Abstract
Background
Staphylococcus aureus is a major cause of food poisoning due to their ability to produce enterotoxins which if ingested in sufficient amounts results in sickness. Food handlers carrying enterotoxin-producing S. aureus in their noses or hands can contaminate food leading to food poisoning. We characterized 200 S. aureus obtained from food handlers in different restaurants for antibacterial resistance and the carriage of virulence genes.
Findings
Susceptibility to antibacterial agents was determined by disk diffusion and Etest. PCR was used to detect genes for accessory gene regulator (agr); capsular polysaccharide (cap) 5 and 8, staphylococcal enterotoxins (SE), toxic shock syndrome toxin-1 (TSST-1) and Panton-Valentine leukocidin (PVL). Isolates were typed using pulsed-field gel electrophoresis. In total 185 (92.5%) of the 200 isolates expressed resistance to antibacterial agents. They were resistant to penicillin G (82.0%), tetracycline (19.0%), erythromycin (2.5%), clindamycin (2.0%), trimethoprim (7.5%), kanamycin (2.5%), streptomycin (1.5%), ciprofloxacin (1.5%), fusidic acid (1.0%) and cadmium acetate (68.0%). Seventy-six (38.0%) and 114 (57.0%) isolates had type 5 and type 8 capsular polysaccharides respectively. The agr types I, II and III alleles were detected in 50.5%, 20.0% and 23.5% of the isolates respectively. They contained genes for SEI (38.5%), SEG (24.0%), SEC (23.0%), SEB (12.5%), SEH (21.5%), SEA (11.0), SED (1.5%), SEE (1.5%), TSST-1 (4.0%) and PVL (9.0%).
Conclusion
This study revealed a high prevalence of antibacterial resistance and virulence determinants in S. aureus from food handlers in Kuwait restaurants justifying the screening of food handlers to detect and treat carriers and protect restaurant customers from staphylococcal food poisoning.
Background
Staphylococcus aureus can cause localized and invasive infections in humans. This is attributed to its ability to produce a variety of virulence factors such as capsular polysaccharides, staphylococcal enterotoxins (SEs), toxic shock syndrome toxin 1 (TSST-1) [1,2], panton – valentine leukocidin (PVL) [3] and accessory gene regulators (agr) [1,4-6]. Although S. aureus isolates produce one of 11 serotypes of capsular polysaccharides, most clinical isolates belong to serotypes 5 and 8 [2]. Enterotoxin-producing S. aureus are common causes of food poisoning in many parts of the world. The ingestion of the preformed toxins in food often leads to the development of food poisoning. The symptoms typically have a rapid onset (2–6 h) and may include nausea, vomiting, diarrhea and abdominal pain [1]. Nasal and hand carriage of enterotoxigenic S. aureus by food handlers is an important source of staphylococcal food contamination in restaurants and fast food outlets [7]. Therefore it is important to detect S. aureus carriage among food handlers to prevent possible food contamination by them resulting in food poisoning.
Food poisoning outbreaks result in huge financial losses to restaurants, in addition to the loss of reputation and confidence among the public. Staphylococcal food-borne diseases are estimated to cause 6 – 81 million illnesses and up to 9000 deaths, and accounts for 14–20% of outbreaks involving contaminated food in the USA [8].
Most of the studies on S. aureus associated with food poisoning have focused on screening of the isolates for enterotoxins [1,9-13] with only sparse data on the carriage of other virulence factors and antimicrobial resistance among S. aureus obtained from food handlers [14-17] especially in the Arabian Gulf countries. Although we previously studied the prevalence of staphylococcal enterotoxins in S. aureus isolated from food handlers in Kuwait city [10], their susceptibility to antibacterial agents were not investigated. Therefore in the present study, we characterized S. aureus isolated from food handlers in Kuwait City restaurants for their susceptibility to antibacterial agents and the carriage of virulence associated genes.
Methods
S. aureus isolates
In total, 200 S. aureus isolates were recovered from food handlers working in 50 Kuwait City restaurants from 2003 to 2005. They consisted of 133 (102 isolates from nasal and 31 hand swabs) of 500 swabs from 250 adult male workers in 50 restaurants, during screening of food handlers as demanded by the City Council, yielding a carriage rate of 53.2%. The Kuwait Municipal Council mandated a compulsory screening of restaurant workers to detect S. aureus carriers. Only one sample was investigated from the same food handler. The study also included 67 isolates obtained from 31 stool samples, 9 throat swabs and 27 nasal samples obtained from food handlers during routine investigations of suspected cases of food poisoning in different restaurants. Bacteria were isolated and identified using standard bacteriological methods including cultural characteristics, Gram stain, catalase, tube coagulase and DNase tests [10]. The isolates were preserved in skimmed milk at -80°C.
Susceptibility to antibacterial agents
Susceptibility to antimicrobial agents was determined by the disk diffusion method [18] Disks containing cadmium acetate (50 μg), propamidine isethionate (100 μg), and mercuric chloride (109 μg) were prepared in the laboratory [19]. A zone diameter of 6–10 mm indicated resistance to cadmium acetate, propamidine isethionate and mercuric chloride. The minimum inhibitory concentrations (MICs) of methicillin, vancomycin and teicoplanin were determined with Etest strips (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. S. aureus strain ATCC25923 was used for quality control.
Isolation of DNA for PCR amplification
DNA for PCR was extracted as described previously [3]. The extracted DNA was used for PCR immediately or stored at 4°C and used within three months.
Detection of genes for staphylococcal enterotoxin (SE) and toxic shock syndrome toxin (TSST-1)
PCR amplification was performed for genes encoding staphylococcal enterotoxins SEA (sea), SEB (seb), SEC (sec), SED (sed), SEE (see), SEG(seg), SEH (seh), and SEI (sei) and toxic shock syndrome gene TSST-1(tst) with primers published previously [13,20] in three multiplex (MT) assays as follows: MT1 (sec, sei and sed), MT 2 (seg, seh and see) and MT 3 (sea, seb and tst). Based on optimization experiments, the primer concentrations used were 0.1 μM for sea, sec, and tst, and 0.2 μM for seb, see, sed, seg and seh. Amplifications comprised an activation step at 95°C for 15 min followed by 37 cycles of 94°C for 2 min, 51°C for 1 min and 72°C for 1 min with a final extension at 72°C for 7 min. Control strains used were S. aureus; ATCC13565 (SEA), ATCC14458 (SEB), ATCC23235 (SED), ATCC27664 (SEE), ATCC51811 (SEH), WBG525 (SEC, TSST-1), C90 (SEI) [16], C29 (SEG). The 16S rRNA primers, 5'-GGA GGA AGG TGG GGA TGA CG-3' and 5'-ATG GTG TGA CGG GCG GTG TG-3' [21] were used as internal controls. All tests were considered positive if the positive controls and the internal controls yielded the expected results. Negative controls for all PCR consisted of the PCR mixes without template DNA. PCR products (5 μl) were loaded in 2.0% (w/v) molecular biology grade agarose (BioRad, Hercules, USA) gels. PCR products were analyzed by agarose gel electrophoresis.
Detection of genes for accessory gene regulators (agr), capsular polysaccharides (cap) and Panton-Valentine Leucocidin (lukS-lukF)
All isolates were tested for the presence of genes for PVL, agr types I, II, III and IV and cap type 5 and type 8 in PCR assays as described previously [3,22,23]. The 16S rRNA primer [21] was used as internal control in PCR amplifications.
Pulsed-field gel electrophoresis (PFGE)
Counter-clamped homogeneous electric field (CHEF) electrophoresis of Sma I-digested chromosomal DNA was performed as described previously [19].
Statistics
Differences in the distribution of virulence determinants between groups were assessed by Chi square test. A P-value of = 0.05 was considered significant.
Results
One hundred and eighty five (92.5%) of the 200 S. aureus isolates were resistant to antibacterial agents. The majority, 164 (82.0%) and 136 (68.0%) of them were resistant to penicillin G and cadmium acetate respectively. Forty-two isolates (21.0%) were resistant to tetracycline, 15 isolates (7.5%) were resistant to trimethoprim and six isolates (3%) were resistant to kanamycin. Five and four isolates were resistant to erythromycin and streptomycin respectively. Two isolates were resistant to fusidic acid and only one isolate was resistant to methicillin and gentamicin. They were all susceptible to mupirocin, rifampicin, vancomycin, teicoplanin and linezolid. Sixty-five percent of them were resistant to two or more antibacterial agents and 23% were multiresistant with resistance to three or more classes of antibacterial agents.
The distribution of genes for capsular types and agr allotypes is summarized in Table 1. Whereas 114 isolates had gene for cap8, 76 isolates had gene for cap5. Ten isolates yielded negative results for both capsular polysaccharides. Only one agr type was detected in each isolate. One hundred and one isolates were agr type I, 47 isolates were type III, and 40 isolates were type II. None was positive for agr type IV. Twelve isolates yielded negative results for the four agr types tested.
Table 1.
Distribution of genes for virulence factors in 200 S. aureus isolated from food handlers.
| Number (%) of isolates positive for virulence genes | ||||||||
| Sources of S. aureus (#) | SE | cap5 | cap8 | agr I | agr II | agrIII | lukS-lukF | tst |
| Nose (129) | 93 (72.1) | 49 (38.0) | 72 (55.8) | 67 (51.9) | 24 (18.6) | 28 (21.7) | 11 (8.5) | 7 (5.4) |
| Stool (31) | 21 (67.7) | 15 (48.4) | 15 (48.4) | 17 (54.8) | 6 (19.3) | 7 (22.5) | 4 (12.9) | 1 (3.2) |
| Hands (31) | 22 (70.9) | 9 (29.0) | 21 (67.7) | 13 (41.9) | 6 (19.3) | 11 (34.0) | 3 (9.7) | ND |
| Throats (9) | 6 (66.6) | 3 (33.3) | 6 (66.6) | 4 (44.4) | 4 (44.4) | 1 (11.1) | ND | ND |
| Total (200) | 142 (71.0) | 76 (38.0) | 114 (57.0) | 101 (50.5) | 40 (20.0) | 47 (23.5) | 18 (9.0) | 8 (4.0) |
Abbreviations: cap, genes for capsular polysaccharide, agr, genes for accessory gene regulator, luKS-lukF genes for PVL, tst, genes for TSST-1. ND, Not detected
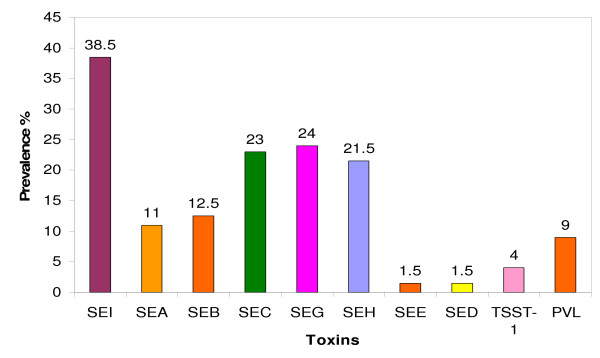
The distribution of genes for SE, TSST-1 and PVL is summarized in Figure 1 and Table 2. In total, 142 (71.0%) isolates were positive for SE, 8 (4.0%) were positive for TSST-1 and, 18 isolates, including the only MRSA isolate, were positive for PVL. Gene for SEI was the most common SE gene. It was detected in 77 isolates. Fifty isolates contained single SE genes, 54 isolates carried two SE genes, 28 and 10 isolates contained three and four SE genes respectively.
Figure 1.
Distribution of toxins in 200 S. aureus isolated from food handlers.
Table 2.
Enterotoxin gene profiles of the 142 enterotoxin-positive S. aureus from food handlers.
| Enterotoxin profiles | |
| 1 |
S aureus isolates with single SE genes (50) sea (6), seb (9), sec (13), seg (9), seh (6), sei (7) |
| 2 |
S. aureus isolates with two SE genes (54) seg, sei (13); seh, sei (17); sea, sec (4); sec, seg (4); sec, seh (4); seb, seg (3); sea, sei (2); seb, sei (2); seg, sed (1); seb, sec (1); sec, sei (1); see, sei (1); seb, see (1). |
| 3. |
S. aureus isolates with three SE genes (28) sec, seg, sei (9); sec, seh, sei (5); sea, seg, sei (4); seb, seg, sei (3); sea, seh, sei (3); seb, seh, sei (2);; sec, sed; seg (1); seb, sec, seg (1). |
| 4 |
S. aureus isolates with four SE genes (10) sea, sec, seg, sei (3); sea, sec, seh, sei (1); seb, seg, seh, sei (1); seb, seg, seh, sei (1); seb, see, seh, sei (1); sea, seg, seh, sei (1); sea, sec, sed, seh(1); seb, sec, seh, sei (1). |
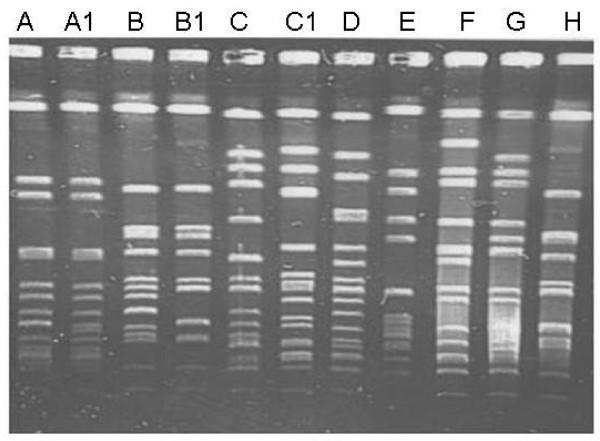
The characteristics of the 18 lukS-lukF positive isolates are presented in Table 3. They had different resistance patterns, belonged to different agr and cap types and harbored different SE genes but none carried gene for TSST-1. When typed by PFGE to determine if they were related, they yielded eight PFGE patterns designated types A to H (Figure 2, Table 3). Eight isolates obtained from stool, hand and nasal samples had the same PFGE pattern (type A) and 10 isolates belonged to seven PFGE patterns.
Table 3.
Characteristics of 18 S. aureus isolates containing lukS-lukF
| Isolates | Source | Resistance pattern | agr class | cap type | SE genes | PFGE pattern |
| 33N | Nose | Pc, Tc, Tp, Cd | III | 8 | sea, sei | A |
| 78N | Nose | Mc, Pc, Km, Tc, Fd, Cd | III | 8 | ND | A |
| 157N | Nose | Pc, Km, Sm | III | 8 | Seg | A |
| 342N | Nose | Cd | III | 8 | Sei | A1 |
| 515N | Nose | Cd | III | 8 | ND | H |
| 42N | Nose | Pc, | I | 5 | seb, seh, sei | B |
| 46N | Nose | Pc, Sm | I | 5 | seh, sei | C |
| 268N | Nose | Cd | I | 8 | ND | C |
| 298N | Nose | Pc | I | 5 | seh | C |
| 489N | Nose | Pc, Cd | I | 5 | seh, sei. | E |
| 666N | Nose | Pc, Tc, Tp, Cd | I | 5 | sec, seg, sei. | G |
| 9S | Stool | Pc, Tp, Fd | III | 8 | sea, seh, sei | A |
| 10S | Stool | Pc, Tp | III | 8 | sea, seg, sei | A |
| 58S | Stool | Pc, Tp, Cd | III | 8 | sea, sec, seg, sei | A |
| 76S | Stool | None | III | 8 | Seb | D |
| 4S | Hand | Cd | III | 8 | ND | F |
| 515S | Hand | Pc, Tc, Cd | III | 8 | seh, sei | A |
| 666S | Hand | Pc, Tc, Tp, Cd | I | 5 | sec, seg, sei. | B |
Abbreviations: Cd, cadmium acetate, Fd. fusidic acid. Km, kanamycin, Mc, methicillin, Pc, benzyl penicillin, Sm, streptomycin, Tc, tetracycline, Tp, trimethoprim, ND, not detected, agr, accessory gene regulator class, cap, capsular polysaccharide serotype type.
Figure 2.
PFGE of representatives of 18 PVL positive S. aureus from food handlers. Lanes A: PFGE type A, Lane A1: PFGE type A1, Lane B: PFGE type B, Lane B1: PFGE type B1, Lane C: PFGE type C, Lane C1: PFGE type C1, Lane D: PFGE type D, Lane E: PFGE type E, Lane G: PFGE type G, Lane H: PFGE type H.
When the isolates obtained during the screening exercise (screening samples) were compared with those obtained during routine investigation of food poisoning cases (routine samples), the results were not significantly different for the distribution of genes for agr and capsular polysaccharides (data not shown). Similarly, 72.9% of the screening sample isolates (97/133) and 67.1% (45/67) of the routine samples isolates contained genes for SEs showing no significant difference in the distribution of genes for SE between the two sets of isolates (P value = 0.50). Furthermore tst was detected in 5.2% (7/133) of the screening samples isolates and in 1.5% (1/67) of the routine sample isolates (P value = 0.4)
Discussion
This study has demonstrated a high prevalence of antibacterial resistance and diversity in the carriage of virulence genes in S. aureus obtained from food handlers employed in restaurants in Kuwait City. Our results that 92.5% of the isolates expressed resistance to antibacterial agents is similar to the prevalence of antibacterial resistance in methicillin-susceptible S. aureus obtained from patients in Kuwait hospitals [24] and to that found in S. aureus isolates from food handlers in Chile [11] and Botswana [17]. Although only one isolate was methicillin-resistant, the finding is significant because food handlers carrying MRSA had initiated outbreaks in hospitals in the Netherlands [14], USA [25] and Brazil [15]. As food handlers represent a section of the healthy population in the community, besides working in restaurants, the detection of high prevalence of antibiotic resistance in S. aureus isolated from them also highlights the growing problem of antibiotic resistance in the community.
The ability of S. aureus to colonize or infect its host is related to its ability to express virulence factors that facilitate their adherence to surfaces, cause damage or its ability to evade host's immune system [1,2]. Our isolates carried genes for a range of virulence factors including three of the four accessory gene regulators that regulate staphylococcal virulence factors [6]. Our result that 50.5% of the isolates were agr type I concurs with results of other studies that the majority of S. aureus from clinical sources belong to agr type I [4,23]. Similarly, our results that 57% of the isolates belonged to serotype 8 and 38% belonged to serotype 5 are in agreement with previous studies that showed that the majority of S. aureus obtained from clinical samples belonged to capsular polysaccharide serotypes 5 or 8 [2,23].
With regards to the carriage of SE determinants, the results of this study differ from that of a previous study in Kuwait [10]. The 71.0% prevalence of SE genes in this study was lower than the 86.6% prevalence detected previously in S. aureus from food handlers in Kuwait restaurants [10]. However, it was similar to the prevalence of 74.1% obtained from S. aureus isolated from food poisoning cases in Taiwan [26] and higher than the prevalence of SE reported in S. aureus from food handlers in Chile [19%, [11]] and from humans, food and animal sources in Malaysia [20.8%, [27]]. In addition, the demonstration of SEI as the most common SE gene in this study contrasted with the results of a previous study of SE in S. aureus isolated from food handlers in Kuwait restaurants [10] and other studies [9,11,12] where SEA was the most common SE. Furthermore, whereas 64.7% of the SE positive isolates in this study carried genes for two to four SEs, only 8.6% of S. aureus in the previous study from Kuwait carried more than one SE gene [10]. This is consistent with recent reports in the literature that also document the carriage of multiple SE gene in enterotoxigenic S. aureus [9,12,13]. The detection of multiple SE genes in recent S. aureus isolates could be due to the improvement in detection methods following the application of PCR technology and the discovery of new SE genes which were not tested in the previous study. It could also be due to frequent horizontal transmission of phage-mediated toxin genes among the staphylococcal populations [21].
PVL is widely distributed among some clones of community-associated MRSA (CA-MRSA) isolated in Kuwait and other countries [3,28,29] and was thought to contribute to the increased virulence of CA-MRSA isolates [3,28]. However, PVL has recently also been detected in healthcare-associated MRSA and in methicillin-susceptible S. aureus from patients [29]. Our report is the first in S. aureus obtained from food handlers in Kuwait and elsewhere. Its detection in 9.0% of the isolates in this study supports the suggestion that PVL-bearing phages are widespread among S. aureus of different genetic backgrounds and are not a specific characteristic of CA-MRSA [19,29]. The observation that eight of the 18 lukS-lukF positive isolates had same PFGE patterns suggested a transmission of a common strain among these workers. The detection of genes for PVL in S. aureus obtained from food handlers is of public health interest since these food handlers can serve as sources of transmission of PVL producing S. aureus in the community especially among family members.
Given that the majority of the isolates in this study contained genes for an array of virulence factors, they are potential causes of serious S. aureus infections. Therefore food handlers carrying these strains can contaminate food that can lead to food poisoning [7]. Thus, our results support the current practice of screening restaurant workers to identify S. aureus carriers and referring them to the appropriate health authorities for decolonization. Although the S. aureus carriage rate and their antibacterial resistance in this study may be similar to those found among healthy individuals in the general population [30], it is important to decontaminate food handlers carrying S. aureus because their exposure to, and possible contamination of food prepared and served in restaurants is a public health concern.
Conclusion
This study has provided updated data on the carriage of SE and other virulence genes, and initial information on the prevalence of antibacterial resistance in S. aureus obtained from food handlers in restaurants in Kuwait and in the Gulf region. Our results should contribute to better management of S. aureus carriers among the food handlers and enhance the safety of restaurant customers.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors conceived the study. SAM isolated and performed initial characterization of the isolates. EEU and MJA performed molecular characterization of the isolates. All authors participated in the preparation of the manuscript.
Acknowledgments
Acknowledgements
This study was supported by Kuwait University Research Grant No. MI 05/05. We thank Nadia Al Bania, B. Noronha and B. Mathew for technical assistance.
Contributor Information
Edet E Udo, Email: edet@hsc.edu.kw.
Siham Al-Mufti, Email: Sihmufti@kuwait.net.
M John Albert, Email: John@hsc.edu.kw.
References
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/CMR.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- Jarraud S, Mougel C, Thiouluse J, Lina G, Meunier H, Forey F, Nesme X, Etienne J, Vandenesch F. Relationship between Staphylococcus aureus genetic background, virulence factors, agr type (alleles), and human disease type. Infect Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan SJ, Novick RP. The molecular basis of pathogenicity. In: Crossley KB, Archer GL, editor. The staphylococci in human disease. Churchill Livingstone, New York, N.Y.; 1997. pp. 55–81. [Google Scholar]
- Jarraud S, Lyon GJ, Figueiredo AM, Lina G, Vandenesch F, Etienne J, Muir TW, Novick RP. Exfoliatin-producing strains define a four agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/JB.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombari V, Mayer MD, Laicini ZM, Mamizuka E, Franco BD, Destro MT, Landgrave M. Foodborne outbreak caused by Staphylococcus aureus: phenotypic and genotypic characterization of strains of food and human sources. J Food Prot. 2007;70:489–493. doi: 10.4315/0362-028x-70.2.489. [DOI] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JO, Lee JK, Jung YH, Yoo JI, Park YK, Kim BS, Lee YS. Molecular analysis of Staphylococcal food poisoning in South Korea. J Appl Microbiol. 2006;101:864–871. doi: 10.1111/j.1365-2672.2006.02957.x. [DOI] [PubMed] [Google Scholar]
- Al-Bustan MA, Udo EE, Chugh TD. Nasal carriage of enterotoxin-producing Staphylococcus aureus among restaurant workers in Kuwait City. Epidemiol Infect. 1996;116:319–322. doi: 10.1017/S0950268800052638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa G, Navarrete P, Caro M, Troncoso M, Faundez G. Carriage of enterotoxigenic Staphylococcus aureus in food handlers. Rev Med Chil. 2002;130:859–864. [PubMed] [Google Scholar]
- Holeckova B, Holoda E, Fotta M, Kalinacova V, Gondol J, Grolmus J. Occurrence of enterotoxigenic Staphylococcus aureus in food. Ann Agric Environ Med. 2002;9:179–182. [PubMed] [Google Scholar]
- Omoe K, Ishikawa M, Shimoda Y, Hu D-L, Ueda S, Shinagawa K. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxins productivities of S. aureus isolates harboring seg, seh, or sei genes. J Clin Microbiol. 2002;40:857–862. doi: 10.1128/JCM.40.3.857-862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluytmans J, van Rost Leeuwen W, Goessens W, Hollis R, Messer S, Herwaldt L, Bruining H, Heck MJ, van Leeuwen N, van Belkum A, Verbrugh H. Food-initiated outbreak of methicillin-resistant Staphylococcus aureus analyzed by phenol-and genotyping. J Clin Microbiol. 1995;33:1121–1128. doi: 10.1128/jcm.33.5.1121-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MJ, Tokumaru-Miyazaki NH, Noleto AL, Figueiredo AM. Enterotoxin production by Staphylococcus aureus clones and detection of Brazilian epidemic MRSA clone (III::B:A) among isolates from food handlers. J Med Microbiol. 1997;46:214–221. doi: 10.1099/00222615-46-3-214. [DOI] [PubMed] [Google Scholar]
- Udo EE, Al-Bustan MA, Jacob LE, Chugh TD. Enterotoxin production by coagulase-negative staphylococci in restaurant workers from Kuwait City may be a potential cause of food poisoning. J Med Microbiol. 1999;48:819–823. doi: 10.1099/00222615-48-9-819. [DOI] [PubMed] [Google Scholar]
- Loeto D, Matsheka MI, Gashe BA. Enterotoxigenic and antibiotic resistance determination of Staphylococcus aureus strains isolated from food handlers in Gaborone, Botswana. J Food Prot. 2007;70:2764–2768. doi: 10.4315/0362-028x-70.12.2764. [DOI] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards Performance for disk diffusion susceptibility tests. Approved Standards. National Committee for Clinical Laboratory Standards. Wayne, Pa. 2000.
- Udo EE, Farook VS, Mokadas EM, Jacob LE, Sanyal SC. Molecular fingerprinting of mupirocin-resistant Staphylococcus aureus from a Burn unit. Int J Infect Dis. 1999;3:82–87. doi: 10.1016/S1201-9712(99)90014-0. [DOI] [PubMed] [Google Scholar]
- Becker K, Roth R, Peters G. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxins genes, exfoliative toxins, and toxic shock syndrome toxin 1 gene. J Clin Microbiol. 1998;36:2548–2553. doi: 10.1128/jcm.36.9.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monday SR, Bohach GA. Use of multiplex PCR to detect classical and newly described pyrogenic exotoxins in Staphylococcal isolates. J Clin Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PCL, Lindsay JA. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J Clin Microbiol. 2001;39:2760–2767. doi: 10.1128/JCM.39.8.2760-2767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, Al-Sweih N, Dhar R, Dimnitrov TS, Mokaddas EM, Johny M, Al-Obaid IA, Gomaa HH, Mobasher LA, Rotimi VO, Al-Asar A. Surveillance of antibacterial resistance in Staphylococcus aureus isolated in Kuwait hospitals. Med Princ Pract. 2008;17:71–75. doi: 10.1159/000109594. [DOI] [PubMed] [Google Scholar]
- Jones TF, Kellum ME, Porter SS, Bell M, Schaffner W. An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2002;8:82–84. doi: 10.3201/eid0801.010174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YC, Lin CW, Yang CY, Tsen HY. PCR primers for the detection of staphylococcal enterotoxin K, L, and M and survey of staphylococcal enterotoxin types in Staphylococcus aureus isolates from food poisoning cases in Taiwan. J Food Prot. 2006;69:1072–1079. doi: 10.4315/0362-028x-69.5.1072. [DOI] [PubMed] [Google Scholar]
- Lim YS, Jegathesan M, Koay AS. Enterotoxin production by Staphylococcus aureus strains isolated from humans, food and animals in Malaysia. Southeast Asia J Trop Med Public Health. 1982;13:133–137. [PubMed] [Google Scholar]
- Udo EE, O'Brien FG, Al-Sweih N, Noronha B, Mathew B, Grubb WB. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J Clin Microbiol. 2008;46:3514–3516. doi: 10.1128/JCM.00966-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S, Stickers P, Ellington MJ, Kearns AM, Ehricht R. High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus. Clin Microbiol Infect. 2007;13:1157–1164. doi: 10.1111/j.1469-0691.2007.01833.x. [DOI] [PubMed] [Google Scholar]
- Mainous AG, Hueston WJ, Everet CJ, Diaz VA. Nasal carriage of Staphylococcus aureus and methicillin-resistant S. aureus in the United States, 2001–2002. Ann Fam Med. 2006;4:132–137. doi: 10.1370/afm.526. [DOI] [PMC free article] [PubMed] [Google Scholar]