Summary
The Nuclear Pore Complex (NPC) mediates nucleocytoplasmic transport in all eukaryotes and is among the largest cellular assemblies of proteins, collectively referred to as nucleoporins (nups). Nups are organized into distinct subcomplexes. We optimized the isolation of a putative membrane-coating subcomplex of the NPC, the heptameric Nup84 complex, and analyzed its structure by electron microscopy (EM). Our data confirm the previously reported Y-shape. We discerned additional structural details, including specific hinge regions at which the particle shows great flexibility. We determined the three-dimensional structures of two conformers, mapped the localization of two nups within the subcomplex and docked known crystal structures into the EM maps. The free ends of the Y-shaped particle are formed by beta-propellers; the connecting segments consist of alpha-solenoids. Strikingly, the same organizational principle is found in the clathrin triskelion, which was proposed to share a common evolutionary origin with the heptameric complex.
The nuclear pore complex (NPC) is an assembly of multiple copies of ∼30 different proteins called nucleoporins (nups), with an estimated total mass of ∼50 MDa in budding yeast and an even greater mass in metazoan cells (for a recent review, see1). It is localized in a 100 nm wide circular opening in the nuclear envelope that is formed by circumscribed fusion of the outer and inner membrane of the nuclear envelope, yielding a sharply bent “pore membrane” domain. The pore membrane domain contains three specific integral membrane proteins called poms, which presumably function in anchoring the core of the NPC. The NPC core has eight-fold and two-fold axes of symmetry, perpendicular to the plane and in the plane of the nuclear envelope, respectively. Peripheral nups are attached asymmetrically to the cytoplasmic and nucleoplasmic sides of the core. The core NPC contains a central channel of about 40 nm diameter, through which selective transport between nucleus and cytoplasm occurs. Besides transport, NPCs or subcomplexes thereof are involved in several other functions, such as a chromatin organization or mitotic spindle assembly.
Cryo-electron tomography studies have revealed that the NPC undergoes dramatic conformational changes during the transport of cargo, indicating a flexible structure2,3. Likewise, differences in the arrangement of some nups in various crystal forms suggest a dynamic interaction of nups within the NPC4-6
In higher eukaryotes with open mitosis, the NPC undergoes phosphorylation-dependent disassembly into distinct subcomplexes. One of the best characterized of these is a nonameric complex in vertebrate cells, the Nup107-160 complex7-9. The nonameric complex could also be isolated from interphase cells, using detergent and salt7,8. The nonamer is required for NPC formation10-12 and is targeted to kinetochores during mitosis, where it functions in spindle assembly8,13,14.
An equivalent heptameric complex, the Nup84 complex, was isolated from budding yeast, also using non-ionic detergent and salt15. The heptameric complex consists of Nup133, Nup84, Nup145C, Sec13, Nup85, Seh1 and Nup120 and has a predicted molecular mass of 576 kDa. The heptamer could also be reconstituted from proteins that were recombinantly expressed in E. coli16. Both reconstituted complex and native complex were shown by negative-stain electron microscopy to form a Y-shaped structure15,16. Based on reconstitution and negative-stain electron microscopy of various nup modules of the heptamer, the position of these modules within the two-dimensional Y-shaped structure was suggested16. While several crystal structures of heptameric complex nups are known6,17,18, the three-dimensional structure of the entire heptamer has not been previously determined.
Interestingly, the heptamer shares a protein subunit, Sec13, with the COPII complex, which coats vesicles for transport from the ER to the Golgi apparatus. Furthermore, the two principal folds of the heptamer nups, β-propellers and α-solenoids, are also found in coat complexes for vesicular transport. These facts have led to the hypothesis that the heptameric complex, as well as vesicle coats, have evolved from a “protocoatamer”, which played a crucial role in the evolution of eukaryotic cells19. The formation of the nuclear envelope and the endoplasmic reticulum in the evolution of prokaryotic to eukaryotic cells was envisaged to occur by invagination of specific domains of the prokaryotic plasma membrane20. A protocoatamer was suggested to stabilize the sharp membrane bends generated by this process. During the evolution of eukaryotic cells, this protocoatamer would have given rise to present-day membrane coat structures, including the heptameric subcomplex of the NPC19.
Consistent with a membrane-coating function, the heptamer is localized close to the pore membrane in vivo 21. Moreover, several of the heptameric complex nups contain a predicted membrane-curvature sensing motif, and in the case of Nup133, this motif was shown to mediate selective binding to highly curved liposomes22.
We set out to elucidate the architecture of the heptameric complex. In this paper, we describe the structure and flexibility of this NPC subcomplex from budding yeast based on single-particle electron microscopy of negatively stained specimen. We determined the three-dimensional structure of the heptamer by random conical tilt reconstruction23. In addition, we mapped the localization of tagged nups and docked crystal structures of known nups into the three-dimensional heptamer structure. We found that the arrangement of β-propeller and α-solenoid folds within the heptamer resembles the arrangement within the clathrin triskelion.
Results
Purification and EM of the Nup84 complex from budding yeast
We set out to determine the three-dimensional structure of native Nup84 complex from budding yeast by single-particle EM. Our objectives were the elucidation of the architectural principles of the heptameric complex, which would provide further insights into the proposed evolutionary relationship between the NPC subcomplex and vesicle coats, and a detailed characterization of the observed flexibility of the heptamer. Moreover, docking of nup crystal structures into an EM structure of the entire subcomplex should be an important step towards bridging the gap between high-resolution structures of individual nups and low-resolution structures of the entire NPC.
Purification protocols for affinity-tagged Nup84 complex from budding yeast have been reported15,24-26. The published protocols produced particle populations either missing individual proteins (namely Nup133)15,24 or containing additional nups or other proteins25,26. Furthermore, most of the published protocols15,24,25 used lysis buffer containing 1% Triton X-100, well above the critical micelle concentration. Potential incorporation of the protein complex into micelles would likely compromise structure determination by EM.
We therefore developed a protocol for affinity-purification of the heptameric Nup84 complex followed by size-exclusion chromatography, with the following objectives: (i) recovery of intact complexes, (ii) purity, and (iii) minimization of detergent use.
Based on previously published comparisons of different nup-tagging strategies24,26, we decided to purify the heptameric complex from a yeast strain in which the C terminus of Nup85 was genomically tagged with a TAP-tag. To minimize protein degradation, we disrupted cells by cryogenic grinding25, and carried out all further steps on ice or at 4°C while shortening all incubation times as much as possible. We tested NaCl concentration from 0 to 700 mM in the lysis buffer and found that 500 mM NaCl reduced contamination below the limit of detectability without affecting heptameric complex recovery or integrity. During the washing steps, NaCl concentrations were lowered stepwise to 200 mM. We also tested various detergents (Triton X-100, Tween-20, CHAPS, NP-40, octyl β-D-glucopyranoside and combinations thereof at different concentrations) and found that the presence of 0.05% CHAPS throughout the purification procedure was sufficient to prevent heptameric complex aggregation. Using CHAPS at concentrations below 0.05% reduced the recovery of soluble subcomplex.
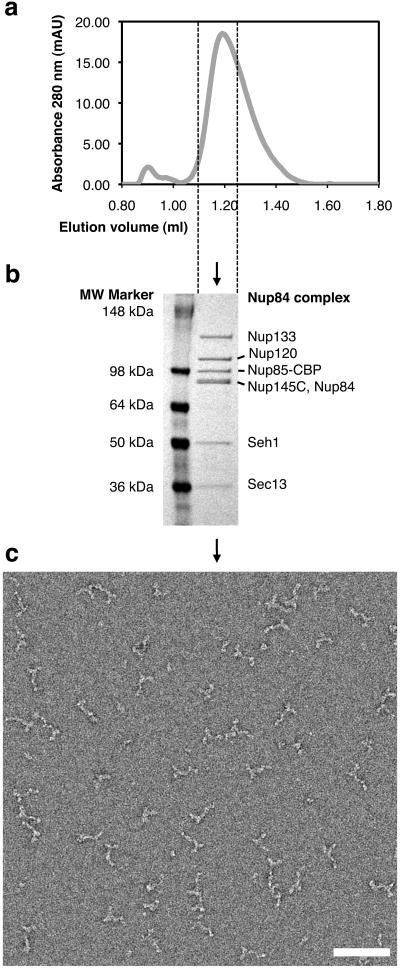
During size-exclusion chromatography, the heptameric complex eluted as one slightly asymmetric peak (Fig. 1a). On-line multi-angle light scattering indicated a molecular weight of ∼0.5 MDa for the peak, consistent with a single heptamer (data not shown). The chromatography fraction indicated by dashed lines in Fig. 1a was analyzed by SDS-PAGE and the identity of each band was confirmed by mass spectrometry (Fig. 1b). An aliquot of this material was applied to glow-discharged carbon-coated grids, negatively stained with uranyl formate, and subjected to EM (Fig. 1c). As previously described15,16, the heptamer appeared as a Y-shaped particle.
Figure 1.
Purification and electron microscopy (EM) of the heptameric Nup84 complex. (a) Size exclusion chromatography profile of affinity-purified Nup84 complex released from IgG-beads by TEV protease cleavage. The fraction indicated by dashed lines contains Nup84 complex and was used for EM. (b) Coomassie-stained SDS-PAGE of the fraction from size exclusion chromatography indicated in (a). All bands were identified by mass spectrometry. Nup85-CBP is Nup85 C-terminally tagged with the calmodulin-binding peptide moiety of the TAP-tag, which was cleaved from the protein A moiety by TEV protease. (c) Negative-stain EM of heptameric complex particles. A field of particles adsorbed to glow-discharged carbon film and stained with 2% uranyl formate is shown. Scale bar, 100 nm.
Two-dimensional analysis of particle structure and flexibility
EM images of individual particles are noisy; the signal-to-noise ratio can be increased by averaging images. Only images representing a highly similar view and conformation of the particle should be averaged, particle images therefore need to be classified into different groups representing distinct particle views or conformations. Alignment of particle images is required prior to classification and averaging. Computational methods for automatic reference-free alignment27 and classification28 of particle images are available.
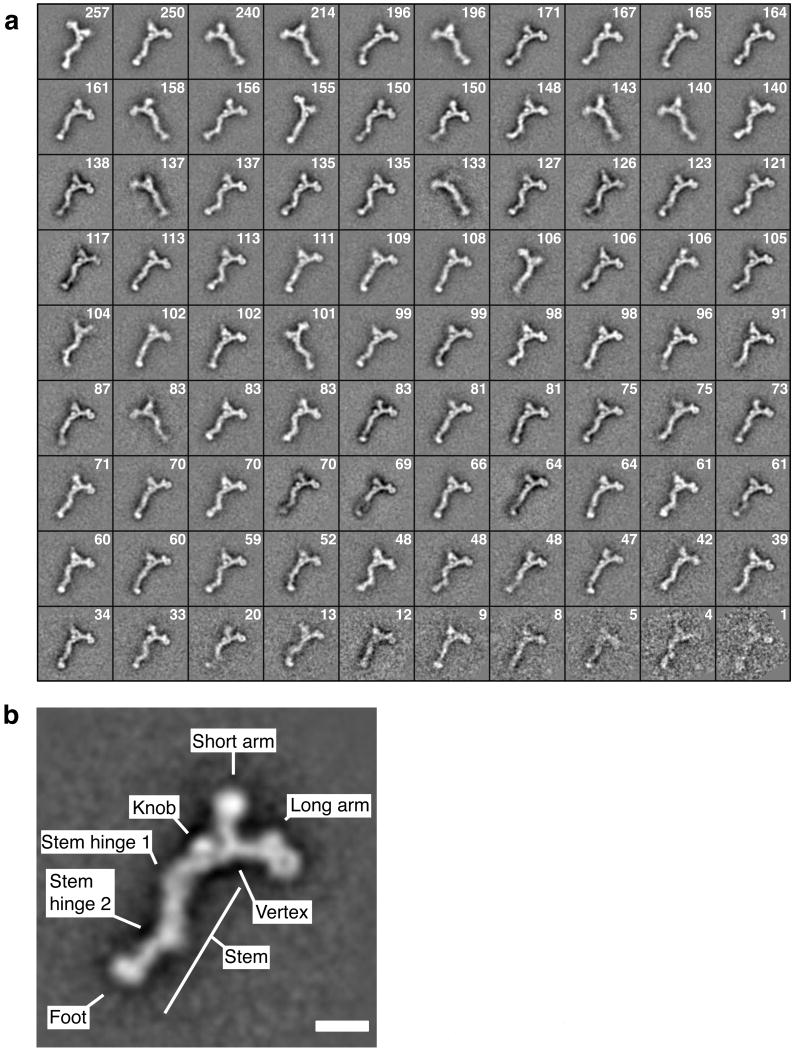
EM images of 9,028 individual particles were subjected to automated reference-free alignment. Aligned particles were then computationally clustered into 90 classes, and particles within each class were averaged. The resulting class averages are shown in Figure 2a.
Figure 2.
Alignment, classification and averaging of particle images reveals structural details of the heptameric complex. (a) Reference-free alignment and k-means classification of 9,028 particle images into 90 classes resulted in the depicted class averages. The number of particles constituting each class is indicated. (b) A well-defined class average is shown and prominent features are named. Scale bar, 100 Å.
Inspection of the class averages revealed characteristic features of the heptameric complex (Fig. 2b). The heptamer forms a ∼45 nm long, branched structure with a short arm, a long arm and a stem meeting at a vertex. In most class averages, the stem appears kinked at two regions, referred to as stem hinge 1 (the vertex-proximal hinge) and stem hinge 2 (the vertex-distal hinge). Four globular regions with a diameter of ∼5 nm are present at the end of the long and short arms, at the foot of the stem and as a knob next to the vertex. Strikingly, the globular region at the tip of the long arm shows a central hole or depression and thus resembles a β-propeller in top view. The present 2D structure is consistent with the structure described by Lutzmann and colleagues16 but reveals more details since averaging of aligned particle images increased the signal-to-noise ratio.
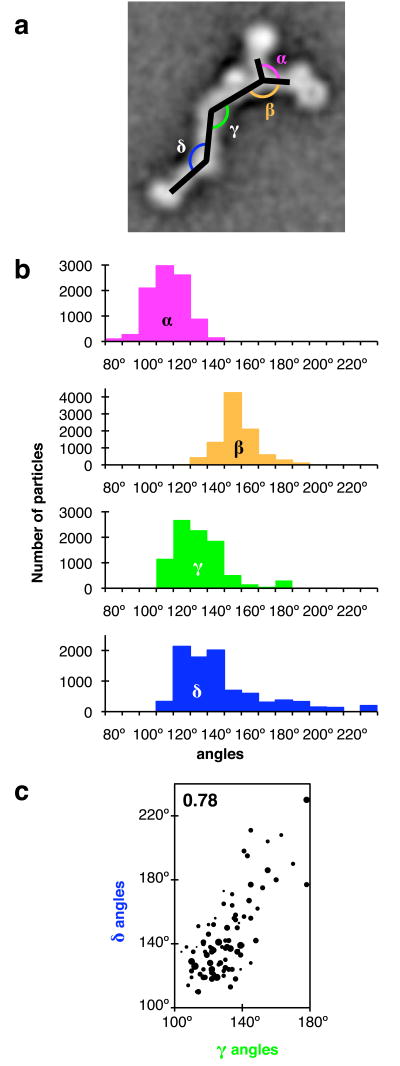
The 2D class averages show a spectrum of different appearances (Fig. 2a), differing mainly in the angles between different segments of the particle. To analyze this variability, we determined angles between particle segments for the 90 class averages (Fig. 2a). Four angles were measured (Fig. 3a): α (the angle between the two arms), β (the angle between the long arm and the vertex-proximal stem segment), γ (the angle at stem hinge 1), and δ (the angle at stem hinge 2).
Figure 3.
Heterogeneity of particle appearance. (a) Definition of angles between particle segments for the two-dimensional view (2D) of particles. α, angle between long and short arms; β, angle between long arm and vertex-proximal stem segment; γ, angle at stem hinge 1; δ, angle at stem hinge 2. (b) Distribution of angles for the 90 classes shown in Figure 2. Angles were measured for 2D class averages and assigned to the number of particles constituting each class. (c) Correlation between γ and δ. Each marker indicates γ and δ for a particle class. Marker area is proportional to class size. Particle-based correlation coefficient: 0.78 (shown in purple); Class-based correlation coefficient: 0.72. All other combinations of angles α, β, γ and δ are plotted in Supplementary Figure 1; the corresponding correlation coefficients are between −0.44 and −0.0006.
Analysis of the 90 classes revealed a continuum of angles between particle segments (Fig. 3b). The highest variability between 2D class averages was observed at the two stem hinges, especially stem hinge 2. Angles γ and δ at the stem hinges varied from ∼110° to ∼180°, and from ∼110° to ∼230°, respectively (Fig. 3b), causing the stem appearance to vary from straight to kinked. The angles around the vertex, α and β, also vary considerably: from ∼80° to ∼140°, and from ∼130° to ∼190°, respectively (Fig. 3b).
The angles at the stem hinges, γ and δ, are positively correlated with a correlation coefficient of 0.78 (Fig. 3c): most particles appear to have either a straight stem or a stem kinked at both hinges; stems kinked at one hinge and straight at the other hinge were not commonly observed. This suggests either that a large stem region undergoes a rigid-body movement, or that conformational changes at the two stem hinges are coupled. Other pairs of angles between particle segments do not correlate strongly (Supplementary Fig. 1), and principal component analysis indicates that correlated changes of the stem hinge angles γ and δ explain 73% of the measured angular variance (Supplementary Fig. 2).
The foot region also shows conformational flexibility, as evidenced by the varying position of the foot with respect to the adjacent stem segment, and the fuzzy appearance in several class averages, which is indicative of in-class variation (Fig. 2a). Since the foot appears as a globular shape without discernible internal features, we could not reliably determine its orientation with respect to the stem and therefore abstained from quantitative analysis of variability in the foot region.
3D structures of the heptameric complex
The method of choice for the de novo determination of 3D structures by EM is random-conical tilt reconstruction (RCT)23: A field of particles is imaged once after tilting the sample by a known angle, in our case 50°, and a second time after returning the sample to an untilted position, and tilt pairs of images of the same particle from the tilted and untilted sample are picked, as illustrated in Supplementary Figure 3. Since the in-plane orientation of particles on the carbon film is random, the images of the tilted specimen show a range of different particle views. Particle images from the untilted sample are used to classify particles and determine their in-plane orientation, and particle images from the tilted sample are used to reconstruct the 3D structure of the particle.
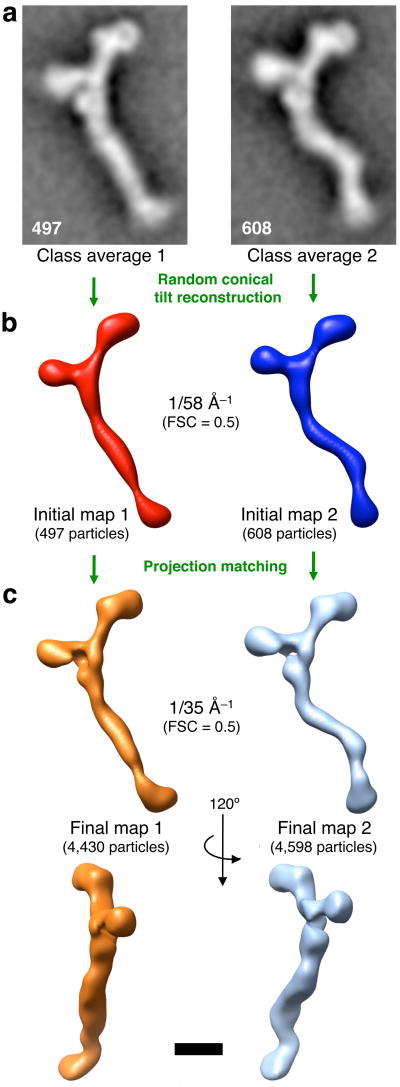
The 9,028 particle images of untilted specimen analyzed above were all taken in conjunction with images of the same particles after tilting. The continuum of different conformations present in the heptamer population poses a challenge to 3D reconstruction. Ideally, a conformationally homogeneous class of particles should be used for reconstruction. At the same time, a class comprising a large number of particles is desirable to improve signal-to-noise ratio and angular coverage for RCT. The number of particles chosen for a 3D reconstruction is thus a trade-off between homogeneity and signal-to-noise ratio. We approached the problem as follows: Particle images from the untilted sample were grouped into larger classes by hierarchical clustering, and two relatively homogeneous classes encompassing 497 and 608 particles, respectively, were chosen for RCT (Fig. 4a). 3D maps were generated by RCT (Fig. 4b). The reproducible resolution of these initial 3D maps is ∼58 Å as determined by the 0.5 Fourier-Shell correlation (FSC) criterion (Supplementary Fig. 4).
Figure 4.
Three-dimensional structures of the heptameric complex (a) 9,028 untilted particle images were grouped by hierarchical ascendant classification and the two depicted classes, comprising 497 and 608 particles, respectively, were chosen for random-conical tilt reconstruction (RCT). (b) Initial maps obtained by RCT from the classes shown in (a) are depicted as isodensity contour surfaces that were low-pass filtered beyond the reproducible resolution (Fourier shell correlation (FSC) = 0.5 at 1/58 Å−1, see Supplementary Fig. 3). (c) The initial maps were used as references for projection matching of all 9,028 tilted particle images (see text for details). 4,430 particles aligned to initial map 1, and 4,598 particles aligned to initial map 2. Final maps 1 and 2 were obtained by the simultaneous iterative reconstruction technique, and are shown in two views as isodensity contour surfaces low-pass filtered beyond the reproducible resolution (FSC = 0.5 at 1/35 Å−1, see Supplementary Fig. 3). All structures depicted to scale; scale bar, 100 Å.
Our next goal was to increase the signal-to-noise ratio of the initial maps by incorporating more particle images from tilted specimen. We hypothesized that some of the variation in particle appearance is likely to be caused by slight differences in the particle orientation with respect to the carbon support. Therefore, additional particles are likely to have a similar conformation to the particles constituting the two initial maps. We proceeded as follows: Projection images were created from the two initial maps, and all particle images from tilted specimen were matched to one of these projections based on the highest correlation coefficient. This approach is discussed in more detail in the Supplementary Discussion section. The simultaneous iterative reconstruction technique29 was used to obtain the final 3D maps 1 and 2 (Fig. 4c). The reproducible resolution of the final maps is ∼35 Å (Supplementary Fig. 4). The overall resolution of the particle is likely limited by the averaging of conformationally divergent particles, and the true resolution of invariant features may be better than 35 Å.
The 3D structures confirm that the isolated heptameric complex is a flexible particle that can exist in different conformations (Fig. 4b,c): map 1 corresponds to a “straight-stem” conformation and map 2 to a “kinked-stem” conformation. In both conformations, the particle lies relatively flat on the carbon support film. It is possible that binding of the particle to the planar support restricts its conformational flexibility, and that the complex displays an even greater conformational variability in solution.
We attempted to characterize the 3D structure of the heptameric complex in solution by collecting cryo-EM images of vitrified samples. Unfortunately, we were not able to detect the particle by either cryo-EM, probably due to the low contrast provided by the thin, extended shape of the particle, nor could we obtain suitable cryo-negative stain images using ammonium molybdate.
Nup positions within the heptameric complex
Several aspects of Nup84 complex architecture have been established previously based on protein-protein interactions and 2D EM16. The two arms of the heptamer are formed by Nup120 and the Nup85·Seh1 heterodimer, the vertex-proximal stem segment is formed by the Nup145·Sec13 heterodimer, the mid-stem is formed by Nup84 and the distal stem segment is formed by Nup133. The experimental evidence for this assignment is reviewed in the Supplementary Discussion section.
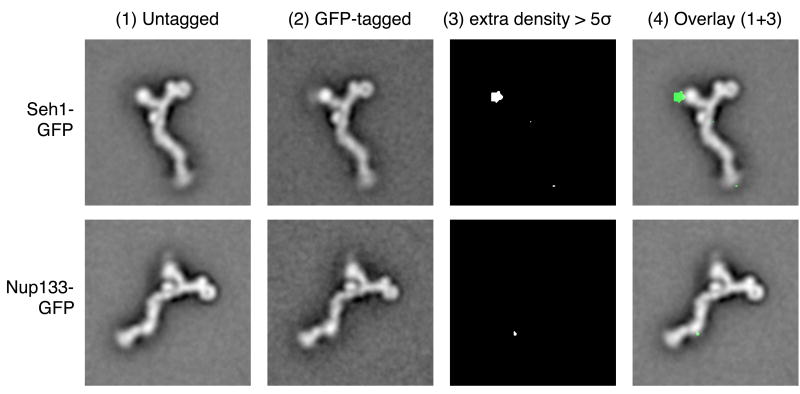
We described here the asymmetric appearance of the two arms of the heptameric complex. Since previous data did not allow us to deduce which arm corresponds to Nup120 and which to Nup85·Seh1, we addressed this question experimentally. We genomically tagged the C terminus of Seh1 with green fluorescent protein (GFP) and purified the heptameric complex from this modified yeast strain. GFP tagging did not interfere with nuclear envelope localization in vivo or with complex purification. EM images were collected for GFP-tagged complexes, and aligned class averages of GFP-tagged and untagged particles were compared. An area of additional density corresponding to the GFP tag could be localized to the short arm (Fig. 5). The GFP density distribution is also visible directly in the class average image of the GFP-tagged particle; however, it appears fuzzy, presumably due to a flexible localization of the attached GFP.
Figure 5.
Mapping of nup localization. Heptameric complexes were purified from yeast strains in which one protein of the subcomplex was genomically tagged with green fluorescent protein (GFP): the C terminus of Seh1 (first row), or the C terminus of Nup133 (second row). Aligned class averages of untagged and GFP-tagged particles are shown in columns (1) and (2). The significance map column (3) shows extra density for the GFP-tagged particles above the five-fold pixel-based standard deviation of the class averages. Column (4) shows an overlay of columns (1) and (3).
In a separate experiment, GFP was attached to the C terminus of Nup133, which is known to interact with the C-terminal domain of Nup84. The additional density was mapped as before, and found to co-localize with the stem hinge 2 region of the heptamer (Fig. 5). The GFP density appears even more fuzzy and delocalized, and the area of statistically significant additional mass is much smaller than for Seh1-GFP, likely because of even greater flexibility of the attached GFP in this case.
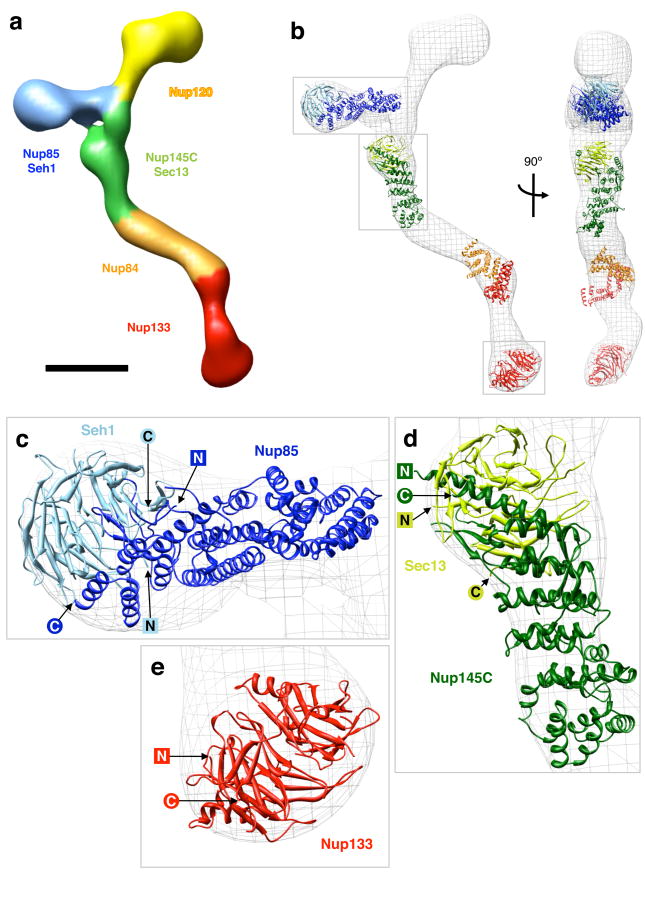
These localization maps, in conjunction with established interactions between members of the heptameric complex5,6,16-18, enabled us to segment the heptameric complex as shown in Figure 6a. Some details, in particular the exact nature of the interfaces between the Nup120, Nup145C and Nup85 regions at the vertex, are currently unknown and displayed tentatively.
Figure 6.
Protein arrangement within the heptameric complex (a) Segmentation of the particle (map 2) based on mapped nup localizations and previously established biochemical interactions. The particle surface is color-coded to represent the regions of the particle corresponding to different modules. Boundaries between regions are approximate. Scale bar, 100 Å. (b) Docking of available crystal structures (ribbon representation) into map 2 (isodensity contour mesh representation). Two views related by a 90° rotation around a vertical axis are shown. The crystal structures are of: yeast Nup85 (amino acids 1-570 of 744, dark blue) in complex with yeast Seh1 (full length, light blue)6, yeast Nup145C (amino acids 125-555 of 711, dark green) in complex with human Sec13 (amino acids 1-316 of 322, light green)18, human Nup107 (the homologue of yeast Nup84, amino acids 658-925 of 925, orange) in complex with human Nup133 (amino acids 934-1156 of 1156, red)5, and Nup133 (amino acids 76-478 of 1156, red)30. The conformation of the Nup107·Nup133 fragment is likely to differ from the actual Nup84·Nup133 conformation in map 2, as evidenced by the poor fit, and the structure is included for illustrative purposes only. Empty regions in the particle map correspond to proteins and protein domains for which no crystal structure is available yet. Grey boxes indicate the regions of the map shown in the subsequent panels. (c-e) Detailed views of crystal structures docked into map 2; N and C termini of the crystallized nup domains are indicated.
Nup crystal structures docked into the EM map
We proceeded to dock known crystal structures of heptameric complex components into the 3D EM structure. Docking was carried out independently for heptamer maps 1 and 2, and the resulting structures are shown in Figure 6b, Supplementary Figure 7 and Supplementary movies 1 and 2. Details of the docked structures in map 2 are shown in Figure 6c-e and Supplementary movie 3. Empty regions in the EM maps correspond to nup domains for which no crystal structures are currently available.
The Nup85·Seh1 dimer6,17 fit into the short arm of the heptamer (Fig. 6c); the Seh1 β-propeller corresponds to the thicker globular region at the end of the short arm; the α-solenoid regions of Nup85 localize to the thinner arm region. The Nup145C·Sec13 dimer18 localizes to the vertex-proximal stem region (Fig. 6d); the Sec13 β-propeller forms the knob close to the vertex. The middle part of Nup145 extends to the stem hinge 1 region of the heptamer, where it interacts with Nup84. This suggests that stem hinge 1 is formed at the interface between Nup145C and Nup84.
Since structures of yeast Nup133 and Nup84 have not been determined, we used the partial crystal structures of their human homologues Nup133 and Nup107 for docking. The N-terminal Nup133 β-propeller30 forms the globular foot region of the heptamer. The heterodimerization of Nup107 and Nup133 is mediated by the C-terminal domains of both nups, which have been crystallized as a complex5. Since the C terminus of Nup133 maps to stem hinge 2 (Fig. 5), the Nup133·Nup107 crystal structure can be expected to be situated in this segment of the heptamer. However, the crystal structure does not fit neatly into the EM map (Fig. 6b). The likely explanation is that since stem hinge 2 is a conformationally flexible region, the conformation of human Nup133·Nup107 in the crystal structure is somewhat different from the conformation of the homologous yeast Nup133·Nup84 in the particle conformation of map 2. Thus, the docking of the Nup133·Nup107 fragment is tentative, and mainly shown for illustrative purposes. Further details of the crystal structure arrangement within the particle are examined in the Supplementary Discussion section.
Discussion
We have presented 3D structures of an entire NPC subcomplex, the heptameric Nup84 complex from budding yeast. Our data confirm the overall architecture that was proposed previously based on 2D EM16. Averaging of images allowed us to discern additional details, such as four globular regions and the asymmetry of the two arms of the particle, and to characterize the conformational heterogeneity of the particle. The 3D maps, in combination with protein labeling experiments, enabled us to dock available nup crystal structures into the heptamer structure.
The EM structures (Fig. 4) do not necessarily represent the conformation of the heptameric complex in the context of the NPC since (i) interactions with other nups may affect the conformation of the heptameric complex, and (ii) the present structures are of the particle bound to a planar support film, whereas in the context of the NPC, the heptameric complex coats a highly curved surface. Distortions of the particle structure caused by negative staining and by missing-cone effects due to incomplete angular coverage of particle views are a potential concern, but they are unlikely to be dramatic in the present study, since the particle is not very extended in the direction perpendicular to the carbon support film. The simultaneous iterative reconstruction technique was used to minimize missing-cone effects. The docking of nups into the EM map (Fig. 6) represents the best possible fit given the current data; future higher-resolution EM maps and additional crystal structures may lead to a refinement of nup positions and orientations.
Despite these caveats, the present 3D structures yield fundamental insights into the architecture of the heptameric complex. The main architectural principle of the heptameric complex is that the globular domains at the ends of the arms and the stem are formed by β-propeller domains, whereas the thinner connecting segments are formed mainly by α-solenoid folds. While the crystal structure of Nup120 is not yet available, we expect Nup120 to conform to this principle: the shape of the long arm strongly suggests that the predicted Nup120 β-propeller localizes to the thick, globular end of the arm, whereas the predicted α-helical regions form the thinner connection to the vertex. This arrangement is supported by the 2D class averages (Fig. 2b): the long arm ends in a round shape ∼5 nm in diameter with a central hole or depression, compatible with a β-propeller in top view. Intriguingly, the same architectural principle of α-solenoid arms ending in β-propeller domains is also found in the clathrin triskelion31, thus lending further support to the hypothetical evolutionary relationship between vesicle coats and the heptameric complex.
β-propellers occur in many biological contexts, frequently acting as platforms for interactions with other proteins. The structural basis for this function is their rigid fold and the availability of several highly variable interaction surfaces.32 Remarkably, the surfaces of the four β-propellers in the heptamer are mostly exposed, and thus available for interactions with other proteins. As suggested previously6,17,18, the β-propellers may be involved in higher-order interactions between heptameric complexes within the NPC. Such an arrangement occurs in the COPII vesicle coat33. Moreover, a scaffold formed by heptameric complexes is likely to form a platform that organizes other nups within the NPC.
Whereas the β-propellers are rigid structural units, the connecting regions formed by α-solenoids and, possibly, by unstructured regions are likely to account for the conformational flexibility of the heptameric complex. Flexibility of α-solenoid arms was described for both COPII coatamers33 and clathrin triskelia34, where it is thought to allow the formation of vesicle coats in different sizes.
The flexibility of the heptameric complex is also potentially of physiological relevance. Flexibility of the entire NPC was described2,3,35 and may reflect conformational changes that accompany active transport. In particular, dilation of the NPC may be required to allow passage of large cargoes, such as ribosomal subunits. Molecular sliding of nups located near the central channel of the NPC was suggested to form the basis for NPC dilation4. It is likely that conformational changes of these central nups would occur in concert with conformational changes of the more peripheral nups, including the heptameric complex. A further requirement for flexibility may apply to the vertebrate homologue of the heptameric subcomplex, which has additional functions outside the NPC during mitosis1, and may adopt distinct conformations in different cellular contexts.
The heptameric complex was reported to play an essential role in the formation of NPCs, both post-mitotically and during interphase. Immunodepletion of the vertebrate homologue of the heptamer from nuclear assembly reactions leads to the formation of a continuous nuclear envelope devoid of pores10,11. Similarly, the heptamer is required for de novo insertion of NPCs into the interphase nuclear envelope12. While the mechanism of NPC assembly is currently unknown, a specific structural role for the heptameric complex in this process can be envisaged based on its structure and its affinity for highly curved membrane surfaces.
Formation of new NPCs during interphase requires the formation of a fusion pore between the outer and inner nuclear membranes. In other biological contexts, membrane fusion was shown to occur by a stepwise process: apposition of two membranes, hemifusion between the inner leaflets of the two lipid bilayers, reversible formation of a small fusion pore, stabilization and expansion of the fusion pore36. In different biological processes, such as exocytosis or viral membrane fusion, these steps are catalyzed by specific proteins that interact with the membranes to overcome the inherent energy barriers of each step along the fusion pathway, and to control the geometry of fusion.
The heptameric complex may function in one or several phases during the formation of nuclear envelope pores. The formation of the initial fusion pore is likely catalyzed by integral membrane proteins, possibly by the poms, which are components of the mature NPC. The heptamer may then stabilize initial fusion pores, by binding to the sharply bent membrane lining the pore. The following step of fusion pore expansion is particularly interesting in the case of nuclear envelope pores: whereas in other biological contexts, such as vesicle fusion, fusion pores expand maximally to integrate the vesicle membrane into the target membrane, the nuclear envelope fusion pore expands to a defined diameter of ∼100 nm to accommodate the NPC. A scaffold formed by several heptamers may control the final size of the fusion pore, and thus act as a molecular ruler. An octagon with a diameter of 100 nm has edges that are ∼38 nm long. Such an octagon could be formed by a previously suggested18 head-to-tail arrangement of eight heptameric complexes in a ring: The heptameric complex is ∼45 nm long (Fig. 4), allowing for some overlap at the head-to-tail interface.
Once the heptamer scaffold has stabilized the 100 nm pore, it can serve as a platform for the recruitment of other nups. The eightfold symmetry of the NPC may be dictated by the eightfold symmetry of the initial scaffold formed by heptameric complexes. Interestingly, the length of the heptameric complex was constant in the different conformations we observed (Fig. 2). This means that a ring of heptamers could maintain a fixed size based on head-to-tail interactions, while the flexibility of the heptamer would buffer conformational changes of other parts of the NPC that are anchored to the heptamer ring.
Methods
Yeast strains
We purified Nup84 complex from budding yeast in which the C terminus of Nup85 was genomically tagged with a tandem affinity purification (TAP) tag37. We derived C-terminally GFP-tagged strains from this strain using standard methods for genomic tagging38, including a seven-amino-acid spacer with the sequence (Gly)2-Ser-Gly-Ser-(Gly)2 between GFP and the tagged nup, and verified the strains by PCRs on genomic DNA, fluorescence microscopy and SDS-PAGE.
Nup84 complex purification
Yeast cells were grown in YPD medium at 30°C, harvested in mid-exponential phase (OD600 = 0.4–0.6), washed in water and frozen in liquid nitrogen. We ground frozen cell pellets cryogenically in a Retsch MM301 bead mill, following the method by Cristea and colleagues25. 4.5 l of cell culture yielded ∼3 g of cell powder. 1.5 g cell powder was thawed in 13.5 ml lysis buffer (500 mM NaCl, 110 mM KOAc, 2 mM MgCl2, 20 mM HEPES pH 7.5, 0.05% (w/v) CHAPS, 1 mM DTT, 1 mM PMSF, 5 μM pepstatin A and 1/100 volume of protease inhibitor cocktail, Sigma catalogue number P8340), homogenized in a Dounce homogenizer (40 strokes, tight pestle) and the lysate was clarified by centrifugation for 15 minutes at 3,200 g. We incubated clarified lysate for 1 hour at 4°C with 109 magnetic beads (Dynal) freshly coated with rabbit IgG (MP Biomedicals). A twofold dilution series of lysis buffer into elution buffer (200 mM NaCl, 110 mM KOAc, 2 mM MgCl2, 20 mM HEPES pH 7.5, 0.05% (w/v) CHAPS, 1 mM DTT) was prepared, and the beads were washed with five 1ml aliquots of sequentially diluted buffer. The beads were then resuspended in 500 μl of elution buffer, 100 units of AcTEV protease (Invitrogen) were added, and the sample was incubated for 1 hour at 4°C. The eluate was concentrated tenfold using Amicon Ultra filters (Millipore, 50 kDa molecular weight cutoff) and aggregates were removed from the concentrated sample by centrifugation for 15 minutes at 18,000 g. We subjected the soluble fraction to size-exclusion chromatography in elution buffer on a Superose-6 column (2.4 ml column volume). We analyzed eluted fractions by SDS-PAGE, stained with Coomassie, and each individual protein band was identified by mass spectrometry.
During optimization of the purification protocol, we estimated recovery and loss of heptameric complex at different steps by quantifying signals from Western blotting using an antibody against the calmodulin-binding peptide moiety of the TAP-tag (Open Biosystems). The optimized procedure recovered ∼20% of total cellular heptameric complex and the amount of beads was limiting for recovery.
Electron microscopy
For EM, we applied 3 μl of the 150 μl fraction containing heptameric complex (elution volume 1.1–1.25 ml, see Fig. 1a) directly to a glow-discharged carbon-coated copper grid and stained with three drops of 2% (w/v) uranyl formate. We collected electron micrographs of negatively stained samples with a defocus of −1.0 μm and doses not exceeding 10 e− per Å2 per exposure on a JEOL2100 field emission gun transmission electron microscope at 200 kV and recorded them with a CCD camera at a calibrated magnification of 40,641, corresponding to 5.91 Å per pixel. Micrograph pairs were taken at tilt angles of 50° and 0° in a semi-automated manner using SerialEM39. All images for a given construct were taken from a single grid over several days.
Image processing and analysis, 3D reconstruction
We applied low-pass filtering of the micrographs at (15 Å)−1, the spatial frequency corresponding to the first node of the contrast transfer function (CTF), instead of CTF-correction. Tilt pairs of particles were selected interactively in the program WEB40. Reference-free alignment and classification of images of untilted specimen was carried out using the program EMAN41. Angles between particle segments in 90 class averages were measured in ImageJ42. Principal component analysis was carried out in Matlab (The Mathworks). We implemented all remaining steps in SPIDER40, based on procedures described in the SPIDER documentation. The global set of aligned particles was subjected to correspondence analysis and hierarchical ascendant classification to group particles. For two well-defined groups, initial 3D reconstructions were obtained from images of tilted specimen using random conical tilt reconstruction23. These initial maps were used as references for projection matching. Reference projections were created in 5° increments for tilt angles from 40° to 55°, resulting in 205 projections per initial map. Each of the 9,028 images of tilted specimen was matched to the projection from either of the two references that resulted in the highest cross-correlation coefficient. We obtained final maps by the simultaneous iterative refinement technique (SIRT) to minimize missing-cone effects.
Mapping of GFP tags
To localize GFP densities for tagged complexes, we pooled images of 9,028 untagged particles either with 3,016 images of Seh1-GFP-tagged particles or with 2,114 images of Nup133-GFP-tagged particles, and subsequently aligned and classified them as before. We selected well-defined classes and calculated separate class averages for tagged and untagged particles from the same class. The class selected from the Seh1-GFP set contained 1,423 particles, 36% of which were GFP-tagged. The class selected from the Nup133-GFP set contained 1,143 particles, 31% of which were GFP-tagged. We calculated significant differences between the two class averages using the method by Wagenknecht and colleagues43; pixels with intensity values below the average value in both tagged and untagged class average images were not included in the significance map.
Map visualization and crystal structure fitting
We prepared all figures of crystal structures and EM maps using Chimera44. Structures with the PDB codes 3F3F (Nup85·Seh1), 1XKS (Nup133) and 3BG0 (Nup145C·Sec13) were docked independently as rigid bodies into particle maps 1 and 2 by exhaustive cross-correlation based search in Situs45,46. From the list of likely positions and orientations within the EM map generated by the algorithm, we accepted the highest-scoring fit that localized the crystal structure to the correct segment of the heptamer (as defined in Fig. 6a). PDB code 3CQG (Nup107·Nup133) was docked locally based on cross-correlation in Chimera, since docking in Situs did not place the crystal structure in the correct segment of the EM map.
Supplementary Material
Acknowledgments
We thank Ruben Diaz-Avalos and William Rice (New York Structural Biology Center, NYSBC) and other NYSBC staff for their excellent support and stimulating discussions. We thank David King (Howard Hughes Medical Institute Mass Spectrometry Facility) for mass spectrometry. We thank Erik Debler, Vivien Nagy, Claire Atkinson and Alexa Mattheyses for critical reading of the manuscript and Daniel Andor, Kuo-Chiang Hsia and other members of the Blobel lab for helpful discussions. M.K. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
References
- 1.Lim RY, Ullman KS, Fahrenkrog B. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol. 2008;267:299–342. doi: 10.1016/S1937-6448(08)00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–90. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 3.Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–5. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 4.Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–32. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–31. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debler EW, et al. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Glavy JS, et al. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107-160 subcomplex. Proc Natl Acad Sci USA. 2007;104:3811–6. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–60. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loïodice I, et al. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–44. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walther TC, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 11.Harel A, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–64. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 12.D'Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–3. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 13.Orjalo AV, et al. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–18. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuccolo M, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–64. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siniossoglou S, et al. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–97. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–73. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–26. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devos D, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci U S A. 1980;77:1496–500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rout MP, et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–46. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 23.Radermacher M, Wagenknecht T, Verschoor A, Frank J. A new 3-D reconstruction scheme applied to the 50S ribosomal subunit of E. coli. J Microsc. 1986;141:RP1–2. doi: 10.1111/j.1365-2818.1986.tb02693.x. [DOI] [PubMed] [Google Scholar]
- 24.Siniossoglou S, et al. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–75. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 25.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–41. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Lutzmann M, et al. Reconstitution of Nup157 and Nup145N into the Nup84 complex. J Biol Chem. 2005;280:18442–51. doi: 10.1074/jbc.M412787200. [DOI] [PubMed] [Google Scholar]
- 27.Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 28.van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–94. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert P. Iterative methods for the three-dimensional reconstruction of an object from projections. J Theor Biol. 1972;36:105–17. doi: 10.1016/0022-5193(72)90180-4. [DOI] [PubMed] [Google Scholar]
- 30.Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–7. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fotin A, et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–9. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 32.Paoli M. Protein folds propelled by diversity. Prog Biophys Mol Biol. 2001;76:103–30. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 33.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–36. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 34.Ferguson ML, et al. Clathrin triskelia show evidence of molecular flexibility. Biophys J. 2008;95:1945–55. doi: 10.1529/biophysj.107.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–93. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- 36.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–83. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 38.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 41.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 42.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 43.Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. Direct localization of the tRNA--anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J Mol Biol. 1988;203:753–60. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–84. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 46.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–95. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.