SUMMARY
Early in the development of animal embryos, superficial cells of the blastula form a distinct lineage and adopt an epithelial morphology. In different animals, the fate of these primary superficial epithelial (PSE) cells varies, and it is unclear whether pathways governing segregation of blastomeres into the PSE lineage are conserved. Mutations in the gene encoding Interferon Regulatory Factor 6 (IRF6) are associated with syndromic and non-syndromic forms of cleft lip and palate, consistent with a role for Irf6 in development of oral epithelia, and mouse Irf6 targeted null mutant embryos display abnormal differentiation of oral epithelia and skin. In Danio rerio (zebrafish) and Xenopus laevis (African clawed frog) embryos, zygotic irf6 transcripts are present in many epithelial tissues including the presumptive PSE cells and maternal irf6 transcripts are present throughout all cells at the blastula stage. Injection of antisense oligonucleotides with ability to disrupt translation of irf6 transcripts caused little or no effect on development. By contrast, injection of RNA encoding a putative dominant negative Irf6 caused epiboly arrest, loss of gene expression characteristic of the EVL, and rupture of the embryo at late gastrula stage. The dominant negative Irf6 disrupted EVL gene expression in a cell autonomous fashion. These results suggest Irf6 translated in the oocyte or unfertilized egg suffices for early development. Supporting the importance of maternal Irf6, we show that depletion of maternal irf6 transcripts in X. laevis embryos leads to gastrulation defects and rupture of the superficial epithelium. These experiments reveal a conserved role for maternally-encoded Irf6 in differentiation of a simple epithelium in X. laevis and D. rerio. This epithelium constitutes a novel model tissue in which to explore the Irf6 regulatory pathway.
Keywords: Interferon Regulatory Factor 6, enveloping layer, superficial epithelium, gastrulation, epiboly, trophectoderm, keratin
INTRODUCTION
One of the first examples of cell diversification during embryogenesis occurs during the blastula stage, when superficial cells adopt an epithelial morphology and become a distinct lineage. These cells form a tight, simple epithelium around the embryo that serves to protect it, to regulate osmotic balance, and to provide mechanical support necessary for early morphogenesis (Regen and Steinhardt, 1986; Zalik et al., 1999). In D. rerio this layer is referred to as the enveloping layer (EVL), in X. laevis as the primary or superficial epithelium (SE), and in mammals as trophectoderm (TE). We refer to this layer in general as the primary superficial epithelium (PSE). In each of these taxa, PSE layers form a distinct lineage, but the fate of PSE cells varies among species. In D. rerio the sole fate of the EVL is to become periderm (Kimmel et al., 1990), an embryonic skin that is shed as the underlying epidermis matures (shown in trout) (Bouvet, 1976). In X. laevis, ventral PSE cells contribute to the superficial layer of the skin that may also eventually be shed (Chalmers et al., 2006), while dorsal PSE cells are incorporated into the neural tube as secondary neuronal precursors (Hartenstein, 1989). In mammals the PSE contributes to the placenta and other extra-embryonic membranes, which are structures that do not form in D. rerio and X. laevis. Thus PSE cells in different species share the general features of having a simple epithelial architecture and of segregating early from other lineages, but yet have distinct fates.
Because PSE cells adopt different fates in different species, it is unclear whether mechanisms governing segregation of the PSE lineage from underlying blastomeres is conserved among animals. In X. laevis, cell division planes predict which daughters of a PSE cell will remain in the PSE and which will join deeper layers of ectoderm, arguing that unequal segregation of cell components contributes to the choice between PSE and deep cell fates (Chalmers et al., 2003). The polarity of X. laevis SE cells is regulated by atypical Protein Kinase C (aPKC), partition defective 1 (PAR1), Lethal giant larvae and Scribble, all members of the widely-conserved PAR/aPKC polarity pathway (Chalmers et al., 2005; Dollar et al., 2005; Suzuki and Ohno, 2006). aPKC is localized to the egg cortex and later to the apical side of PSE cells (Chalmers et al., 2003; Nakaya et al., 2000). Forced expression of aPKC can drive deep cells to express markers of the PSE fate, which suggests inheritance of aPKC is a determinant of PSE fate (Ossipova et al., 2007). PAR1 is basally localized in PSE cells and likely serves as a determinant of the deep blastomere fate (Ossipova et al., 2007). In mouse embryos, after an initial period where cells can change fate after rearrangement, cells become polarized, and, as in X. laevis, apically-localized aPKC appears to promote the trophectoderm fate (Johnson and McConnell, 2004). In D. rerio it is unknown whether the aPKC/PAR pathway contributes to commitment of cells to the PSE. However, lineage studies revealed that after about 4 hpf, deep layer blastomeres do not intercalate into the PSE, and conversely daughters of EVL cells remain within the EVL (Kimmel et al., 1990). Transplant studies indicated that PSE cells become committed to an epithelial morphology at around 5 hours post fertilization (hpf) because they will do so even when transplanted into the deep layer (DEL) of blastomeres (Ho, 1992). When EVL and DEL of animal pole explants are separated, DEL cells do not gain expression of EVL markers, or do so at a very low level (Sagerstrom et al., 2005). Together these findings are consistent with the possibility that inheritance of egg membrane components is required to specify PSE fate in D. rerio.
The transcription factors that act downstream of aPKC, or other inherited determinants, to promote PSE development have not been well characterized. A candidate transcription factor to regulate PSE development is Interferon Regulatory Factor 6 (Irf6) because it is implicated in differentiation of several other epithelial tissues. IRFs are a family of transcription factors with a shared structure including an amino-terminal DNA binding domain and a carboxy-terminal domain that mediates homo and heterodimerization called the Interferon Association Domain (IAD). For most IRF family members, a pathogenic stimulus leads to phosphorylation of the IAD, stimulating nuclear translocation, dimerization, and activation of transcription from target genes including interferon genes (reviewed in Paun and Pitha, 2007). IRF6 by contrast plays no role in innate immunity but is essential for embryonic development. Genetic variation in IRF6 is associated with syndromic and non-syndromic forms of cleft lip and palate (Kondo et al., 2002a; Zucchero et al., 2004), possibly reflecting a role in the epithelia covering palatal shelves, where it is expressed at high levels (Knight et al., 2006; Kondo et al., 2002b). Mouse embryos homozygous for targeted mutations in Irf6 display defects in differentiation of keratinocytes, and exhibit adhesions between the tongue and maxillary prominence, suggesting a defect in differentiation of oral epithelia (Ingraham et al., 2006; Richardson et al., 2006). IRF6 is expressed in breast epithelial cells where it may regulate the decision to divide vs. differentiate (Bailey et al., 2008a; Bailey et al., 2005). However it is unknown whether Irf6 function in epithelial development extends to the PSE.
Here we report that in D. rerio and X. laevis embryos at blastula stage, irf6 mRNA is present in both deep and superficial layers of blastomeres, and that interfering with maternal Irf6 causes epiboly arrest concomitant with a gross failure of PSE development. Thus, expressing a dominant negative variant of Irf6 in D. rerio, or depleting maternal irf6 transcripts in X. laevis embryos, causes PSE cells to lose characteristic gene expression. We show that the requirement for Irf6 for PSE cell differentiation is cell autonomous. Overall, these data demonstrate a critical role for Irf6 in simple epithelia, and underscore the utility of studying PSE as a model epithelium to explore the Irf6 regulatory pathway.
MATERIALS AND METHODS
Danio rerio maintenance
Breeding D. rerio were maintained as described (Westerfield, 1993) in the University of Iowa Animal Care Facility. Embryos were staged according to Kimmel et al. at 28.5°C by hours or days post fertilization (hpf or dpf) (Kimmel et al., 1995).
Isolation of irf6 clones and plasmid construction
The D. rerio cDNA clone for an EST corresponding to irf6 (GenBank accession number AW422721. Full length cDNA, AY725802, Ben et al., 2005) was purchased from Research Genetics (Invitrogen). Bi-directional DNA sequencing was performed across the entire cDNA insert using gene specific primers. Plasmids for mRNA production were prepared as follows: irf6 cDNA corresponding to the full open reading frame (Irf6ORF, for rescue experiments), or to only the DNA binding domain, i.e. amino acids 1-115, (for construction of Irf6DBD), was inserted into the pCS2+ vector or the pCS2+MT vector, respectively. For use in testing the efficacy of translation blocking MO, irf6 cDNA corresponding to most of the 5’ untranslated region (excluding the initial 85 bases of 279 bases total in order to remove two start codons in an alternative open reading frame) plus the irf6 open reading frame were inserted in the C2+MT vector, yielding a transcript that encodes Irf6 with 6 Myc epitopes on the carboxy terminus. Two X. laevis irf6 orthologues have been identified which we refer to as irf6.1 (previously called irf-6, GenBank accession number D86492, Hatada et al., 1997), and irf6.2 (GenBank accession number BC071111, Klein et al., 2002). An irf6.2 plasmid was obtained commercially (Open Biosystems, Huntsville, AL) and verified by sequencing. The coding sequence was amplified by PCR, cloned into pCR8-GW-TOPO and sub-cloned into a custom Gateway-compatible (Invitrogen) pCS2+ vector for expression studies.
Morpholino and mRNA injections – Danio
Three translation blocking morpholinos (MO), targeting the start codon or the 5’ untranslated region (5’ UTR) of D. rerio irf6 (ZGC Clone BC056772.1, zgc.nci.nih.gov) and two splice-blocking morpholinos, targeting splice sites were ordered (Gene Tools, Philomath, OR) (Splice sites from Ensembl gene ENSDARG00000043296). Sequence of MO as follows: irf6 AUG: ATGAGACGACATCACACCGGCTGAG; irf6 5’utr: GGCTCAAAGCAGGAAAAGGCAGCAC; irf6 5’utr-b: GCCTTTGCCGCCAAAACGAACGGTT; irf6 exon 2/intron 2 (e2/i2): TTCTGACCTTAAAGATGGTGTTCTC; irf6 intron 3/ exon 4 (i3/e4): GAACTGAACCTGAAGTAGAGCATTA. To test the ability of irf6 splice-blocking MO to disrupt normal splicing of irf6 mRNA, RNA was collected with Trizol, following manufacturers instructions (Invitrogen), at 20 hpf from embryos injected at the 2-cell stage with 5 ng of MO. First strand cDNA was generated with random hexamers as described previously (O’Brien et al., 2004). To check efficacy of e2i2 MO by PCR, a forward PCR primer in exon 1 and a reverse primer in exon 3 were used, for i3/e4 MO, primers in exon 1 and exon 5 were used. An MO targeting p53 and the standard negative control MO from Gene Tools were injected at 5 ng/embryo. MO were diluted to 5 mg/ml in Danieau solution (Nasevicius and Ekker, 2000) for storage at -20°C and diluted before use to 1 mg/ml in 0.2 M KCl. Embryos were injected with MO into the yolk beneath the blastomeres at the 1-4 cell stage. Template DNA was prepared by linearization and cleaned of histones by Proteinase K digestion followed by phenol extraction. Capped RNA synthesis, using SP6, was carried out using the mMessage mMachine (Ambion, Austin, TX) transcription. RNA was recovered with Microcon microconcentrators (Millipore, Billerica, MA).
In Danio experiments, mRNA was injected into the cytoplasm at the 1-cell stage for global injections, into the cytoplasm of a single cell at the 16-32-cell stage for mosaic injections, and into the yolk after sphere stage (4 hpf) for YSL injections. Sytox green (Invitrogen) was co-injected in YSL injections for labeling yolk syncytial nuclei. The high dose, used in all experiments herein except as noted was 3 ng mRNA/embryo in global injections. Low dose, total of 1.2 ng RNA/embryo. These RNA doses are about 10 times higher than those required to observe specific phenotypes with another dominant negative transcription factor in D. rerio (dlx3, Kaji and Artinger, 2004), although in the latter case, a constitutive repressor domain was fused to the DNA binding domain, which may have improved its efficacy as a dominant negative. Importantly, embryos injected with 3 ng of control RNAs encoding GFP or beta galactosidase developed normally.
Histology
An anti-cMyc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used in whole-mount immunohistochemistry as previously described (O’Brien et al., 2004). This antibody was also used in combination with DIG-labeled zfk8 RNA probes in whole mount double labeling as previously described (Cornell and Eisen, 2000). Cell death by apoptosis was monitored by terminal transferase dUTP nick-end labeling in whole embryos as previously described (Li and Cornell, 2007). To label polymerized actin, embryos were fixed at the shield stage for 48 hours in 4% PFA, rinsed in PBS + 0.1% Tween 20 (PBST), incubated for 1 hour in 0.165 M Phalloidin Alexa 546 (U. Iowa Central Microscopy Research Facility, Iowa City, IA), washed with PBST, and visualized with a Texas Red filter. Sytox green (Invitrogen) was used to label yolk syncytial nuclei and was injected into the yolk of live embryos after the sphere stage (4 hpf) as described (D’Amico and Cooper, 2001). Alcian green was used to label cartilage in 4 dpf embryos that were fixed in 4% PFA for 24 hours as described (Kimmel et al., 1998). For thick sections, embryos fixed in 4% PFA were embedded in 1.5% agar, soaked in 30% sucrose overnight, and sectioned on a Jung Frigocut 2800 cryostat (Leica, Wetzlar, Germany) to a thickness of 14-16 microns.
Probes for in situ hybridization were generated for irf6, cebpb and gsc genes by RT-PCR (Primer sequences are available upon request). Fragments were inserted into the pCR4TOPO vector (Invitrogen). Plasmids were linearized, and DIG-labeled antisense RNA probes transcribed (Roche Diagnostics, Mannheim, Germany) as follows: irf6, (PstI/T7); cebpb, (NotI/T3); gsc, (NotI/T3). Alternatively, probes were linearized and transcribed from existing plasmids as follows: tfap2a (O’Brien et al., 2004), NotI/T7; zfk8 (Imboden et al., 1997), Asp718/SP6; gata2 (Detrich et al., 1995), XbaI/SP6; ntl (Schulte-Merker et al., 1994), SalI/T7; sox2 (Li and Cornell, 2007); krt18 (Pei et al., 2007), NotI/SP6; k4, (Pei et al., 2007), Not1/SP6.
Immunostaining and whole-mount in situ hybridization-Xenopus
Embryos for immunostaining were devitellined and fixed in methanol at -20°C. Samples were embedded, sectioned and immunostained with mAb LP3K (a gift from Birgitte Lane, see Lane et al., 1985) as described (Godsave et al., 1984). Secondary antibodies were Alexa-488 conjugated goat anti-mouse antibodies (Invitrogen/Molecular Probes). Azan staining was done as previously described (Houston and King, 2000).
Whole-mount in situ hybridization was performed essentially as described (Sive et al., 2000), with replacement of the triethanolamine and acetic anhydride washes by a single 20 minute wash in 0.1% active DEPC in PBS (after Braissant and Wahli, 1998). The probe for intelectin 2 (XL020e14) was identified through the work of Hayes et al. (2007) and obtained through the NIBB/NIG/NBRP Xenopus laevis EST project (Hayes et al., 2007). Antisense probes were made by linearizing plasmid templates with EcoRI, followed by transcription with T7 RNA polymerase.
In situ hybridization on paraffin sections was performed essentially as described (Butler et al., 2001). Irf6 antisense probes were prepared from a full-length clone in pCMVSPORT6 by linearizing with SalI and transcribing with T7.
Microscopy -Danio
D. rerio images were documented on a DMRA 2 compound microscope or a MZ FLIII stereo-microscope (Leica, Wetzlar, Germany). Images were captured with a Retiga 1300 camera (Q-Imaging, Burnaby, Canada) and Openlab software (Improvision, Lexington, MA). Labeling and contrast adjustments were done in Photoshop 7.0 (Adobe, San Jose, CA).
Xenopus oocytes, embryos and explants
Ovary was isolated from mature Xenopus laevis females, subdivided into small segments, and stored at 18°C in oocyte culture medium (OCM; 70% L-15, 0.04% BSA, 1 mM Glutamax (Invitrogen), 1.0 μg/ml gentimicin, pH 7.6-7.8), modified from Heasman et al (1991). Oocytes were isolated by manual defolliculation and cultured in OCM and oocyte maturation was induced by the addition of 2.0 μM progesterone. Ovulated eggs from females induced with human chorionic gonadotropin (hCG) were fertilized in vitro using a sperm suspension and placed in 0.1× MMR (1× MMR: 0.1 M NaCl, 1.8 mM KCl, 2.0 mM CaCl2, 1.0 mM MgCl2, 15.0 mM HEPES, pH 7.6). Jelly coats were removed in cysteine (2% in 0.1× MMR, pH 7.8). For isolation of superficial and deep layers, animal caps were dissected at stage 9 in calcium-magnesium-free MMR, transferred to 67 mM phosphate buffer (pH 7.2) for 3 minutes. The caps were then transferred to fresh calcium-magnesium-free MMR + 0.1% BSA and incubated for 10 minutes, after which the superficial layer was gently teased away from deep cells using a tungsten needle. Blastomeres in the deep cell layer became loosely adhering, but were never fully disaggregated during the procedure. The layers were cultured separately in OCM, which contains calcium, until the desired stage.
Antisense oligos and host-transfer -Xenopus
An MO was designed to complement the Xenopus laevis irf6 mRNA 5’ UTR and start codon. This sequence is expected to target both identified irf6 orthologues. Although there is a one-base mismatch with irf6.2, this difference is not predicted to alter the binding of the MO. (irf6-MO; 5’-GTGCATAGCCATGACCTCTGCCTGG -3’, Gene-Tools). The predicted start codon is underlined. Morpholinos were dissolved in nuclease-free water to 1 mM and stored at -80°C. Manually defolliculated stage VI oocytes were injected animally with 24 ng of irf6-MO and maintained in OCM at 18°C for 24-48 hours prior to maturation with progesterone. Oocytes were stimulated to mature approximately 10-12 hours prior to implantation into laying host females (Heasman et al., 1991). Oocytes were injected while in OCM; embryos were injected in Ficoll solution (2% Ficoll, 0.5× MMR, pH 7.8).
RT-PCR -Xenopus
Embryos and explants for RT-PCR analysis were frozen on dry ice and stored at -80°C. Samples were homogenized in 200 μl of RNA lysis buffer per embryo, DNase I treated, subjected to RT-PCR and analyzed either by gel or by quantitative PCR, essentially as described (Houston and King, 2000; Houston and Wylie, 2005). Real time PCR was performed on a LightCycler 480 machine (Roche Applied Science). Relative expression values for quantitative RT-PCR were determined by comparison to a standard curve of serially diluted cDNA from uninjected controls, followed by normalization to ornithine decarboxylase (odc). The data are shown as representative individual experiments using unreplicated samples, but were repeated at least twice with similar results. Primer sequences and detailed protocols are available upon request.
Western blots
60 embryos at the 1-2 cell stage were injected with 1 ng of irf6mt RNA. At the 2-4 cell stage, 20 of the previously RNA injected embryos were injected a second time with approximately 5 ng of irf6 AUG MO, and 20 with approximately 6 ng of irf6 5’ UTR MO. At the shield stage (6hpf) embryos were frozen at -80°C. Frozen embryos were thawed and diluted in 5 ml loading buffer per embryo. Three embryo equivalents were run on a pre-cast 10% Tris HCL protein gel (Bio-Rad Laboratories) for 105 minutes at 100V. The protein was transferred to a nitrocellulose membrane for 1 hour at 400mA. The membrane was blocked in milk blocking buffer for 1 hour and then probed with mouse anti-Myc antibody (9E10, Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:1500 in blocking buffer (Phosphate buffered solution (PBS), 0.15 molar NaCl, 0.1% Tween 20, 0.04 g/ml Dried Milk) overnight at 4 degrees. The membrane was washed in blocking buffer and then probed with anti-mouse IR800 secondary antibody (Rockland Immunochemicals, Gilbertsville, PA) diluted 1:10,000 in blocking buffer for 1 hour at RT. The membrane was washed with blocking buffer and then PBS and imaged with the Odyssey system (LI-COR Biosciences, Lincoln, NE).
RESULTS
D. rerio irf6 expression
To investigate the function of Irf6 in D. rerio, we searched D. rerio cDNA and genomic DNA sequence databases and detected a single orthologue of mammalian IRF6. By whole mount in situ hybridization we observed irf6 expression in cleavage-stage embryos (Fig. 1A) and confirmed the presence of maternal irf6 message with semi-quantitative PCR on cDNA generated from fertilized eggs (data not shown). Sections of 6 hpf embryos revealed diffuse irf6 transcripts in cells of the deep layer, and higher level expression in EVL and dorsal forerunner cells (DFC) (Fig. 1C). Diffuse expression remained throughout the embryo at 13 hpf but was no longer detectable by 24 hpf (Fig. 1D-F). Starting at 13 hpf, irf6 expression was observed in many epithelial structures including, nasal and otic placodes, semi-circular canals, and oral epithelium (Fig. 1D-J). Expression of irf6 was not detected in embryonic skin (Fig. 1G). Our independent analysis of D. rerio irf6 expression matches an earlier published report (Ben et al., 2005).
Figure 1. Danio rerio irf6 expression by in situ hybridization.
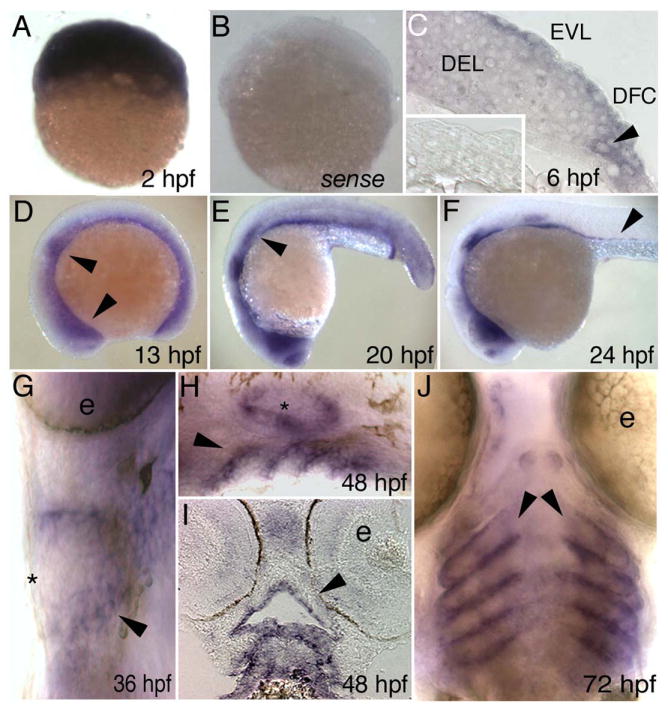
Lateral views, except as indicated, of D. rerio embryos fixed at the indicated stage and processed with an antisense irf6 probe, except B and inset in C. A) A cleavage stage embryo, revealing high-level maternal expression. B) A cleavage stage embryo, processed with an irf6 sense probe, revealing very little background staining. C) Section at shield stage showing low-level expression in deep cells (DEL) and higher level expression in EVL and dorsal frontrunner cells (DFC). Inset, section of a shield stage embryo processed with a sense probe. D) Diffuse expression throughout the embryo, with increased expression in the nasal and otic placodes (arrowheads). Head is to the left in panels D-F. E) Decreasing diffuse expression, expression present in nasal and otic placodes and lateral line primordium (arrowhead). F) Expression as in E but now including putative gut precursor cells (arrowhead). G) Ventral view, showing expression in pharyngeal pouches (arrowhead). Expression is not detectable in skin at this stage (asterisk) (e, eye). H) Expression in pharyngeal pouches (arrowhead) and ear (asterisk). I) Horizontal section, showing expression in oral epithelium (arrowhead). J) Ventral view, expression in pharyngeal pouches (arrowheads). (e, eye).
A putative dominant negative Irf6 disrupts gastrulation
To examine the role of zygotically-expressed Irf6 in early D. rerio development we injected embryos with a standard dose (5 ng, Nasevicius and Ekker, 2000) of antisense morpholino oligonucleotides (MO) targeting one or another of two splice sites of the irf6 transcript. We harvested RNA from MO-injected embryos at 22 hours post fertilization (hpf), synthesized first-strand cDNA, and performed PCR with primers flanking the targeted splice site. Gel analysis followed by sequencing of the MO-dependent PCR products revealed that both MO altered splicing of a fraction of irf6 RNA in a manner that would destroy the encoded protein (Fig 2A and not shown). Because of defects in skin and craniofacial defects in mouse Irf6 mutants (Ingraham et al., 2006; Richardson et al., 2006), we anticipated similar problems in irf6 MO-injected D. rerio embryos. However in MO-injected embryos the skin morphology at 2 days post fertilization (dpf) and craniofacial cartilage at 4 dpf, and the overall morphology of the embryos, were grossly normal (n >100 embryos). Embryos injected with high doses of these MO (20 ng/embryo) still developed grossly normally, without defects in skin, pectoral fins, or craniofacial cartilage (n > 100 embryos). A possible explanation for these unexpected findings is the existence an Irf6 homologue that can compensate for the reduction of zygotic Irf6 expression; alternatively, maternally-encoded Irf6 protein serves this function.
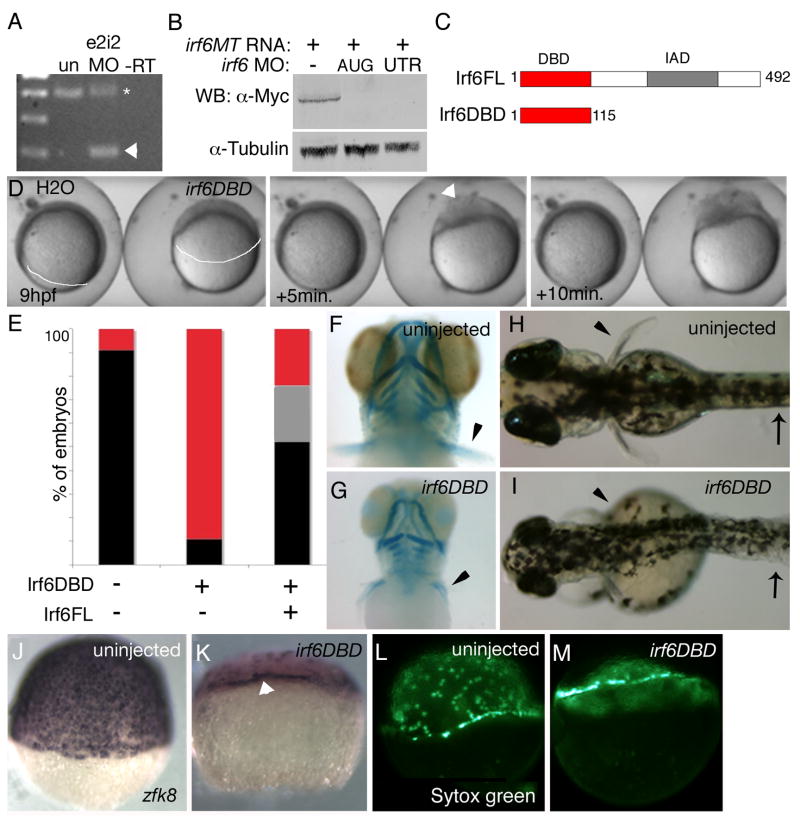
Figure 2. In D. rerio, expression of a putative dominant negative Irf6 causes severe gastrulation defects and late defects in the pectoral fin and skin.
A) RT-PCR analysis of efficacy of splice blocker MO targeting D. rerio irf6. RNA was harvested from 22 hpf embryos that were either uninjected or injected at the single cell stage with 5 ng of irf6 exon 2/intron 2 MO. First strand cDNA was synthesized and PCR carried out with forward primer in exon 1 and reverse primer in exon 3, yielding, from uninjected embryos (un), a single band of expected size, and from MO-injected embryo a second, smaller band (arrowhead). The MO-dependent PCR product was excised and sequenced and found to reflect loss of the first coding exon, which encodes the first 60 amino acids, including half of the predicted DNA binding domain. In e2i2 MO injected embryos, a substantial fraction of irf6 mRNA, possibly corresponding to maternal stores, is spliced correctly (Asterisk). -RT, negative control lacking reverse transcriptase B) Western blot analysis of efficacy of MO targeting translation of Irf6. All lanes show lysates from 16 hpf embryos injected at the two-cell stage with in vitro transcribed irf6 RNA altered to include most of the 5’ UTR and to encode full length Irf6 with a carboxy-terminal Myc tag. -, embryos not injected with MO. AUG, RNA-injected embryos subsequently with 5 ng of irf6 AUG MO; 5’UTR, RNA-injected embryos subsequently with 5 ng of irf6 5’UTRb MO. In a similar Western blot experiment, embryos injected with 20 ng of a 5-base pair mismatch variant of the AUG MO were comparable to embryos not injected with MO (not shown). C) Schematics of full-length Irf6 and the DNA binding domain variant, which contains the amino terminal 115 amino acids. IAD, Interferon Association Domain (Taniguchi et al., 2001). D) Frames from a time-lapse video of a water-injected control, left, and an irf6DBD-injected embryo, right, taken at the indicated times. In the latter, thinning of the DEL is delayed (most obvious at the animal pole), as is vegetal migration of the DEL (white line, extent of DEL migration). Finally, when siblings are at about 80% epiboly (9 h), the irf6DBD-injected embryo ruptures near the animal pole. Arrowhead indicates first area of embryo rupture. Staging was analyzed in 10 embryos in three separate experiments with equivalent results. E) Histogram showing percentage of embryos at 24 hpf of the indicated class that were normal (black), or dead (red), or with head and tail defects (grey). F,G) Ventral views of 4 dpf embryos stained with alcian-green. F) Uninjected embryo. G) irf6DBD-injected embryo showing pharyngeal cartilage elements are present but smaller than normal and pectoral fins (arrowheads) are reduced. H,I) Dorsal views of live, H) uninjected embryo and, I) irf6DBD-injected embryos at 4 dpf. The later shows blistered skin (arrows) and abnormal fins (arrowheads). J-M) Lateral views of, J,L) uninjected and, K,M) irf6DBD-injected embryos at shield stage. J,K) Embryos processed to reveal expression of zfk8, a marker of the EVL. In the irf6DBD-injected embryo, the vegetal limit of the EVL (arrowhead) is at the position appropriate for dome stage. L,M) YSL nuclei labeled with sytox green, revealing that in, M) the irf6DBD-injected embryo vegetal migration of the YSL has stalled at dome stage.
To attempt to block translation of maternal irf6 transcripts, we ordered an MO complementary to the start codon of irf6 mRNA (irf6 AUG MO). To test efficacy of this MO, we injected it into embryos previously injected with RNA containing the irf6 5’UTR and encoding a carboxy-terminal Myc-tagged variant of Irf6 (i.e., irf6MT RNA). Injection of 5 ng or more of this MO efficiently blocked translation of the irf6MT RNA as monitored by Western analysis, while injection of 20 ng of a negative control MO did not block translation (Fig 2B and not shown). Surprisingly, embryos injected with 5 ng of irf6 AUG MO developed grossly normally (not shown). To confirm these findings, we ordered two additional irf6 MO, complementary to the 5’ UTR or start codon, and found they gave comparable results to the irf6 AUG MO (not shown). Of note, it is possible that irf6 MO do not effectively block maternal Irf6 expression because Irf6 protein is translated in the oocyte, as has been proposed to explain the limited effectiveness of MO targeting other maternal transcripts (Bruce et al., 2003).
We reasoned that the function of maternally-expressed Irf6 might be blocked by injection of RNA encoding a dominant inhibitory Irf6 protein. To create such a reagent, we engineered D. rerio irf6 cDNA to encode solely the amino-terminal 115 amino acids, containing the presumed DNA binding domain (Irf6DBD) (Fig. 2C, Kondo et al., 2002a). Preliminary gel shift experiments with human IRF6DBD indicated that it indeed binds a subset of previously identified Interferon Stimulated Regulatory Elements (BCS, unpublished findings). Injection of a high dose (3 ng) of irf6DBD mRNA into 1-cell stage embryos led to death, with embryonic disintegration, by 90% epiboly (Fig. 2D) (82% affected, n=55). Importantly, co-injection of 3 ng of irf6 mRNA (irf6ORF) was able to partially rescue the phenotype of death at gastrulation, arguing for specificity of Irf6DBD (Fig. 2E, 24% affected, n=70). Embryos injected with 3 ng of irf6ORF RNA alone underwent gastrulation normally (not shown). A fraction of embryos injected with a lower dose of irf6DBD mRNA survived gastrulation and at 3 days post fertilization (dpf) displayed short pectoral fins and blistered skin (Fig. 2G) (19%, n=36). In these embryos, cartilage elements of the craniofacial skeleton are present but reduced in size and variably disorganized (Fig. 2H and not shown). Of note, these phenotypes are reminiscent of the truncated forelimbs, abnormal skin, and disorganized craniofacial skeleton of mice deficient for Irf6 (Ingraham et al., 2006; Richardson et al., 2006). These results suggest that maternally-expressed Irf6 is necessary during epiboly in D. rerio embryos. However it is possible that the relevant Irf6-type activity is instead provided by an Irf6 homologue whose activity is inhibited by Irf6DBD. We provide evidence in favor of the first model later in this work.
Irf6 is required for epiboly of all three cell layers
In D. rerio, gastrulation movements include epiboly, internalization, and convergence and extension (Solnica-Krezel, 2006). During epiboly the three cell layers of D. rerio blastula, i.e. the deep layer (DEL), the enveloping layer (EVL), and the yolk syncytial layer (YSL), undergo characteristic behaviors, and the motions of these layers can be affected differentially in mutants. For instance, in the ecad mutant epiboly of the DEL is primarily affected (Kane et al., 1996; McFarland et al., 2005), while in poky mutants, epiboly of all three layers is slower than normal (Wagner et al., 2004) (molecular characterization of the poky locus has not yet been reported). We sought to assess which layers were affected in irf6DBD- injected embryos. In control embryos at 80% epiboly stage (8.3 hpf), the DEL thinned to just a few layers thick (Fig. 2D and Supplemental movie) (behavior of DEL cells during epiboly described in Kane et al., 2005). However, in irf6DBD-injected embryos from the same clutch, thinning of the DEL stopped at approximately dome stage (4.3 hpf) (Fig. 2D panels and Supplemental movie). The upward doming of the yolk, caused by YSL migration, was profoundly delayed and asymmetric in irf6DBD-injected animals (Fig. 2D and Supplemental movie). To aid in visualization of EVL cells, we stained embryos to reveal expression of a keratin 8 homologue, zfk8 (Imboden et al., 1997). In irf6DBD-injected embryos the number of zfk8 expressing cells is highly reduced (discussed below), but the leading edge of the EVL is still discernible and does not extend past the position found in control embryos at about sphere stage (4.3 hpf) (Fig. 2 J,K). To monitor YSL migration we labeled YSL nuclei by injecting SYTOX green into the yolk (D’Amico and Cooper, 2001). When uninjected sibling embryos were at 75% epiboly stage, YSL nuclei in irf6DBD-injected embryos remained at the margin adjacent to the DEL cells (88% affected, n=16) (Fig. 2 L,M). Ultimately, when control embryos are at approximately 90% epiboly, irf6DBD-injected embryos rupture near the animal pole and cells spill out (82% affected, n=55) (Fig. 2D and Supplemental movie). Thus, morphogenesis of all three cell layers is affected in irf6DBD-injected embryos, similar to the maternal effect mutant poky (Wagner et al., 2004).
Markers of EVL are lost in irf6DBD-injected embryos
The rupture of irf6DBD-injected embryos during epiboly suggests a failure of EVL integrity. To explore this further, we examined expression of zfk8 (Imboden et al., 1997), a keratin18 orthologue (krt18) (Pei et al., 2007), a keratin 4 orthologue (k4) (Pei et al., 2007), and CCAAT/enhancer binding protein, beta (cebpb) (Lyons et al., 2001). These genes are strongly expressed in EVL of uninjected control embryos at 50% epiboly (Fig. 3A,C,E, and not shown). In irf6DBD-injected embryos at 50% epiboly, the normally continuous expression of these genes becomes patchy and, in strongly affected embryos, is almost absent (Fig. 3B,D,F, and not shown) (for all panels in Fig. 3, ≥ 60% affected, n ≥ 25). EVL cells in control embryos at 50% epiboly are characterized by a cortical ring of filamentous actin (F-actin) revealed by phalloidin stain (Fig. 3G)(Koppen et al., 2006). In high-dose irf6DBD-injected embryos this pattern is lost, with phalloidin stain in superficial cells either diffuse or absent (Fig. 3H).
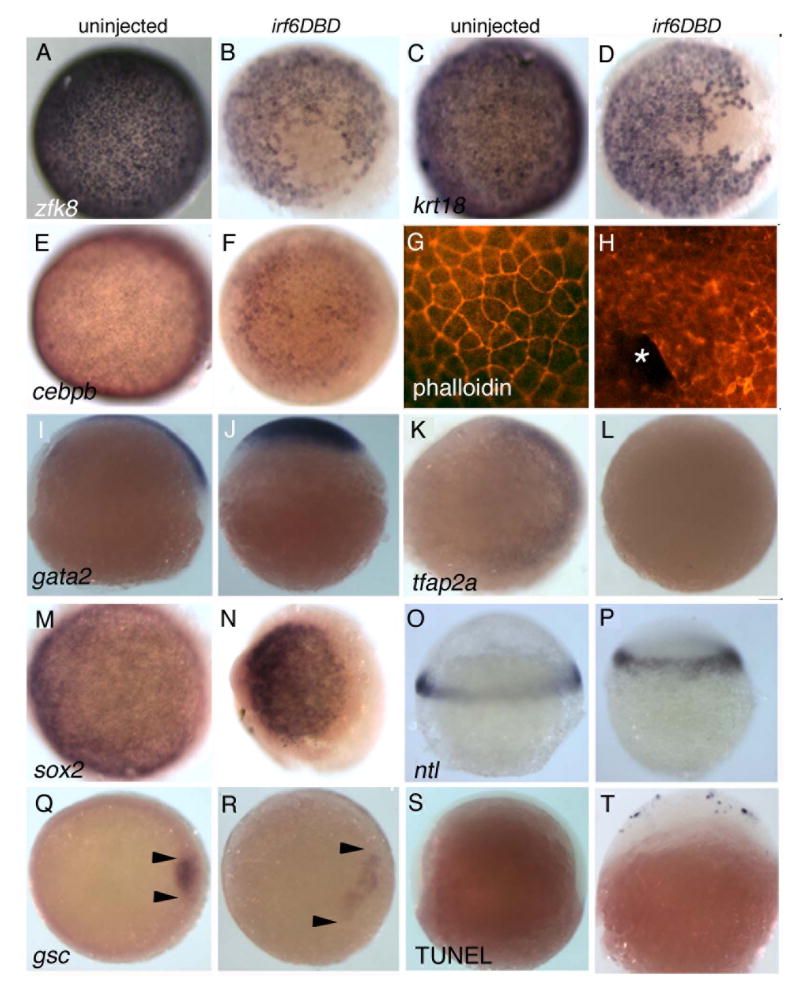
Figure 3. Irf6DBD-mediated changes in gene expression in shield-stage D. rerio embryos.
Dorsal views, except as indicated, of uninjected and irf6DBD-injected embryos, as indicated by the column header, fixed when uninjected sibling embryos reached shield stage. Embryos processed for in situ hybridization to reveal expression of the indicated gene, except as noted. Images portray the phenotype displayed by 60%-80% of embryos in the experimental group, n = 25 embryos or greater. A-F) Expression of zfk8, krt18, cebpb, are expressed in all EVL cells of uninjected embryos but absent in large patches in uninjected embryos. G) Uninjected and, H) irf6DBD-injected embryos stained with phalloidin, revealing polymerized actin (i.e., F-actin). G) In the uninjected embryo, F-actin is visible in a cortical ring within every EVL cell. H) In the irf6DBD-injected embryo, there is loss (*) or disorganization of F-actin in superficial cells. I,J) Lateral views, showing gata2 expression in non-neural epiblast is present but displaced the irf6DBD-injected embryo. K,L) In contrast to gata2, expression of tfap2a, a marker of non-neural epiblast, is highly reduced in the irf6DBD-injected embryo. M,N) sox2 expression, a marker of neural epiblast, is present in the irf6DBD-injected embryo. O,P) Lateral views, showing that ntl expression, a marker of hypoblast, is present in the irf6DBD-injected embryo. Q,R) gsc expression is present the shield hypoblast of the uninjected embryo, and in an expanded arc in the irf6DBD-injected embryo, consistent with developmental delay in the latter (arrowheads show extent of gsc expression arc). S,T) Lateral views of embryos processed to reveal cells undergoing apoptosis by TUNEL. TUNEL-positive cells are absent from this uninjected embryo. In the irf6DBD-injected embryo, many TUNEL-positive cells are visible, primarily within the EVL.
We sought to determine if there was abnormal gene expression in other cell layers. The DEL is composed of epiblast, which will give rise to neural and non-neural ectoderm, and hypoblast, which will give rise to mesoderm and endoderm (Warga and Kimmel, 1990). At 60% epiboly gata2 and tfap2a are expressed in non-neural epiblast (Fig. 3 I,K), and sox2 is expressed in neural epiblast (Fig. 3M). In irf6DBD-injected embryos, when control sibling embryos have reached 60% epiboly, gata2 and sox2 are expressed at high levels on one side of the embryo (Fig. 3 J,N), but tfap2a expression is markedly reduced (Fig. 3L). It possible the differential effect on tfap2a reflects overall delay in irf6DBD-injected embryos, because tfap2a initiates expression later than gata2 (RAC unpublished). Expression of the pan-hypoblast marker ntl is comparable in control and irf6DBD-injected (Fig. 3O,P), implying no defect in hypoblast induction. However, in irf6DBD-injected embryos, expression of gsc in dorsal hypoblast, the shield, was expanded to a larger arc, suggesting reduced convergence movements in the hypoblast (Fig. 3Q,R). Thus in irf6DBD-injected embryos we observed strong reduction in levels of four makers of the EVL, while we detected evidence of developmental delay but not of gross mispatterning of the DEL.
Effect of irf6DBD on cell survival
To determine if epiboly arrest in irf6DBD-injected embryos reflected a role for Irf6 in cell survival, we assessed apoptosis. At 90% epiboly (8.5 hpf), control embryos had few or no TUNEL-positive cells (0% with >5 TUNEL-positive cells, n=55; Fig 3S), while high-dose irf6DBD-injected embryos fixed at the same time had many TUNEL-positive cells (100% with ≥ 5 TUNEL-positive cell, n=45), most prominently in the EVL (Fig. 3T). Because cell death pathways are not active in D. rerio until 7 hpf (Ikegami et al., 1999) but Irf6DBD-injected embryo appear to stall at about 4.3 hpf, we doubted that control of cell survival was the principle function of Irf6. To test this prediction, we co-injected embryos with p53 MO and high-dose irf6DBD mRNA. We processed a portion of these embryos at 90% epiboly and observed that the number of TUNEL-positive cells was highly reduced (0% with ≥ 5 TUNEL-positive cells, n=42). However, the majority of the remaining embryos ruptured at around 90% epiboly (84%, n=40), and had reduced k4 expression (95% n =22). Together these results argue against the possibility that loss of gene expression in the EVL in Irf6DBD results from such cells entering a cell death pathway; instead they suggest Irf6 directly regulates EVL cell differentiation.
Irf6 function in the EVL is cell autonomous
To determine whether loss of gene expression in the EVL of irf6DBD-injected embryos reflected a cell-autonomous requirement of Irf6 in the EVL, we generated cDNA encoding Irf6DBD with a Myc-epitope tag at the C-terminal end (irf6DBD-MT). Embryos injected at the 1-cell stage with irf6DBD-MT mRNA displayed the same phenotype as embryos injected with irf6DBD (data not shown). We injected irf6DBD-MT mRNA into one blastomere of 16-cell stage embryos, fixed the embryos at 70% epiboly, and processed them to reveal zfk8 and anti-Myc immunoreactivity (IR). EVL cells with prominent nuclear anti-Myc IR did not express zfk8, although adjacent cells did (Fig. 4A). These results imply a cell-autonomous function of Irf6 within EVL cells.
Figure 4. The Irf6DBD effect is cell autonomous to the EVL.
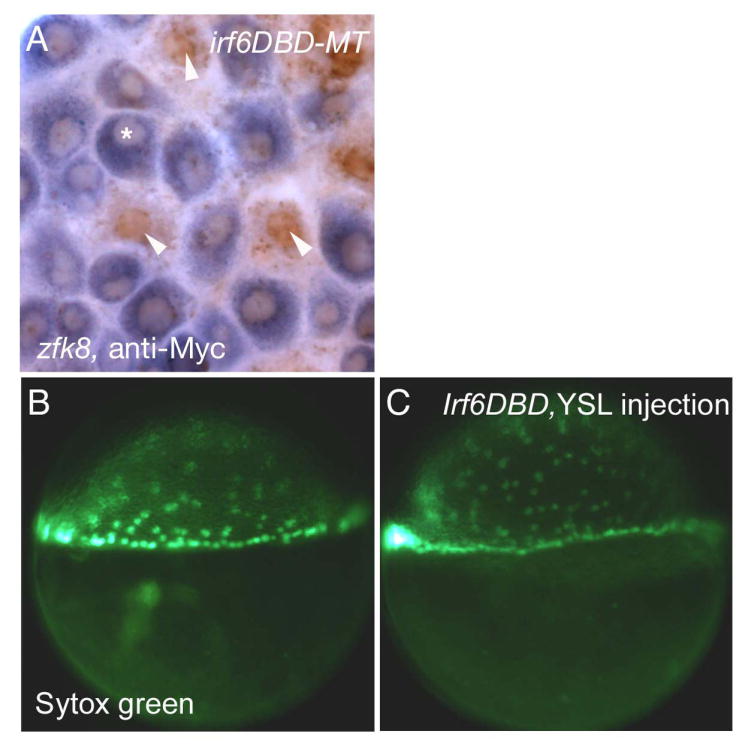
A) Animal pole view of an embryo injected with irf6DBD-MT mRNA into a single blastocyst at 16-32 cell stage, processed at shield stage for zfk8 mRNA (purple) and anti-cMyc IR (brown). Superficial cells which contain anti-Myc IR (arrowheads) have highly reduced zfk8 expression in comparison to cells that do not contain anti-Myc IR (*). (6 cells clearly containing nuclear anti-Myc IR and also clearly in the same plane at the EVL observed in 3 embryos, 0/6 displayed detectable zfk8 signal) B,C) Lateral views of embryos at shield stage which had been injected with B) SYTOX green or C) SYTOX green and irf6DBD into the yolk at sphere stage (4 hpf). There is normal progression of epiboly of yolk syncytial nuclei in both embryos.
Because Irf6DBD blocks epiboly, and epiboly requires the YSL, we tested for a cell-autonomous requirement for Irf6 in the YSL. We injected irf6DBD mRNA and SYTOX green either into 1-cell stage embryos or into the yolk after the sphere stage (4 hpf) (D’Amico and Cooper, 2001). Embryos injected at the 1-cell stage almost all died by 90% epiboly (88%, n=16) as expected; by contrast, the embryos with YSL-targeted injections developed with normal YSL migration and normal morphology (86%, n= 28) (Fig. 4B,C). These findings argue against a cell-autonomous requirement for Irf6 activity in the YSL, although an important caveat is that mRNA injected at sphere stage has less time to be translated prior to epiboly than RNA injected into early blastomeres.
Identification and characterization of Xenopus Irf6 genes
The strong effect of Irf6DBD in D. rerio, along with the lack of effect using irf6 MOs and the presence of maternal irf6 mRNA, all suggested that the Irf6DBD interferes with DNA binding of the maternal pool of Irf6 protein. Alternatively, Irf6DBD might block the activity of one or more Irf6-related proteins. To distinguish these models, we turned to another vertebrate model, Xenopus laevis, where it is possible to deplete maternal stores of mRNA (Heasman et al., 1991). In X. laevis, we identified two irf6 ESTs that share 92% identity at the nucleotide level (irf6.1 and irf6.2) (Hatada et al., 1997; Klein et al., 2002). Although these transcripts may represent different alleles of irf6 or separate paralogues, two irf6 paralogues are present in the sequenced genome of a related species, X. tropicalis (DWH, unpublished observations), providing evidence for the latter possibility. A previous study in X. laevis showed that irf6 (irf6.1) was maternally expressed, with later expression surrounding the blastopore and in the tailbud blastema (Hatada et al., 1997). Using primers specific for irf6.2 we detected maternal and zygotic expression by RT-PCR (Fig. 5A), and using a probe derived from irf6.2, we detected a pattern similar to that reported for irf6.1 (Fig. 5B-G). Because irf6.1 and irf6.2 probes may cross-hybridize, this pattern likely represents a combination of the expression of both genes. Alternatively, irf6.1 and irf6.2 may have identical expression patterns. The pattern of irf6 expression in X. laevis shares many similarities with irf6 expression in D. rerio, including prominent expression at gastrula stage, in the otic vesicle and in facial epithelium.
Figure 5. Expression of irf6 in Xenopus laevis.
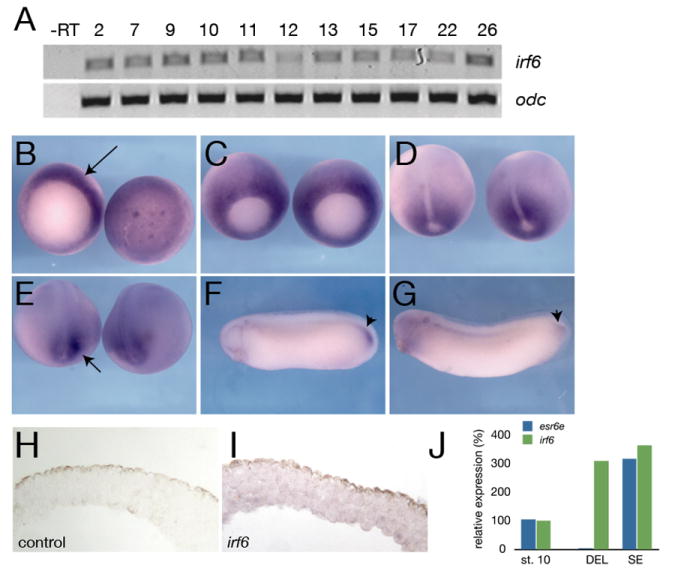
A) RT-PCR analysis of irf6 expression at different stages of development. Developmental stages are indicated at the top (Nieuwkoop and Faber, 1956). -RT, negative control reaction without reverse transcriptase. B-G) Whole mount in situ hybridization for irf6. B) Stage 10.5, left is vegetal view, arrow indicates expression in the dorsal marginal zone. Right is animal pole view. C) Stage 11, expression is throughout marginal zone, but beginning to be lost from dorsal midline. D) Stage 12.5, expression around blastopore, absent from prospective notochord. E) stage 18, expression flanking prospective tailbud region, arrow. F) Stage 28 and G) stage 30, notable expression in tailbud, arrowheads. H) Section of animal cap hybridized with an irrelevant control probe (xnr3). I) Section of animal cap hybridized with an antisense irf6 probe. Diffuse expression is found in both superficial and deep layers. J) Quantitative RT-PCR of irf6 (green) and esr6e (blue) expression in ectodermal explants; st. 10 (stage 10 whole embryo), DEL (deep layer explant), SE (superficial layer explant).
Because blocking Irf6 function in D. rerio caused EVL defects, we more closely examined irf6 expression in the SE and deep layers of early X. laevis embryos. We performed in situ hybridization for irf6 on sectioned late blastulae and found expression in both layers (Fig. 5I). These data were confirmed by quantitative RT-PCR, using RNA isolated from explanted superficial and deep epithelial layers. Levels of irf6 were similar in the two layers, whereas a marker of the superficial layer, enhancer of split related 6-epidermis (esr6e, Deblandre et al., 1999) was detected only in the SE layer (Fig. 5J). Thus, irf6 is not restricted to specific layers of the early ectoderm, at least at the level of mRNA expression, which is consistent with the inheritance of irf6 transcripts from the egg.
Injection of irf6DBD and depletion of maternal Irf6 disrupts SE formation in Xenopus
We found that injection of the D. rerio irf6DBD mRNA into X. laevis embryos caused exogastrulation and eventual rupture of the embryo near the animal pole, consistent with a possible defect in the superficial epithelium (Fig. 6 D,F). Supporting this possibility, by RT-PCR we detected greatly reduced expression of esr6e, a marker of the superficial epithelium, in animal explants harvested from X. laevis embryos injected with irf6DBD. A second marker, grainyhead-like 3 (grhl3), is normally expressed at higher levels in the SE than in DEL (Fig 6G, Chalmers et al., 2006). We found that expression ghrl3 was reduced but not absent in animal explants from irf6DBD-injected embryos, and by separating SE and deep blastomere layers we observed that ghrl3 expression is preferentially lost from the SE of such explants (Fig. 6G). Of note, SE layers isolated from irf6DBD injected embryos could be cultured for 24 hours without disintegration or signs of widespread cell death, which are easily recognized (Hensey and Gautier, 1997). This suggests that cell death is unlikely to be the primary cause of loss of gene expression characteristic of the SE. Thus, in X. laevis as in D. rerio, Irf6DBD inhibits development of the superficial epithelium, suggesting a conserved requirement for Irf6 in simple epithelial formation.
Figure 6. Defects in development of superficial epithelium in irf6DBD-injected X. laevis embryos.
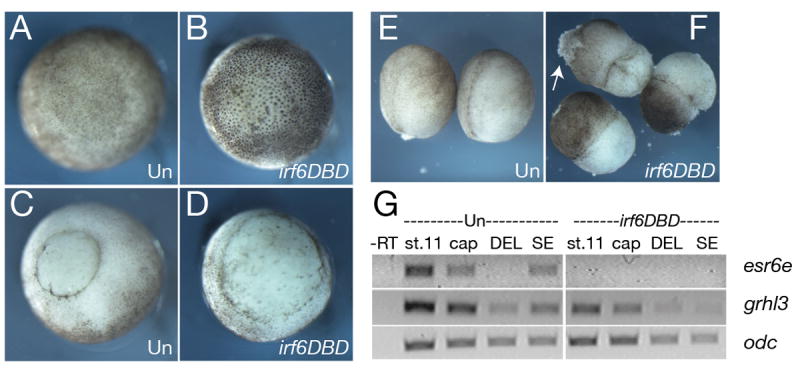
A) Animal and, C) vegetal views of stage 11 uninjected controls (Un). B) Animal and D) vegetal views of stage 11 embryos injected with 2 ng D. rerio irf6DBD mRNA. E) Uninjected (Un) and F) irf6DBD-injected stage 14 embryos. Arrow in F) indicates lesion in the ectoderm. G) RT-PCR analysis of SE markers, esr6e and grhl3 in control (Un) and irf6DBD-injected embryos and explants. St. 11, whole embryo stage 11; cap, stage 11 animal cap; DEL, deep layer explants; SE, superficial layer explants.
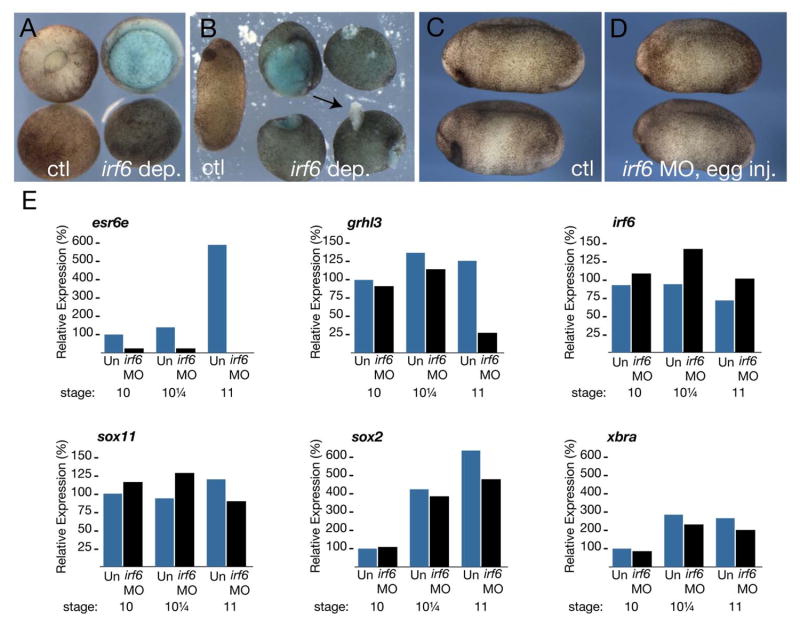
To further test whether inhibition of maternally-encoded Irf6 was sufficient to disrupt superficial epithelium formation, we blocked the translation of maternal irf6 transcripts in X. laevis oocytes with a morpholino (MO) designed to complement both irf6.1 and irf6.2 transcripts (see Methods). We harvested oocytes, injected them with 24 ng of irf6 MO, and cultured them in vitro for 24 hours (Heasman et al., 1991). We then matured the oocytes with progesterone, transferred them into the body cavity of egg-laying host females, and fertilized the resulting eggs to generate Irf6-depleted embryos. Depleted embryos developed normally through cleavage and blastula stages, but were delayed in gastrulation and formed open blastopores (Fig. 7A, right hand embryos), closely resembling X. laevis embryos injected with irf6DBD (Fig. 6B). Irf6-depleted embryos failed to elongate fully and subsequently developed epidermal lesions (Fig. 7B, right hand embryos; 90% affected, n=50). Like irf6DBD-injected embryos, irf6-depleted embryos lacked visible heads (discussed further below). Interestingly, fertilized eggs injected with the same dose of MO (24 ng) developed normally, with a minor proportion showing blastopore closure defects (Fig. 7C-D, 5% affected, n=30). The contrast in the effects of Irf6 depletion before and after fertilization strongly suggests that the maternal store of Irf6 that is critical for normal early development.
Figure 7. Maternal Irf6 is required for SE specification in X. laevis.
A) Control (left side) and Irf6-depleted (right side) embryos at stage 12. Upper row is a vegetal view; lower row is an animal view. B) Control (left embryo) and Irf6-depleted (right four embryos) embryos at stage 24. Arrow indicates epidermal lesion in an Irf6-depleted embryo. C) Control uninjected embryos. D) embryos injected with 24 ng irf6 MO at the 1-2 cell stage. E) Quantitative RT-PCR of esr6e, grhl3, sox11, irf6, sox2 and xbra expression in uninjected embryos (Un) and Irf6-depleted embryos (irf6 MO) at the indicated stages. Graphs represent data from a single PCR run.
To test for alterations in SE fate in maternal irf6-depleted embryos, we analyzed the expression of SE markers, esr6e and grainyhead-like-3 (grhl3), as well as a deep layer marker, sox11, over several gastrula stages by quantitative RT-PCR (Chalmers et al., 2006; Chalmers et al., 2002). Expression of esr6e was nearly eliminated in Irf6-depleted embryos (Fig. 7E). Whereas grhl3 was unaffected at early gastrula stages, it was reduced to 25% of control levels by midgastrula stage (stage 11) (Fig. 7E). In embryos where irf6 MO was injected after fertilization, esr6e was reduced to 50% of control levels at stage, and grhl3 was unaffected, reflecting the reduced effectiveness of this depletion method (data not shown). By contrast, levels of the deep layer marker sox11 in whole embryos were not significantly different from controls (Fig. 7E). Additionally, irf6 levels were not greatly altered, indicating a lack of compensatory regulation (Fig. 7E). Expression of the presumptive neural precursor marker sox2, the mesodermal marker xbra, and two organizer markers (nodal related 3 and goosecoid) (not shown) were also not severely affected (Fig. 7E and not shown). SE layers isolated from Irf6-depleted embryos could be cultured for 24 hours without disintegration or signs of widespread cell death (not shown), suggesting the loss of SE markers is not due to loss of the cells themselves. These results suggest that defects in Irf6-depleted embryos during gastrula stages are primarily within the SE layer. The later defects in these embryos, including anterior defects, most likely result from abnormal morphogenesis.
Consistent with reduced expression of early SE markers in Irf6-depleted tissue, we observed reduced expression of markers of cell types that are derived from the SE. Control and Irf6-depleted embryos were analyzed by whole-mount in situ hybridization for intelectin 2 (intel2), a marker of SE-derived mucosecretory cells (Hayes et al., 2007). We also stained sections from control and Irf6-depleted embryos with the monoclonal antibody LP3K, which was raised against mouse cytokeratins and labels the apical surface of SE cells (Lane et al., 1985). In control embryos at stage 14, intel2 robustly labeled cells in the SE layer and LP3K stained the surface of the superficial epithelium (10/10 cases each; Fig. 8A, C). In sibling Irf6-depleted embryos, intel2 and LP3K staining were absent (0/10 embryos each; Fig. 8B, D). We further assessed overall development of pre-larval skin in tailbud stage embryos by histological staining. Azan staining of control embryos showed thinned ectodermal layers with apically localized embryonic pigment granules, a hallmark of mature epidermis (3/3 cases; Fig. 8E)(Nieuwkoop and Faber, 1956). Irf6-depleted embryos at this stage displayed thicker than normal epidermis with basally localized pigment granules (3/3 cases; Fig. 8F), suggesting that the epidermis is in a less differentiated state than the controls. These results demonstrate that maternal Irf6 is necessary for the normal differentiation of superficial epithelial cells in Xenopus.
Figure 8. Maternal Irf6 is required for SE differentiation in X. laevis.
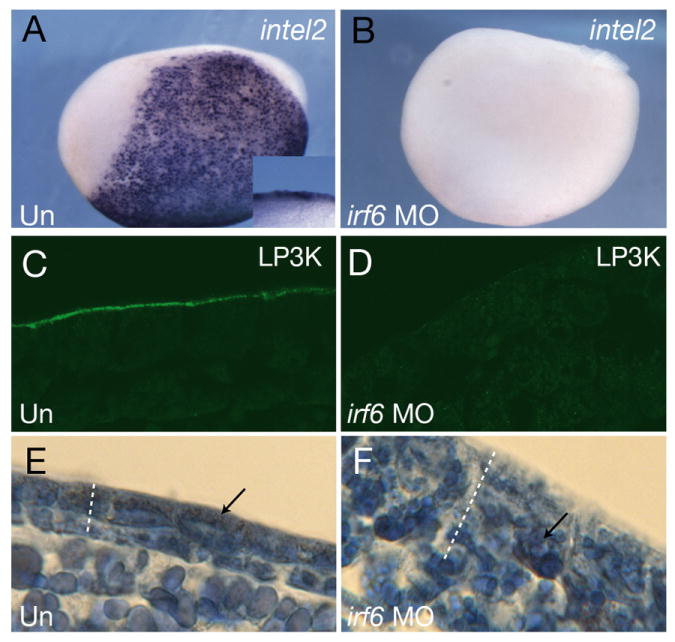
A,B) Whole mount in situ hybridization of stage 14, control A) uninjected (Un) and B) Irf6-depleted (irf6 MO) embryos for intelectin 2 (intel2). Lateral view is shown, anterior is to the left. Inset shows close up views of the epidermis in a bisected embryo. C,D) Sections of C) uninjected and D) Irf6-depleted stage 13 embryos stained with mAb LP3K. E,F) Sections of, E) control and, F) Irf6-depleted stage 22 embryos stained with Azan. Dotted lines indicate the thickness of the ectoderm; arrows indicate the location of embryonic pigment.
Specificity of Irf6 depletion
To confirm the specificity of phenotypes seen in Irf6-depleted embryos, we performed rescue experiments using an irf6.2 mRNA lacking the full MO-binding sequence. MO-injected oocytes were cultured for 24 hours and then injected with irf6.2 mRNA, fertilized by host transfer and analyzed. Interestingly, injection of a wide range of irf6 mRNA amounts, from 20 pg to 500 pg, into Irf6-depleted embryos, exacerbated the embryonic phenotype rather than rescuing it (data not shown). However, in RT-PCR analysis gastrula stage embryos, injection of 50 pg of X. laevis irf6.2 mRNA partially reversed the inhibition of esr6e and grhl3 expression (Fig. 9A), suggesting that the effects of irf6 MO injection are indeed specific to the depletion of Irf6. In separate experiments, injection of a higher dose (250 pg) of irf6.2 mRNA was able to almost completely rescue esr6e and grhl3 expression (Fig. 9B), but was also unable to rescue the exogastrulation phenotype (data not shown). We also assessed the degree of rescue in whole embryos by in situ hybridization for intel2 expression. Controls robustly expressed intel2 at stage 14 (8/8; Fig. 9C), and expression was eliminated in Irf6-depleted embryos (0/8; Fig. 9D). However, patches of intel2-expressing cells were evident in a subset of Irf6-depleted embryos injected with irf6 (2/8 with patchy expression; Fig. 9E), even though the embryo morphology was grossly abnormal. Thus, these instances of partial rescue provide evidence that the effects of MO injection are due to inhibition of Irf6. They further suggest that levels of Irf6 activity must be precisely controlled for normal development to occur.
Figure 9. Specificity of Irf6 depletion in Xenopus.
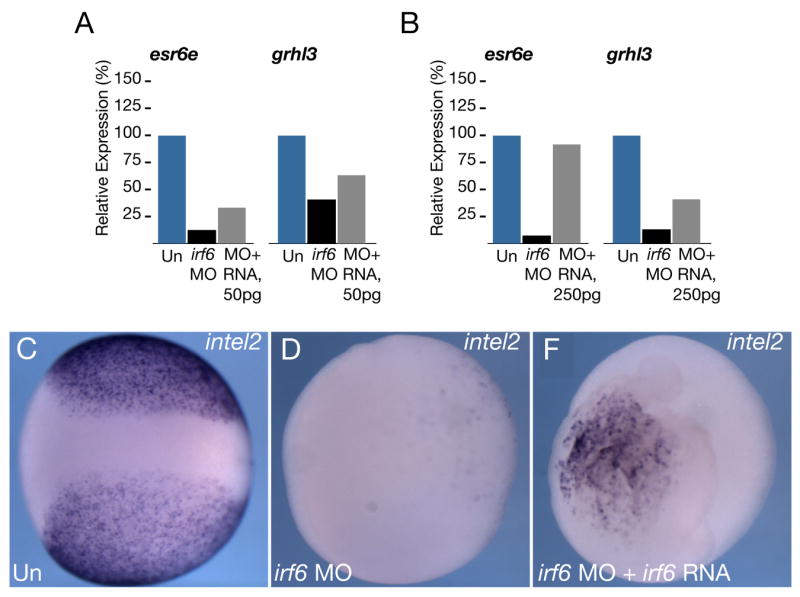
A) Quantitative RT-PCR of esr6e and grhl3 expression in controls (Un), Irf6-depleted embryos (irf6 MO) and depleted embryos injected with 50 pg irf6.2 mRNA (MO+RNA), or B) with 250 pg irf6.2 mRNA (MO+RNA). C-E) Whole mount in situ hybridization for intelectin 2 at stage 14, C) control uninjected (Un), D) Irf6-depleted (irf6 MO) and E) rescued embryos (irf6 MO+ irf6 RNA).
DISCUSSION
Irf6 depleted embryos die at gastrula stage
These findings suggest an Irf6-like activity is required for gastrulation in D. rerio and X. laevis, and several lines of evidence indicate that this activity is provided by maternally-encoded Irf6. First, maternal irf6 mRNA is readily detected by RT-PCR or in situ hybridization in both organisms. Second, injection of antisense MO after fertilization, which should target zygotic irf6 transcripts efficiently but maternal ones less well or not at all, does not affect gastrulation in either species. Third, forced expression of Irf6DBD causes D. rerio and X. laevis embryos to stall in epiboly. The effect of Irf6DBD in both species is partially rescued by co-injection of RNA encoding full-length Irf6. The simplest interpretation of these findings is that maternal irf6 transcripts are translated too early to be effectively targeted by MO, while maternal Irf6 protein is displaced from its normal chromatin binding sites by Irf6DBD. However, X. laevis embryos derived from oocytes injected with irf6 MO display a phenotype that is remarkably similar to embryos injected with irf6DBD. Thus maternal Irf6 protein may largely substitute for the loss of zygotic Irf6, at least over the period when morpholinos are effective. While most maternally supplied mRNAs are degraded prior to or about the time of mid blastula transition, a microarray study identified 142 maternal transcripts that are stable until segmentation stages and beyond (Mathavan et al., 2005). Moreover, many maternal-effect mutants have been isolated whose phenotype is not discernible until after zygotic transcription begins, and one such mutant, pollywog, appears to develop normally until at least 10 hpf (Wagner et al., 2004). It is an intriguing possibility that there is a role for maternal Irf6 in mammals, perhaps in formation of the trophectoderm. Unfortunately, this possibility cannot be readily tested because mouse Irf6 null mutants die shortly after birth (Ingraham et al., 2006; Richardson et al., 2006), precluding examination of pups derived from Irf6 null females.
An alternative model consistent with these data is that there is an Irf6 homologue whose binding to DNA is inhibited by Irf6DBD. Indeed, given the profound defects in skin and limb development in mouse Irf6 mutant embryos (Ingraham et al., 2006; Richardson et al., 2006), we were surprised that defects did not emerge in post-gastrula stage D. rerio embryos that had been injected with splice-blocking irf6 MO. In addition, the observation that embryos derived from irf6-MO-injected oocytes have a less severe phenotype than embryos injected with Irf6DBD is consistent with the possibility that Irf6DBD interferes with activity of a protein other than Irf6 (although the difference may reflect incomplete knockdown of Irf6 levels in MO-injected oocytes). In teleost fishes there are duplicated copies of many genes that are present in single copy in mammals (Postlethwait et al., 1998). However, our database searches yielded only a single D. rerio irf6 orthologue, on chromosome 22. The most closely related gene is a clear irf5 orthologue on chromosome 4 (peptide sequence is 46% identical/60% similar). While we detect expression of irf5 at embryonic stages, our preliminary double knock down experiments did not support the model that Irf5 and Irf6 act coordinately (J.S., C.d’A. and RAC, unpublished observations). At this point we cannot rule out the possibility of redundant activity of an Irf6 homologue, sensitive to inhibition by Ird6DBD, during early development in D. rerio. Creation of a maternal-zygotic irf6 null mutant embryos, which may be possible in D. rerio (Ciruna et al., 2002; Doyon et al., 2008; Meng et al., 2008), would allow definitive resolution of these models.
Why do irf6DBD-injected D. rerio embryos stall in epiboly? Experiments conducted in Fundulus and D. rerio suggest the engines driving epiboly include microtubule-dependent vegetal migration of the YSL, which pulls the firmly-attached EVL behind it (Betchaku and Trinkaus, 1978; Solnica-Krezel, 2006), contraction of microtubules at the margin (Strahle and Jesuthasan, 1993), and possibly actively crawling behavior of leading-edge EVL cells (Keller and Trinkaus, 1987; Zalik et al., 1999). Epiboly movements of the YSL are blocked in Irf6DBD injected animals. It is unclear whether this reflects a cell autonomous requirement for Irf6 within the YSL, however our finding that injection of irf6DBD mRNA into the YSL at 2 hpf has no effect on epiboly does not support this possibility. There is high level expression of irf6 in dorsal forerunner cells (DFC), a cluster of ingressing EVL cells present just vegetal to the shield (Cooper and D’Amico, 1996; Oteiza et al., 2008). Perhaps failure of epiboly in irf6DBD-injected embryos in part reflects loss of a DFC-mediated role in epiboly. However, any such role is presumably confined to the dorsal side of the embryo, while Irf6DBD appears to block epiboly at all radial positions. Instead, we favor the model that a cell-autonomous requirement for Irf6 in the EVL largely or entirely accounts for the stall in epiboly of irf6DBD-injected D. rerio embryos. There is precedent for such a possibility, because knockdown of a set of keratin genes expressed only in EVL prevents epiboly in D. rerio (Pei et al., 2007), and depletion of maternal ker8 disrupts gastrulation in X. laevis (Torpey et al., 1992). We envision that in irf6DBD-injected animals, as in normal embryos, a vegetal-pole directed force is generated in the YSL by microtubule-based mechanisms (Solnica-Krezel and Driever, 1994; Strahle and Jesuthasan, 1993). When this force is transmitted to the EVL, the abnormal differentiation of EVL cells prevent them from spreading during epiboly. Consequently, force builds up until the EVL ruptures and deep cells spill out. In X. laevis embryos, epiboly is driven by outward radial intercalation of deep blastomeres, although not into the superficial epithelium (Keller, 1980). Similar to the model for D. rerio, exogastrulation in Irf6-depleted X. laevis embryos may reflect a loss of mechanical strength in the superficial epithelium.
Irf6 governs differentiation of epithelial tissues
These present results extend earlier studies indicating Irf6 is required for differentiation of a stratified epithelium. In D. rerio embryos and X. laevis embryos with reduced Irf6, all epithelial features of the PSE analyzed were lost, and a mosaic experiment indicates the requirement for Irf6 is cell autonomous to the PSE. However, the principle function of Irf6 in superficial epithelial cells is unclear. It is possible that the primary requirement for Irf6 is to maintain cell adhesiveness between PSE cells, because expression of simple epithelium keratins in PSE depends on cell contact (Jones and Woodland, 1986; Sagerstrom et al., 2005). Could the primary function of Irf6 be to promote survival of PSE cells? We feel this is unlikely because previous studies have shown treating embryos with inducers of apoptosis (gamma irradiation, camptothecin, alpha-amanitin) does not result in cell death until 7 hpf in D. rerio (Ikegami et al., 1999) and stage 10.5 in X. laevis (Hensey and Gautier, 1997). This critical period is later than the time that we detect changes in gene expression or morphology in Irf6 knockdown embryos. Moreover, reagents that block cell death induced by Irf6DBD, neither prevented the morphological phenotype, nor the loss of expression of an EVL marker. It is also possible that Irf6 governs specification of PSE cells. A prediction of this model is that superficial cells deprived of Irf6 will be competent to join the deep layer; experiments to test this prediction are underway. The strongest conclusion from the present data is that Irf6 is required for superficial cells to adopt simple epithelial architecture (i.e. simple squamous in the case of D. rerio PSE and simple cuboidal in X. laevis PSE). This adds to evidence that Irf6 promotes exit from the cell cycle and differentiation of keratinocytes (Ingraham et al., 2006; Richardson et al., 2006) and breast epithelium (Bailey et al., 2008a; Bailey et al., 2008b).
The Irf6 regulatory pathway governing PSE development
Which gene products act in concert with Irf6 to promote PSE development? Excellent candidates are those that, like Irf6, have been implicated in differentiation of epidermal keratinocytes, including AP-2 family members (Guttormsen et al., 2008; Wang et al., 2006), p63 (Truong and Khavari, 2007), Ikka (Hu et al., 1999; Li et al., 1999), 14-3-3sigma (Li et al., 2005), and aPKC (Helfrich et al., 2007). aPKC is an especially intriguing candidate because not only is it required for differentiation of basal keratinocytes in mouse (Helfrich et al., 2007), inhibition of aPKC disrupts SE development (Ossipova et al., 2007), similar to knock down of Irf6. Activity of other IRF family members is stimulated by phosphorylation (reviewed in Paun and Pitha, 2007), and Irf6 is regulated by phosphorylation in breast epithelium (Bailey et al., 2005). An interesting avenue for future work will be to test whether Irf6 is directly activated by aPKC-mediated phosphorylation. Which genes act downstream of Irf6? Loss of adhesion among X. laevis blastomeres is observed upon misexpression of the small GTPases Rac, Cdc42, and Rnd1 (Choi and Han, 2002; Hens et al., 2002; Wunnenberg-Stapleton et al., 1999) and also after disruption of p21-activated kinase (PAK) (Faure et al., 2005); genes encoding these proteins are all potential targets of Irf6. In irf6DBD-injected embryos, EVL cells lack the cortical band of F-actin normally associated with adherens junctions (Nagafuchi, 2001). In D. rerio embryos disruption of actin-polymerization with cytochalasin D causes a phenotype of embryonic rupture that is morphologically indistinguishable to that observed in irf6DBD-injected embryos (Zalik et al., 1999). Moreover, in X. laevis embryos, cytochalasin D blocks expression of an epidermal marker and the cell cycle (Jones and Woodland, 1986). Thus, crucial transcriptional targets of Irf6 in PSE cells may include genes that govern organization of actin into filaments, of which a large number are known (Uribe and Jay, 2007). Finally, genes disrupted in D. rerio maternal-effect mutants with abnormal epiboly are intriguing candidates for members of the Irf6 regulatory pathway (Wagner et al., 2004).
Supplementary Material
Timelapse movie of water-injected (left) and irf6DBD-RNA-injected (right) embryos from about 5hpf to about 9hpf. The water-injected embryo develops normally but the irf6DBD-injected embryo has delayed epiboly including stalled EVL movement, YSL doming, and DEL thinning which eventually leads to eruption.
Acknowledgments
We thank Gregory Bonde, Wei Li, Nick Rorick and Margaret Malik for expert technical assistance; David Kimelman, Diane Slusarski and the NIBB for plasmids; and Chris Wylie and Birgitte Lane for LP3K supernatant. This work was supported by: The Roy J. Carver Charitable Trust (DWH); Research Grant No. 1-FY05-922 from the March of Dimes Foundation (RAC); the Interdisciplinary Graduate Program in Genetics training grant (JLS); NIH/NIDCR DE13513 (BCS); and NIH/NIGMS GM067841 (RAC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey CM, Abbott DE, Margaryan NV, Khalkhali-Ellis Z, Hendrix MJ. Interferon regulatory factor 6 promotes cell cycle arrest and is regulated by the proteasome in a cell cycle-dependent manner. Mol Cell Biol. 2008a;28:2235–43. doi: 10.1128/MCB.01866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CM, Abbott DE, Margaryan NV, Khalkhali-Ellis Z, Hendrix MJ. IRF6 Promotes Cell Cycle Arrest and is Regulated by the Proteasome in a Cell Cycle-dependent Manner. Mol Cell Biol. 2008b doi: 10.1128/MCB.01866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CM, Khalkhali-Ellis Z, Kondo S, Margaryan NV, Seftor RE, Wheaton WW, et al. Mammary serine protease inhibitor (Maspin) binds directly to interferon regulatory factor 6: identification of a novel serpin partnership. J Biol Chem. 2005;280:34210–7. doi: 10.1074/jbc.M503523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben J, Jabs EW, Chong SS. Genomic, cDNA and embryonic expression analysis of zebrafish IRF6, the gene mutated in the human oral clefting disorders Van der Woude and popliteal pterygium syndromes. Gene Expr Patterns. 2005;5:629–38. doi: 10.1016/j.modgep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Betchaku T, Trinkaus JP. Contact relations, surface activity, and cortical microfilaments of marginal cells of the enveloping layer and of the yolk syncytial and yolk cytoplasmic layers of fundulus before and during epiboly. J Exp Zool. 1978;206:381–426. doi: 10.1002/jez.1402060310. [DOI] [PubMed] [Google Scholar]
- Bouvet J. Enveloping layer and periderm of the trout embryo (Salmo trutta fario L.)20U. Cell Tissue Res. 1976;170:367–82. doi: 10.1007/BF00219418. [DOI] [PubMed] [Google Scholar]
- Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha, -beta, and -gamma during rat embryonic development. Endocrinology. 1998;139:2748–54. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- Bruce AE, Howley C, Zhou Y, Vickers SL, Silver LM, King ML, Ho RK. The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development. 2003;130:5503–17. doi: 10.1242/dev.00763. [DOI] [PubMed] [Google Scholar]
- Butler K, Zorn AM, Gurdon JB. Nonradioactive in situ hybridization to xenopus tissue sections. Methods. 2001;23:303–12. doi: 10.1006/meth.2000.1142. [DOI] [PubMed] [Google Scholar]
- Chalmers A, Lachani K, Shin Y, Sherwood V, Cho K, Papalopulu N. Grainyhead-like 3, a transcription factor identified in a microarray screen, promotes the specification of the superficial layer of the embryonic epidermis. Mech Dev. 2006;123:702–718. doi: 10.1016/j.mod.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Pambos M, Mason J, Lang S, Wylie C, Papalopulu N. aPKC, Crumbs3 and Lgl2 control apicobasal polarity in early vertebrate development. Development. 2005;132:977–86. doi: 10.1242/dev.01645. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Strauss B, Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development. 2003;130:2657–68. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev Cell. 2002;2:171–82. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Choi SC, Han JK. Xenopus Cdc42 regulates convergent extension movements during gastrulation through Wnt/Ca2+ signaling pathway. Dev Biol. 2002;244:342–57. doi: 10.1006/dbio.2002.0602. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Weidinger G, Knaut H, Thisse B, Thisse C, Raz E, Schier AF. Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc Natl Acad Sci U S A. 2002;99:14919–24. doi: 10.1073/pnas.222459999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MS, D’Amico LA. A cluster of noninvoluting endocytic cells at the margin of the zebrafish blastoderm marks the site of embryonic shield formation. Dev Biol. 1996;180:184–98. doi: 10.1006/dbio.1996.0294. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–82. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- D’Amico LA, Cooper MS. Morphogenetic domains in the yolk syncytial layer of axiating zebrafish embryos. Dev Dyn. 2001;222:611–24. doi: 10.1002/dvdy.1216. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 1999;126:4715–28. doi: 10.1242/dev.126.21.4715. [DOI] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–7. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollar GL, Weber U, Mlodzik M, Sokol SY. Regulation of Lethal giant larvae by Dishevelled. Nature. 2005;437:1376–80. doi: 10.1038/nature04116. [DOI] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–8. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Cau J, de Santa Barbara P, Bigou S, Ge Q, Delsert C, Morin N. Xenopus p21-activated kinase 5 regulates blastomeres’ adhesive properties during convergent extension movements. Dev Biol. 2005;277:472–92. doi: 10.1016/j.ydbio.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Godsave SF, Wylie CC, Lane EB, Anderton BH. Intermediate filaments in the Xenopus oocyte: the appearance and distribution of cytokeratin-containing filaments. J Embryol Exp Morphol. 1984;83:157–67. [PubMed] [Google Scholar]
- Guttormsen J, Koster MI, Stevens JR, Roop DR, Williams T, Winger QA. Disruption of epidermal specific gene expression and delayed skin development in AP-2 gamma mutant mice. Dev Biol. 2008;317:187–95. doi: 10.1016/j.ydbio.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3:399–411. doi: 10.1016/0896-6273(89)90200-6. [DOI] [PubMed] [Google Scholar]
- Hatada S, Kinoshita M, Takahashi S, Nishihara R, Sakumoto H, Fukui A, et al. An interferon regulatory factor-related gene (xIRF-6) is expressed in the posterior mesoderm during the early development of Xenopus laevis. Gene. 1997;203:183–8. doi: 10.1016/s0378-1119(97)00512-x. [DOI] [PubMed] [Google Scholar]
- Hayes JM, Kim SK, Abitua PB, Park TJ, Herrington ER, Kitayama A, et al. Identification of novel ciliogenesis factors using a new in vivo model for mucociliary epithelial development. Dev Biol. 2007;312:115–30. doi: 10.1016/j.ydbio.2007.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Holwill S, Wylie CC. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 1991;36:213–30. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- Helfrich I, Schmitz A, Zigrino P, Michels C, Haase I, le Bivic A, et al. Role of aPKC isoforms and their binding partners Par3 and Par6 in epidermal barrier formation. J Invest Dermatol. 2007;127:782–91. doi: 10.1038/sj.jid.5700621. [DOI] [PubMed] [Google Scholar]
- Hens MD, Nikolic I, Woolcock CM. Regulation of Xenopus embryonic cell adhesion by the small GTPase, rac. Biochem Biophys Res Commun. 2002;298:364–70. doi: 10.1016/s0006-291x(02)02459-2. [DOI] [PubMed] [Google Scholar]
- Hensey C, Gautier J. A developmental timer that regulates apoptosis at the onset of gastrulation. Mech Dev. 1997;69:183–95. doi: 10.1016/s0925-4773(97)00191-3. [DOI] [PubMed] [Google Scholar]
- Ho RK. Cell movements and cell fate during zebrafish gastrulation. Dev Suppl. 1992:65–73. [PubMed] [Google Scholar]
- Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–56. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- Houston DW, Wylie CW. Maternal Xenopus Zic2 negatively regulates Nodal-related gene expression during anteroposterior patterning. Development. 2005;127:447–56. doi: 10.1242/dev.02066. [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284:316–20. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Hunter P, Yager TD. Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev Biol. 1999;209:409–33. doi: 10.1006/dbio.1999.9243. [DOI] [PubMed] [Google Scholar]
- Imboden M, Goblet C, Korn H, Vriz S. Cytokeratin 8 is a suitable epidermal marker during zebrafish development. C R Acad Sci III. 1997;320:689–700. doi: 10.1016/s0764-4469(97)84816-0. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–40. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–97. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Jones EA, Woodland HR. Development of the ectoderm in Xenopus: tissue specification and the role of cell association and division. Cell. 1986;44:345–55. doi: 10.1016/0092-8674(86)90769-5. [DOI] [PubMed] [Google Scholar]
- Kaji T, Artinger KB. dlx3b and dlx4b function in the development of Rohon-Beard sensory neurons and trigeminal placode in the zebrafish neurula. Dev Biol. 2004;276:523–40. doi: 10.1016/j.ydbio.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Hammerschmidt M, Mullins MC, Maischein HM, Brand M, van Eeden FJ, et al. The zebrafish epiboly mutants. Development. 1996;123:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- Kane DA, McFarland KN, Warga RM. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development. 2005;132:1105–16. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- Keller RE. The cellular basis of epiboly: an SEM study of deep cell rearrangement during gastrulation in Xenopus laevis. J Embryol Exp Morphol. 1980;60:201–234. [PubMed] [Google Scholar]
- Keller RE, Trinkaus JP. Rearrangement of enveloping layer cells without disruption of the epithelial permeability barrier as a factor in Fundulus epiboly. Dev Biol. 1987;120:12–24. doi: 10.1016/0012-1606(87)90099-6. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203:245–63. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–94. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–91. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–7. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002a;32:285–9. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002b;32:285–9. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Fernandez BG, Carvalho L, Jacinto A, Heisenberg CP. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 2006;133:2671–81. doi: 10.1242/dev.02439. [DOI] [PubMed] [Google Scholar]
- Lane EB, Bártek J, Purkis PE, Leigh IM. Keratin antigens in differentiating skin. Ann N Y Acad Sci. 1985;455:241–58. doi: 10.1111/j.1749-6632.1985.tb50415.x. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Estepa G, Verma IM. Identification of 14-3-3sigma mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proc Natl Acad Sci U S A. 2005;102:15977–82. doi: 10.1073/pnas.0508310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, Verma IM. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 1999;13:1322–8. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol. 2007;304:338–54. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SE, Shue BC, Lei L, Oates AC, Zon LI, Liu PP. Molecular cloning, genetic mapping, and expression analysis of four zebrafish c/ebp genes. Gene. 2001;281:43–51. doi: 10.1016/s0378-1119(01)00774-0. [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–76. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland KN, Warga RM, Kane DA. Genetic locus half baked is necessary for morphogenesis of the ectoderm. Dev Dyn. 2005;233:390–406. doi: 10.1002/dvdy.20325. [DOI] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A. Molecular architecture of adherens junctions. Curr Opin Cell Biol. 2001;13:600–3. doi: 10.1016/s0955-0674(00)00257-x. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Fukui A, Izumi Y, Akimoto K, Asashima M, Ohno S. Meiotic maturation induces animal-vegetal asymmetric distribution of aPKC and ASIP/PAR-3 in Xenopus oocytes. Development. 2000;127:5021–31. doi: 10.1242/dev.127.23.5021. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. taf/DynaPage.taf?file=/ng/journal/v26/n2/full/ng1000_216.html taf/DynaPage.taf?file=/ng/journal/v26/n2/abs/ng1000_216.html. [DOI] [PubMed]
- Nieuwkoop P, Faber J. Normal tables of Xenopus laevis. New Holland: Publishing Co., Amsterdam; 1956. [Google Scholar]
- O’Brien EK, d’Alencon C, Bonde G, Li W, Schoenebeck J, Allende ML, et al. Transcription factor Ap-2alpha is necessary for development of embryonic melanophores, autonomic neurons and pharyngeal skeleton in zebrafish. Dev Biol. 2004;265:246–61. doi: 10.1016/j.ydbio.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Tabler J, Green J, Sokol S. PAR1 specifies ciliated cells in vertebrate ectoderm downstream of aPKC. Development. 2007;134:4297–306. doi: 10.1242/dev.009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteiza P, Koppen M, Concha ML, Heisenberg CP. Origin and shaping of the laterality organ in zebrafish. Development. 2008;135:2807–13. doi: 10.1242/dev.022228. [DOI] [PubMed] [Google Scholar]
- Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–53. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Noushmehr H, Costa J, Ouspenskaia MV, Elkahloun AG, Feldman B. An early requirement for maternal FoxH1 during zebrafish gastrulation. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–9. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Regen CM, Steinhardt RA. Global properties of the Xenopus blastula are mediated by a high-resistance epithelial seal. Developmental Biology. 1986;113:147–154. [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38:1329–34. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Sagerstrom CG, Gammill LS, Veale R, Sive H. Specification of the enveloping layer and lack of autoneuralization in zebrafish embryonic explants. Dev Dyn. 2005;232:85–97. doi: 10.1002/dvdy.20198. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, van Eeden FJ, Halpern ME, Kimmel CB, Nusslein-Volhard C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 1994;120:1009–15. doi: 10.1242/dev.120.4.1009. [DOI] [PubMed] [Google Scholar]
- Sive H, Grainger RM, Harland RM. Early Development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Laboratory Press.; New York: 2000. [Google Scholar]
- Solnica-Krezel L. Gastrulation in zebrafish -- all just about adhesion? Curr Opin Genet Dev. 2006;16:433–41. doi: 10.1016/j.gde.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120:2443–55. doi: 10.1242/dev.120.9.2443. [DOI] [PubMed] [Google Scholar]
- Strahle U, Jesuthasan S. Ultraviolet irradiation impairs epiboly in zebrafish embryos: evidence for a microtubule-dependent mechanism of epiboly. Development. 1993;119:909–19. doi: 10.1242/dev.119.3.909. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. Journal of Cell Science. 2006:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–55. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Torpey N, Wylie CC, Heasman J. Function of maternal cytokeratin in Xenopus development. Nature. 1992;357:413–5. doi: 10.1038/357413a0. [DOI] [PubMed] [Google Scholar]
- Truong AB, Khavari PA. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 2007;6:295–9. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- Uribe R, Jay D. A review of actin binding proteins: new perspectives. Mol Biol Rep. 2007 doi: 10.1007/s11033-007-9159-2. [DOI] [PubMed] [Google Scholar]
- Wagner D, Dosch R, Mintzer K, Wiemelt A, Mullins M. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell. 2004;6:781–90. doi: 10.1016/j.devcel.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Wang X, Bolotin D, Chu DH, Polak L, Williams T, Fuchs E. AP-2alpha: a regulator of EGF receptor signaling and proliferation in skin epidermis. J Cell Biol. 2006;172:409–21. doi: 10.1083/jcb.200510002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108:569–80. doi: 10.1242/dev.108.4.569. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene, OR: 1993. [Google Scholar]
- Wunnenberg-Stapleton K, Blitz IL, Hashimoto C, Cho KW. Involvement of the small GTPases XRhoA and XRnd1 in cell adhesion and head formation in early Xenopus development. Development. 1999;126:5339–51. doi: 10.1242/dev.126.23.5339. [DOI] [PubMed] [Google Scholar]
- Zalik SE, Lewandowski E, Kam Z, Geiger B. Cell adhesion and the actin cytoskeleton of the enveloping layer in the zebrafish embryo during epiboly. Biochem Cell Biol. 1999;77:527–42. [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–80. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Timelapse movie of water-injected (left) and irf6DBD-RNA-injected (right) embryos from about 5hpf to about 9hpf. The water-injected embryo develops normally but the irf6DBD-injected embryo has delayed epiboly including stalled EVL movement, YSL doming, and DEL thinning which eventually leads to eruption.