Abstract
Through the classic study of genetics, much has been learned about the regulation and progression of human disease. Specifically, cancer has been defined as a disease driven by genetic alterations, including mutations in tumor-suppressor genes and oncogenes, as well as chromosomal abnormalities. However, the study of normal human development has identified that in addition to classical genetics, regulation of gene expression is also modified by ‘epigenetic’ alterations including chromatin remodeling and histone variants, DNA methylation, the regulation of polycomb group proteins and the epigenetic function of non-coding RNA. These changes are modifications inherited both during meiosis and mitosis, yet they do not result in alterations of the actual DNA sequence. A number of biological questions are directly influenced by epigenetics, such as how does a cell know when to divide, differentiate or remain quiescent, and more importantly, what happens when these pathways become altered? Do these alterations lead to the development and/or progression of cancer? This review will focus on summarizing the limited current literature involving epigenetic alterations in the context of human cancer stems cells (CSCs). The extent to which epigenetic changes define cell fate, identity, and phenotype are still under intense investigation, and many questions remain largely unanswered. Before discussing epigenetic gene silencing in CSCs, the different classifications of stem cells and their properties will be introduced. This will be followed by an introduction to the different epigenetic mechanisms Finally, there will be a discussion of the current knowledge of epigenetic modifications in stem cells, specifically what is known from rodent systems and established cancer cell lines, and how they are leading us to understand human stem cells.
Keywords: epigenetics, embryonic stem cells, normal and cancerous adult stem/progenitor cells
Introduction
Embryonic, Adult and Cancer Stem Cells
It is first important to define and understand the difference between embryonic, adult and cancer stems cells. The mammalian zygote, or fertilized egg, represents the beginning of life for an organism. The zygote demonstrates totipotency, meaning it has the potential to develop into a complete organism from a single cell, and can divide and produce all the differentiated cells in an organism, including extra-embryonic tissues (Allis, 2007; Keller, 2005). In a developing embryo, the first visible cell differentiation event to occur is the formation of the blastocyst, which contains the trophoblast stem cells (TS), the inner cell mass cells (ICMs) and the blastocyst cavity. Embryonic stem cells (ESCs) are derived from the ICMs, and unlike the blastocyst are pluripotent in nature (Allis, 2007; Keller, 2005). Pluripotency is defined as having the ability to generate stem cells and the subsequent differentiated cells of all three of the germ layers (ectoderm, mesoderm, and endoderm), with the exception of the extra-embryonic tissues (Allis, 2007). As the germ layers of the early embryo develop, however, asymmetric division begins and the process of differentiation starts (Sell, 2004). If maintained properly in vitro, ESCs are an attractive model system to investigate proprieties of stem cells since they do not undergo senescence, and carry the potential to regenerate all types of cells and organs present in an adult organism (Allis, 2007). However, maintenance of ESCs pluripotency still remains largely unknown, as well as how normal development proceeds when every cell has the same genetic content, yet some cells are able to follow different developmental patterns.
In contrast to ESCs are adult (somatic) stem cells (ASCs). ASCs have the ability to regenerate cells of the specific tissue where they reside in response to dying or damaged tissue, and unlike ESCs, they are not pluripotent in character. These cells divide asymmetrically meaning one daughter cell remains as an ASCs and will continue the process of self-renewal, whereas the other daughter cell starts the process of differentiation and is referred to as a ‘transient amplifying cell’ (Sell, 2004). One hypothesis regarding ASCs is that these cells are a unique reservoir that is not only responsible for the normal reparative and regenerative processes, but are the prime target for genetic and epigenetic changes culminating in many abnormal conditions, including cancer (Allis, 2007). Within the last 5 years it has been well documented that only a small fraction of epithelial tumor cells have the ability to form colonies in vitro or to initiate a new tumor upon injection into a host in vivo (Al-Hajj et al., 2003; Graziano et al., 2008; Cariati and Purushotham, 2008; Kasper, 2008; Takaishi et al., 2008; Lee et al., 2008). These cells have been termed the cancer stem cells (CSCs) within the tumor. However, as early as 1994, Lapidot et al. showed that after using fluorescence-activated cell sorting (FACS) of cells based on their expression of the cell surface markers CD34 and CD38, the CD34+CD38− cells could be identified as potential stem cells of acute myeloid leukemia (AML) (Lapidot et al., 1994). This ‘stem-cell’ phenotype was assigned since nonobese diabetes/severe combined immunodeficiency (NOD/SCID) mice injected with low numbers of CD34+CD38− cells developed leukemia, whereas those injected with even larger numbers of more mature cells (CD34−CD38+) did not (Lapidot et al., 1994).
Similar to a true stem cell, CSCs require the ability to undergo self-renewal, are highly proliferative and can differentiate (Allis, 2007). However, it is becoming more evident that CSCs are not governed by the same type of genetic regulation as normal stem cells (Clarke, 2005). Understanding the current evidence that supports epigenetic silencing as a regulating mechanism between normal and cancer stem cells will increase our chances of better targeting them in specific therapies.
Chromatin Structure and Methods of Epigenetic Regulation
In order for a cell to efficiently package the entire genome into its nucleus, the DNA is organized into a complex referred to as chromatin (consisting of DNA and protein). Nucleosomes are the core repeating unit of chromatin. They are comprised of 147 base pairs of DNA tightly wound around a highly basic protein octamer containing two molecules of each core histone (H2A, H2B, H3, and H4) (Kornberg, 1974). The core histone contains a globular domain with a flexible amino-terminal histone tail that protrudes from the surface. Chromatin is then further classified as either being euchromatic or heterochromatic. Euchromatic usually refers to chromatin that is decondensed and active, whereas heterochromatic DNA is highly condensed and silenced (Adkins et al., 2004). Changes in the chromatin can occur when the histones themselves are modified, resulting in either an altered structure or charge (referred to as a cis-effect) or an altered affinity for chromatin-associated proteins themselves (trans-effect) (Ehrlich, 2002).
As previously mentioned, the core histones contains a globular domain with a flexible amino-terminal histone tail that protrudes from the surface. This tail is highly susceptible to post-translational modification including methylation, acetylation, phosphorylation, ubiquitination, sumoylation, citrullination, and ADP-ribosylation (Jenuwein and Allis, 2001). This introduces the idea that a ‘histone code’ exists, and it may considerably extend the information potential of the genetic (DNA) code by regulating DNA at the epigenetic level. These epigenetic marks can provide either an ON or OFF signature which result in the tight regulation of gene expression (Table 1). For example, methylation of lysine 4, 36, or 79 on H3 (H3K4, H3K36, and H3K79 respectively), lysine 20 of H4 (H4K20), and lysine 5 of K2B (H2BK5) results in activation of gene transcription (Barski et al., 2007; Steger et al., 2008). However, trimethylation on H3K9, H3K27, or H4K20 represents repression of gene expression (Barski et al., 2007). Additionally, acetylation of H3K9 and H3K14 results in gene activation (Koch et al., 2007).
Table 1.
Examples of Epigenetic Histone Code modification and function
| Epigenetic modification | Effect on Transcription | Ref. |
|---|---|---|
| H3K4, H3K36, H3K36, H3K79, H4K20 and H2BK5 methylation | Active |
Barski, 2007 Steger, 2008 |
| H3K9, H3K27, and H4K20 Tri-methylation | Repressed | Barski, 2007 |
| H3K9 and H3K14 acetylation | Active | Koch, 2007 |
H: histone number identification, K: lysine residue identification.
Histone modifications are established by a number of chromatin-associated enzyme systems. Methylation of histones are carried out by two enzymes, protein arginine methyltransferases (PRMTs) (Pal and Sif, 2007) and histone lysine methyltransferases (HKMTs) (Shi, 2007). In addition, two enzymes work together to regulate histone acetylation, histone acetylases (HATs) and histone deacetylases (HDACs) (Hadnagy et al., 2008). The regulation of these enzyme networks tightly controls histone modification within differentiating cells.
A second mechanism of epigenetic modification is DNA methylation. The process of DNA methylation involves covalent modification of cytosine nucleotides at the C5 position in specific areas of CpG dinucleotides. The notion that methylation of DNA could represent a mechanism of memory within cells was discovered in 1975 by two independent labs when they determined that patterns of DNA methylation were replicated semiconservatively similar to DNA (Riggs, 1975; Holliday and Pugh, 1975). The majority of methylated CpG dinucleotides are present in heterochromatic regions, and thus are unexpressed in the genome (Jones and Takai, 2001). However, there are islands of GC dinucleotides, termed CpG islands, which are usually found within promoter regions of a gene. Interestingly, during the mapping of the genome most individual gene promoters were found to have GC-rich sequences, and currently 60% of all known human genes contain them (Jones and Takai, 2001; Bird et al., 1985). The process of methylating DNA is controlled by a set of enzymes called DNA Methyltransferases (Dnmts). The first Dnmt identified, termed Dnmt1, was discovered in 1983 by Bestor and Ingram, and subsequently was shown to prefer hemi-methylated DNA as its substrate (Bestor and Ingram, 1983). The de novo methylation of DNA, however, depends on Dnmt3a and Dnmt3b. These two enzymes are developmentally regulated and encode catalytically active proteins, unlike Dnmt1 (Xie et al., 1999). Although Dnmt2 and Dnmt3L (Dnmt3-like) have been characterized, they do not play a substantial role in the enzymatic process (Okano et al., 1998; Aapola et al., 2000).
Methylation in mammals evolved as a method of silencing genes when their expression is not required. For example, a number of CpG islands on one X chromosome are methylated during a process called X-chromosome inactivation (Wolf et al., 1984). This process ensures an equal amount of gene expression between males and females. Additionally, the process of genomic imprinting involves DNA methylation where one allele of a gene, either maternal or paternal, is silenced (Reik et al., 1987). This process only affects a few hundred genes within the genome, most of which encode for genes that regulate embryonic and neonatal growth (Rugg-Gunn et al., 2007).
Finally, another set of genes which demonstrate epigenetic regulation during differentiation and development are members of the Polycomb group (PcG) of proteins. The function of PcG proteins is to maintain the silenced state of developmental regulators such as the HOX genes. These genes were first identified in studies of Drosophila homeotic (Hox) genes which encode homeodomain transcription factors that specify the identities of body segments in the fly (Gellon and McGinnis, 1998). The majority of these genes were characterized by mutations that cause homeotic transformations due to the failure to maintain these repressed states. The majority of PcG genes encode subunits which are a part of the Polycomb Repressive Complex (PRC) 1 and 2 (Otte and Kwaks, 2003). Mammalian PRC1 contains homologous proteins to the complex discovered in Drosophila; human polycomb (HPC), human polyhomeotic (HPH), BMI1 and RING (Otte and Kwaks, 2003). The mammalian PRC2 is composed of three proteins, Enhancer of Zeste protein-2 (Ezh2), Suppressor of Zeste 12 (Suz12) and the histone methyltransferase embryonic ectoderm development (Eed) (Otte and Kwaks, 2003). PRC3 is similar to PRC2 expect that it contains two different and smaller isoforms of Eed, Eed3 and 4, while PRC2 contains the largest, Eed1 (Kuzmichev et al., 2002). In 2005, PRC4 was characterized and was found to contain Eed2, a protein which only appears to be expressed in undifferentiated pluripotent cells (Kuzmichev et al., 2005). Furthermore, the PcG genes are closely associated with coordinated regulation of histone modification and methylation, thus tying a number of epigenetic mechanisms together.
Also, during the past several years RNA interference (RNAi) as a method of epigenetic regulation has been investigated with detail in plants. One specific example involves RNA-directed DNA methylation (RdDM) in recombinant viroid-infected plants. This occurs when double-stranded RNA (dsRNA) is directed at the promoter region of a gene resulting in de novo cytosine methylation at homologous DNA sequences, and thus causes stable silencing (Pelissier and Wassenegger, 2000). In addition to the short interfering RNAs used in RNAi, many other non-coding RNAs which alter transcription have been discovered. These RNAs include microRNAs (miRNAs) (Chuang and Jones, 2007), small nucleolar RNAs (snoRNAs), repeat-associated small interfering (rasiRNA), and P-element induced wimpy testis in Drosophila (PIWI) interacting RNA (piRNA) (Kawaji and Hayashizaki, 2008). PIWI proteins are coded by a class of genes that was originally identified as encoding regulatory proteins responsible for maintaining incomplete differentiation in stem cells and maintaining the stability of cell division rates in germ line cells (Cox et al., 2000).
Now that the epigenetic mechanisms including chromatin remodeling, DNA methylation, polycomb group proteins and the epigenetic function of non-coding RNA (Figure 1) have been well defined, their contribution to each area of stem cell regulation will be further examined.
Figure 1.
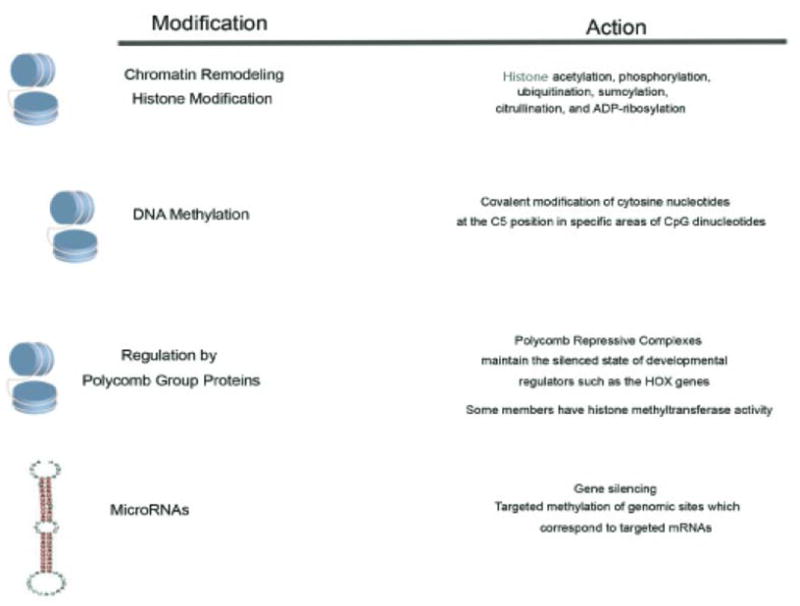
Known mechanisms of epigenetic gene regulation
Epigenetic Regulation in ESCs
The majority of the research investigating epigenetic mechanisms of gene silencing in ESCs to date has been conducted in mouse lines as a result of the controversial issues around acquiring human embryos. However, in the early 1980s it was found that ESCs explanted from mouse blastocysts could be maintained in culture for extended periods of time (Evans and Kaufman, 1981). A working hypothesis in relation to ESCs is that during differentiation the genome undergoes alterations shifting it from being rich in euchromatin, to a more compact heterochromatic structure, reviewed in (Atkinson and Armstrong, 2008). Determination of global histone modification patterns in mouse ESCs have demonstrated that the genome has a generalized pattern of histone acetylation and methylation at lysine 4 of H3 (reviewed in Atkinson and Armstrong, 2008) allowing for permissive gene expression. In addition, a correlation has been found between the chromatin environment and the DNA sequence itself, suggesting that it can dictate patterns of histone modification (Bernstein et al., 2005). For example, the methylation of lysine 27 on H3 is associated with the binding components PRC1 and PRC2 PcG complexes (Boyer et al., 2006). The PcG complexes then act to repress key developmental regulators in ESCs which are expressed only upon differentiation cues.
One key observation is that the maintenance of pluripotency within ESCs requires the expression of a number of genes, including three key genes, Oct4, Nanog, and Sox2, reviewed in (Atkinson and Armstrong, 2008). In 2008, using murine fibroblasts and human primary cells it was shown that these cells regain pluripotency through ectopic expression of four transcription factors (Oct4, Sox2, Klf4 and Myc) (Park et al., 2008). These genes are not normally expressed in differentiated cells, yet when reintroduced resulted in ‘induced pluripotent stem (iPS) cells’ (Park et al., 2008). Mechanistically, it was also shown that the reprogrammed iPS cells had a decrease in methylation at the Oct4 and Nanog promoters (Park et al., 2008). In addition, when comparing histone modifications at specific gene loci in iPS cells and mouse ESCs, an identical pattern of histone 3 K4 and K27 trimethylation was found (Maherali et al., 2007). Recently, it has also been shown that in mESCs, Nanog and Oct4 physically interact with each other as well as other proteins from multiple repression complexes (Liang et al., 2008).
All of these observations are in murine cells, however, and few labs have truly investigated the role of epigenetic gene silencing in human ESCs (hESCs). These cells have been isolated from early-stage embryos collected from in vitro fertilization clinics. In 2007, Pan et al. conducted a whole-genome analysis of trimethylation of lysine 4 and 27 of H3 (H3K4me and H3K27me3) in hESCs (Pan et al., 2007). Previous analyses suggest that H3K4me3 modification associates with active promoters, while H3K27me3 with silenced ones. It was found that Oct4, Nanog, and Sox2 are initially marked only with H3K4me3, yet as the cells differentiate, become marked with H3K27me3 as well. However, in the same year Rodriguez et al. further demonstrated that reduced expression of Oct4 in human ESCs using siRNA promoted upregulation of markers indicative of mesoderm and endoderm differentiation, and unlike mouse ESCs, when Oct4 levels were elevated, it also promoted upregulation of markers of endoderm derivatives (Rodriguez et al., 2007). This is not the first incidence where differences are seen between human and mouse ESCs since LIF signaling through gp130/STAT3 supports undifferentiated cell growth in mESCs, yet has no importance in hESCs (Rodriguez et al., 2007). This analysis, however, was only conducted in one hESC line, the HSG-6 line from the National Institutes of Health. Yet in a previous study in 2006, Lagarkova et al. demonstrated the promoters of Oct4, Nanog, and two Oct4 related genes, DppA3 and DppA5, are in fact unmethylated in three different hESC lines (Lagarkova et al., 2006).
With regards to additional mechanisms of epigenetic silencing in hESCs the literature is limited. Methyltransferase activity (unlike expression) of the de novo Dnmt3b has been shown to be high in several hESC lines (Allegrucci and Young, 2007). Also, a subset of the short RNAs referred to as microRNAs (miRNAs) have been found to be specifically expressed in hESCs, yet downregulated during development into embryoid bodies (Suh et al., 2004). The hope is that this methylation and miRNA signature will be used to better identify undifferentiated hESCs.
Thus, differences in the epigenetic regulation of ESCs between mice and humans do exist, which is further compounded by differences in individual established cell lines. Such differences include overall growth rates and genetic as well as epigenetic stability in long-term culture. True patterns of epigenetic regulation will have to be further defined in either short term culture systems or in primary patient samples. Currently, efforts are being made to identify a critical set of genes in hESCs that represent a global methylation signature, and not just differences between lines (Allegrucci et al., 2005). It is clear, however, that a network of tightly regulated epigenetic signals exist to ensure proper maintenance of pluripotent ESCs, and upon differentiation, this network undergoes vast rearrangement to induce development of somatic cells.
Epigenetic gene silencing of adult stem cells: Do errors lead to the development of cancer stem cells?
ASCs are undifferentiated cells found throughout the body after embryonic development that divide to replenish dying cells and regenerate damaged tissues (Allis, 2007). Unlike ESCs they are not pluripotent and are usually lineage restricted to the tissue in which they reside, reviewed in (Preston et al., 2003). Multipotent stem cells, however, have been characterized for a number of systems. Hematopoietic stem cells (HSCs), which give rise to all blood cells, have been characterized in the bone marrow (Preston et al., 2003). In addition, stem cells of the central nervous system (neuronal stem cells: NSCs) have trilineage ability and can generate neurons, oligodendrocytes, and astrocytes (Preston et al., 2003). Mesenchymal stem cells (MSCs) have the capacity to differentiate, both in vivo and in vitro, into osteoblasts, chondroblasts, and adipocytes, and more recently, into beta-pancreatic islet cells when exposed to the appropriate stimuli (Preston et al., 2003). There are many questions about ASCs that still remain unanswered. For instance, do CSCs originate from misregulation or errors in epigenetic mechanisms from ASCs? Since organ stem cells are long-lived, during the process of aging they might acquire genetic mutations and/or more deleterious epigenetic modifications resulting in altered signaling and function (Feinberg, 2007).
Hematopoietic stem cells
ASCs were first studied and characterized in the hematopoietic system. Indeed, the hierarchical organization of the hematopoietic system has been hypothesized since the early 20th century (Metcalf, 2007). This model predicts that a small number of undifferentiated stem cells generates a hierarchy of progenitor populations with progressively restricted developmental potential, eventually leading to the production of eight different lineages of mature blood cells. HSCs were first characterized by Baum and coworkers as a subset of human bone marrow cells containing multipotent progenitors (Baum et al., 1992). These cells were subsequently shown to rescue lethally irradiated hosts by reconstituting the entire repertoire of hematopoietic cells in the recipients (Uchida et al., 1994).
HSCs show an impressive self-renewal capacity, evidenced by the continuous supply of hematopoietic cells throughout a person’s lifetime. HSCs are rare, occurring at a frequency of about 1 in 105 bone marrow cells (Sieburg et al., 2002), and they are generally quiescent (non-cycling) (Rossi et al., 2007). It is likely that at steady state only a small fraction of HSCs enter into the cell cycle to generate differentiating cells. To the contrary, hematopoietic stress (e.g. bleeding, infection and chemotherapy) recruits a larger number of HSCs into the cell cycle (Metcalf, 2007). The phenotypic characterization of murine HSCs has been firmly established: a single bone marrow cell that is CD34−/lo/Kit+ (receptor for stem cell factor), and negative for granulocytic, lymphoid and erythroid markers (Lin−) is multipotent and capable of self-renewing (Ema et al., 2006). In humans, the HSC phenotype is not conclusively defined. However, the CD34−/Lin− bone marrow fraction seems to be enriched of quiescent HSCs, while CD34+/Lin− cells may be enriched of activated precursor cells with enhanced clonogenic potential (Engelhardt et al., 2002).
Hematopoiesis is tightly regulated by the interplay between external stimuli and the activation of signal transduction pathways and transcription factor networks within the differentiating cell (Rice et al., 2007). Depending on the functional needs of the organism, niche cells (e.g. fibroblasts, osteoblasts, osteoclasts and adipocytes) produce extracellular mediators that sustain HSC self-renewal or induce HSC differentiation (Porter and Calvi, 2008). This eventually leads to the activation of specific transcription factors determining HSC fate. For example, Hox transcription factors are essential for HSC self-renewal (Abramovich and Humphries, 2005), while PU.1 is required for myeloid development and GATA-1 supports the differentiation of erythroid and megakaryocytic cells, reviewed in (Rice et al., 2007). Two main mechanisms regulate the specificity of transcription factor-induced gene expression: binding of the transcription factor to defined DNA sequences and epigenetic gene silencing. It is well known that GATA-1 and PU.1 mutually inhibit their expression during hematopoiesis. PU.1 functions by binding to the PU-box, a purine-rich DNA sequence (5′-GAGGAA-3′). As an example, PU.1 overexpression in G1ER cells (proerythroblast cells) blocked the DNA binding of GATA-1, as well as the erythroid differentiation of these cells (Zhang et al., 2000). PU.1 has been shown to bind to GATA-1 on its target genes, thereby recruiting a repression complex comprised of pRb, the heterochromatin protein HP1a and the histone methyltransferase Suv39h (Stopka et al., 2005). This leads to H3K9 methylation (a marker of inactive genes) in nucleosomes near the GATA-1 binding site. PU.1-dependent gene silencing plays a physiological role during myeloid lineage specification. On the other hand, inappropriate PU.1 expression in non-myeloid cells may block differentiation and cause leukemic transformation (Matushansky et al., 2000). In the absence of PU.1, GATA-1 recruits the histone acetyltransferase CBP to its target genes, thereby producing transcriptionally active chromatin (Stopka et al., 2005). During erythroid differentiation, GATA-1 directly represses PU.1 transcriptional activity, by preventing c-Jun binding to PU.1 (Zhang et al., 1999). Recently, it has become apparent that these two antagonistic transcription factors are involved in the earliest steps of hematopoiesis. Indeed, the activation of one transcription factor, coupled to the inhibition of the other, occurs during HSC commitment to myelo-lymphoid or erythro-megakaryocytic differentiation (Arinobu et al., 2007).
It is evident that DNA methylation plays a crucial role in the activity of lineage-specific transcription factors, both in physiological and in pathological context. For example, Suzuki et al. demonstrated that PU.1 forms complexes with Dnmt3a and 3b, thereby allowing the methylation of specific genes, such as p16 (a gene that is frequently silenced in human leukemias) (Suzuki et al., 2006). The PU.1 locus itself is subject to epigenetic control. For example, it is expressed in HSC and differentiated B cells, but is silenced by DNA methylation in T cells (Ivascu et al., 2007). It is noteworthy that PU.1 over expression or demethylation has been linked with T- and B cell lymphomas, respectively (Mahadevan et al., 2005; Ivascu et al., 2007). As will be discussed below, PU.1 seems to play an opposite role in myeloid leukemias, where it functions as a differentiating agent.
Leukemias are neoplastic diseases arising from impaired differentiation of HSCs or committed cells re-acquiring self-renewal potential. Leukemia classification is mainly based on differentiation status and clonogenic potential of the leukemic blast. Disease-initiating cells have been isolated for both acute and chronic, as well as lymphoid and myeloid leukemias (Bonnet and Dick, 1997; Sirard et al., 1996; Kong et al., 2008). This work was based on the identification of specific human leukemia subpopulation, called leukemia stem cells (LSCs). When LSCs are injected into NOD/SCID mice, they are capable of producing a disease that is identical to that of the donor. Based on these results, two main hypotheses have been addressed about leukemogenesis (Passegue and Weisman, 2005). One model suggests that leukemogenic events affect only the HSC compartment. Depending on the nature and timing of these molecular alterations, transformed HSCs may give rise to different leukemia phenotypes. This view is supported by the fact that since HSCs have an endless self-renewal potential, they can accumulate a sufficient number of genetic and epigenetic alterations to initiate leukemia (Rossi et al., 2007). According to the second hypothesis, the leukemogenic process may occur at any stage of hematopoiesis, as a result of an alteration of the normal developmental program. In this case, the degree of commitment of target cells determines the characteristics of leukemic blasts. Although the origin of LSCs is still a matter of debate, current evidence support both views, suggesting that leukemias may arise from either HSCs or committed cells. Epigenetic modulation of gene silencing plays a crucial role in both processes.
Acute myeloid leukemia (AML) was the first neoplasm with a defined subset of cancer initiating cells. Bonnet and Dick (Bonnet and Dick, 1997) clearly showed that this leukemia is initiated by transformed HSCs. When injected into NOD/SCID mice, only the CD34+CD38− fraction of leukemic cells was able to generate a heterogeneous population of leukemic-blasts, recapitulating the human disease. Based on these observations, the authors proposed a hierarchical organization for AML. However, subsequent work questioned this assumption. One of the genes involved in AML initiation and progression is mixed lineage leukemia (MML). MML is a human homologue of Drosophila melnogaster trithorax (Trx), and is known to control the expression of many genes during human development. In HSCs, Trx and PcG complexes serve as chromatin remodeling driving forces, with opposite roles (Krivtsov and Armstrong, 2007). Indeed, H3K4 methylation by some members of Trx group, including MML, is a positive regulator of Hox genes expression, while H3K27 methylation by PcG proteins mediates the silencing of these genes. Hox genes are important for HSC self-renewal, but their hyperactivation leads to leukemia (Abramovich and Humphries, 2005). The regulation of Hox genes expression by MLL is affected by several factors, including the activity of PcG proteins, the appropriate binding of MLL cofactors to DNA, and the occurrence of chromosomal translocations producing MML fusion proteins. More than 50 different MML translocation fusion partners have been identified, although five of them (AF4, AF9, AF10, ENL, ELL) account for 80% of leukemia associated MML rearrangements. These translocations are frequent in pediatric and therapy-related leukemia, and occur in 10% of adult AML (Krivtsov and Armstrong, 2007). The mechanisms of MML-driven leukemogenesis are not completely understood, although some recent studies shed new light on this issue. It has been shown that one of the most frequent rearrangements, the MML-ENL fusion protein, results in loss H3K4 methyltransferase activity, and acquisition of H3K79 methyltransferase activity (Milne et al., 2005). H3K79 is a marker of active chromatin: it is broadly distributed across promoter and open reading frame regions, and is thought to be a very efficient activator of DNA polymerase II (Guenther et al., 2007). It is conceivable that the acquisition of this novel methyltransferase activity confers to MML oncogenic properties, probably through the abnormal activation of Hox genes. Mouse models of leukemogenesis, (Cozzio et al., 2003)showed that MML-ENL fusion protein is able to transform both HSC and committed myeloid progenitors, thereby questioning the Bonnet and Dick paradigm. This phenomenon could be linked to the ability of MML fusion protein to confer self-renewal proprieties to committed cells (Krivtsov and Armstrong, 2007). However, HSCs are the more efficient target of leukemogenesis since the induction of AML in mice required 10- times more myeloid progenitors than HSC. Moreover, these observations still wait to be confirmed by studies on human AML samples.
In contrast with AML, chronic myeloid leukemia (CML) is a paradigm of HSC-driven neoplasm. Both human and mouse data follow this hypothesis, and key pathways have been identified for CML initiation and progression (Kavalerchik et al., 2008). The first step of this process is the expression of a BCR-ABL fusion protein by an HSC. BCR-ABL usually originates from a chromosomal translocation t(9q34:22q11), and activation of this aberrant protein in HSC leads to preferential expansion of myeloid progenitors (Chalandon et al., 2005). BCR-ABL expression is associated with an uncontrolled ABL tyrosine kinase activity, leading to the activation of several anti-apoptotic proteins, reviewed in (Clarkson et al., 2003). These oncogenic functions are mainly mediated by the mitogen–activated protein kinase (MAPK) pathway (Faderl et al., 1999). However, BCR-ABL expression per se is not sufficient for HSC leukemic transformation, since this aberrant protein was found in the blood of 22/73 healthy adults and 1/22 children (Clarkson et al., 2003). Moreover, it has been shown that BCR-ABL does not confer self-renewal ability to committed progenitors, thus contributing only to HSC transformation (Huntly et al., 2004). Additional leukemogenic events are required for CML development. Among these, epigenetic silencing of the JunB transcription factor was shown to occur in BCR-ABL positive CML patients (Yang et al., 2003). A murine model of CML showed that JunB acts as a tumor suppressor gene during myeloid development, probably through its anti-proliferative and proapoptotic activity (Passegue and Weissman, 2005). Interestingly, microarray data showed that during CML progression to more clinically advanced phases, BCR-ABL-driven MAPK activation is progressively lost, and this phenomenon is coupled to the activation of the Wnt pathway (Radich et al., 2006). Imatinib, a BCR-ABL inhibitor employed for the treatment of CML, principally targets the MAPK pathway. It has been suggested that resistance to this drug in advanced CML is due to the emergence of clones less addicted to the MAPK pathway and more dependent on Wnt signaling. Moreover, JunB epigenetic silencing and Wnt pathway activation may be mediated by MDFI (MyoD Family Inhibitor), a transcription factor inhibitor influencing both Wnt and Jun signaling (Kusano and Raab-Traub, 2002). Thus, epigenetic control of gene silencing seems to play a role in both the initiation and progression of CML. Keeping with this hypothesis is the observation that the PcG protein Bmi-1 is essential for the proliferative capacity of LSCs (Lessard and Sauvageau, 2003). Moreover, Bmi-1 expression in CML was correlated with disease progression and prognosis: patients showing higher Bmi-1 expression progressed more rapidly to an advanced CML phase (Mohty et al., 2007).
Overall, epigenetics clearly plays a significant role in gene silencing in HSCs. The information to date will only allow us to better understand epigenetics and the regulation of tissue specific adult stem cells.
Breast and Prostate Stem Cells
Some of the most investigated systems with regards to normal progenitor cells are those from the mammary and prostate glands. The notion that normal adult stem cells exist in the mammary gland has been under investigation for a number of years, reviewed in (Cariati and Purushotham, 2008). As early as 1959, researchers found that mouse epithelium isolated from different regions of a mammary gland at various stages in postnatal development was able to generate fully functional mammary epithelial outgrowths (Deome et al., 1959). The development of mammary gland is a dynamic process where growth and differentiation occur during neonatal and prepubertal periods. The gland itself originates from thickening of the embryonic endoderm which forms into a closely packed knob of cells that lengthens to form a solid chord of epithelial cells. This chord then branches into the adult mammary gland (Hogg et al., 1983). Once sexual maturity is reached, branching continues, and alveoloar buds form (Hovey et al., 2002). The normal human breast is composed of two cell layers, an inner luminal cell population and a distinct outer cell layer, juxtaposed to the basement membrane, termed the ‘basal’ layer (Anbazhagan et al., 1998). During pregnancy, the gland undergoes another process of differentiation when lobulalveolar development occurs, leading to lactation, nursing and finally involution of the weaned gland (Brisken, 2002). In 1988, using transplantation studies, Smith and Medina discovered what they called a latent stem cell in the mammary epithelium (Smith and Medina, 1988). In addition, mammary stem cells have been found to respond specifically to their microenvironment and hormonal milieu (Tsai, 2004). Recently it has been thought that the stem cells of the mammary gland reside in the basal layer, and recently more aggressive forms of breast cancer have been originating from this population of cells (Stein et al., 2005).
In the mammary gland, the vertebrate homeobox genes Msx1 and Msx2 facilitate proper development, and are found at sites where mesenchymal and epithelial tissue interact (Jowett et al., 1993). It is not surprising then that misexpression in adult tissues has been implicated in mammary tumorigenesis (Wang et al., 1994). In the mouse mammary gland, the PcG protein Bmi1, which commonly acts as an epigenetic silencer in adult tissue to maintain cellular identity, is crucial for outgrowth of the ductal epithelium and maintains premature lobuloalveolar differentiation of mammary stem cells (Pietersen et al., 2008). Thus, Bmi1 also has plays a role in the development of breast cancer, and this will be discussed later in further detail.
Similar observations have been made in the prostate. Specifically, it was found that recombining adult epithelium with embryonic urogenital mesenchyme (UGM) induced epithelial proliferation and ductal branching morphogenesis (Kinbara et al., 1996), indirectly demonstrating the presence of stem/progenitor cells. The molecules produced by the UGM which induce the differentiation of the prostate epithelium are not entirely characterized, and neither are the regulatory events occurring. However, one epigenetic fingerprint, expression of the homeobox gene Nkx3.1, which only appears in males, is the earliest prostate marker expressed when epithelial identity is occurring (Sciavolino et al., 1997). The mature prostate contains three distinct cell layers, secretory luminal, basal, and neuroendorcrine (Bonkhoff and Remberger, 1996), and the traditional model proposes that prostate stem cells (PSCs) exist in the basal cell compartment (De Marzo et al., 1998). During development, the basal cells express cytokeratin (K) K5 and K15, and are negative for luminal markers K8 and K18, as well as in androgen receptor (AR) (Prins and Birch, 1995). As development proceeds, basal cells differentiate into luminal cells and then express K8 and AR. The epigenetic regulation of these events, however, has yet to be fully described in humans.
In the last 40 years there have been a number of approaches used to identify and enrich for true mammary stem cells. Applying the theory that only a small population of tumor cells have the ability to proliferate and form new tumors, in 2003 Al-Hajj et al. was successful at identifying and isolating this subset of human mammary progenitor cells (Al-Hajj et al., 2003) termed mammary CSCs. Using cell surface adhesion markers, it was demonstrated that cells positive for CD44 (CD44+), negative for CD24 (CD24−), and negative for specific cell lineage markers (CD2, CD3, CD10, CD16, CD18, CD31, CD64, and CD140b) were highly tumorigenic, where a sub-population of only 100 injected cells could form tumors in mice (Al-Hajj et al., 2003).
Similar to breast, CSCs have been identified in the prostate (Tang et al., 2007), and furthermore, human prostate cancer cells (LNCaP) expressing CD44+/CD24− have been shown to form prostatospheres in vitro (Hurt et al., 2008), similar to mammospheres cultured from mammary progenitor cells. What is less well understood is how the normal stem cells within these glandular tissues transform to become CSCs, or if the cancer is able to develop from differentiated cells which have reactivated the self-renewal process. There is more evidence supporting the first hypothesis and furthermore, that it requires regulation of epigenetic pathways. The epigenetic regulatory events which might be controlling the differentiation of these progenitor cells are currently under intense investigation.
With regards to CSCs in these tissues, it is well known that the PcG protein Bmi1 maintains the self-renewal of hematopoietic and neuronal stem cells, and recently it was shown that the Bmi1 oncogene-driven pathway is one of the key regulatory mechanisms of the ‘stemness’ function of normal and CSCs (Glinsky, 2008). It was determined that Bmi1 regulates the proliferation of stem cells and the differentiation to more committed cells in the mammary gland (Pietersen et al., 2008). The authors state that Bmi1 overexpression in breast cancer leads to a block in terminal differentiation, thus leading to an increase in the cells that are susceptible for further oncogenic transformation (Pietersen et al., 2008). The involvement of Bmi1 has been further supported by the identification of a gene expression signature (GES) in human metastatic prostate cancer cells that is similar to expression patterns active in ESCs (Glinsky, 2008). Specifically, in CD44+/CD24− metastatic PC3-32 cells, quantitative colocalized immunofluorescence demonstrated an increase in expression of Bmi1/Ezh2 positive cells compared with cells isolated from the parental PC3 line. A similar analysis was carried out where a ‘Polycomb repression signature’ was identified in LNCaP cells and three metastatic prostate cancer tissues from independent patients (Yu et al., 2007b). It was found that the samples share a common set of H3K27me3-marked genes and those patients expressing this PcG signature actually have a higher risk and less favorable survival outcome. The PcG signature was also investigated in estrogen-receptor positive breast cancer patients and a similar pattern was found where patients expressing the signature demonstrated an increased risk of relapse and lower probability of survival (Yu et al., 2007b). Since PcG proteins are normally expressed at a very low level in differentiated cells and highly expressed in stem cells, it is likely that the metastatic breast and prostate cells expressing the PcG signature are either transformed normal stem cells which have progressed to CSCs, or have undergone dedifferentiation resulting in a stem-cell like phenotype. Furthermore, when breast progenitor cells are exposed to estrogen, the differentiated progeny of epithelial based cells demonstrated a unique ‘methylome’ (Cheng et al., 2008). Methylation was enriched in genes related to nucleic acid and cellular metabolism, however 23% of the genes are known PcG targets. A similar analysis with regards to androgen/AR expression and (PSCs) has not been conducted, but could reveal some interesting results since 12–33 months after androgen deprivation therapy in patients a population of cells (which could be stem cells) become resistant (Chung et al., 2003). It is known that the promoter for the AR is methylated in a few prostate cancer cell lines and this can be reversed by addition of the DNA methylation inhibitor 5-aza-2′ deoxycytidine (Jarrard et al., 1998). What is also known is that expression of the PcG protein, Ezh2, is regulated by Rb1 in a cell-cycle related fashion (Bracken et al., 2003), and this increase may be reflective of the increase in cell proliferation in advanced prostate cancers. Recall that expression of Ezh2 (a member of the Polycomb Repressive Complex 2 (PRC2)) is normally found in ESCs, it is not surprising to see it misregulated in cancer. A microarray studying examining Ezh2 overexpression in metastatic hormone-refractory prostate cancer demonstrated downregulation of approximately 100 genes, and increased expression of a smaller subset (Varambally et al., 2002). The actual contribution of the PcG group of proteins however is not known, and overall they seem to be reactivated or expressed at a higher level in a population of cancer cells within the prostate and mammary gland. Exactly how this increased expression is occurring still requires more investigation, and it will need to be determined if these cells truly are progenitor cells.
Turning to another method of epigenetic regulation, much less is known about how miRNAs regulate the stem cells of the breast and the prostate. In one study, it was found that breast progenitor cells isolated from human tumors have much less of the miRNA let-7, yet when overexpressed using a lentivirus, the cells had an increase in proliferation, mammosphere formation, and the proportion of undifferentiated cells in vitro and tumor formation and metastasis in NOD/SCID mice (Yu et al., 2007a). Additionally, it was found that in these cells let-7 resulted in a decrease in Ras and HMG2A expression, and shifted the cells toward differentiation (Yu et al., 2007a). Overall, much more is known about their contribution to maintenance and control of ‘stemness’ in different cell lineages, including hematopoietic cells, cardiomyocytes, myoblasts, and neural cells (Foshay and Gallicano, 2007), yet still very little is known specifically with regard to epigenetics, thus leaving much open for investigation.
In addition, direct target genes are affected by methylation in both breast and prostate cancer. For example, the Wnt signaling pathway is regulated by epigenetic gene silencing in breast cancer, reviewed in (Klarmann et al., 2008), as well as prostate cancer (Robinson et al., 2008). The Wnt signaling pathway is involved in the regulation of cell differentiation and proliferation, as well as cell movement and polarity, reviewed in (Mohinta et al., 2007). It also regulates the maintenance and self-renewal of hematopoietic stem cells and alterations in this pathway contribute to tumorigenesis. In addition, aberrant activation of Wnt signaling has been shown to induce mammary tumors from stem/progenitor cells (Lindvall et al., 2007). A number of genes involved in Wnt signaling have been identified as being methylated in breast cancer, reviewed in (Klarmann et al., 2008). These include the pathway components WIF1, SFRP1-5, APC and DKK1. Wnt signals are conferred through the Frizzled (FRZ) family of transmembrane proteins and the low-density lipoprotein receptor-related protein (LRP5/6) coreceptor, thus resulting in the stimulation of β-catenin. β-catenin interacts with the TCF/LEF-1 transcription factors within the nucleus to activate downstream Wnt signals. If Wnt signaling is not induced, β-catenin is targeted for destruction by the proteosome involving CK1, Axin, GSK-3β and APC. Since Wnt is a secreted ligand, many proteins regulate its expression by binding either Wnt itself or the receptor to regulate signaling. Wnt inhibitory factor-1 (WIF1) and secreted frizzled-related proteins (SFRPs) prevent Wnt from binding to its FRZ receptor, while the DICKKOPF (DKKs) proteins block activation by targeting LRP5/6 for degradation. Thus, epigenetic regulation of these critical proteins within the pathway greatly influences expression of Wnt target genes. What is less known, however, is how the epigenetic regulation of Wnt influences the regulation of the stem cell pool within the breast and the prostate. Using mouse ESCs as well as C17.2 cells (which have neural stem cell-like properties) and C2C12 cells (which are myogenic cells) it was recently shown that Wnt responsive TCF-bound states correlate with DNA hypomethylation, histone H3 hyperacetylation and H3K4 trimethylation (Wohrle et al., 2007). Clearly, this data supports that the Wnt pathway significantly contributes to the epigenetic regulation of stem cells.
Another target gene which is commonly methylated in prostate cancer is glutathione S-transferase pi 1 (GSTP1), reviewed in (Perry et al., 2006). The normal function of this protein is to detoxify potential carcinogens within a cell and prevent lesions from occurring within the genome that could lead to tumor development (Perry et al., 2006). Alterations in GSTP1 have been reported in 75–100% of all prostate carcinomas and up to 70% of high-grade prostatic intraepithelial neoplasia (HGPIN) (Perry et al., 2006) making it an extremely interesting target for further investigation. In rats, GSTP1 has been found to be expressed in oligodendrocyte progenitor cells (Tamura et al., 2007), and more recently, upon examining tissue specimens from patients with proliferative inflammatory atrophy (PIA), it has been found that cells expressing similar levels of GSTP1 also express the basal marker p63 (Kuzmanov et al., 2007). The authors suggest that these cells might represent the putative human prostate carcinoma stem cells and that prostate cancer may arise from this immature cell type. Additional genes, including APC, Ras-association domain family 1A (RASSF1A) and retinoic acid receptor β2 (RARβ2) also demonstrate increased promoter methylation as disease progression occurs in the prostate, yet, similar to GSTP1 their methylation analysis has not been investigated in progenitor cells of either the human prostate or breast.
Clearly the breast and the prostate have been well studied systems in the field of CSC research, and in addition, the epigenetic regulation of solid tumors originating from these organs, yet the direct analysis of epigenetics and human CSCs still remains largely unknown.
Neuronal Stem Cells
Cells derived from the central nervous system (CNS) originate from the neuroepithelium, which composes the neural plate (Morest and Silver, 2003). Embryonic neuroepithelial (NEP) cells are undifferentiated stem cells, capable to generate committed progenitors, which in turn produce three different lineages of fully differentiated cells (neurons, astrocytes and oligodendrocytes). Evidence suggests that this process is not restricted to pre-natal development, but can occur also during adult life, reviewed in (Lederer and Santama, 2008). NEP cells can be isolated from several regions of the CNS, with the highest densities in the subventricular and subgranular zones. Adult neurogenesis is thought to recapitulate the hierarchical organization of embryonic neurogenesis, and to occur mainly after stress (ischemia, bleeding) (Jensen and Parmar, 2006). The lack of an established in vivo assay makes normal NEP cell isolation and characterization difficult. The most accepted assay for NEP expansion is the in vitro propagation of neurospheres in serum-replacement medium. In addition, cells isolated from the subventricular zone can be enriched in NEP cells by FACS based on the expression of nestin, neurotrophin R (p75) or prominin-1 (CD133) (Frederiksen and McKay, 1988; Morrison et al., 1999; Nakafuku et al., 2008). At present, neither marker has been shown to be unequivocally associated with NEP cells, thus it is likely that NEP cells are identified by a combination of different markers. Consistent with this evidence, human embryonic CD133+/CD24− cells showed neural stem cell (NSC) features, including the ability to form neurospheres and to differentiate in CNS cells when injected into immunodeficient mice (Uchida et al., 2000).
The process of differentiation in embryonic NEPs in relation to epigenetic mechanisms has been well investigated in mice, reviewed in (Namihira et al., 2008). In cultures of NEPs prepared from mouse telencephalon at embryonic day 11 (E11, mid-gestation) and 14 (E14, late gestation) there is a significant change in the methylation status of STAT-3 binding element within the glial fibrillary acidic protein (GFAP) promoter (Namihira et al., 2008). Further experiments revealed that the STAT3-binding site was hypermethylated in E11 cells (which do not respond to the JAK-STAT3 pathway-activating cytokine LIF), yet are barely methylated in E14 cells which do express GFAP in response to LIF stimulation. This lack of methylation in the E14 cells allows for expression of astrocytic genes. Furthermore, it was shown that another astrocyte-inducing cytokine, bone morphogenic protein 2 (BMP2), increases histone acetylation around the CpG site in E14 cells and not E11 cells (Namihira et al., 2008). However, much less is known about the differentiation of adult neuronal stem cells. Again in mice, it has been shown that in NEPs lacking Mbd1 (methyl-CpG binding protein-1), the promoter of fibroblast growth factor-2 (Fgf-2) is hypomethylated and the number of stem cells increase (Li et al., 2008).
One key regulator of neurogenesis is the neuronal repressor REST (RE1-silencing transcription factor, also called NRSF) (Ballas and Mandel, 2005). In mouse ESCs, the REST/NRSF complex is bound to the RE1 site within neuronal-specific genes such as NeuroD and SCG10/stathmin and the chromatin is maintained along with HDAC1 and methyl CpG binding protein-2 (MeCP2) to allow only basal expression (Ballas and Mandel, 2005; Schoenherr et al., 1996). However, as mouse ESCs differentiate, the complex disappears from these promoter regions (triggered by unknown activation of proteosome degradation) and the cells become activated neurons. Recently, it has also been shown that REST maintains self-renewal and pluripotency in mouse ESCs through the suppression of microRNA 21 (miR-21), and futhermore Rest+/− cells express reduced levels of Oct4, Sox2, Nanog, and c-Myc (Singh et al., 2008). In addition, another miR, miR-124a, is regulated by the REST/NRSF complex (Conaco et al., 2006). Expression of miR-124a is repressed in neuronal progenitor cells expressing this complex. Once the complex disappears, the miR-124a gene is induced, and differentiation occurs. This demonstrates that REST is not only important in maintaining the pluripotency of NSCs, but is also involved in regulating self-renewal of mouse ESCs. Although most research involving REST has been conducted in mouse lines, one study demonstrated the connection in humans. Using medulloblastoma tumor samples and immunohistochemistry, it was found that 17 of 21 stained positively for REST/NRSF complex compared to almost no expression in normal samples (Su et al., 2006). Considering that medulloblastoma is thought to arise from undifferentiated neural stem/progenitor cells present in the external granule layer of the cerebellum, this evidence along with that from mouse systems warrants it as a target for future study with regard to its epigenetic modification and regulation of neuronal differentiation.
In addition to methylation-specific regulation of NEPs, histone modifications have been shown to dramatically affect their differentiation patterns. For example, the differentiation of adult rat hippocampal neural progenitors is blocked by the well known anti-epileptic, valproic acid (VPA) (Hsieh et al., 2004). VPA is classified as an HDAC inhibitor and can also inhibit glial differentiation of NEPs even under conditions that favor lineage-specific differentiation (Hsieh et al., 2004). Recently, in patients, it has been shown that VPA treatment can decrease the differentiation and markers associated with the progression of acute myeloid leukemia (Mahlknecht and Schonbein, 2008), suggesting that the regulation of histone modification can truly impact the progression of human disease.
On the contrary to normal NSCs, brain cancer stem cells seem to be identified by the expression of only the CD133 surface protein. Indeed, only CD133+ cells isolated from brain pediatric tumors (medulloblastoma and pilocytic astrocytoma) were capable to form neurospheres in serum replacement medium (Singh et al., 2003). Subsequently, the same group demonstrated that CD133+ cells are much more tumorigenic than CD133− cells (Singh et al., 2004). When injected into NOD/SCID mice, as few as 100 CD133+ cells formed a tumor that was a phenocopy of the patient’s tumor, however, injection of 105 CD133− cells did not form a tumor. In this case, CD133+ cells were isolated from both pediatric medulloblastoma and adult glioblastoma. Regardless of patient’s age and tumor histology, all CD133+ cells displayed stem cell properties in vivo, prompting the hypothesis of a common CSC phenotype for brain cancers. The simplicistic dichotomy between CD133+ and CD133− cells has been questioned by more recent findings. Indeed, the CD133− fraction proved to retain a minimal tumorigenic activity, and thus based on this observation, it has been proposed that not all glioblastoma CSCs express CD133 (Beier et al., 2007). Interestingly, the CD133 gene is subject to epigenetic gene silencing in brain tumors, as it can be methylated (Yi et al., 2008). Thus, it is conceivable that the majority of glioblastoma cells do not express CD133 because of promoter methylation, while glioblastoma CSCs harbor an unmethylated promoter. The clinical relevance of CD133+ population has been confirmed by the finding that high CD133 expression is associated with a worse prognosis glioblastoma (Pallini et al., 2008). On the other hand, the chemotherapic temozolomide has been shown to selectively deplete CD133+ cells, and could be employed in future trails to target brain tumor CSCs (Beier et al., 2008).
Finally, in human-and mouse-established glioma cell lines, a side population (SP) expressing CD133 has been found (Wu et al., 2008). The tumors from these cells are characterized as glioblastoma multiforme (GBM), which is a lethal primary brain tumor that is comprised of a phenotypically diverse population of cells (Wu et al., 2008). The mouse lines have been characterized as being highly tumorgenic, capable of forming tumor spheres with high frequency, and are able to differentiate into glial-and neuronal-like cells. The identification of neuronal progenitor cells expressing CD133 will lead to further experiments characterizing these cells and an increased understanding of what epigenetic mechanisms might be altered.
It is clear, however, that the majority of our understanding comes from the murine models of NSCs. Hopefully as labs begin to culture human NSCs more efficiently (most likely as neurospheres) and from different sources, a better understanding of their epigenetic fingerprint will be available.
Liver and pancreatic stem cells
It is well known that the liver has regenerative capacity following partial hepatectomy or chemical injury and this can occur either by proliferation/differentiation of hepatic stem cells. With regards to the normal development, the liver and the pancreas develop from a common multipotent population of endoderm cells, reviewed in (Zaret, 2008). The differentiation of the two organs is specified by a unique set of genetic cues within the endoderm cells, and following this activation, the hepatic and pancreatic endoderm cells begin to invade the local mesenchyme and form tissues buds representative of their unique organ (Zaret, 2008). The earliest markers of hepatic differentiation are the expression of albumin, transthyretin and α-fetoprotein (AFP), while markers of early pancreatic differentiation include duodenal homeobox gene 1 (Pdx1), reviewed in (Zaret, 2008).
Due to the explosion of stem cell research and the identification of specific markers of CSCs, this regenerative capacity was investigated in hepatic and pancreatic lines as well. In rats, Sigal et al. were the first to identify hepatic stem/progenitor cells (Sigal et al., 1994) using FACS. These cells have been characterized as expressing c-Met+ CD49f+/low c-Kit− CD45− TER119− and demonstrate an increased enrichment in their ability to from hepatic colony-forming units in culture (H-CFU-Cs) (Suzuki and Nakauchi, 2002). Less characterization of these hepatic progenitor cells has occurred in humans, but nevertheless, similar cells have been isolated since it was noted after bone marrow transplantation in adults, hematopoietic stem cells migrate into the liver and give rise to oval cells (hepatic stem/progenitor cells in rodents), and then hepatocytes and biliary epithelial cells (Crosby et al., 2001). This concept of transdifferentiation, the idea that a somewhat differentiated stem cell creates cells outside its already established lineage (Udani, 2006; Liu and Rao, 2003) is discussed further in the future prospects. Interestingly, H-CFU-Cs derived from fetal rat livers have the ability to differentiate in vitro into cells of the hepatocyte, bile-duct, pancreas, gastric-epithelium and intestinal-epithelium, and thus following transplantation, can develop into the liver, pancreas, and intestine, respectively (Suzuki et al., 2002). More recently, normal hepatic stem cells (HpSCs) have been isolated using epithelial specific antigen (ESA), commonly called epithelial cell adhesion molecule (EpCAM), and AFP (Yamashita et al., 2008), the earlier maker of hepatic differentiation.
Human pancreatic stem cells have been isolated using FACS analysis and a diverse group of cell surface adhesion markers, reviewed in (Mimeault and Batra, 2008). Specifically, cells expressing CD133 were found to be endocrine progenitor cells further expressing makers of stemness; telomerase, efflux pump protein ABCG2, Oct-3/4, nanog and Rex-1 (Koblas et al., 2008). The cells were able to differentiate into insulin-producing cells in vitro, and further secreted C-peptide in a glucose dependent manner (Koblas et al., 2008).
Pancreatic cancer stem cells (termed PaCSCs) have been isolated using bulk cancer cells derived from patient primary tumors or primary tumors established from xenografts in NOD-SCID mice. It was found that the PaCSCs expressed CD44, CD24, and ESA, only representing about 0.2–0.8% of the total cell population, and these cells also had the highest tumorigenic potential (Lee et al., 2008). In a separate study, a similar result was seen when as few as 500 CD133+ pancreatic cancer cells were injected into immunocompromised mice (Hermann et al., 2007). Two situations are possible here; there exist separate populations of PaCSCs, where both have the same potency for tumor formation and represent distinct CSC populations, or PaCSCs expressing all four markers are the most highly enriched for true PaCSC function (Lee et al., 2008). Additional experiments need to be performed in order to determine the true nature of these cells.
These adult PaCSCs are able to differentiate into insulin-, glucagon-, and somatostatin-positive cells in vitro in the presence of certain growth factors (Mimeault and Batra, 2008). More recently, PaCSCs have also been isolated from acinar pancreatic tissue (meaning islet tissue has been removed) using CXCR4 and express the stem cell makers nestin, ABCG2, CD177, neurogenin-3, Nanog, CD133 and Oct-4 (Koblas et al., 2007). Likewise, as previously mentioned, CD133 itself can be used to isolate PaCSCs, and these cells also express similar markers of stemness (Koblas et al., 2008). There remains much debate as to which cells are really PaCSCs due to this large list of cell surface makers used in their isolation.
A number of groups, however, have recently begun studying the epigenetic regulation of PaCSCs. An interesting study by Mutskov et al. investigated the epigenetic control of human islet-derived precursor cells (hIPCs) (Mutskov et al., 2007). These mesenchymal cells are derived in vitro from the adult pancreas, and although they proliferate independent of insulin, they can be differentiated to epithelial cells which are able to secrete it (Gershengorn et al., 2004). Overall, within the actual isolated human islets, the insulin gene was found to have high levels of histone modifications typical of active genes (H4 hyperacetylation and dimethylation of H3 lysine 4) (Mutskov et al., 2007). The hIPCs cells, however, demonstrated half the level of active chromatin modification and had no measurable H3 lysine methylation. The authors conclude that although hIPCs do not express insulin, they demonstrate unique epigenetic marks that could enable them to undergo activation of insulin expression in response to certain pressures or alterations in programming (Mutskov et al., 2007).
With regard to genetic differences within these cells, a four-fold increase in sonic hedgehog (Shh) mRNA was observed in bulk pancreatic cancer cells, yet a forty-six fold increase was seen in CD44+CD24+ESA+ sorted cells (Lee et al., 2008). Much less is known about the epigenetic contribution to PaCSCs, however a great deal is known about epigenetic alteration in pancreatic cancer in general. For example, it is known that genes such as cyclin-D2, cyclin-dependent kinase inhibitors 1A and 1C and retinoic acid receptor-beta are hypermethylated in pancreatic cancer cell lines, as well as primary or xenografted pancreatic cancer, reviewed in (Sato and Goggins, 2006). Additionally, tumor suppressor genes such as 14-3-3-sigma (stratifin) and maspin are also hypomethylated in pancreatic cancer (Sato et al., 2004).
Much less is known with regard to hepatic cancer stem cells (HpCSCs), yet when CD133+ cells were isolated from human hepatocellular carcinoma cells as cancer stem/progenitor cells, it was found that the expression of glutamine synthetase and cytochrome P450 was decreased (Suetsugu et al., 2006) yet methylation was not defined as the mechanism of silencing. A recent review has summarized the expression of different markers and commonalities of human liver progenitor cells, different lineages (cholanglocytic and hepatocytic), expression in hepatocellular carcinoma (HCC), and then putative HpCSCs (Mishra et al., 2009). Although this list nicely summarizes the expression on certain makers on a variety of liver cell types, it does not functionally characterize any of the lines listed as stem cells.
Thus, an increased understanding of the epigenetic networks at hand in both of these tissues will allow for further understanding of the disease in general, as well as any therapeutic interventions involving epigenetics which could be applied.
Kidney stem cells
In addition to the discovery of adult stem cells in glandular tissues like the breast, prostate, liver and pancreas, adult stem cells have recently been identified in the kidney, reviewed in (Gupta and Rosenberg, 2008). The developing kidney differentiates from the metanephric mesenchyme and ureteric bud into 26 different types of tissue. A number of genes have been identified that are specifically expressed in the undifferentiated metanephric mesenchyme and are required for proper differentiation of the kidney, reviewed in (Metsuyanim et al., 2008). Although no true stem/progenitor cell markers exist for the kidney, recent studies using the Hoechst 33348 efflux method to isolate side populations of cells enriched in stem-like characteristics has been successful and determined that 1.3% of all cells are stem-like (Inowa et al., 2008). Side populations are characterized as specific cells in a population that can efflux or pump out dyes such has Hoechst, yet are not fully considered to be stem cells (Scharenberg et al., 2002). In another paper, these cells were isolated at a slightly higher percentage, 3.8% from normal kidneys, and 5.9% from kidneys of patients with renal cell carcinoma (RCC), yet they demonstrate an enriched expression of the putative stem cells markers β-catenin, CD133 and the mesenchymal stem cell marker Pax2 (Addla et al., 2008) compared to the total population (Addla et al., 2008). The population is still heterogeneous, however, and the authors found that the terms side-population (SP) and stem cell cannot be used interchangeably.
With regards to epigenetic regulation, it has recently been shown that the PcG proteins are key players in regulating normal and malignant renal progenitors. First using a murine model where differentiation of the metanephric mesenchyme to the nephrogenic mesenchyme was carried out it was found that the regulation of only Ezh2 was important for maintenance of the normal renal progenitor population (Metsuyanim et al., 2008). In the adult kidney, however, when ischemia/reperfusion injury was induced it was found that Bmi1, Eed and Suz12 expression was increased and Ezh2 was actually silenced. Additionally, in tumor studies using Wilms’ tumor samples (pediatric kidney cancer) in a xenograft model, it was determined that the same genes Suz12, Bmi1, and Eed (although significance was not achieved) along with Ezh2 were all elevated, as well as an increase in the stem cell marker CD133. Clearly, these PcG genes are regulated differently in the progenitor cells of the kidney depending on the developmental stage the organ is in.
In human RCC cell lines and patient samples, the methylation status of a number of genes has been examined. For example, the heptatocyte growth factor activator HAI-2/SPINT2, and four other genes (PLAU, CDH1, IGFBP3 and MT1G) have previously been shown to undergo promoter methylation (Morris et al., 2008). Additionally, methylation analysis of RCC cell lines and primary tumors demonstrated that CK19 and CXCL16 were methylated. Less is known about the methylation of the progenitor cells within the kidney. Clearly the knowledge about kidney stem cells is very limited and will require more intense investigation into adequate surface markers, as well as which epigenetic programs might be altered in their cancer stem cells pools.
Reversibility of epigenetic changes
The question that still remains, do errors in epigenetic regulation lead to the development of cancer stem cells? There is emerging evidence to support that this hypothesis is true since reversal of these changes can reverse the overall phenotype of the cell. For example, when the colorectal carcinoma cell line RKO and the leukemia cell line KG1a were treated with a low dose of 5-aza-2′ deoxycytidine and trichostatin A, a specific inhibitor of histone deacetylase activity, it resulted in robust re-expression of MLH1, TIMP3, CDKN2A CDKN2B, p15, and p16 (Cameron et al., 1999). Additionally, this same treatment in the AR-negative prostate cancer line DU145 resulted in restoring functional expression of AR and downstream targets such as prostate specific antigen (PSA) (Nakayama et al., 2000). With regards to Ezh2, it has recently been shown that loss of miR-101 leads to in increase in Ezh2 expression (Varambally et al., 2008). Conversely, the authors found that restoration miR-101 expression in DU145 cells led to a decrease in Ezh2 expression, and reduced the invasiveness of these cells (Varambally et al., 2008). Although this evidence strongly supports that errors in epigenetic patterning could lead to the development of CSCs, the limitation of this data, however, still remains that these changes were investigated using the entire cancer cell line, and not the sub-population of CSCs we are addressing here. To truly determine if epigenetic changes are altering the normal stem cell pool, future experiments will have to be designed to target these cells in particular.
Experimental challenges
Currently, sorting of cells for specific makers of ‘stemness’ seems to be the most widely accepted method for the isolation of CSCs from bulk populations of tumor cells, reviewed in (Fabian et al., 2009). The cell surface makers CD44 and CD133 have been used to isolate CSCs from colorectal, head and neck squamous cell carcinoma, liver, ovarian, and prostate cancer, while expression of CD133 has been used in these tissues as well as in the brain, lung and pancreas (Fabian et al., 2009). CD24 positive cells on the other hand have been used to identify CSCs in the pancreas, yet CD24 negative cells identify the CSCs in prostate and breast cancer tissues (Hurt et al., 2008; Fabian et al., 2009). Overlapping and non-overlapping populations do seem to exist, however. For example, two different non-overlapping populations of breast CSCs were identified in cells isolated from BRAC1 deficient tumors in mice, one being CD44+CD24−, and the other CD133+(Wright et al., 2008). Additional methods of isolation include side population analysis for efflux of Hoescht dye and staining with ALDH1 (which result in high levels of toxicity), and enrichment in vitro to form tumorspheres in stem cell culture media (Fabian et al., 2009). The later method has had some debate since the stem cell media could alter the cell surface expression from the original tissue over time. Also, one must considering the handling/preparation of samples and alteration to epigenetic changes that occur over time. In order to ensure differences in epigenetic signatures are not solely due to differences in the amount of time it takes to harvest stem cells from a piece of tissue or how long the cells are maintained in culture conditions, standardized protocols will need to be implemented.
A relatively new method that has been investigated in our lab uses a Matrigel invasion to isolate CSCs. We characterized differences in non-invasive/invasive cells from human prostate cancer cell lines (LNCaP and DU145), as well as clonally isolated human primary prostate cancer stem cells (PCSCs). We observed that only a subpopulation of cells within all established cell lines could invade the Matrigel-coated membranes through an EMT-mediated process (Klarmann et al., 2009). These cells were found to express much higher levels of CD44 and exhibit gene expression profiles consistent with those of previously characterized CD44+CD24− prostate CSCs isolated from the LNCaP cell line (Hurt et al., 2008). While in contrast, non-invasive cells do not express high levels of ‘stemness’ genes. Moreover, purified CD44+ cells, and not CD44− cells, are invasive. The invasive cell subpopulation was also tumorigenic in NOD/SCID mice whereas the non-invasive cells were only weakly tumorigenic. Thus, these data strongly suggest that the stem cell-like component of cancer cells is responsible for invasion, the first step in metastasis.
Regardless, we know that the CSCs exist and that they represent a highly tumorigenic population within a bulk tumor. One hypothesis is that CSCs themselves represent a heterogeneous population with different abilities to form tumors and metastasize, and that overtime the CSCs themselves undergo their own clonal evolution (Fabian et al., 2009). The idea must be considered when studying the epigenetic changes that are occurring in these populations of cells. Also, when thinking about comparing normal tissue specific stem cells to those isolated from tumor tissue, will the same makers be employed? The original concept of CSCs is that they are mutated versions of tissue-derived stem cells, and overtime, they are more likely to accumulate the genetic changes required for malignant transformation (Wicha et al., 2006). Thus, the current methods of isolating the transformed population should in theory isolate the parent cells as well.
Unfortunately the idea of investigating the epigenetic changes of these cells is much easier said on paper than carried out in the laboratory. The majority of experiments performed by molecular biologists require adequate amounts of RNA, DNA, and protein, as well as additional cells for repeat analyses or new questions that arise from previous data. This is challenging when studying the cancer stem cell population from either an established cell line or a solid tumor since the number of cells is quite low. However, in response to these challenges companies have begun to develop assays where, for example, RNA can be isolated from as little as 10 cells and real-time QPCR can be performed using TaqMan® Gene Expression Cells-to-CT™ Kit (Ambion). In addition, Ambion’s MessageAmp™ II aRNA Amplification Kit allows for RNA amplification for downstream experiments from very small amounts of isolated RNA. With regards to studying epigenetic changes, chromatin immunoprecipitation (ChIP) assays coupled with Affymetrix high-density tiling arrays (called ChIP-on-chip) is a powerful technique for identifying transcription factor binding sites, sites of histone modification, DNA methylation and other regulatory elements. In our lab we have developed such an assay that requires only 2 μg of total genomic DNA which can be obtained from only 300,000 cells (each cell contains 6 pg of DNA) (Alberts, 2004), while the ChIP-IT express from Active Motif only requires 100,000 cells per reaction. Coupling such experiments will further support that the observed epigenetic changes are truly having an effect on the proposed target genes. Since epigenetic methods of regulation indirectly regulate target genes this is crucial to prove the effects are directly related. In addition, using our Matrigel invasion system we were able to complete microarray analyses, QRT-PCR, immunocytochemistry, and xenograft studies with the subpopulation of invaded cells (Klarmann et al., 2009). Overall, assays are being developed in order to adequately analyze such as small population of cells.
Finally, the collection of such epigenetic data will lead to the development of software able to collate these epigenetic signatures (similar to those used in microarray data). Currently, the field of computational epigenetics is very new, yet genome-wide mapping of epigenetic information (mostly from ChIP-on-chip, ChIP-seq and bisulfite sequencing data) has lead to the development of specific epigenetic fingerprints of a variety of different tissues, including global changes which occur in normal verses cancer tissue. Using a similar approach with data collected from CSCs, we can determine differences in their epigenetic fingerprint from their normal counterpart.
Future Prospects
Transdifferentiation and stem cells
Many concepts have emerged about the true definition of cancer stem cells. Recently, the concept of transdifferentiation, the idea that a cell can differentiate across lineage boundaries, (i.e. that a non-stem cell transforms into a different type of cell) or an already differentiated stem cell creates cells outside its already established lineage (Udani, 2006; Liu and Rao, 2003). The best known evidence for this process exists in hematopoietic stem cells (HSCs) which have been shown to give rise to skin, respiratory, intestinal, and renal epithelium, liver parenchyma, pancreas, skeletal muscle, vascular endothelium, myocardium, and neurons of the central nervous system, reviewed in (Udani, 2006). There are a number of criteria that must be present to support that transdifferentiation has occurred.
First of all, the transdifferentiated stem cell must acquire the morphology of the native cells within the tissue it now resides in (Wagers et al., 2002). Secondly, the transdifferentiated stem cell must display the appropriate tissue specific markers demonstrating that it has in fact adopted a new cell fate (Wagers et al., 2002), and must also not express the markers that once identified it as a progenitor cell. These transdifferentiated cells have further been characterized as nonhematopoietic tissue committed stem cells (TCSC), and are enriched for a population expressing CXCR4+, CD34+, AC133+, lin−, CD45− for humans and CXCR4+, Sca-1+, lin−, CD45− in mice (Kucia et al., 2005).
The most supported models involving transdifferentiation of HSCs occurs either after a tissue is injured, or in early neoplasia. For example, mouse models have demonstrated that progenitor cells from adult mouse bone marrow are mobilized into the circulation by transient renal ischemia and home specifically to injured regions of the renal tubule (Kale et al., 2003). Once localized, they differentiate into renal tubular epithelial cells and appear to constitute the majority of the cells present in the previously necrotic tubules (Kale et al., 2003). Thus, transdifferentiation could be a process which occurs only under certain circumstances, such as injury or inflammation. This process, however, is not well understood; yet, it is thought to involve epigenetic changes in cells exposed to the external stimuli mentioned above (Kucia et al., 2005). One concept is that transdifferentiation occurs by cell fusion where the donor derived HSCs may fuse with differentiated cells in recipient tissues (Kucia et al., 2005). In more defined systems, for example in myofibroblast transdifferentiation, the conversion of rat hepatic stellate cells into hepatic myofibroblasts is triggered at sites of injury. Interestingly, treatment of these hepatic stellate cells with 5-aza-2′-deoxycytidine blocked this transdifferentiation from occurring (Mann et al., 2007). Similarly, the DNA methylation inhibitor, zebularine, induced the morphological transformation of C2C12 mouse myoblasts into smooth muscle cells accompanied by de novo synthesis of smooth muscle markers such as smooth muscle alpha-actin and transgelin (Lee and Kim, 2007).
The idea of transdifferentiation is a very intriguing model for the regulation of stem cell pools within adult tissues. Clearly, this process must be tightly regulated and certain cues must notify cell populations when to change. The current literature does suggest that epigenetic regulation is involved in the process of transdifferentiation, however, to what extent and the complete details remain largely unknown.
Overall, there is very limited literature published about transdifferentiation and human stem cells, and the data becomes even more sparse when the concept of epigenetic regulation is added. This concept should, however, be considered when investigating the role that epigenetics has on regulating human stem cells.
Therapeutic implications
Understanding the origin of leukemia stem cells and their epigenetic characteristics is particularly important for the rational design of leukemia treatment. Gene expression studies have demonstrated that HSC express higher levels of anti-apoptotic and drug efflux pumps than committed cells (Terskikh et al., 2003). Thus, it is conceivable that leukemias derived from HSC may be more prone to drug-resistance. As it has been shown in this section, epigenetic modulation of gene expression is crucial for the control of anti-apoptotic and drug resistance pathways. Moreover, epigenetic changes are mainly reversible through the administration of drugs such as Dnmt inhibitors and histone deacetylase inhibitors (HDACis). Such drugs have been developed for targeting total cell populations and not specifically stem cells. Azacitidine (5-aza-cytidine), has already been approved by the US Food and Drug Administration (FDA) for the treatment of all subtypes of myelodysplastic syndromes (MDS) (Kaminskas et al., 2005). This drug acts as a Dnmt inhibitor, and thus clinical efficacy depends on its ability to reverse promoter methyaltion of specific genes, many of which are unknown. A second agent that is very similar in structure, decitabine (5-aza-2′-deoxycytidine), has also received approval by the FDA for clinical use in patients with MDS (Kantarjian et al., 2006). One HDACi that has been FDA approved is vorinostat for the treatment of cutaneous T-cell lymphoma (Zelent et al., 2008). Treatment with an HDACi would permit re-acetylation of histone lysine residues required for transcriptionally active chromatin and reactivate gene expression. Clearly these treatments are effective in targeting abnormalities in MDS and AML, but what about the treatment of solid tumors? One group recently tested the ability of two HDACis, hydralazine and magnesium valproate, to sensitize tumor cells to chemotherapy in patients with advanced and refractory solid tumors (Candelaria et al., 2007). After treatment with hydralazine and magnesium valproate the patients demonstrated a reduction in global DNA methylation, histone deacetylase activity, and promoter demethylation, and an overall subsequent clinical response when re-challanged to the same chemotherapy they once received (Candelaria et al., 2007). Overall the authors state that this ‘lends support to the epigenetic-driven tumor-cell chemoresistance hypothesis’ (Candelaria et al., 2007).
An emerging concept is that classical anti-leukemia drugs may also act through the modulation of epigenetically silenced genes in leukemic cells. For example, all-trans retinoic acid (ATRA) has being used for years for the treatment of acute promyelocytic leukemia (APL). ATRA was known to promote the full differentiation of APL clones, leading to the production of postmitotic neutrophils. However, the molecular mechanism underlying this process was unknown until Mueller and coworkers showed that ATRA restores PU.1 expression in APL cells (Mueller et al., 2006). As it has been previously described, PU.1 is a required gene during myeloid development, and its epigenetic silencing is frequent in some hematological malignancies.
Although studies have begun to understand the effects of epigenetic modification in cancer, the specifics of these effects in stem cells remains a mystery and a sample of what is known to date is summarized in Table 2. This review has further demonstrated the lack of epigenetic data in human stem cells, yet has highlighted what information we do have from rodent studies, and how it can lead us in the right direction. Future research will have to investigate the mechanisms of epigenetic regulation/silencing in both normal and cancer stem cells isolated from humans, as well as the changes that occur as a normal stem cell progresses to a cancerous one. There are epigenetic cues that must occur for normal development/maturation of cells to thrive, however there are also aberrant cues that led to mis-regualtion (Figure 2). Exactly what these cues are in different cell types remains unknown. With the emerging technologies of systems biology/informatics, a vast amount of information can be acquired from samples with minimal materials required. In addition, using these systems we can ask what inhibitors or small molecules might alter these changes and restore normal function within the stem cell population.
Table 2.
Epigenetically controlled genes in tissue-specific stem cells
| Gene | Tissue Type | Epigenetic modification | Comments | Ref. |
|---|---|---|---|---|
| GFAP BMP2 |
Murine NEP | DNA met. Histone acetyl. |
GFAP and BMP2 are silenced in early NEP | Namihira, 2008 |
| FGF-2 | Murine NSC | DNA met. | Mbd1 dependent silencing | Li, 2008 |
| Axin2 Cdx1 T/Bra |
Murine NSC and myogenic cells | DNA met. H3 acetyl. H3K4 met. |
Wnt responsive genes | Wohrle, 2007 |
| INS | Human IPCS |
H4 acetyl. H3K4 met. |
Gene inactivated in IPCS | Mutskov, 2007 |
| GATA-1 | Murine erythroleukemia | H3K9 met. | Gene involved in normal and malignant hematopoiesis | Zhang, 2008 |
| PU.1 | Human hematopoietic cells | DNA met. | PU.1 is expressed in HSCs and B cells. Repressed in T cells | Ivascu, 2007 |
| HoxA7 HoxA9 HoxA10 |
Human/murine hematopoietic cells | H3K4 met. H3K27 met. |
PcG/Trx balance: regulates Hox gene expression | Krivtsov, 2007 |
met: methylation, acetyl: acetylation, NEP: neuroepithelial cells, NSC: neural stem cells, IPCS: islet-derived precursor cells, HSCs: hematopoietic stem cells, PcG: Polycomb group, Trx: Trithorax group.
Figure 2.
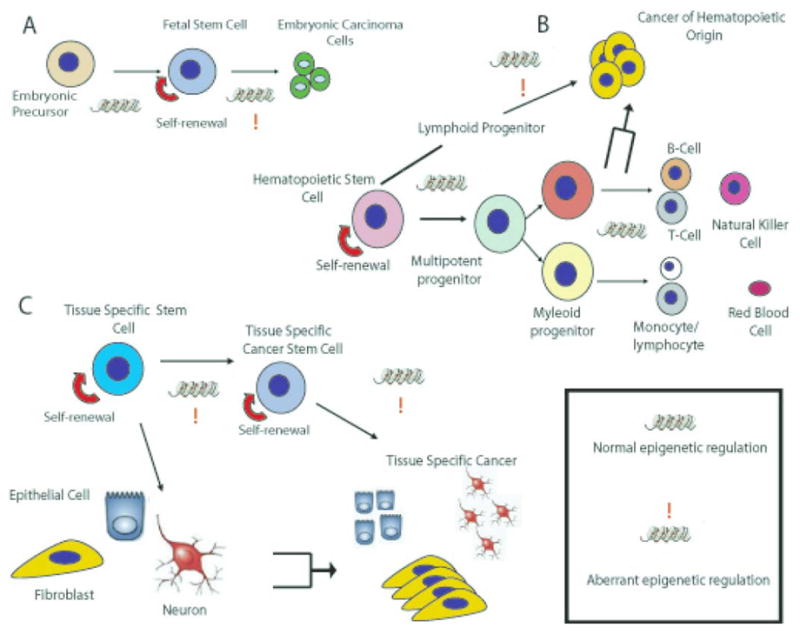
Overview of how epigenetic modifications can regulate differentiation of embryonic precursor cells (A), hematopoietic stem cells (B) or tissue specific stem cells (C). The histone symbol identifies normal epigenetic changes that occur to regulate either differentiation or self-renewal, whereas the symbol with the exclamation point identifies patterns of aberrant epigenetic modifications.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400, and supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We would also like to thank Dr. Elaine M. Hurt for her extensive editing and scientific perspective throughout the generation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Kawasaki K, Minoshima S, Krohn K, Antonarakis SE, Shimizu N, Kudoh J, Peterson P. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Current opinion in hematology. 2005;12:210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- Addla SK, Brown MD, Hart CA, Ramani VA, Clarke NW. Characterization of the Hoechst33342 side population from normal and malignant human renal epithelial cells. Am J Physiol Renal Physiol. 2008 doi: 10.1152/ajprenal.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins NL, Watts M, Georgel PT. To the 30-nm chromatin fiber and beyond. Biochim Biophys Acta. 2004;1677:12–23. doi: 10.1016/j.bbaexp.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. Essential cell biolog. Garland Science Pub; New York, NY: 2004. [Google Scholar]
- Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129:137–149. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- Allegrucci C, Young LE. Differences between human embryonic stem cell lines. Hum Reprod Update. 2007;13:103–120. doi: 10.1093/humupd/dml041. [DOI] [PubMed] [Google Scholar]
- Allis C, Jenuwein T, Reinberg D. Epigenetics. Cold Spring Harbor Laboratory Press; New York: 2007. [Google Scholar]
- Anbazhagan R, Osin PP, Bartkova J, Nathan B, Lane EB, Gusterson BA. The development of epithelial phenotypes in the human fetal and infant breast. The Journal of pathology. 1998;184:197–206. doi: 10.1002/(SICI)1096-9896(199802)184:2<197::AID-PATH992>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell stem cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Atkinson S, Armstrong L. Epigenetics in embryonic stem cells: regulation of pluripotency and differentiation. Cell Tissue Res. 2008;331:23–29. doi: 10.1007/s00441-007-0536-x. [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer research. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Beier D, Rohrl S, Pillai DR, Schwarz S, Kunz-Schughart LA, Leukel P, Proescholdt M, Brawanski A, Bogdahn U, Trampe-Kieslich A, Giebel B, Wischhusen J, Reifenberger G, Hau P, Beier CP. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer research. 2008;68:5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Bonkhoff H, Remberger K. Differentiation pathways and histogenetic aspects of normal and abnormal prostatic growth: a stem cell model. The Prostate. 1996;28:98–106. doi: 10.1002/(SICI)1097-0045(199602)28:2<98::AID-PROS4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C. Hormonal control of alveolar development and its implications for breast carcinogenesis. Journal of mammary gland biology and neoplasia. 2002;7:39–48. doi: 10.1023/a:1015718406329. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature genetics. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Candelaria M, Gallardo-Rincon D, Arce C, Cetina L, Aguilar-Ponce JL, Arrieta O, Gonzalez-Fierro A, Chavez-Blanco A, de la Cruz-Hernandez E, Camargo MF, Trejo-Becerril C, Perez-Cardenas E, Perez-Plasencia C, Taja-Chayeb L, Wegman-Ostrosky T, Revilla-Vazquez A, Duenas-Gonzalez A. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol. 2007;18:1529–1538. doi: 10.1093/annonc/mdm204. [DOI] [PubMed] [Google Scholar]
- Cariati M, Purushotham AD. Stem cells and breast cancer. Histopathology. 2008;52:99–107. doi: 10.1111/j.1365-2559.2007.02895.x. [DOI] [PubMed] [Google Scholar]
- Chalandon Y, Jiang X, Christ O, Loutet S, Thanopoulou E, Eaves A, Eaves C. BCR-ABL-transduced human cord blood cells produce abnormal populations in immunodeficient mice. Leukemia. 2005;19:442–448. doi: 10.1038/sj.leu.2403650. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang TH. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer research. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- Chung LW, Hsieh CL, Law A, Sung SY, Gardner TA, Egawa M, Matsubara S, Zhau HE. New targets for therapy in prostate cancer: modulation of stromal-epithelial interactions. Urology. 2003;62:44–54. doi: 10.1016/s0090-4295(03)00796-9. [DOI] [PubMed] [Google Scholar]
- Clarke MF. A self-renewal assay for cancer stem cells. Cancer Chemother Pharmacol 56 Suppl. 2005;1:64–68. doi: 10.1007/s00280-005-0097-1. [DOI] [PubMed] [Google Scholar]
- Clarkson B, Strife A, Wisniewski D, Lambek CL, Liu C. Chronic myelogenous leukemia as a paradigm of early cancer and possible curative strategies. Leukemia. 2003;17:1211–1262. doi: 10.1038/sj.leu.2402912. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development (Cambridge, England) 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes & development. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby HA, Kelly DA, Strain AJ. Human hepatic stem-like cells isolated using c-kit or CD34 can differentiate into biliary epithelium. Gastroenterology. 2001;120:534–544. doi: 10.1053/gast.2001.21175. [DOI] [PubMed] [Google Scholar]
- De Marzo AM, Meeker AK, Epstein JI, Coffey DS. Prostate stem cell compartments: expression of the cell cycle inhibitor p27Kip1 in normal, hyperplastic, and neoplastic cells. The American journal of pathology. 1998;153:911–919. doi: 10.1016/S0002-9440(10)65632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer research. 1959;19:515–520. [PubMed] [Google Scholar]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- Ema H, Morita Y, Yamazaki S, Matsubara A, Seita J, Tadokoro Y, Kondo H, Takano H, Nakauchi H. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nature protocols. 2006;1:2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- Engelhardt M, Lubbert M, Guo Y. CD34(+) or CD34(−): which is the more primitive? Leukemia. 2002;16:1603–1608. doi: 10.1038/sj.leu.2402620. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fabian A, Barok M, Vereb G, Szollosi J. Die hard: are cancer stem cells the Bruce Willises of tumor biology? Cytometry A. 2009;75:67–74. doi: 10.1002/cyto.a.20690. [DOI] [PubMed] [Google Scholar]
- Faderl S, Talpaz M, Estrov Z, O’Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. The New England journal of medicine. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Foshay KM, Gallicano GI. Small RNAs, big potential: the role of MicroRNAs in stem cell function. Curr Stem Cell Res Ther. 2007;2:264–271. doi: 10.2174/157488807782793781. [DOI] [PubMed] [Google Scholar]
- Frederiksen K, McKay RD. Proliferation and differentiation of rat neuroepithelial precursor cells in vivo. J Neurosci. 1988;8:1144–1151. doi: 10.1523/JNEUROSCI.08-04-01144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon G, McGinnis W. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays. 1998;20:116–125. doi: 10.1002/(SICI)1521-1878(199802)20:2<116::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Gershengorn MC, Hardikar AA, Wei C, Geras-Raaka E, Marcus-Samuels B, Raaka BM. Science. Vol. 306. New York, N.Y: 2004. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells; pp. 2261–2264. [DOI] [PubMed] [Google Scholar]
- Glinsky GV. “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J Clin Oncol. 2008;26:2846–2853. doi: 10.1200/JCO.2008.17.0266. [DOI] [PubMed] [Google Scholar]
- Graziano A, d’Aquino R, Tirino V, Desiderio V, Rossi A, Pirozzi G. The stem cell hypothesis in head and neck cancer. J Cell Biochem. 2008;103:408–412. doi: 10.1002/jcb.21436. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Rosenberg ME. Do stem cells exist in the adult kidney? Am J Nephrol. 2008;28:607–613. doi: 10.1159/000117311. [DOI] [PubMed] [Google Scholar]
- Hadnagy A, Beaulieu R, Balicki D. Histone tail modifications and noncanonical functions of histones: perspectives in cancer epigenetics. Mol Cancer Ther. 2008;7:740–748. doi: 10.1158/1535-7163.MCT-07-2284. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hogg NA, Harrison CJ, Tickle C. Lumen formation in the developing mouse mammary gland. Journal of embryology and experimental morphology. 1983;73:39–57. [PubMed] [Google Scholar]
- Holliday R, Pugh JE. Science. Vol. 187. New York, N.Y: 1975. DNA modification mechanisms and gene activity during development; pp. 226–232. [PubMed] [Google Scholar]
- Hovey RC, Trott JF, Vonderhaar BK. Establishing a framework for the functional mammary gland: from endocrinology to morphology. Journal of mammary gland biology and neoplasia. 2002;7:17–38. doi: 10.1023/a:1015766322258. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, Akashi K, Gilliland DG. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+ CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inowa T, Hishikawa K, Takeuchi T, Kitamura T, Fujita T. Isolation and potential existence of side population cells in adult human kidney. Int J Urol. 2008;15:272–274. doi: 10.1111/j.1442-2042.2007.01984.x. [DOI] [PubMed] [Google Scholar]
- Ivascu C, Wasserkort R, Lesche R, Dong J, Stein H, Thiel A, Eckhardt F. DNA methylation profiling of transcription factor genes in normal lymphocyte development and lymphomas. The international journal of biochemistry & cell biology. 2007;39:1523–1538. doi: 10.1016/j.biocel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Jarrard DF, Kinoshita H, Shi Y, Sandefur C, Hoff D, Meisner LF, Chang C, Herman JG, Isaacs WB, Nassif N. Methylation of the androgen receptor promoter CpG island is associated with loss of androgen receptor expression in prostate cancer cells. Cancer research. 1998;58:5310–5314. [PubMed] [Google Scholar]
- Jensen JB, Parmar M. Strengths and limitations of the neurosphere culture system. Molecular neurobiology. 2006;34:153–161. doi: 10.1385/MN:34:3:153. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Science. Vol. 293. New York, N.Y: 2001. Translating the histone code; pp. 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jones PA, Takai D. Science. Vol. 293. New York, N.Y: 2001. The role of DNA methylation in mammalian epigenetics; pp. 1068–1070. [DOI] [PubMed] [Google Scholar]
- Jowett AK, Vainio S, Ferguson MW, Sharpe PT, Thesleff I. Epithelial-mesenchymal interactions are required for msx 1 and msx 2 gene expression in the developing murine molar tooth. Development (Cambridge, England) 1993;117:461–470. doi: 10.1242/dev.117.2.461. [DOI] [PubMed] [Google Scholar]
- Kale S, Karihaloo A, Clark PR, Kashgarian M, Krause DS, Cantley LG. Bone marrow stem cells contribute to repair of the ischemically injured renal tubule. J Clin Invest. 2003;112:42–49. doi: 10.1172/JCI17856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC, Pazdur R. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, Helmer R, 3rd, Shen L, Nimer SD, Leavitt R, Raza A, Saba H. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- Kasper S. Exploring the Origins of the Normal Prostate and Prostate Cancer Stem Cell. Stem Cell Rev. 2008;4:193–201. doi: 10.1007/s12015-008-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalerchik E, Goff D, Jamieson CH. Chronic myeloid leukemia stem cells. J Clin Oncol. 2008;26:2911–2915. doi: 10.1200/JCO.2008.17.5745. [DOI] [PubMed] [Google Scholar]
- Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kinbara H, Cunha GR, Boutin E, Hayashi N, Kawamura J. Evidence of stem cells in the adult prostatic epithelium based upon responsiveness to mesenchymal inductors. The Prostate. 1996;29:107–116. doi: 10.1002/(SICI)1097-0045(199608)29:2<107::AID-PROS6>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2008;3:59–63. doi: 10.4161/epi.3.2.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarmann GJ, Hurt EM, Mathews LA, Zhang X, Duhagon MA, Mistree T, Thomas SB, Farrar WL. Invasive Prostate Cancer Cells Are Tumor Initiating Cells That Have A Stem Cell-Like Genomic Signature. Clinical and Experimental Metastasis. 2009;3:59–63. doi: 10.1007/s10585-009-9242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblas T, Pektorova L, Zacharovova K, Berkova Z, Girman P, Dovolilova E, Karasova L, Saudek F. Differentiation of CD133-positive pancreatic cells into insulin-producing islet-like cell clusters. Transplantation proceedings. 2008;40:415–418. doi: 10.1016/j.transproceed.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Koblas T, Zacharovova K, Berkova Z, Mindlova M, Girman P, Dovolilova E, Karasova L, Saudek F. Isolation and characterization of human CXCR4-positive pancreatic cells. Folia biologica. 2007;53:13–22. doi: 10.14712/fb2007053010013. [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Yoshida S, Saito Y, Doi T, Nagatoshi Y, Fukata M, Saito N, Yang SM, Iwamoto C, Okamura J, Liu KY, Huang XJ, Lu DP, Shultz LD, Harada M, Ishikawa F. CD34+CD38+CD19+ as well as CD34+CD38−CD19+ cells are leukemia-initiating cells with self-renewal capacity in human B-precursor ALL. Leukemia. 2008;22:1207–1213. doi: 10.1038/leu.2008.83. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Kucia M, Reca R, Jala VR, Dawn B, Ratajczak J, Ratajczak MZ. Bone marrow as a home of heterogenous populations of nonhematopoietic stem cells. Leukemia. 2005;19:1118–1127. doi: 10.1038/sj.leu.2403796. [DOI] [PubMed] [Google Scholar]
- Kusano S, Raab-Traub N. I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Molecular and cellular biology. 2002;22:6393–6405. doi: 10.1128/MCB.22.18.6393-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmanov A, Hayrabedyan S, Karaivanov M, Todorova K. Basal cell subpopulation as putative human prostate carcinoma stem cells. Folia histochemica et cytobiologica/Polish Academy of Sciences, Polish Histochemical and Cytochemical Society. 2007;45:75–80. [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lederer CW, Santama N. Neural stem cells: mechanisms of fate specification and nuclear reprogramming in regenerative medicine. Biotechnology journal. 2008;3:1521–1538. doi: 10.1002/biot.200800193. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26:2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Kim HJ. Inhibition of DNA methylation is involved in transdifferentiation of myoblasts into smooth muscle cells. Mol Cells. 2007;24:441–444. [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. The Journal of biological chemistry. 2008;283:27644–27652. doi: 10.1074/jbc.M804899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Lindvall C, Bu W, Williams BO, Li Y. Wnt signaling, stem cells, and the cellular origin of breast cancer. Stem Cell Rev. 2007;3:157–168. doi: 10.1007/s12015-007-0025-3. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rao MS. Transdifferentiation--fact or artifact. J Cell Biochem. 2003;88:29–40. doi: 10.1002/jcb.10281. [DOI] [PubMed] [Google Scholar]
- Mahadevan D, Spier C, Della Croce K, Miller S, George B, Riley C, Warner S, Grogan TM, Miller TP. Transcript profiling in peripheral T-cell lymphoma, not otherwise specified, and diffuse large B-cell lymphoma identifies distinct tumor profile signatures. Molecular cancer therapeutics. 2005;4:1867–1879. doi: 10.1158/1535-7163.MCT-05-0146. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mahlknecht U, Schonbein C. Histone deacetylase inhibitor treatment downregulates VLA-4 adhesion in hematopoietic stem cells and acute myeloid leukemia blast cells. Haematologica. 2008;93:443–446. doi: 10.3324/haematol.11796. [DOI] [PubMed] [Google Scholar]
- Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- Matushansky I, Radparvar F, Rekhtman N, Skoultchi A. Reprogramming erythroleuekmia cells to terminal differentiation and terminal cell division. Front Biosci. 2000;5:D488–492. doi: 10.2741/matushan. [DOI] [PubMed] [Google Scholar]
- Metcalf D. Concise review: hematopoietic stem cells and tissue stem cells: current concepts and unanswered questions. Stem cells (Dayton, Ohio) 2007;25:2390–2395. doi: 10.1634/stemcells.2007-0544. [DOI] [PubMed] [Google Scholar]
- Metsuyanim S, Pode-Shakked N, Schmidt-Ott KM, Keshet G, Rechavi G, Blumental D, Dekel B. Accumulation of malignant renal stem cells is associated with epigenetic changes in normal renal progenitor genes. Stem Cells. 2008;26:1808–1817. doi: 10.1634/stemcells.2007-0322. [DOI] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer research. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Mimeault M, Batra SK. Recent progress on normal and malignant pancreatic stem/progenitor cell research: therapeutic implications for the treatment of type 1 or 2 diabetes mellitus and aggressive pancreatic cancer. Gut. 2008;57:1456–1468. doi: 10.1136/gut.2008.150052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra L, Banker T, Murray J, Byers S, Thenappan A, He AR, Shetty K, Johnson L, Reddy EP. Liver stem cells and hepatocellular carcinoma. Hepatology (Baltimore, Md. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohinta S, Wu H, Chaurasia P, Watabe K. Wnt pathway and breast cancer. Front Biosci. 2007;12:4020–4033. doi: 10.2741/2368. [DOI] [PubMed] [Google Scholar]
- Mohty M, Yong AS, Szydlo RM, Apperley JF, Melo JV. The polycomb group BMI1 gene is a molecular marker for predicting prognosis of chronic myeloid leukemia. Blood. 2007;110:380–383. doi: 10.1182/blood-2006-12-065599. [DOI] [PubMed] [Google Scholar]
- Morest DK, Silver J. Precursors of neurons, neuroglia, and ependymal cells in the CNS: what are they? Where are they from? How do they get where they are going? Glia. 2003;43:6–18. doi: 10.1002/glia.10238. [DOI] [PubMed] [Google Scholar]
- Morris MR, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Teh BT, Latif F, Maher ER. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br J Cancer. 2008;98:496–501. doi: 10.1038/sj.bjc.6604180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, White PM, Zock C, Anderson DJ. Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell. 1999;96:737–749. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- Mueller BU, Pabst T, Fos J, Petkovic V, Fey MF, Asou N, Buergi U, Tenen DG. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107:3330–3338. doi: 10.1182/blood-2005-07-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V, Raaka BM, Felsenfeld G, Gershengorn MC. The human insulin gene displays transcriptionally active epigenetic marks in islet-derived mesenchymal precursor cells in the absence of insulin expression. Stem Cells. 2007;25:3223–3233. doi: 10.1634/stemcells.2007-0325. [DOI] [PubMed] [Google Scholar]
- Nakafuku M, Nagao M, Grande A, Cancelliere A. Revisiting neural stem cell identity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:829–830. doi: 10.1073/pnas.0711637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Watanabe M, Suzuki H, Toyota M, Sekita N, Hirokawa Y, Mizokami A, Ito H, Yatani R, Shiraishi T. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Laboratory investigation; a journal of technical methods and pathology. 2000;80:1789–1796. doi: 10.1038/labinvest.3780190. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Abematsu M, Nakashima K. Epigenetic mechanisms regulating fate specification of neural stem cells. Philos Trans R Soc Lond B Biol Sci. 2008;363:2099–2109. doi: 10.1098/rstb.2008.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213:306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, De Maria R. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Passegue E, Weissman IL. Leukemic stem cells: where do they come from? Stem cell reviews. 2005;1:181–188. doi: 10.1385/SCR:1:3:181. [DOI] [PubMed] [Google Scholar]
- Pelissier T, Wassenegger M. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. Rna. 2000;6:55–65. doi: 10.1017/s135583820099201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AS, Foley R, Woodson K, Lawler M. The emerging roles of DNA methylation in the clinical management of prostate cancer. Endocrine-related cancer. 2006;13:357–377. doi: 10.1677/erc.1.01184. [DOI] [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, van Lohuizen M. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18:1094–1099. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- Porter RL, Calvi LM. Communications between bone cells and hematopoietic stem cells. Archives of biochemistry and biophysics. 2008;473:193–200. doi: 10.1016/j.abb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston SL, Alison MR, Forbes SJ, Direkze NC, Poulsom R, Wright NA. The new stem cell biology: something for everyone. Mol Pathol. 2003;56:86–96. doi: 10.1136/mp.56.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GS, Birch L. The developmental pattern of androgen receptor expression in rat prostate lobes is altered after neonatal exposure to estrogen. Endocrinology. 1995;136:1303–1314. doi: 10.1210/endo.136.3.7867585. [DOI] [PubMed] [Google Scholar]
- Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, Sawyers C, Shah N, Stock W, Willman CL, Friend S, Linsley PS. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2794–2799. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Zylstra CR, Williams BO. Wnt signaling and prostate cancer. Curr Drug Targets. 2008;9:571–580. doi: 10.2174/138945008784911831. [DOI] [PubMed] [Google Scholar]
- Rodriguez RT, Velkey JM, Lutzko C, Seerke R, Kohn DB, O’Shea KS, Firpo MT. Manipulation of OCT4 levels in human embryonic stem cells results in induction of differential cell types. Exp Biol Med (Maywood) 2007;232:1368–1380. doi: 10.3181/0703-RM-63. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Seita J, Czechowicz A, Bhattacharya D, Bryder D, Weissman IL. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell cycle (Georgetown, Tex. 2007;6:2371–2376. doi: 10.4161/cc.6.19.4759. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16(Spec No 2):R243–251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2006;13:286–295. doi: 10.1007/s00534-005-1057-1. [DOI] [PubMed] [Google Scholar]
- Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99:507–512. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Seftor EA, Brown KM, Chin L, Kirschmann DA, Wheaton WW, Protopopov A, Feng B, Balagurunathan Y, Trent JM, Nickoloff BJ, Seftor RE, Hendrix MJ. Epigenetic transdifferentiation of normal melanocytes by a metastatic melanoma microenvironment. Cancer research. 2005;65:10164–10169. doi: 10.1158/0008-5472.CAN-05-2497. [DOI] [PubMed] [Google Scholar]
- Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Sieburg HB, Cho RH, Muller-Sieburg CE. Limiting dilution analysis for estimating the frequency of hematopoietic stem cells: uncertainty and significance. Experimental hematology. 2002;30:1436–1443. doi: 10.1016/s0301-472x(02)00963-3. [DOI] [PubMed] [Google Scholar]
- Sigal SH, Brill S, Reid LM, Zvibel I, Gupta S, Hixson D, Faris R, Holst PA. Characterization and enrichment of fetal rat hepatoblasts by immunoadsorption (“panning”) and fluorescence-activated cell sorting. Hepatology (Baltimore, Md. 1994;19:999–1006. [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer research. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard C, Lapidot T, Vormoor J, Cashman JD, Doedens M, Murdoch B, Jamal N, Messner H, Addey L, Minden M, Laraya P, Keating A, Eaves A, Lansdorp PM, Eaves CJ, Dick JE. Normal and leukemic SCID-repopulating cells (SRC) coexist in the bone marrow and peripheral blood from CML patients in chronic phase, whereas leukemic SRC are detected in blast crisis. Blood. 1996;87:1539–1548. [PubMed] [Google Scholar]
- Smith GH, Medina D. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. Journal of cell science. 1988;90 (Pt 1):173–183. doi: 10.1242/jcs.90.1.173. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein T, Price KN, Morris JS, Heath VJ, Ferrier RK, Bell AK, Pringle MA, Villadsen R, Petersen OW, Sauter G, Bryson G, Mallon EA, Gusterson BA. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin Cancer Res. 2005;11:6872–6879. doi: 10.1158/1078-0432.CCR-05-0547. [DOI] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. The EMBO journal. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, Snyder E, Eberhart CG, Majumder S. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu A, Nagaki M, Aoki H, Motohashi T, Kunisada T, Moriwaki H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351:820–824. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Nakauchi H. Identification and propagation of liver stem cells. Semin Cell Dev Biol. 2002;13:455–461. doi: 10.1016/s1084952102001349. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Zheng YW, Kaneko S, Onodera M, Fukao K, Nakauchi H, Taniguchi H. Clonal identification and characterization of self-renewing pluripotent stem cells in the developing liver. J Cell Biol. 2002;156:173–184. doi: 10.1083/jcb.200108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Hara E, Tenen DG, Hozumi N, Oikawa T. Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b. Oncogene. 2006;25:2477–2488. doi: 10.1038/sj.onc.1209272. [DOI] [PubMed] [Google Scholar]
- Takaishi S, Okumura T, Wang TC. Gastric cancer stem cells. J Clin Oncol. 2008;26:2876–2882. doi: 10.1200/JCO.2007.15.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Kataoka Y, Cui Y, Takamori Y, Watanabe Y, Yamada H. Intracellular translocation of glutathione S-transferase pi during oligodendrocyte differentiation in adult rat cerebral cortex in vivo. Neuroscience. 2007;148:535–540. doi: 10.1016/j.neuroscience.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Tang DG, Patrawala L, Calhoun T, Bhatia B, Choy G, Schneider-Broussard R, Jeter C. Prostate cancer stem/progenitor cells: identification, characterization, and implications. Mol Carcinog. 2007;46:1–14. doi: 10.1002/mc.20255. [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Miyamoto T, Chang C, Diatchenko L, Weissman IL. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;102:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- Tsai RY. A molecular view of stem cell and cancer cell self-renewal. The international journal of biochemistry & cell biology. 2004;36:684–694. doi: 10.1016/j.biocel.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Uchida N, Aguila HL, Fleming WH, Jerabek L, Weissman IL. Rapid and sustained hematopoietic recovery in lethally irradiated mice transplanted with purified Thy-1.1lo Lin-Sca-1+ hematopoietic stem cells. Blood. 1994;83:3758–3779. [PubMed] [Google Scholar]
- Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udani VM. The continuum of stem cell transdifferentiation: possibility of hematopoietic stem cell plasticity with concurrent CD45 expression. Stem Cells Dev. 2006;15:1–3. doi: 10.1089/scd.2006.15.1. [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002;297:2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer research. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895–1886. [DOI] [PubMed] [Google Scholar]
- Wohrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol Cell Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SF, Jolly DJ, Lunnen KD, Friedmann T, Migeon BR. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:2806–2810. doi: 10.1073/pnas.81.9.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Oh S, Wiesner SM, Ericson K, Chen L, Hall WA, Champoux PE, Low WC, Ohlfest JR. Persistence of CD133+ cells in human and mouse glioma cell lines: detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cells Dev. 2008;17:173–184. doi: 10.1089/scd.2007.0133. [DOI] [PubMed] [Google Scholar]
- Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, Reid LM, Minato H, Honda M, Kaneko S, Tang ZY, Wang XW. EpCAM-Positive Hepatocellular Carcinoma Cells Are Tumor-Initiating Cells With Stem/Progenitor Cell Features. Gastroenterology. 2008;136:1012–24. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Liu TC, Chang JG, Lin PM, Lin SF. JunB gene expression is inactivated by methylation in chronic myeloid leukemia. Blood. 2003;101:3205–3211. doi: 10.1182/blood-2002-05-1598. [DOI] [PubMed] [Google Scholar]
- Yi JM, Tsai HC, Glockner SC, Lin S, Ohm JE, Easwaran H, James CD, Costello JF, Riggins G, Eberhart CG, Laterra J, Vescovi AL, Ahuja N, Herman JG, Schuebel KE, Baylin SB. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer research. 2008;68:8094–8103. doi: 10.1158/0008-5472.CAN-07-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007a;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, Mehra R, Wang X, Ghosh D, Shah RB, Varambally S, Pienta KJ, Chinnaiyan AM. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer research. 2007b;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–340. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- Zelent A, Petrie K, Boix-Chornet M, Melnick AM, Waxman S, Gore SD. Derepression in the desert: the third workshop on clinical translation of epigenetics in cancer therapeutics. Cancer research. 2008;68:4967–4970. doi: 10.1158/0008-5472.CAN-07-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]


