Abstract
The peopling of Europe is a complex process. One of the most dramatic demographic events, the Neolithic agricultural revolution, took place in the Near East roughly 10 000 years ago and then spread through the European continent. Nevertheless, the nature of this process (either cultural or demographic) is still a matter of debate among scientists. We have retrieved HVRI mitochondrial DNA sequences from 11 Neolithic remains from Granollers (Catalonia, northeast Spain) dated to 5500 years BP. We followed the proposed authenticity criteria, and we were also able, for the first time, to track down the pre-laboratory-derived contaminant sequences and consequently eliminate them from the generated cloning dataset. Phylogeographic analysis shows that the haplogroup composition of the Neolithic population is very similar to that found in modern populations from the Iberian Peninsula, suggesting a long-time genetic continuity, at least since Neolithic times. This result contrasts with that recently found in a Neolithic population from Central Europe and, therefore, raises new questions on the heterogeneity of the Neolithic dispersals into Europe. We propose here a dual model of Neolithic spread: acculturation in Central Europe and demic diffusion in southern Europe.
Keywords: ancient DNA, mtDNA, Neolithic, demic diffusion
1. Introduction
One of the main features of the European genetic diversity landscape, especially in classical genetic markers and Y chromosome, is a clinal pattern that has been interpreted as reflecting a population movement from the southeast to the northwest, with a significant demographic impact (Cavalli-Sforza et al. 1994; Rosser et al. 2000). There are currently two processes in the demographic and evolutionary history of Europe, as documented from the archaeological record, that can account for such a cline: the Palaeolithic colonization of Europe (starting around 40 000 years BP) and the Neolithic agricultural diffusion (starting around 10 000 years BP) (Ammerman & Cavalli-Sforza 1984; Barbujani & Goldstein 2004). Unfortunately, clines do not have dates associated with them, and as both population movements followed the same axis through Europe, the attribution of the genetic cline to either process is not straightforward. In addition, the short evolutionary time elapsed between both the processes makes it difficult to disentangle both scenarios. Different authors have tried to distinguish between these two hypotheses from the analysis of genetic data in current European populations, but the conclusions obtained are contradictory (Chikhi et al. 1998; Torroni et al. 1998; Richards et al. 2000; Semino et al. 2000; Simoni et al. 2000; Chikhi et al. 2002; Richards 2003; Barbujani & Goldstein 2004).
To explain the spread of the agriculture through the European continent, two opposite hypotheses (with intermediate scenarios) have been proposed: the cultural diffusion model and the demic diffusion model (also known as the wave of advance). The cultural diffusion model supports the idea that the farmers did not move and the agricultural knowledge was transmitted from the Near East to Europe through the movement of technology and ideas (Whittle 1996). In contrast, the demic diffusion model supports the idea that farmers moved to neighbouring places while expanding from the Near East and spread into Europe taking the agricultural technology with them; thus, this model involves gene flow between the hunter-gatherers who inhabited Europe at that moment and the farmers who arrived in a slow wave through the generations (Ammerman & Cavalli-Sforza 1984).
Ancient DNA (aDNA) could potentially help to resolve between both the hypotheses, since it allows us to directly study the ancient populations that were undergoing these evolutionary processes and not their descendent populations. Recently, Haak et al. (2005) successfully extracted and sequenced the HVRI of the mitochondrial DNA (mtDNA) from 24 out of 57 Neolithic skeletons from various locations in Germany, Austria and Hungary. All human remains were dated to the LBK (also known as linear pottery culture or Linearbandkeramik) period (7000–7500 years ago). They found that 25% (6 out of 24) of the samples are of a distinctive and rare N1a lineage of the mtDNA well-known phylogeny. Furthermore, five of these six individuals display different N1a haplotypes and they were widespread in the LBK area. Europeans today have a 150 times lower frequency (0.2%) of this mtDNA type, suggesting that these first Neolithics did not have a strong impact on the genetic background of the modern European female lineages. They proposed that small pioneer farming groups carried farming into new areas of Europe and, that once the technique had taken root, the surrounding hunter-gatherers adopted the new culture and then outnumbered the original farmers, diluting their N1a frequency to the low modern value. Thus, this result and its interpretation would support the cultural diffusion model, where the farming culture itself spread without the people originally carrying these ideas. They proposed that within the current debate on whether Europeans are genetically of Palaeolithic or Neolithic origin, leaving aside the possibility of significant post-Neolithic migration, their data lend weight to the arguments for a Palaeolithic origin of Europeans. Recently, however, the results of Haak et al. (2005) have been criticized (Ammerman et al. 2006; Barbujani & Chikhi 2006) because, among other things, of the limited sample size, the uniparental mode of inheritance of the mtDNA and the generalization of the results to the whole of Europe. Moreover, aDNA studies on human samples, especially ancient Europeans, have also been put into question owing to the impossibility of distinguishing between potential contaminants and endogenous sequences. Nevertheless, such a problem could be avoided if information on all the putative contaminants present in a particular sample set is available.
In this paper, a Neolithic population from Southern Europe (Granollers, Catalonia and northeast Spain) has been subjected to aDNA genetic analysis, previous to the typing of all people involved in the manipulation of the samples. The putative endogenous sequences obtained do not match those found by Haak et al. (2005) in a sample from Central Europe. This raises new questions on the heterogeneity of the Neolithic dispersals and supports a totally different demographic model for southern Europe, compatible with a demic diffusion model.
2. Material and methods
The site ‘Camí de Can Grau’ (Granollers, Barcelona, Spain) is a necropolis excavated in 1994, which comprised 23 tombs dated by C14 between 3500 and 3000 cal years BC. There were two different funerary typologies with separate geographical locations that corresponded to different periods, spanning several hundreds of years; the older tombs formed squared sepulchral chambers, while the younger ones were hypogean tombs with an access through a vertical well.
A tooth sample was removed from 23 adult individuals for DNA analysis, with the exception of a toothless specimen, from which a bone fragment was obtained. In two well-preserved specimens (specimens 1 and 5), a second tooth was removed for independent replication in Florence. Standard methodological precautions were followed to provide as much support as possible for the authenticity of the results. Contamination by handling has been recognized as a major problem facing aDNA studies that focus on ancient human remains, especially when researchers and remains are from the same geographical area (Handt et al. 1994; Gilbert et al. 2003, 2005; Lalueza-Fox et al. 2005; Malmström et al. 2005, 2007). The authentication criteria proposed by different authors (e.g. Cooper & Poinar 2000) can help in preventing putative intralaboratory contamination, but they are useless for monitoring pre-laboratory contamination. However, what makes Can Grau an exceptional site is that its pre-laboratory history is perfectly recorded. The remains were excavated, handled and washed by the archaeologists R.P. and M.M. in 1994; once dried, they were reconstructed and studied by a physical anthropologist (E.V.) and posteriorly stored in closed plastic boxes for ca 10 years in a local museum until the genetic study was attempted. By typing the mtDNA of all the people involved in the manipulation of the skeletal remains and the laboratory analysis (M.L.S., C.L.-F. and D.C.), we have been able to trace all contaminants present in our samples (Sampietro et al. 2006). As far as we know, this is the first time in the history of aDNA research where it has been possible to control a posteriori the putatively pre-laboratory-derived contaminant DNA sequences and consequently to eliminate them of the cloning dataset. Thus and under such circumstances, the Neolithic remains that we subjected to the genetic analysis were unique in their possibilities of authentication.
(a) DNA extraction
The surface of each sample was cleaned with bleach and then ground to powder. The extraction method has been described elsewhere (Sampietro et al. 2005, 2006). One extraction blank was included for every three Neolithic samples. In brief, 10 ml of EDTA (pH 8; 0.5 M) was added to the powder overnight at 37°C to remove mineral salts; after centrifugation, the EDTA was carefully poured off and the powder was incubated overnight at 50°C in a lysis solution (1 ml SDS 5%, 0.5 ml Tris 1 M, 8.5 ml H2O and 100 μl of 1 mg ml−1 proteinase K). The samples were subsequently extracted three times with phenol, phenol–chloroform and chloroform–isoamylic alcohol and concentrated with centricons (Millipore) up to a 50–100 μl volume.
Extraction procedures were carried out in both Barcelona and Florence in an isolated pre-PCR area exclusively dedicated to aDNA studies, physically isolated from the main laboratory, with positive air pressure, overnight UV light and frequent bench cleaning with bleach. All samples and reagent manipulation were performed in a laminar flow cabinet routinely irradiated with UV light. To help avoid intralaboratory contaminations, aliquoted reagents, filter pipette tips, sterile gloves, sterile pipettes, facemasks and coverall coats were used.
(b) Amino acid racemization analysis
A limit of 0.10 in the stereoisomeric D/L ratio for the aspartic acid has been proposed as compatible with DNA preservation (Poinar et al. 1996). Although the general usefulness of the racemization data has been debated (Collins et al. 1999), we estimated the D/L ratio for three amino acids (aspartic acid, glutamine and alanine) for control purposes related to the general taphonomical conditions of the site, in two randomly chosen specimens, following procedures described in Caramelli et al. (2003).
(c) Amplification, cloning and sequencing
The mtDNA HVRI region (Anderson et al. 1981) was amplified in 21 Neolithic samples in different overlapping fragments with sizes ranging from 98 to 212 bp (excluding primers) combining several primer pairs (table 1). Some additional primer pairs were used to amplify mtDNA coding regions, where diagnostic SNPs that unequivocally define haplogroups in the mtDNA genealogy are located (table 2). PCR amplifications were performed in 25 μl reactions with 1–5 μl of extract (some extracts were subjected to 1 : 3 dilution in order to overcome inhibitors), 1.2 U of taq polymerase (Ecogen), 1X reaction buffer (Ecogen), 1.4 mg ml−1 BSA, 2.1 mM MgCl2, 0.2 mM dNTPs and 1 μM of each primer. The PCRs were subjected to 40 amplification cycles (1 min step at 94°C, 1 min step at 50°C and 1 min step at 72°C) with an initial denaturing step at 94°C for 2 min and a final elongating step for 7 min at 72°C.
Table 1.
Primers used to amplify the HVRI of the mitochondrial genome in the Neolithic samples.
| L primer | sequence (5′→3′) | H primer | sequence (5′→3′) |
|---|---|---|---|
| L16022 | CTAATTTAAACTATTCTCT | H16142 | ATGTACTACAGGTGGTCAAG |
| L16055 | GAAGCAGATTTGGGTACCAC | H16156 | TGTGGATTGGGTTTTTATGTAC |
| L16081 | TTGACTCACCCATCAACAA | H16211 | TAGTTGAGGGTTGATTGCTG |
| L16122 | CATTACTGCCAGCCACCATGAATA | H16218 | TGTGTGATAGTTGAGGGTTG |
| L16131 | CACCATGAATATTGTACGGT | H16247 | CAACTATCACACATCAACTGCAA |
| L16185 | AACCCAATCCACATCAAAACC | H16282 | TTAAGGGTGGGTAGGTTTG |
| L16209 | CCCCATGCTTACAAGCAAGT | H16378 | CAAGGGACCCCTATCTGAGG |
| L16247 | CAACTATCACACATCAACTGCAA | H16401 | TGATTTCACGGAGGATGGTG |
| L16261 | CAACTATCACACATCAACTGCAA | ||
| L16347 | CGTACATAGCACATTACAGT | ||
| L16313 | CCCTTAACAGTACATAATAC |
Table 2.
Primers used to amplify diagnostic SNPs in the coding region of the mtDNA genome.
| L primer | sequence (5′→3′) | H primer | sequence (5′→3′) | haplogroup |
|---|---|---|---|---|
| L12227 | GAAAGCTCACAAGAACTGC | H12341 | GGTTATAGTAGTGTGCATGG | U (12208) |
| L 6999 | CAAACTCATCACTAGACATCG | H 7066 | GAATGAAGCCTCCTATGATGG | H (7028) |
| L13669 | CACCCTTACTAACATTAACG | H13725 | TAGTAATGAGAAATCCTGCG | J (13708) |
| L13257 | AATCGTAGCCTTCTCCACTTCA | H13372 | TTGTTAAGGTTGTGGATGAT | T (13368) |
| L56 | GAGCTCTCCATGCATTTGGT | H131 | GGATGAGGCAGGAATCAAG | Pre-HV (073) |
| L10014 | TTTTAGTATAAATAGTACCG | H10088 | GTAGTAAGGCTAGGAGGGTG | I (10034) |
| L8215 | ACAGTTTCATGCCCATCGTC | H8297 | ATGCTAAGTTAGCTTTACAG | W (8251) |
PCR products were electrophoresed in 1.6% low-melting point agarose gels (Invitrogen) stained with ethidium bromide. Bands with the expected right size were excised from the gel, purified with silica and routinely cloned using pMOS Blue blunt end cloning kit (Amersham Biosciences) following the manufacturer's instructions. In brief, 7 μl of PCR product was treated with pK enzyme mix, incubated at 22°C for 40 min and ligated into pMOS Blue vector overnight. Two microlitres of the ligation product were transformed into 40 μl of competent cells, grown in 160 μl of SOC medium at 37°C for 1 h and plated on IPTG/X-gal agar plates. After 16 h, white colonies were subjected to direct PCR screening using T7 and U-19 universal primers. Inserts that yielded the correct size were identified by agarose gel electrophoresis, purified and sequenced with an ABI 3100 DNA sequencer (Applied Biosystems), following the supplier's instructions.
(d) Uracil-N-glycosylase (UNG) treatment
Hydrolytic deamination of cytosines causes uracil residues that are incorrectly read by the polymerase, resulting in false C→T/G→A changes in the clone sequences (Hofreiter et al. 2001a); this is the most common form of post-mortem damage in aDNA sequences (Stiller et al. 2006). In three extracts (table 3), a UNG treatment was followed in order to eliminate possible miscoding lesions. Ten microlitres of DNA extract were treated with 1 U of UNG for 30 min at 37°C to excise uracil residues in the original template (Hofreiter et al. 2001b). After this treatment, extracts were subjected to the same PCR amplifications described above and subsequently cloned.
Table 3.
Amplification strategy of the Neolithic samples. (R, fragment independently replicated; UNG, fragment with UNG treatment of the extract.)
| specimen | fragments amplified | no. of clones |
|---|---|---|
| Neolithic 1 | 16 022–16 156 (R) | 4 |
| 16 055–16 142 | 3 | |
| 16 083–16 282 (R) | 4 | |
| 16 131–16 211 (UNG) | 9 | |
| 16 185–16 261 | 5 | |
| 16 247–16 378 | 5 | |
| Neolithic 5 | 16 131–16 261 (R) | 5 |
| 16 247–16 378 | 8 | |
| Neolithic 6 | 16 055–16 142 | 2 |
| 16 131–16 211 | 4 | |
| 16 122–16 261 | 2 | |
| 16 247–16 378 | 7 | |
| Neolithic 8 | 16 055–16 142 | 8 |
| 16 122–16 261 | 3 | |
| 16 247–16 378 | 3 | |
| 16 247–16 378 (UNG) | 7 | |
| 16 313–16 401 | 10 | |
| Neolithic 10 | 16 122–16 261 | 6 |
| 16 247–16 378 | 5 | |
| 16 247–16 378 | 5 | |
| Neolithic 11 | 16 055–16 142 | 3 |
| 16 122–16 261 | 7 | |
| 16 131–16 211 | 9 | |
| 16 247–16 378 | 10 | |
| 16 209–16 401 | 8 | |
| Neolithic 12 | 16 055–16 142 | 4 |
| 16 131–16 211 | 10 | |
| 16 185–16 261 | 5 | |
| 16 247–16 378 | 7 | |
| Neolithic 14 | 16 055–16 142 | 5 |
| 16 122–16 218 | 10 | |
| 16 131–16 211 | 9 | |
| 16 247–16 378 | 9 | |
| Neolithic 21 | 16 055–16 142 | 8 |
| 16 122–16 261 | 8 | |
| 16 209–16 401 | 11 | |
| Neolithic 22 | 16 055–16 142 | 11 |
| 16 122–16 218 | 7 | |
| 16 247–16 378 | 6 | |
| Neolithic 23 | 16 022–16 142 (UNG) | 5 |
| 16 122–16 158 | 7 | |
| 16 209–16 378 | 6 |
(e) Statistical analysis
The information obtained from the HVRI sequence (table 4) together with the result of typing different diagnostic coding SNPs allows us to classify each Neolithic sample into its corresponding mitochondrial DNA haplogroup in the well-known mtDNA phylogeny. To explore comparatively the haplogroup composition of the Iberian Neolithics together with current populations from the Iberian Peninsula (Galicians, Cantabrians, Basques, Aragoneses, Catalans, Valencians, Andalucians, Central Spain, North Portugal, Central Portugal and South Portugal), the southeast of Europe (Croatians, Romanians, Bulgarians, Albanians, Italians and Greeks) and the Middle East, the putative place of Neolithic origin (Palestinian, Druze, Jordanians, Syrians, Iraquis and Kurds), a correspondence analysis on haplogroup frequencies was performed with Statistica software (StatSoft, Inc., 2001 v. 6). As the only ancient population from the same historical period available until now was that analysed by Haak et al. (2005), we performed the same corresponding analysis, but included this population in the dataset as well.
Table 4.
mtDNA haplotype and haplogroup composition of the Iberian Neolithic samples. (Specimen 5 could also have 129A (this position has not been typed).)
| specimen | HVRI mtDNA haplotype | haplogroup |
|---|---|---|
| Neolithic 1 | CRS | H |
| Neolithic 5 | (129A) 223T 264T 270T 311C 319A 362C | I1 |
| Neolithic 6 | CRS | H |
| Neolithic 8 | 069T 126C | J1c |
| Neolithic 10 | 223T, 292T, 295T, 304C | W1 |
| Neolithic 11 | 288C 362C | H |
| Neolithic 12 | CRS | H |
| Neolithic 14 | 069T 126C | J1c |
| Neolithic 21 | 126C, 140C, 294T, 296T, 311C | T2 |
| Neolithic 22 | 126C, 140C, 189C, 294, 296T, 311C | T2 |
| Neolithic 23 | 134T | U4 |
3. Results
(a) Characterization of endogenous sequences
Twenty-three Neolithic remains were analysed; two samples yielded no amplification products and were subsequently discarded, nine samples were discarded due to the irreproducible or fragmentary results and four more samples could not be unambiguously attributed to one of the main European mitochondrial DNA lineages. The remaining 11 sequences were considered to be endogenous and included in the posterior population analysis. The amino acid racemization values obtained for two of these samples (numbers 1 and 5) are fully compatible with DNA preservation. In the first sample, the D/L value was 0.0564 (Asp), 0.0086 (Glu) and 0.0048 (Ala); in the second, the D/L value was 0.0756 (Asp), 0.0222 (Glu) and 0.0122 (Ala). The misincorporation ratios in the fragments subjected to UNG treatment were not significantly lower than those generated without UNG from the same extracts (data not shown). Therefore, no significant differences in the damage were expected between samples with and without UNG treatment.
A total of 572 clones were sequenced, from which 98 (17.13%) could be definitely identified as being from the only six people involved in the manipulation and laboratory analysis of the Neolithic remains (Sampietro et al. 2006). Since we were able to monitor all the people who had ever had access to this set of samples, it was possible to track down the pre-laboratory-derived contaminant sequences and, consequently, we could definitely eliminate them from the generated cloning dataset (electronic supplementary material).
However, we faced some problems in particular situations related to the impossibility of working with long DNA fragments. For instance, two Neolithic samples predominantly display the haplotype 069T, 126C and two out of the six handlers also have that haplotype, albeit only in the first part of the HVRI sequence (M.L.S. has the haplotype 069T, 126C, 185T, 189C and R.P. 069T, 126C, 278T, 366T). Therefore, 069T, 126C could potentially be a contaminant, but two results allow us to consider these sequences as endogenous: they reach frequencies up to 85% of the clones in the amplified 055–142 fragment, while up to 85% of the clones for the second half of the HVRI are essentially CRS. Therefore, the alternative hypothesis that the first fragment was totally contaminated while the second one was almost free of contaminants seems less plausible. In addition, we must consider the problem that contaminant sequences with no substitutions in particular fragments will result in a background of CRS sequences (for instance, M.M. has the 129A haplotype and accounts for 20.41% of detected contaminant sequences and will undoubtedly result in CRS sequences in the second half of the HVRI fragments, while E.V. carrying the 298C haplotype will result in CRS sequences in the first HVRI half). Therefore, some fragments display a rather high level of CRS sequences that probably correspond to this unspecific contaminant background. However, the putative endogenous sequences share some characteristics, such as to be reproducible, to be in many cases exclusive of a particular sample and to be present in higher frequencies than the distinguishable contaminants. Moreover, three Neolithic samples probably have CRS as endogenous haplotype, since these sequences are in overwhelming majority and present in higher frequency than the detected contaminants in the other Neolithic samples. Finally, some sporadic background of contamination in the air or the reagents cannot be discarded, and this phenomenon can account for some few distinctive clones.
(b) Neolithic haplotype sequences
Iberian Neolithic sequences showed haplotypes (see table 4) widely distributed throughout Europe when comparing them with a haplotype dataset composed of more than 10 000 individuals from Europe and the Middle East (information compiled by F. Calafell, UPF). Interestingly, one of them carries one haplotype (223T, 292T, 295T, 304C) that is only found in the Middle East while the other two Neolithic samples display haplotypes that are only found in the Iberian Peninsula (223T, 264T, 270T, 311C, 319A, with the addition of 129A, not typed in this specimen) and in Italy (126C, 140C, 189C, 294T, 296T, 311C).
Nevertheless, there are two samples that show a particular haplotype not found in the dataset: one Neolithic individual is assigned to the haplogroup T2, but the haplotype (126C, 140C, 294T, 296T, 311C) does not show the 189C substitution that is always found associated with this haplotype. The most plausible situation is that this mtDNA haplotype is ancestral to the haplotype with 189C. Another Neolithic sample displays a U haplogroup sequence with the 134T substitution that has always been described in association with 356C. Again, the most plausible explanation for the absence of 356C is the ancestrality of the 134T haplotype.
(c) Neolithic haplogroups
The general haplogroup composition of the Neolithic sample is: H (36.4%); T2 (18.2%); J1c (18.2%); I1 (9.1%); U4 (9.1%); and W1 (9.1%) (table 4). Although the sample size is recognized to be small and, consequently, some haplogroups are not represented, the general composition is not significantly different from that obtained from the current Iberian Peninsula dataset when random resamplings of 11 sequences are made (data not shown).
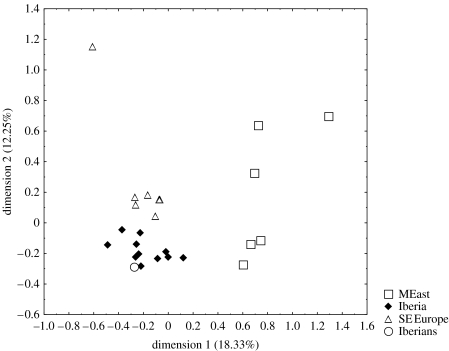
The correspondence analysis shows that the Iberian Neolithic population clusters together with the modern Iberian populations, but not with the Middle East groups (figure 1). This further suggests that the haplogroup composition of the Iberian Neolithic population is not significantly different from that found in the current population from the Iberian Peninsula.
Figure 1.
Correspondence analysis with present populations from Iberian Peninsula (Iberia), Middle East (MEast) and southeast of Europe (SE Europe). The Neolithics from the Iberian Peninsula (Iberians) are also included.
The correspondence analysis performed with the previous populations and the addition of data by Haak et al. (2005; figure 2) indicate that, considering the haplogroup composition, the North Central European Neolithic population analysed by these authors is, in fact, quite different from modern European populations, from the Iberian Neolithic population analysed here and even from those of the Near East, making by itself the second dimension of the correspondence analysis. This can be attributed to the unusually high frequency of N1a haplogroup found in the Central European Neolithic population (Haak et al. 2005) but not in the other populations so far analysed.
Figure 2.
Correspondence analysis with populations from Iberian Peninsula (Iberia), Middle East (MEast) and southeast of Europe (SE Europe), along with Neolithics from North Central Europe (NCEurope) and from the Iberian Peninsula (NIberians).
4. Discussion
There has been a general tendency to try to understand the spread of the Neolithic in Europe as a result of a single, unique and homogeneous process, but in fact there is evidence against this simplistic view. Zvelebil (2004) has shown evidence of geographical stratification in the local adoption of agriculture between Atlantic and Mediterranean models. Archaeologically, two main cultural traditions, marked by two different potteries, can be distinguished in the Early Neolithic: the linear pottery culture (or LBK) that runs along the Danubian route and the impressed ware pottery (also called cardial) that spreads along the Mediterranean. This is not just a question on ceramic decoration. The diffusion of the new economy took two main routes after the colonizations of the Balkans that implied different necessities of adaptation of the agriculture and the farming to specific climatic and ecological conditions. In this sense, it is probable that the Palaeolithic populations should have had lesser demographic numbers in the Mediterranean than in the Atlantic and Central Europe, the former being a less productive area for hunting and gathering (Zvelebil 2004).
Haak et al. (2005) when analysing an older Neolithic population (7000–7500 years BP) from Central Europe found a genetic discontinuity between linear pottery Neolithics and current European populations. Twenty-five per cent (6 out of 24) of the samples were of a distinctive and rare N1a lineage (currently present at 0.2% in the European population) of the mtDNA well-known phylogeny. They discarded the possibility that genetic drift affected the N1a lineages over the last 7500 years by means of demographic models, and proposed that small pioneer farming groups carried farming into new areas of Europe, and therefore the dispersal of the agricultural techniques were through a cultural model; hence, at the end of the process, the frequency of the N1a haplogroup was diluted to the low modern value that is observed today.
The genetic continuity found in this study in the Iberian Peninsula since the Neolithic period sharply contrasts with what was found by Haak et al. (2005). The absence of sequences carrying the N1a haplogroup in the Iberian Neolithic population could be due to the difference in time (approx. 2000 years) and geographical distance (North versus South Europe) with those specimens analysed by Haak et al. (2005). However, if the absence of N1a lineage cannot be explained by genetic drift, as suggested by Haak et al. (2005), other hypotheses (Barbujani & Chikhi 2006) have to be invoked in order to explain such a discrepancy between their results and those observed in the current work.
First, it seems evident that in the impressed ware or cardial culture studied here, there is no evidence for a genetic discontinuity. Second, this implies that the Neolithic spread was neither genetically nor geographically a uniform process in Europe. We hypothesize that the dispersal of agriculture involved both demographic and cultural diffusion, depending on the region where it took place. Whereas the dispersal of the agricultural in Central Europe could follow a cultural diffusion model, in the Mediterranean our results suggest a demic diffusion model. This finding is in agreement with that of most archaeologists who consider the impressed ware complex as representing a cultural and demic intrusion, and essentially not a local development (Zilhao 2000). Thus, the south–north heterogeneity shown by archaeologists could also have a genetic correspondence and, in fact, some studies (Comas et al. 1997; Simoni et al. 2000) have observed a significant east to west clinal variation in the Mediterranean, which was not possible to detect in Central and Northern Europe.
To test this hypothesis, we should also analyse Palaeolithic populations from the Iberian Peninsula and Central Europe. The only previous aDNA analysis on 25 000-year-old European remains (Caramelli et al. 2003) yielded two mtDNA sequences from the pre-HV and N haplogroups that seem compatible with both models. Therefore, the palaeogenetic study of additional specimens is definitively needed to clarify the Neolithic expansion into Europe. Nevertheless, since handling and washing the remains uncovered cause huge amounts of contamination, we strongly recommend future researchers to adhere to our methodological rules, i.e. to analyse only skeletal material from sites where all the people involved in the excavation and analysis are known and can be genetically typed.
Acknowledgments
This research was supported by the Ministerio de Educación y Ciencia of Spain (grant CGL2006-03987), the Institut d'Estudis Catalans and by a fellowship AP2002-1065 to M.L.S. We are grateful to Francesc Calafell (University Pompeu Fabra, Barcelona) for allowing us to use the European mtDNA database.
Supplementary Material
Supporting information: clone sequences for the Neolithic samples
References
- Ammerman A.J, Cavalli-Sforza L.L. The Neolithic transition and the genetics of population in Europe. Princeton University Press; Princeton, NJ: 1984. [Google Scholar]
- Ammerman A.J, Pinhasi R, Banffy E. Comment on “Ancient DNA from the first European farmers in 7500-year-old Neolithic sites”. Science. 2006;312:1875. doi: 10.1126/science.1123984. doi:10.1126/science.1123936 author reply 1875. [DOI] [PubMed] [Google Scholar]
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. doi:10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- Barbujani G, Chikhi L. Population genetics: DNAs from the European Neolithic. Heredity. 2006;97:84–85. doi: 10.1038/sj.hdy.6800852. doi:10.1038/sj.hdy.6800852 [DOI] [PubMed] [Google Scholar]
- Barbujani G, Goldstein D.B. Africans and Asians abroad: genetic diversity in Europe. Annu. Rev. Genomics Hum. Genet. 2004;5:119–150. doi: 10.1146/annurev.genom.5.061903.180021. doi:10.1146/annurev.genom.5.061903.180021 [DOI] [PubMed] [Google Scholar]
- Caramelli D, et al. Evidence for a genetic discontinuity between Neandertals and 24,000-year-old anatomically modern Europeans. Proc. Natl Acad. Sci. USA. 2003;100:6593–6597. doi: 10.1073/pnas.1130343100. doi:10.1073/pnas.1130343100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L.L, Menozzi P, Piazza A. History and geography of human genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Chikhi L, Destro-Bisol G, Bertorelle G, Pascali V, Barbujani G. Clines of nuclear DNA markers suggest a largely Neolithic ancestry of the European gene pool. Proc. Natl Acad. Sci. USA. 1998;95:9053–9058. doi: 10.1073/pnas.95.15.9053. doi:10.1073/pnas.95.15.9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Nichols R.A, Barbujani G, Beaumont M.A. Y genetic data support the Neolithic demic diffusion model. Proc. Natl Acad. Sci. USA. 2002;99:11 008–11 013. doi: 10.1073/pnas.162158799. doi:10.1073/pnas.162158799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M.J, Waite E.R, van Duin A.C. Predicting protein decomposition: the case of aspartic-acid racemization kinetics. Phil. Trans. R. Soc. B. 1999;354:51–64. doi: 10.1098/rstb.1999.0359. doi:10.1098/rstb.1999.0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, Pérez-Lezaun A, Bosch E, Bertranpetit J. Mitochondrial DNA variation and the origin of the Europeans. Hum. Genet. 1997;99:443–449. doi: 10.1007/s004390050386. doi:10.1007/s004390050386 [DOI] [PubMed] [Google Scholar]
- Cooper A, Poinar H.N. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. doi:10.1126/science.289.5482.1139b [DOI] [PubMed] [Google Scholar]
- Gilbert M.T.P, Willerslev E, Hansen A.J, Barnes I, Rudbeck L, Lynnerup N, Cooper A. Distribution patterns of postmortem damage in human mitochondrial DNA. Am. J. Hum. Genet. 2003;72:32–47. doi: 10.1086/345378. doi:10.1086/345378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M.T.P, et al. Biochemical and physical correlates of DNA contamination in archaeological human bones and teeth excavated at Matera, Italy. J. Arch. Sci. 2005;32:785–793. doi:10.1016/j.jas.2005.02.006 [Google Scholar]
- Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- Handt O, Hoss M, Krings M, Pääbo S. Ancient DNA: methodological challenges. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. doi:10.1007/BF01921720 [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Serre D, Poinar H.N, Kuch M, Pääbo S. Ancient DNA. Nat. Rev. Genet. 2001a;2:353–359. doi: 10.1038/35072071. doi:10.1038/35072071 [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, Haeseler A.v, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001b;29:4793–4799. doi: 10.1093/nar/29.23.4793. doi:10.1093/nar/29.23.4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalueza-Fox C, et al. Neandertal evolutionary genetics: mitochondrial DNA data from the Iberian Peninsula. Mol. Biol. Evol. 2005;22:1077–1081. doi: 10.1093/molbev/msi094. doi:10.1093/molbev/msi094 [DOI] [PubMed] [Google Scholar]
- Malmström H, Stora J, Dalen L, Holmlund G, Götherstrom A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol. Biol. Evol. 2005;22:2040–2047. doi: 10.1093/molbev/msi195. doi:10.1093/molbev/msi195 [DOI] [PubMed] [Google Scholar]
- Malmström H, Svensson E.M, Gilbert M.T.P, Willerslev E, Götherstrom A, Holmlund G. More on contamination: the use of asymmetric molecular behaviour to identify authentic ancient human DNA. Mol. Biol. Evol. 2007;24:998–1004. doi: 10.1093/molbev/msm015. doi:10.1093/molbev/msm015 [DOI] [PubMed] [Google Scholar]
- Poinar H.N, Hoss M, Bada J.L, Pääbo S. Amino acid racemization and the preservation of ancient DNA. Science. 1996;272:864–866. doi: 10.1126/science.272.5263.864. doi:10.1126/science.272.5263.864 [DOI] [PubMed] [Google Scholar]
- Richards M. The Neolithic invasion of Europe. Annu. Rev. Anthropol. 2003;32:135–162. doi:10.1146/annurev.anthro.32.061002.093207 [Google Scholar]
- Richards M, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- Rosser Z.H, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 2000;67:1526–1543. doi: 10.1086/316890. doi:10.1086/316890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampietro M.L, Caramelli D, Lao O, Calafell F, Comas D, Lari M, Agusti B, Bertranpetit J, Lalueza-Fox C. The genetics of the pre-Roman Iberian Peninsula: a mtDNA study of ancient Iberians. Ann. Hum. Genet. 2005;69:535–548. doi: 10.1111/j.1529-8817.2005.00194.x. doi:10.1111/j.1529-8817.2005.00194.x [DOI] [PubMed] [Google Scholar]
- Sampietro M.L, Gilbert M.T.P, Lao O, Caramelli D, Lari M, Bertranpetit J, Lalueza-Fox C. Tracking down human contamination in ancient human teeth. Mol. Biol. Evol. 2006;23:1801–1807. doi: 10.1093/molbev/msl047. doi:10.1093/molbev/msl047 [DOI] [PubMed] [Google Scholar]
- Semino O, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. doi:10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G. Geographic patterns of mtDNA diversity in Europe. Am. J. Hum. Genet. 2000;66:262–278. doi: 10.1086/302706. doi:10.1086/302706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller M, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:13 578–13 584. doi: 10.1073/pnas.0605327103. doi:10.1073/pnas.0605327103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, et al. MtDNA analysis reveals a major Late Paleolithic population expansion from southwestern to northeastern Europe. Am. J. Hum. Genet. 1998;62:1137–1152. doi: 10.1086/301822. doi:10.1086/301822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle A. Europe in the Neolithic. The creation of New Worlds. Cambridge University Press; Cambridge, UK: 1996. [Google Scholar]
- Zilhao J. From the Mesolithic to the Neolithic in the Iberian Peninsula. In: Price T.D, editor. Europe's first farmers. Cambridge University Press; Cambridge, UK: 2000. pp. 144–182. [Google Scholar]
- Zvelebil M. Who were we 6,000 years ago? In search of prehistoric identities. In: Jones M, editor. Traces of ancestry: studies in honour of Colin Renfrew. McDonald Institute Monographs; Cambridge, UK: 2004. pp. 41–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information: clone sequences for the Neolithic samples