Google Scholar]
- [86].Ryan KM, Hendren JD, Helander LA, Cripps RM. The NK homeodomain transcription factor Tinman is a direct activator of seven-up in the Drosophila dorsal vessel. Dev Biol. 2007;302:694–702. doi: 10.1016/j.ydbio.2006.10.025. [DOI] [PubMed] [Google Scholar]
- [87].Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech Dev. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- [88].Gajewski K, Choi CY, Kim Y, Schulz RA. Genetically distinct cardial cells within the Drosophila heart. Genesis. 2000;28:36–43. doi: 10.1002/1526-968x(200009)28:1<36::aid-gene50>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- [89].Akasaka T, Klinedinst S, Ocorr K, Bustamante EL, Kim SK, Bodmer R. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc Natl Acad Sci U S A. 2006;103:11999–12004. doi: 10.1073/pnas.0603098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Hendren JD, Shah AP, Arguelles AM, Cripps RM. Cardiac expression of the Drosophila Sulphonylurea receptor gene is regulated by an intron enhancer dependent upon the NK homeodomain factor Tinman. Mech Dev. 2007;124:416–426. doi: 10.1016/j.mod.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chartier A, Zaffran S, Astier M, Semeriva M, Gratecos D. Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development. 2002;129:3241–3253. doi: 10.1242/dev.129.13.3241. [DOI] [PubMed] [Google Scholar]
- [92].Wang J, Tao Y, Reim I, Gajewski K, Frasch M, Schulz RA. Expression, regulation, and requirement of the toll transmembrane protein during dorsal vessel formation in Drosophila melanogaster. Mol Cell Biol. 2005;25:4200–4210. doi: 10.1128/MCB.25.10.4200-4210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ikle J, Elwell JA, Bryantsev AL, Cripps RM. Dev Dyn. 2008. Cardiac expression of the Drosophila transglutaminase gene CG7356 is directly controlled by Myocyte enhancer factor-2. in print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088.0081-0088.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Samsonova AA, Niranjan M, Russell S, Brazma A. Prediction of Gene Expression in Embryonic Structures of Drosophila melanogaster. PLoS Comput Biol. 2007;3:e144. doi: 10.1371/journal.pcbi.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- [97].Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- [98].Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- [99].Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci U S A. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kelly KK, Meadows SM, Cripps RM. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002;110:39–50. doi: 10.1016/s0925-4773(01)00586-x. [DOI] [PubMed] [Google Scholar]
- [101].Stronach BE, Renfranz PJ, Lilly B, Beckerle MC. Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol Biol Cell. 1999;10:2329–2342. doi: 10.1091/mbc.10.7.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Marin MC, Rodriguez JR, Ferrus A. Transcription of Drosophila troponin I gene is regulated by two conserved, functionally identical, synergistic elements. Mol Biol Cell. 2004;15:1185–1196. doi: 10.1091/mbc.E03-09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- [104].Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Tanaka KK, Bryantsev AL, Cripps RM. Myocyte enhancer factor 2 and chorion factor 2 collaborate in activation of the myogenic program in Drosophila. Mol Cell Biol. 2008;28:1616–1629. doi: 10.1128/MCB.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kremser T, Gajewski K, Schulz RA, Renkawitz-Pohl R. Tinman regulates the transcription of the beta3 tubulin gene (betaTub60D) in the dorsal vessel of Drosophila. Dev Biol. 1999;216:327–339. doi: 10.1006/dbio.1999.9425. [DOI] [PubMed] [Google Scholar]
- [107].Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M. Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development. 2006;133:4073–4083. doi: 10.1242/dev.02586. [DOI] [PubMed] [Google Scholar]
- [108].Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].van Rooij E, Olson EN. microRNAs put their signatures on the heart. Physiol Genomics. 2007;31:365–366. doi: 10.1152/physiolgenomics.00206.2007. [DOI] [PubMed] [Google Scholar]
- [110].Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]