Abstract
Mustn1 encodes a small nuclear protein expressed specifically in the musculoskeletal system that was originally identified as a strongly up-regulated gene during bone regeneration, especially in fracture callus proliferating chondrocytes. Further experiments were undertaken to investigate its expression and role during chondrogenesis. Initially, whole mount mouse in situ hybridization was carried out and revealed Mustn1 expression in areas of active chondrogenesis that included limb buds, branchial arches and tail bud. To elucidate its function, experiments were carried out to perturb Mustn1 by overexpression and silencing in the pre-chondrocytic RCJ3.1C5.18 (RCJ) cell line. In these cells, Mustn1 is normally differentially regulated, with a spike in expression 2 days after induction of differentiation. Further, Mustn1 was successfully overexpressed in multiple RCJ cell lines by ~2–6 fold, and reduced to ~32–52% in silenced cell lines as compared to parental Mustn1 levels. Overexpressing, silenced, control, and parental RCJ cell lines were assayed for proliferation and differentiation. No statistically significant changes were observed in either proliferation or proteoglycan production when Mustn1 overexpressing lines were compared to parental and control. By contrast, both proliferation rate and differentiation were significantly reduced in Mustn1 silenced cell lines. Specifically, RNAi silenced cell lines showed reductions in populations of ~55–75%, and also ~34–40% less matrix (proteoglycan) production as compared to parental and random control lines. Further, this reduction in matrix production was accompanied by significant downregulation of chondrogenic marker genes, such as Sox9, Collagen type II (Col II), and Collagen type X (Col X). Lastly, reintroduction of Mustn1 into a silenced cell line rescued this phenotype, returning proliferation rate, matrix production, and chondrogenic marker gene expression back to parental levels. Taken together these data suggest that Mustn1 is a necessary regulator of chondrocyte function.
Keywords: Chondrogenesis, RNAi, Mustn1, RCJ, Sox9
Introduction
Previous work in our laboratory has identified Mustn1 as a small (9.6 kDa) protein that localizes predominantly to the musculoskeletal system [1]. Investigation of Mustn1’s sequence revealed that it is highly homologous in vertebrates and contains a nuclear localization sequence but no other significant motifs, only potential phosphorylation and myristoylation sites. GPF-Mustn1 fusion experiments showed that Mustn1 localizes to the nucleus but not to the nuclear membrane or nucleoli. Our laboratory has also showed that Mustn1 is differentially regulated during fracture repair [1, 2], specifically, in proliferating chondrocytes, osteoblasts and periosteal osteoprogenitor cells within the fracture callus [1]. In addition, we also found Mustn1 expression in the superficial layer of articular cartilage of a knee joint (unpublished observations), as well as during developing intervertebral discs and limbs [1]. Finally, while proliferating chondrocytes showed Mustn1 expression in these tissues, terminally differentiated hypertrophic chondrocytes did not. Further, during fracture repair, Mustn1’s peak expression correlates to a time point when mesenchymal precursor cells are migrating to the fracture site, differentiating into chondrocytes, and proliferating within the callus prior to becoming terminally hypertrophic. Based on these data, it was speculated that Mustn1 possibly functions in a transcriptional complex specific to chondrocyte differentiation [1].
In an effort to further characterize this gene, its promoter was identified and analyzed. Within the 1512bp region flanking Mustn1’s 5′ end, several transcription factor binding sites were identified that belong to the Activator Protein (AP) family [3]. It was also showed that AP-1 family members (JunD, c-Fos, and Fra-2), known as key transcriptional regulators of musculoskeletal specific genes [3], bind to Mustn1’s promoter during both proliferation and differentiation of myogenic C2C12 cells. Knockout studies of these genes showed varying degrees of phenotypic deficiency within the musculoskeletal system including osteoporosis, osteosarcoma, and chondrosarcoma as well as sterility, embryonic lethality, and ocular malformations [4]. However, because AP family members function within multiple signaling pathways, additional analyses are needed to determine within which one(s) Mustn1 functions.
Based on the aforementioned data, we reasoned that functional perturbation of Mustn1 during chondrocyte proliferation and differentiation would help to clarify its role during chondrogenesis. To this end, this study sought to modulate Mustn1 expression in the pre-chondrogenic RCJ cell line. The RCJ line represents a heterogeneous cell population capable of differentiation from proliferating chondrocytes to ultimately terminally differentiated collagen X producing hypertrophic chondrocytes [5]. Also, since Mustn1 is highly expressed in the early stages of fracture repair and this process is known to recapitulate embryonic development [2,6,7,8], we decided to further investigate Mustn1 expression during embryogenesis. This combination of in vivo and in vitro approaches revealed that Mustn1 is abundantly expressed in developing cartilaginous structures (limb buds, branchial arches and tail bud) and is necessary for both chondrocyte proliferation and differentiation.
Materials and Methods
Materials
The Anti-DIG Antibody, dexamethasone and β-glycerophosphate, bovine calf serum (BCS), Dulbecco’s modified Eagle medium (DMEM) and antibiotics (penicillin/streptomycin), and G418 were purchased from Roche (Indianapolis, IN), Sigma-Aldrich (St. Louis, MO), Gibco BRL (Gaithersburg, MD), and Invitrogen (Carlsbad, CA), respectively. Oligonucleotides primers were designed against the coding regions of β-2 Microglobulin, Mustn1, Col II, Col X, and Sox9 (Table 1).
Table 1.
List of primers and conditions used for qPCR.
| Target Gene | Accession Number | Primer Sequence | Amplicon Size | Tm(oC) |
|---|---|---|---|---|
| β-2microglobulin | NM 012512 | 5′ - TGGTGTGCTCATTGCTATTC 3′ - CTCTGAAGGAGCCCAAAAC |
152 | 58 |
| Mustn1 | NM 181390 | 5′ - GCTTTTCCTCTGCCACCTC 3′ - ATTCCCCGACCCACCTC |
129 | 58 |
| Collagen II | NM 012929 | 5′ - TGTGCTTCTTCTCCTTGCTC 3′ - GACCTGAAACTCTGCCACC |
187 | 58 |
| Collagen X | AJ131848 | 5′ - ACCTGGGGCAACTTAGAAAA 3′-CAGTGGAATAGAAGGCACACA |
179 | 58 |
| Sox9 | AB073720 | 5′ - CCGACACGGAGAACACAC 3′ – CAGTCATAGCCCTTCAGCAC |
98 | 58 |
Whole mount embryo in situ hybridization
All methods and animal procedures were reviewed and approved by the university’s Institutional Animal Care and Use Committee and met or exceeded all federal guidelines for the humane use of animals in research. Riboprobes were created by using T7 and Sp6 primers to amplify sense and antisense RNA probe sequences. These products were then purified and labeled using the DIG RNA labeling Kit (Roche). The original PCR product was degraded using RNase free DNase 1 (Qiagen). This reaction was stopped using 0.2M EDTA (pH 8.0). The final riboprobes were then purified using sephadex G-50 (Qiagen) quick spin columns. The in situ analysis was completed following the protocol as described by Hsieh et. al. [9]. Briefly, embryos were dissected out on 9.5, 10.5, and 11.5dpc and fixed with 4% paraformaldehyde for 1hr at room temperature (RT). Larger embryos (10.5 and 11.5dpc) were then permeabilized with proteinase K and all embryos were then bleached with H2O2 and stored in 100% methanol at −20°C. Embryos were then rehydrated through a methanol gradient and hybridized overnight at 65°C in a probe concentration of 250ng/ml., followed by a wash with MABT, blocked in MABT+20% Goat serum + 2% Boehringer Blocking Reagent (BM 1096 176) and incubated overnight in 1:2000 dilution anti-DIG 2° antibody. Finally, the embryos were again washed with MABT and expression was visualized with a solution of NTMT/BCIP. Photomicrographs were taken under bright field using a Zeiss Discovery. V8 SteREO microscope with an AxioCam MRc digital camera.
Cell culture
RCJ cells were maintained in DMEM supplemented with 10% BCS, 1% penicillin/streptomycin, and 10−7M dexamethasone. Cells were fed every 3–4 days and passaged prior to confluence.
RNA interference
Three 19-mer RNAi target sequences were designed (5′-AAGGAAGAAGACCTGAAGG -3′, 5′ – CCTGTGAAGGAAGAAGACC – 3′, and 5′ – TATTCAGCCGCAACCGCAC – 3′) which correspond to three different regions of the Mustn1 coding sequence (first and second sequence overlap). Oligonucleotides of hairpins containing these sequences (M2, M3, and M4 respectively) were generated (Invitrogen) and cloned into the pSuppressorRetro plasmid (Imgenex, San Diego, CA). A random control plasmid was generated by cloning a 19-mer sequence (5′ – GACTCCAGTGGTAATCTAC – 3′), which shows no similarity to any sequence within the mouse genome, into the same expression plasmid. A BLASTN (Basic Local Alignment Search Tool Nucleotide http;//www.ncbi.nlm.nih.gov/BLAST/) search was conducted to ensure that this random control sequence showed no genomic homology. Using a Retroviral GeneSuppressor System kit (Imgenex) these plasmids were co-transfected along with a packaging vector designed to create an ectotropic retrovirus protein coat into 293 cells. Virus containing media was harvested and used to infect proliferating RCJ cells. After a 24hr infection period, RCJ cells were selected using G418 (.5–.75mg/ml) for one week. Selected cells were plated at low density and colony forming units were allowed to grow up over several weeks. Homogeneous colonies were isolated and expanded. Five clones for each RNAi sequence were assayed for Mustn1 expression and three clones (M2-2, M3-5, and M4-4) showing the most effective reduction in Mustn1 levels were used in subsequent proliferation and differentiation experiments.
Overexpression
The Mustn1 coding sequence was cloned into the pCDNA vector downstream of the CMV promoter (Invitrogen). RCJ cells were transfected with this plasmid using Fugene 6 Transfection Reagent (Roche). As a control, an empty pCDNA plasmid was also transfected. After a 24hr transfection period, RCJ cells were selected using G418 (0.5–0.75mg/ml) for one week. Selected cells were plated at low density and colony forming units were allowed to grow up over several weeks. Homogeneous colonies were isolated and expanded. Eight clones were assayed for Mustn1 expression and the three clones (OE3, OE4, and OE6) showing the highest Mustn1 levels were used in subsequent proliferation and differentiation experiments.
Site-directed mutagenesis
Oligonucleotides were designed with alterations to the 3′ nucleotide in each codon within the M2 RNAi sequence (M2 RNAi sense sequence 5′ AAGGAAGAAGACCTGAAGG 3′, Rescue sense sequence 5′ AAAGAGGAGGATCTAAAAG 3′). These oligonucleotides were then used to amplify the original Mustn1 pCDNA plasmid using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). The original plasmid was then degraded using Dpn1 restriction digestion and the rescue plasmid was transformed into XL1-Blue Supercompetent Cells. Following Ampicillin selection, the rescue plasmid was isolated, verified and then used to transiently transfect the RCJ M2-2 cell line prior to proliferation and differentiation analyses.
Cell Proliferation
RCJ cells were seeded in 96-well plates (Falcon) at a density of ~2,000 cells/well. The regular RCJ media was removed, cells were washed with PBS, and 2.5%BCS RCJ media was added at the 0hr time point. Triplicate wells for each cell line were assayed during a 72hr time course (at 2hr, 24hr, 48hr, and 72hr) via MTS reagent (Promega). 5μl of MTS reagent was added to each well containing 100μl media. The plates were then incubated at 37°C for 1hr in the dark. The MTS/media solution was then diluted in 400ul dH2O and analyzed by spectrophotometer (BioRad) at 490nm. Values from control wells (MTS/media solution that did not contain cells) were subtracted from each experimental value. Values were further normalized to the 2hr time point to reflect population doublings. The triplicate values for each cell line at each time point were then averaged and standard deviation (sd) was determined.
Differentiation/Matrix production analysis
RCJ cells from each cell line were seeded in 24-well plates (Falcon) at a density of ~20,000cells/well (for the experiments with the overexpressing cell lines). Since Mustn1 silencing reduces the proliferative rate of RCJ cells, we conducted preliminary experiments with our silenced cell lines to ensure that all lines reach ~90% confluency (based on manual counts) concurrently. As such, we plated different concentrations, 20,000 – 30,000cells/well at the beginning of our experiments. For all cell lines, at near confluence (~90%) the regular RCJ media was removed and replaced with differentiation media (DMEM with 10% BCS, 1% penicillin/streptomycin, 10−7M dexamethasone, 50μg/ml ascorbic acid, and 10mM β-glycerophosphate) on Day 0 and was replaced every three days. Samples were taken throughout a 14 day time course (on Day 0, 2, 5, 7, 10, and 14). At each time point triplicate wells for each cell line were fixed for 4 min with 10% Buffered Formalin Phosphate (Fisher) and stored in PBS at 4°C. On Day 14, all plates were stained with a 1% Alcian blue solution for 45min at room temperature. Wells were washed with PBS and the Alcian blue was eluted in 4%Guanidine Hydrochloride in PBS for 30min at RT. Samples were then diluted 1:2–1:4 in dH2O and analyzed at 600nm via spectrophotometer (BioRad) and normalized to the Guanidine Hydrochloride/dH2O solution. The triplicate values for each cell line at each time point were then averaged and the sd determined. Values were then further normalized to the Day 0 time point to reflect fold change. Also, prior to Alcian blue elution, all wells were examined with a Zeiss Axiovert 200 microscope under bright field light and random fields from each well were captured with a CCD camera (Zeiss).
RNA isolation
RCJ cells were seeded in 24-well plates (Falcon) at a density of ~20,000cells/well. At near confluence the regular RCJ media was removed and replaced with differentiation media (DMEM with 10% BCS, 1% penicillin/streptomycin, 10−7M dexamethasone, 50μg/ml ascorbic acid, and 10mM β-glycerophosphate) on Day 0 and was replaced every three days. Samples were taken throughout a 14 day time course (on Day 0, 2, 5, 7, 10, and 14). At each time point triplicate wells for each cell line were lysed using TriZol reagent (Invitrogen), pooled, and stored at −80°C. Upon completion of the time course RNA was extracted using chloroform and 100% isopropanol using the recommended protocol (Invitrogen). The RNA was then resuspended and diluted in dH2O in preparation for quantitative real-time RT-PCR (qPCR).
Quantitative real-time RT-PCR (qPCR)
RNA isolated from Day 0, 2, 5, 7, 10, and 14 of the differentiation time course was treated with DNase 1 (Qiagen) and quantified using a ND-1000 Spectrophotometer (Nano Drop). The qPCR experiments were carried out using a QuantiTect SYBR Green RT-PCR Kit (Qiagen) on a LightCycler system (Roche) as previously described by our laboratory [8,10,11,12]. Primer pairs for Mustn1, Col II, Col X, and Sox9 were designed (Table 1) to anneal at 58°C. Each run consisted of mRNA (4–20ng/reaction) from Day 0, 2, 5, 7, 10, and 14 (for Mustn1) or Day 0, 5, 10, 14 (for Col II, Col X, and Sox9) and assayed together with the housekeeping gene β2-microglobulin, as well as 5 point calibration curves (1.25–20ng/reaction). The calculated concentration value for each gene was normalized to its corresponding β2-microglobulin value. All qPCR products were checked via agarose gel electrophoresis to assess amplification. Each run was replicated three times and results are reported as expression level ± sd.
Statistical analysis
All quantitative data are reported as the average results of three or more independent experiments with sd indicated by error bars. Significance was determined in all analyses by one way ANOVA followed by a Holm-Sidac Post-hoc when comparing to initial (Day 0 or 2hr) time points and Mann-Whitney tests when values from a single time point were compared.
Results
Mustn1 expression during development
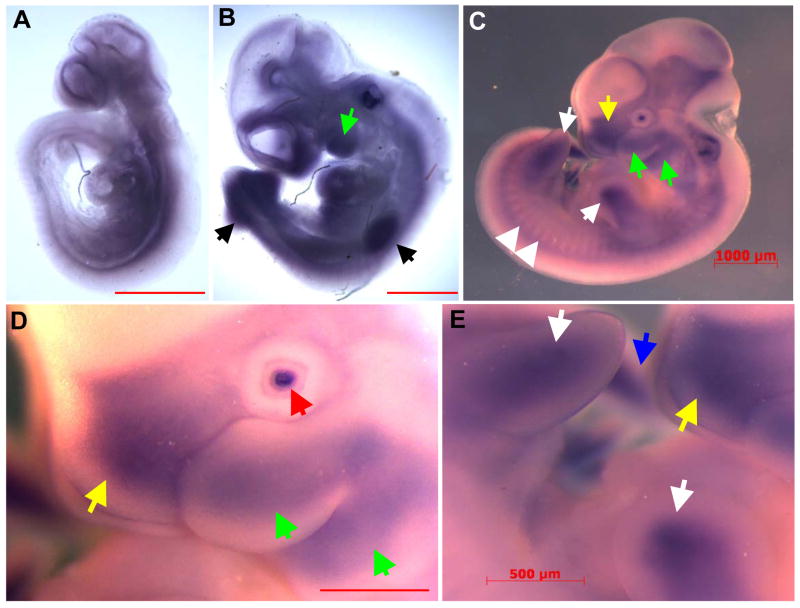
In order to further elucidate Mustn1 expression pattern during mouse development, we performed whole mount in situ hybridization. Mouse embryos incubated with the antisense Mustn1 riboprobes showed staining in several developing tissues at different time points. In a 9.5dpc embryo, staining was present throughout the embryo, possibly in musculoskeletal tissues such as the paraxial mesoderm on either side of the unstained neural tube as well as craniofacial structures (Fig. 1A). At 10.5dpc, hybridization was localized to several areas of cartilage and bone formation. For example, the craniofacial region showed intense staining, especially the branchial arches (Fig. 1B, green arrows), as do the fore and hind limb buds (Fig. 1B, black arrows), and somites in the typical array pattern perpendicular to the spinal column (Fig. 1B). As development proceeds to 11.5dpc, staining was again observed along the entirety of both the fore and hind limb buds (Fig. 1C and E, white arrows). Mustn1 expression was also observed in the craniofacial region, particularly the developing first branchial arch that is beginning to divide into the maxillary and mandibular components (Fig. 1C and D, green arrows). In addition, hybridization is also detected in the frontonasal process (Fig. 1C and D, yellow arrows), the lens of the eye (Fig. 1D, red arrow) and posterior tip of the tail (Fig. 1E, blue arrow). Mouse embryos incubated with a Mustn1 sense control riboprobe showed no staining at any time point (data not shown).
Figure 1. Mustn1 expression localizes to areas of chondrogenesis and myogenesis during mouse embryogenesis.
Whole mount embryos at 9.5dpc (A), 10.5dpc (B) and 11.5dpc (CDE) were hybridized with Mustn1 antisense riboprobes. 9.5dpc embryos stain broadly for Mustn1 throughout mesodermal tissues, while those at 10.5dpc and 11.5dpc embryos show distinct staining in fore and hind limbs (black and white arrows, respectively), branchial arches (green arrows), frontonasal process (yellow arrows), somites (white arrowheads), lens (red arrow) and posterior tail bud (blue arrow). Scale bars = 1000 μm (ABC) and 500μm (DE).
Mustn1 and chondrogenic marker gene expression in differentiating RCJ cells
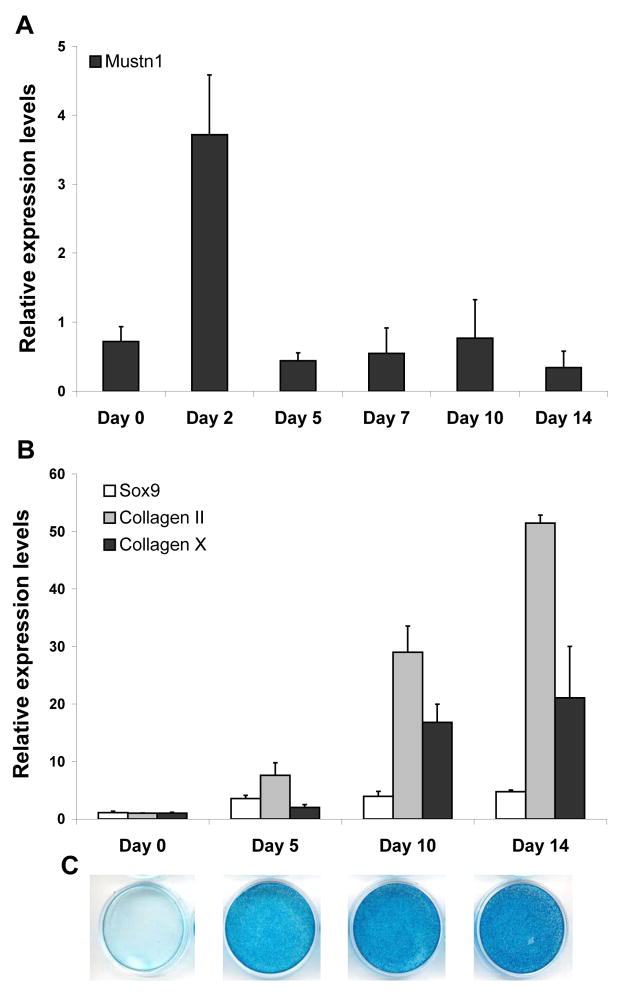
In order to get a baseline expression pattern for Mustn1 and three cartilage marker genes, Sox9, Col II, and Col X during RCJ differentiation, we assayed mRNA levels via qPCR. Mustn1 expression in RCJ cells showed a peak of ~5.2 fold two days after the induction of differentiation as compared to Day 0, followed by a decline to Day 0 levels for the remainder of the time course (Fig. 2A). In contrast, Sox9 showed a steady increase throughout the time course assayed and culminated in a peak of ~4.7 fold over its Day 0 expression value on Day 14 (Fig. 2B). Similarly and as expected, both Col II and Col X showed a similar expression pattern, steadily increasing to a large peak of ~51.4 and ~21.1 fold over Day 0 values on Day 14, respectively (Fig. 2B). Lastly, to compliment the molecular data and verify histologically that these cells do indeed undergo differentiation and produce extracellular matrix, we show the Alcian blue stained cultures from the same time points (Fig. 2C).
Figure 2. Mustn1 and chondrogenic marker gene expression is differentially regulated during RCJ cell differentiation.
Expression of Mustn1 (A), and Sox9, a pre-chondrocyte cell marker, Collagen II, a proliferating chondrocyte cell marker, and Collagen X, a hypertrophic chondrocyte cell marker (B) as assayed via qPCR during RCJ differentiation. Confluent RCJ cells were stimulated to differentiate at Day 0 and mRNA was isolated and assayed at the days indicated. All bars represent average values of raw gene expression normalized to β2-microglobulin in pooled mRNA (n=3). Error bars indicate standard deviation (st. dev.) between qPCR runs (n=3). C. Photomicrographs of tissue culture dishes showing respective Alcian stained cell lines to indicate degree of differentiation on Day 0, 5, 10, and 14.
Modulation of Mustn1 mRNA levels
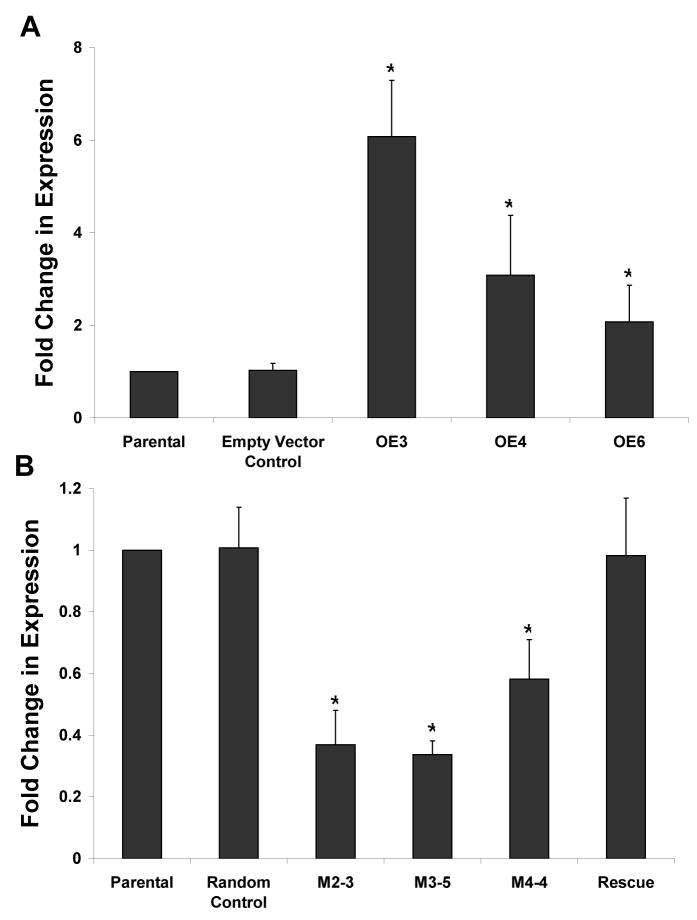
Next we sought to alter Mustn1 expression in RCJ cells via functional perturbation, overexpression and silencing. Overexpression of Mustn1 by stable transfection resulted in multiple homogeneous clones. Three clones were selected and showed significant increased Mustn1 expression al levels of ~6.1, 3.1, and 2.1 fold as compared to those of the parental cell line (OE3, OE4, and OE6, respectively, Fig. 3A). Transfection of the empty expression vector (control) into the parental RCJ cell line caused no significant alteration in Mustn1 expression levels as expected (Fig. 3A). Based on these data, OE3 and OE6 (highest and lowest overexpression levels) were chosen as representative cell lines for investigating the effects of Mustn1 overexpression on cell differentiation.
Figure 3. Modulation of Mustn1 expression via overexpression and silencing.
A.Mustn1 was overexpressed in RCJ cell lines, OE3, OE4 and OE6 at ~6, 3, and 2 fold level of that of the parental cells, respectively. B. Mustn1 was silenced in RCJ cell lines, M2-2, M3-5 and M4-4 at ~0.4, 0.3, and 0.6 fold level of that of the parental cells, respectively. A random sequence with no homology to the rat genome was used as a control (Random Control). The Rescue cell line was created by transiently transfecting the M2-2 cell line with a Mustn1 containing plasmid. Mustn1 expression was assayed by qPCR in proliferating RCJ cells and normalized to β2-microglobulin in pooled mRNA (n=3). Fold change was then determined by normalizing values to those of parental cells. Error bars indicate st. dev. (n=3). *p<.01 determined by Mann-Whitney test vs. Parental.
Silencing of Mustn1 by RNAi infection also resulted in multiple homogeneous cell clones. Three clones, M2-2, M3-5, and M4-4, one resulting from each of the three different RNAi sequences used, were selected and their Mustn1 expression was also analyzed via qPCR. Mustn1 expression was significantly reduced to ~0.4, 0.3, and 0.6 fold of the parental cell line expression levels in M2-2, M3-5, and M4-4, respectively (Fig. 3B). The introduction of the random control sequence showed no significant change in Mustn1 expression (Fig. 3B).
Based on these data, M2-2 was chosen as a representative silenced cell line for investigating the effects of Mustn1 silencing on differentiation, and as such we also targeted it for the rescue experiments. To verify that the rescue worked in terms of restoring Mustn1 levels back to those of parental cells we measured mRNA levels and results from this experiment showed that our strategy work and the rescue sequence did indeed restore Mustn1 levels back to parental levels (Fig. 3B).
Functional perturbation of Mustn1
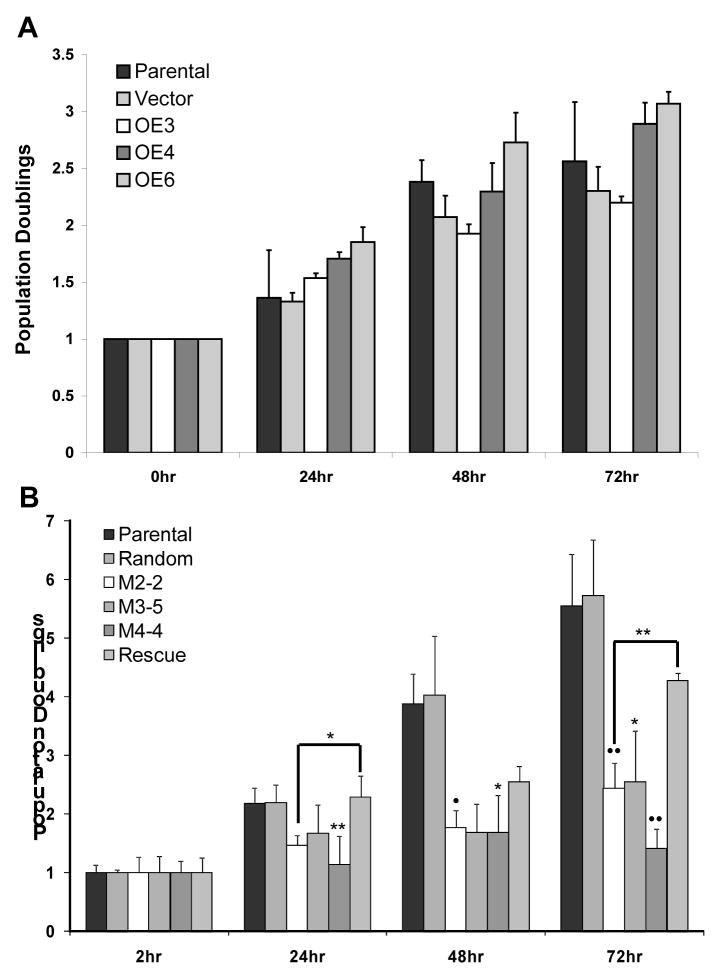
To identify any functional effects caused by overexpressing or silencing Mustn1 on chondrocyte proliferation, we assayed RCJ population growth in low density cell cultures. When the proliferation rate of the overexpressing cell lines, OE3, OE4, and OE6 were compared to both the parental cell line and the empty vector control over 72 hrs, there were no statistically significant differences found at any time point (Fig. 4A). In contrast, when the proliferation rate of the silenced cell lines, M2-2, M3-5, and M4-4 were compared to both the parental and the random control cell lines, significant differences were observed at several time points (Fig. 4B). Specifically, at the 24hr time point, random control, M2-2, M3-5, and rescue cell lines with averages of ~2.2, 1.5, 1.7, and 2.3 population doublings, respectively, showed no significant differences from parental (avg. ~2.2). However, the M4-4 cell line showed significant reduction in population level (~1.1) when compared to parental (p<.01). At 48hrs, the random control, M3-5, and rescue cell lines with population averages of ~4.0, 1.7, and 2.5, respectively, showed no significant differences from parental (~3.9). However, the M2-2 and M4-4 cell lines with averages of ~1.8 and 1.7, respectively, showed significant reduction in population levels when compared to parental (p<.05, Fig. 4B). Interestingly, at 72hrs, only the random control and rescue cell lines with averages of ~5.7, and 4.3, respectively, showed no significant differences from parental (~5.6). However, all three silenced lines, M2-2, M3-5, and M4-4 with averages of ~2.4, 2.5, 1.4, respectively, showed significant reduction in populations when compared to parental (p<.001 for M2-2 and M4-4 and p<0.05 for M3-5, Fig 4B). Furthermore, there was a significant difference in population size between the M2-2 and rescue cell lines at 24hr (p<.05) and 72hr (p<.01, Fig. 4B).
Figure 4. Cell proliferation is unchanged when Mustn1 is overexpressed but is reduced in silenced cells.
A.Cell proliferation measurements in Parental, Vector (control), OE3, OE4 and OE6 cell lines via MTS over a 3 day time course. There was no statistical significance between any cell line at any time point. B. Similar analysis of Parental, Random (control), silenced cell lines, M2-2, M3-5, M4-4 and Rescue. There was no statistical difference between Vector, Random, or Rescue and Parental cell lines at any time point. Each data point represents the average of triplicate MTS experiments (n=3). The value of each cell line was normalized to the 0hr time point to show population doublings. Error bars represent st. dev. All statistical significance was determined by ANOVA on ranks with Tukey post-hoc. Significant difference was only observed in between Parental and silenced cell lines, as well as M2-2 and Rescue. *p<.05, **p<.01, •p<.006, ••p<.001.
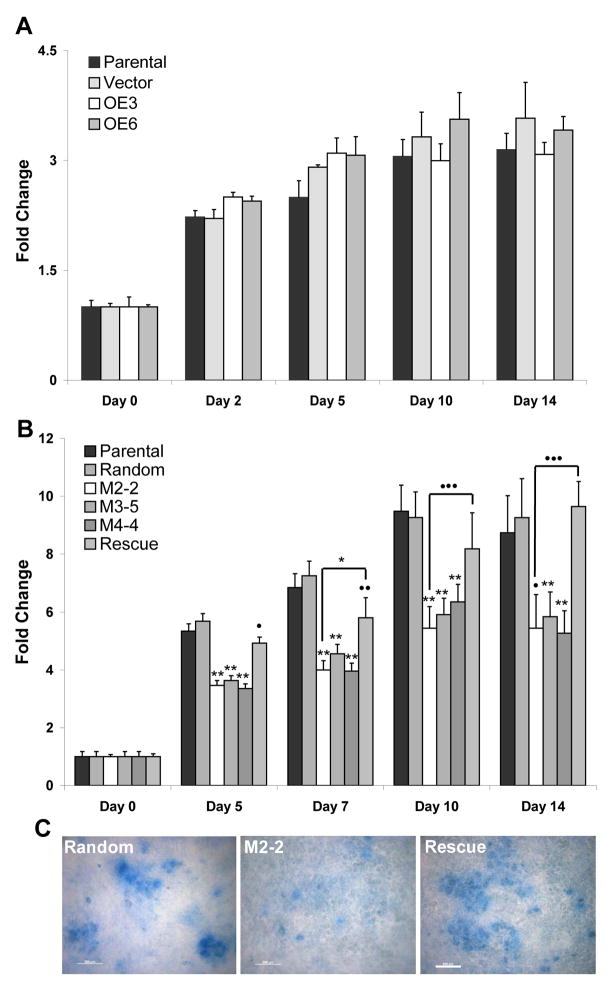
To determine if altering Mustn1 expression affected chondrogenic differentiation, we monitored proteoglycan production (as a function of cartilage matrix) via Alcian blue staining. Using this assay, the two overexpressing cell lines, OE3 and OE6, showing the highest and lowest up-regulation of Mustn1, respectively, demonstrated no significant difference at any time point as compared to both the parental cell line and empty vector control (Fig. 5A). In contrast, when Alcian blue binding was quantified in the silenced cell lines, M2-2, M3-5, and M4-4, and compared to both the parental and the random control cell lines significant differences were found at all time points (Fig. 5B). Specifically, on Day 5, the random control cell line with a value of ~5.7 showed no significant difference from the parental cell line (~5.3). However, the M2-2, M3-5, M4-4 and rescue cell lines, with values of ~3.5, 3.6, 3.7, and 4.9 fold, respectively, over Day 0 levels, showed significant differences in matrix production (p<.001 for M2-2, M3-5, and M4-4 and p<.03 for the rescue cell line when compared to parental). On Day 7, the random control cell line (~7.3 fold change) showed no significant difference from the parental cell line (~6.8). However, the M2-2, M3-5, M4-4, and rescue cell lines with averages of 4.0, 4.6, 4.0, and 5.8 fold, respectively, showed significant differences in matrix production (p<.001 for M2-2, M3-5, and M4-4 and p<.02 for the rescue line when compared to parental). On Day 10, the random control and rescue cell lines with averages of ~9.3 and 8.2 fold change, showed no significant difference from the parental cell line (~9.5). However, the M2-2, M3-5, and M4-4 cell lines with averages of 5.4, 5.9, and 6.4 fold change, respectively, showed significant reduction in matrix production (p<.001 for all three cell lines when compared to parental). Finally, on Day 14, the random control and rescue cell lines with averages of ~9.3 and 9.7 fold change, showed no significant difference from the parental cell line (~8.7). And again, the M2-2, M3-5, and M4-4 cell lines with averages of ~5.4, 5.8, and 5.3 fold change, respectively, showed significant reduction in Alcian blue staining (p<.001 for M3-5 and M4-4 and p<.03 for M2-2 when compared to parental. A significant difference was also found when M2-2 was compared to the rescue cell line on Day 7 (p<.05) as well as Days 10 and 14 (p<.01, Fig 5B). Lastly, Figure 5C shows representative Alcian blue stained cultures at Day 14 to visualize these decreases in matrix production, particularly in the lack of nodule formation and size (degree of differentiation) in the representative silenced M2-2 cell line as compared to Random control and Rescue.
Figure 5. Matrix production is unaffected when Mustn1 is overexpressed, but is reduced in silenced cells.
Confluent cells from each line were stimulated to differentiate on Day 0. At specific times during differentiation all cell lines were fixed and stained with Alcian blue. The dye was then eluted and measured via spectrophotometry to quantitatively determine the amount of matrix produced at each time point. A. Parental, OE 3, OE6 and empty vector cell lines were assayed at Day 0, 2, 5, 10, and 14. B. Parental, Random, M2-2, M3-5, M4-4, and Rescue cell lines on Day 0, 5, 7, 10 and 14. C. Photomicrographs showing a random region from the respective Alcian stained cell line to indicate nodule formation and size (degree of differentiation). All bars indicate average of triplicate experiments ± st. dev. Significance was determined by ANOVA on ranks with a Tukey Post-hoc between the silenced cell lines vs. Parental levels or Mann-Whitney to compare M2-2 to Rescue. *p<.05, **p<.001, •p<.03, ••p<.02, •••p<.01.
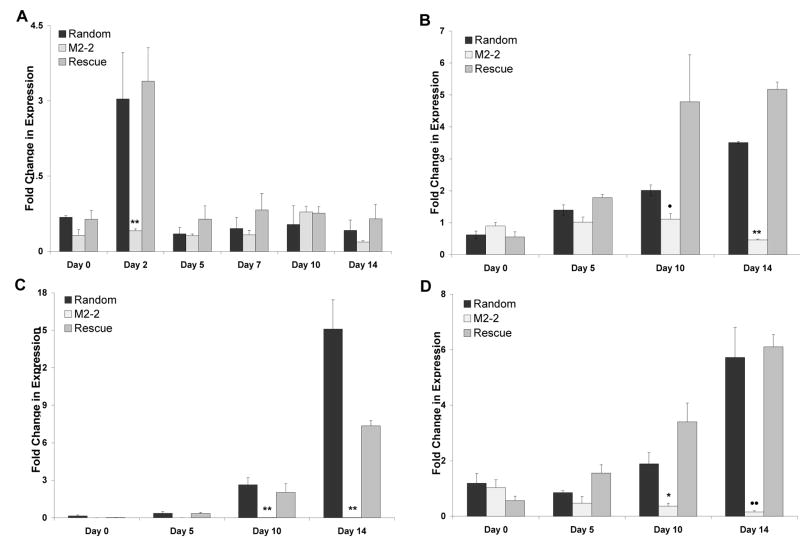
Since we observed inhibitory effects on the differentiation of the cells with reduced Mustn1 levels and since we speculate that Mustn1 functions as a part of a transcriptional complex, we wanted to investigate if this reduction in matrix production is accompanied by molecular changes, especially in chondrogenic gene marker expression (Col II, Col X and SOX9). Thus, Mustn1 and marker gene mRNA levels were determined via qPCR and compared to Random control. When Mustn1 mRNA levels were analyzed in the silenced cell line, M2-2, they were reduced to ~47% as compared to random control at Day 0 (Fig. 6A). In contrast, the rescue cells showed 89% expression when compared to random levels at the same time point (Fig. 6A). No significant differences were found between the Random and Rescue cell lines at any time point and each peaked on Day 2 with increases of ~4.5 and 5.3 fold over Day 0, respectively (consistent with the data shown in Fig. 2A). However, in the silenced cell line, Mustn1 mRNA expression remained suppressed. This was the case at all time points except at Day 10, where M2-2 Mustn1 levels reached those of Random and Rescued cell lines. While the only significant difference between the Random control and M2-2 cell lines was on Day 2, it was indeed very dramatic (p<.001, Fig. 6A).
Figure 6. Mustn1 and chondrogenic marker gene expression is reduced in Mustn1 silenced cells.
Confluent RCJ cells were stimulated to differentiate at Day 0 and mRNA was isolated and assayed via qPCR (normalized to β2-Microglobulin) at Day 0, 2, 5, 7, 10 and 14 for Mustn1 (A) and Day 0, 5, 10 and 14 for Sox9 (B), Collagen type II (C), and Collagen type X (D). All bars represent average raw gene expression values of pooled mRNA (n=3). Error bars indicate st. dev. between qPCR runs (n=3). Significance was determined by Mann-Whitney test vs. Random Expression levels. *p<.05, **p<.001, •p<.01, ••p=.002.
When Sox9 mRNA levels were measured, the Random and Rescue cell lines showed steady increases culminating in peaks on Day 14 with ~5.6 and 9.4 fold increases over Day 0, respectively. The M2-2 cell line did not show this increase in expression and instead Sox9 levels remained suppressed at Days 5 (~1.0), 10 (~1.1) and especially 14 (~0.5, Fig. 6B). The Sox9 values for M2-2 were significantly different on Day 10 (p=0.012) and Day 14 (p<.001) as compared to Random values on those days. The results with Col II were even more dramatic. While both Random and Rescue cell lines showed steady increases in expression culminating in peaks on Day 14 of ~15 and ~7 over Day 0 values respectively, the Col II levels in the M2-2 cell line were completely suppressed (Fig. 6C). The Col II values for M2-2 were significantly different on Day 10 (p<.001) and Day 14 (p<.001) as compared to Random values on those days. Finally, Col X mRNA levels also showed steady increases in the Random and Rescue cell lines culminating in peaks on Day 14 of ~5.7 and ~6.1, respectively, while in the M2-2 cell line they remained severely reduced (Figure 6D). In contrast, Col X expression for M2-2 was remained significantly suppressed on both Day 10 (p<.05) and Day 14 (p=.002) as compared to Random values on those days. There were no significant differences between random and rescue values at any time point in any analysis.
Discussion
The expression pattern of Mustn1 during embryogenesis suggests that this gene is active in areas of early cartilage, bone and muscle formation, especially in limb buds, branchial arches, vertebrae, and somites, all tissues containing differentiating mesenchymal cells, as observed in this and our previous study [1]. Although this correlation does not necessarily reveal its function, it does suggest that Mustn1 is involved in these processes.
Mustn1’s involvement in the early phases of chondrogenesis is further supported by in vitro analyses on cell proliferation and differentiation presented herein. The fact that overexpressing Mustn1 did not cause any significant alteration in either proliferation or matrix (proteoglycan) production can be explained by the notion that the modest increases in Mustn1 expression in RCJ cells (~2 – 6 fold) that were achieved in this study were not enough to alter these processes (when compared to the parental or control cell lines). Unfortunately, we were unable to achieve higher levels of experimental overexpression and thus the possibility remains that higher levels of Mustn1 may be required for inducing changes in cell behavior, although this is highly unlikely given our hypothesis that Mustn1 functions as a co-activator or co-regulator of transcription and as such it is necessary but not sufficient for transcriptional activation. This suggests that increasing the amount of Mustn1 within each cell would not cause any effect(s) as the other molecules in the transcriptional complex would also be required. Lastly, in vivo overexpression on Mustn1 in a model of Xenopus laevis development has no effect (unpublished observations). In contrast, the silencing experiments indicate that Mustn1 is necessary for both proliferation and differentiation. In the silenced cell lines where Mustn1 expression was downregulated to ~33 – 58% expression compared to parental cells, both proliferation and matrix production were clearly suppressed. This decrease was also accompanied by a dramatic reduction in the expression of the three chondrogenic marker genes, Sox9, Col II and Col X, assayed.
While the proliferation and matrix production analyses do not completely elucidate the role of Mustn1, the suppression of Sox9, Col II and Col X mRNA expression does provide a greater understanding of Mustn1’s function during chondrocyte differentiation. Further, the observation that matrix production is reduced in Mustn1 silenced cell lines, could simply be a result of reduced proliferation rate. However, matrix production was tested on visually normalized confluent cells, ensuring consistent cell number across all cell lines tested at initiation of differentiation. Further all cell lines were fully confluent by Day 2 of the time course suggesting that the cells had ceased proliferation and begun the differentiation process concurrently. In addition, the suppression of Col X, a well known marker of hypertrophy [13,14,15] observed in the silenced cells suggests that the lack of matrix production is caused by the inability of these cells to differentiate into terminal hypertrophic chondrocytes and not reduced proliferation rate. Moreover, the parallel and complete suppression seen in Col II, as well as the dramatic reduction in Sox9 expression indicate that even the earlier stages of differentiation [14,16,17] are affected in these silenced cells as a result of Mustn1 downregulation.
Despite these results, low level Alcian blue staining is still present in the Mustn1 silenced cells indicating the presence of glycosaminoglycans (GAGs) within cartilage [18]. One reason for this maybe that even pre-chondrocytes produce minimal levels of proteoglycans, but more probable is the fact that we do not completely eliminate Mustn1 expression in the silenced cells. The residual lower levels of Mustn1 (~30–50%) as detected by qPCR in these cells supports this idea. Even though GAGs are still being produced, the main component of the cartilage extracellular matrix is collagen. Collagen makes up two-thirds of cartilage dry weight with over 75% of that comprised of collagen II [19,20]. The dramatic suppression of Col II mRNA suggests that the matrix produced by the silenced cell lines is probably very different and to some extent structurally deficient when compared to that produced by the parental or random cell lines. although this remains to be experimentally verified.
The most interesting result of these molecular analyses is the suppression of Sox9 observed in the silenced cell lines, which is subsequently rescued after restoring Mustn1 levels into the cells (rescue experiment). These data suggest that Mustn1 functions at an earlier phase during chondrogenic differentiation, as Sox9 is responsible for the transcriptional activation of Col II in multiple chondrogenic and osteogenic tissues [21,22,23]. Thus, it is this reduction in Sox9 expression that likely causes the subsequent suppression of Col II levels. Sox9 activation itself has been shown to be both required and sufficient for chondrogenesis alone or with Sox5 and Sox 6 [22,24,25]. The vital role of Sox9 during chondrocyte differentiation makes Mustn1’s affect on this gene’s expression an interesting avenue to explore. Although these data implicate Mustn1 in the activation of Sox9, they do not indicate which signaling pathway in involved with Mustn1 function as several pathways affect Sox9 expression. Lastly, in vivo downregulation of Mustn1 in a model of Xenopus laevis development using an antisense morpholino oligonucleotide approach resulted in gross morphological defects that included small or lack of eyes, shortened body axis, and tail/body kinks and this was accompanied by an ~40% downregulation of Sox9 mRNA in the developing tadpoles (unpublished observations).
When these data are taken in conjunction with what is previously known about Mustn1, i.e. its expression restricted to the musculoskeletal system, its promoter, and the fact that it is a nuclear protein [1,2], a clearer picture of Mustn1’s function begins to emerge. These data suggest that Mustn1 works within the nucleus, possibly as part of a transcription initiation complex. The lack of a DNA binding motif within its sequence precludes Mustn1 from being a transcription factor per se and thus supports the notion that Mustn1 possibly functions as a musculoskeletal co-activator or co-regulator, although more conclusive experiments have yet to validate this hypothesis. Further, results from the experiments reported herein support the notion that Mustn1 is a co-activator of a transcriptional complex involved in both the proliferation and differentiation of chondrocytes as both processes are negatively affected when Mustn1 is silenced in vitro. However, that Mustn1 helps to inhibit a repressor involved in the transcriptional regulation of these processes cannot be ruled out. Further, if Mustn1 is indeed a transcriptional co-activator, the identification of its target genes holds much promise in the elucidation of chondrogenesis. Because Mustn1 is active very early in chondrocyte differentiation, cartilage and bone development, as well as bone regeneration, finding the direct targets of this gene will aid in identifying the early initiators of each of these vital processes. While Mustn1 itself is not sufficient for chondrogenic differentiation, it is possible that its target gene(s) may be. And although these results are promising and further provide evidence of Mustn1’s critical regulatory role in chondrogenic differentiation, additional research is required to conclusively elucidate Mustn1’s role in the overall process of chondrogenesis.
Acknowledgments
The authors would like to thank Dr. Bernadette Holdener, Christina Leonard, and Janet Chang for technical help and guidance with the whole mount in situs. Additionally, we would like to thank Cheng Liu for help with tissue culture. Finally, we would like to thank Drs. Jane Aubin and William Horton for providing the RCJ cells. This project was funded by a grant from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lombardo F, Komatsu D, Hadjiargyrou M. Molecular cloning and characterization of Mustang, a novel nuclear protein expressed during skeletal development and regeneration. FASEB J. 2004;18:52–61. doi: 10.1096/fj.03-0521com. [DOI] [PubMed] [Google Scholar]
- 2.Hadjiargyrou M, Lombardo F, Zhao S, Ahrens W, Joo J, Ahn H, et al. Transcriptional profiling of bone regeneration. Insight into the molecular complexity of wound repair. J Biol Chem. 2002;277:30177–82. doi: 10.1074/jbc.M203171200. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Hadjiargyrou H. Identification and characterization of the Mustang promoter: Regulation by AP-1 during myogenic differentiation. Bone. 2006;39:815–24. doi: 10.1016/j.bone.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis. 2002;61 (Suppl 2):40–42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellows CG, Sodek J, Yao KL, Aubin JE. Phenotypic differences in subclones and long-term cultures of clonally derived rat bone cell lines. J Cell Biochem. 1986;31:153–69. doi: 10.1002/jcb.240310207. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson C, Alpern E, Miclau T, Helms JA. Does adult fracture repair recapitulate embryonic skeletal formation? Mech Dev. 1999;87:57–66. doi: 10.1016/s0925-4773(99)00142-2. [DOI] [PubMed] [Google Scholar]
- 7.Vortkamp A, Pathi S, Peretti GM, et al. Recapitulation of signals regulating embryonic bone formation during postnatal growth and in fracture repair. Mech Dev. 1998;71(1–2):65–76. doi: 10.1016/s0925-4773(97)00203-7. [DOI] [PubMed] [Google Scholar]
- 8.Gersch RP, Lombardo F, McGovern SC, Hadjiargyrou M. Reactivation of Hox gene expression during bone regeneration. J Orthop Res. 2005;23:882–90. doi: 10.1016/j.orthres.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh J-C, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines M, Rosenquist T, Holdener BC. Mesd Encodes an LRP5/6 Chaperone Essential for Specification of Mouse Embryonic Polarity. Cell. 2003;112:355–67. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu DE, Hadjiargyrou M. Activation of the transcription factor HIF-1 and its target genes, VEGF, HO-1, iNOS, during fracture repair. Bone. 2004;34:680–88. doi: 10.1016/j.bone.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Zhong N, Gersch PR, Hadjiargyrou M. Wnt signaling activation during bone regeneration and the role of Dishevelled in chondrocyte proliferation and differentiation. Bone. 2006;39:5–16. doi: 10.1016/j.bone.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu D, Bosch-Marce M, Semenza GL, Hadjiargyrou M. Enhanced Bone Regeneration Associated with Decreased Apoptosis in Mice with Partial HIF-1α Deficiency. J Bone Miner Res. 2007;22:366–74. doi: 10.1359/JBMR.061207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington EK, Lunsford LE, Svoboda KKH. Chondrocyte Terminal Differentiation, Apoptosis, and Type X Collagen Expression Are Downregulated by Parathyroid Hormone. Anat Rec A Discov Mol Cell Evol Biol. 2004;281:1286–95. doi: 10.1002/ar.a.20129. [DOI] [PubMed] [Google Scholar]
- 14.Shen G, Rabie AB, Zhao Z, Kaluarachchi K. Forward deviation of the mandibular condyle enhances endochondral ossification of condylar cartilage indicated by increased expression of type X collagen. Arch Oral Biol. 2006;51:315–24. doi: 10.1016/j.archoralbio.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Tchetina EV, Kobayashi M, Yasuda T, Meijers T, Pidoux I, Poole AR. Chondrocyte hypertrophy can be induced by a cryptic sequence of type II collagen and is accompanied by the induction of MMP-13 and collagenase activity: Implications for development and arthritis. Matrix Biol. 2007;26:247–58. doi: 10.1016/j.matbio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Sen M, Cheng Y, Goldring MB, Lotz MK, Carson DA. WISP3-Dependent Regulation of Type II Collagen and Aggrecan Production in Chondrocytes. Arthritis Rheum. 2004;50:488–97. doi: 10.1002/art.20005. [DOI] [PubMed] [Google Scholar]
- 17.Uchihashi T, Kimata M, Tachikawa K, Koshimizu T, Okada T, Ihara-Watanabe M, Sakai N, Kogo M, Ozono K, Michigami T. Involvement of nuclear factor I transcription/replication factor in the early stage of chondrocytic differentiation. Bone. 2007;41:1025–35. doi: 10.1016/j.bone.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Scott J, Dorling J. Differential staining of acid glycosaminoglycans (mucopolysaccharides) by alcian blue in salt solutions. Histochemie. 1965;1(5):221–33. doi: 10.1007/BF00306130. [DOI] [PubMed] [Google Scholar]
- 19.Eyre D. Collagens and Cartilage Matrix Homeostasis. Clin Orthop Relat Res. 2004;427(Suppl):S118–22. doi: 10.1097/01.blo.0000144855.48640.b9. [DOI] [PubMed] [Google Scholar]
- 20.Johns D, Athanasiou K. Design characteristics for temporomandibular joint disc tissue engineering: learning from tendon and articular cartilage. Proc Inst Mech Eng [H] 2007;221:509–26. doi: 10.1243/09544119JEIM158. [DOI] [PubMed] [Google Scholar]
- 21.Akiyamaa H, Kamitanic T, Yanga X, Kandyila R, Bridgewaterd LC, Fellouse M, Mori-Akiyamaa Y, de Crombrugghe B. The transcription factor Sox9 is degraded by the ubiquitin–proteasome system and stabilized by a mutation in a ubiquitin-target site. Matrix Biol. 2005;23:499–505. doi: 10.1016/j.matbio.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–9. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 23.Rabie ABM, She TT, Harley VR. Forward Mandibular Positioning Up-regulates SOX9 and Type II Collagen Expression in the Glenoid Fossa. J Dent Res. 2003;82:725–30. doi: 10.1177/154405910308200913. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung U. The Combination of SOX5, SOX6, and SOX9 (the SOX Trio) Provides Signals Sufficient for Induction of Permanent Cartilage. Arthritis Rheum. 2004;50:3561–73. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 25.Kim J-H, Do H-J, Yang H-M, Oh J-H, Choi S-J, Kim D-K, Kwang-Yul Cha K-Y, Chung H-M. Overexpression of SOX9 in mouse embryonic stem cells directs the immediate chondrogenic commitment. Exp Mol Med. 2005;37:261–68. doi: 10.1038/emm.2005.35. [DOI] [PubMed] [Google Scholar]