Abstract
Population stratification (PS) can lead to an inflated rate of false positive findings in genome-wide association studies (GWAS). A commonly used approach is to adjust for a fixed number of principal components (PCs) in GWAS but this approach could have a deleterious impact on power when the cases and controls are equally distributed along selected PCs, or if the adjustment of certain covariates, such as self-identified ethnicity or recruitment center, already included in the association analyses, correctly map to major axes of genetic heterogeneity. We propose a computationally efficient procedure, PC-Finder, to identify a minimal set of PCs while permitting an effective correction for PS. A general pseudo F statistic, derived from a non-parametric multivariate regression model, can be used to assess whether PS exists or has been adequately corrected by a set of selected PCs. Empirical data from two GWAS conducted as part of the Cancer Genetic Markers of Susceptibility (CGEMS) project demonstrate the application of the procedure. Furthermore, simulation studies show the power advantage of the proposed procedure in GWAS over currently used PS correction strategies, particularly when the PCs with substantial genetic variation are distributed similarly in cases and controls and therefore do not induce PS.
INTRODUCTION
Genome-wide association studies (GWAS) have emerged as an effective approach to identify common polymorphisms underlying complex traits [Hunter et al., 2007; The Wellcome Trust Case Control Consortium, 2007; Yeager et al., 2007; Manolio et al., 2008; Pearson and Manolio, 2008]. GWAS frequently employ a case-control design because of the efficiency in investigating a large number of common variants in unrelated cases and controls with sufficient statistical power to detect small-to-moderate effects. However, multi-stage population stratification (PS) can lead to a high fraction of putative associations that are spurious, particularly when many SNPs are tested in follow-up but in actuality, only a few are closely related to disease and alpha level is low. This is particularly problematic in multi-stage GWAS, which have been based on selecting a subset of SNPs from a GWAS underpowered to reliably detect low penetrance alleles (estimated odds ratios of < 1.3) so that only a small percentage of notable variants are carried through subsequent stages [Skol et al., 2006; Yu et al., 2007].
Because the vast majority of single nucleotide polymorphisms (SNPs) genotyped in a GWAS are not associated with the disease under study, it is feasible to use SNPs measured throughout the genome for the detection and correction of PS. Principal component analysis (PCA) [Zhu et al., 2002; Patterson et al., 2006; Price et al., 2006; Li and Yu, 2008] uses SNPs measured throughout the genome to uncover hidden population substructure by detecting axes of large genetic variation and, if necessary, to adjust for ancestral background differences between cases and controls along several major axes. Although adjust for PS in a GWAS based on PCA is becoming routine, little evidence on adequacy of adjustment of a subset of principal components (PCs) for correction s for PS or how to best select relevant principal components is available. One commonly used PCA approach adjusts simultaneously for a fixed number of top-ranked PCs according to the size of eigenvalues [Price et al., 2006]; another approach selects PCs with significant large genetic variation according to the Tray-Wisdom test [Patterson et al., 2006]. However, both approaches may include some unnecessary PCs and could have a deleterious impact on the power if adjustment for one or more of the PC is unnecessary because the PC is equally distributed among cases and controls or because the adjustment of certain covariates (such as self-identified ethnicity, or recruitment center), which correctly map to major axes of genetic heterogeneity, have already been included in the association analyses. Previously, we [Yu et al., 2008] presented an example to demonstrate that the unnecessary adjustment of population substructure by even one PC could lead to a significant loss in power, and proposed a permutation procedure to identify the minimal number of PCs while allowing an effective correction of the confounding effect. To apply this procedure, two sets of SNPs are required, one for PCA, the other for the evaluation of type I error inflation in the permutation steps. Selection of relevant PCs using this procedure can be computationally intensive if it is necessary to calculate the association test statistic on a large number of markers in order to have an accurate estimation of the inflation level in type I error. Here, using techniques derived from the distance-based regression model, we propose a computationally efficient procedure to evaluate and correct, when necessary, for PS.
The distance-based regression model was originally proposed by McArdle and Anderson [2001] for the analysis of ecological data, and can be thought as a non-parametric version of the traditional multivariate regression model. The multivariate regression model is commonly used for the study of the relationship between a set of predictors X and a multivariate outcome Y. The pseudo F statistic [McArdle and Anderson, 2001] can be applied to test the null hypothesis of no effect of X on Y. Recognizing that the pseudo F statistic can be calculated in term of the Euclidean distance between the outcomes of two subjects, McArdle and Anderson [2001] proposed the distance-based regression model for the analysis of pair-wise distance (or similarity) measured among a group of subjects by a chosen distance (similarity) metric, and suggested using the similar pseudo F statistic to assess the effect of predictors X on the pair-wise distance (or similarity). Recently, this method has been used for genetic analyses, such as the comparison of microarray gene expression patterns [Zapala and Schork, 2006], a multilocus test for genetic association studies [Wessel and Schork, 2006], and assessment of genetic background diversity [Nievergelt et al., 2007].
As suggested by Nievergelt et al. [2007], the pseudo F statistic derived from the general distance-based regression model can be used to detect PS in a GWAS if an appropriate metric is used for the measurement of the genetic background similarity between two subjects. Here we extend the pesudo F statistic considered by McArdle and Anderson [2001] and Nievergelt et al. [2007] to allow for the adjustment of covariates. The extended F statistic can evaluate the adequacy of PS correction when potential ancestral confounding factors, such as self-identified ethnicity or selected principal components [Patterson et al., 2006; Price et al., 2006], have already been included in the adjustment. Built upon this pseudo F statistic, a computationally efficient PC selection procedure, called PC-Finder, is proposed to identify relevant PCs for the correction of PS. Empirical data from two GWAS in the Cancer Genetic Markers of Susceptibility (CGEMS) project were used to demonstrate the application of the proposed methods. We also conducted simulation studies to evaluate the performance of the proposed methods.
METHODS
THE PSEUDO F STATISTIC WITH ADJUSTMENT FOR THE COVARIATE’S EFFECT
Suppose that there are n subjects in the sample and S = (sij)n×n is the similarity [what does similarity mean?] matrix with sij describing the similarity between subject i and j, 1≤ i, j ≤ n. Let X = (xij)n×m = (X1 : X2) be the design matrix where X1 and X2 are matrices with dimension of n × m1 and n × m2, respectively, and m1 + m2 = m. We try to obtain the test statistic for testing whether the pair-wise similarity is influenced by the covariates in X2, while adjusting for the effect of X1. McArdle and Anderson [2001] provided the pseudo F statistic for testing the effect of X2 on the pair-wise similarity level when there is no other covariate to be adjusted for, i.e., X1 is a column of 1’s. For the general case, we can use the following more general pseudo F statistic for the adjustment of X1,
| (1) |
where C = In-n-1JJT is the centering matrix, with In and J being the n × n identity matrix and the n-dimensional column vector of 1’s, respectively; ΗX = X(XTX)-1XT and be the projection matrices onto the subspace spanned by columns in X = (X1 : X2) and columns in X1, respectively. The derivation of (1) is given in the Appendix I. Also shown in Appendix I, the pseudo F statistic can be expressed as
| (2) |
where Q = (In-HX1)CSC(In-HX1). This representation is useful in the permutation procedure described below.
If there is no covariate for adjustment when testing whether the pair-wise similarity matrix is influenced by the covariates in X2, we have X1 = J. Since in this case HX1C = 0, the pseudo F statistic in (1) can be simplified as
which becomes the statistic given by McArdle and Anderson [2001].
In applications where a dissimilarity (distance) matrix D = (dij)n×n is available, we can simply transform the dissimilarity matrix into the following similarity matrix S = (sij)n×n, with , i,j = 1,2,⋯,n. Then the pseudo F statistic defined by (1) can be obtained.
EVALUATING THE SIGNIFICANCE LEVEL OF THE PSEUDO F STATISTIC
Since the theoretical asymptotic distribution for the pseudo F statistic is unknown, a permutation procedure has to be used for the assessment of the associated p value. When there are no other covariates to be adjusted for (i.e., X1 = J), a random permutation of rows and corresponding columns in the matrix CSC, while keeping X1 and X2 unchanged, can be used to generate datasets under the null hypothesis [McArdle and Anderson, 2001]. When covariate adjustment is needed, the standard permutation procedure that does not maintain the relationship between the pair-wise similarity matrix and X1 during the permutation is not appropriate for generating datasets under the null hypothesis. Instead, we propose to use a residual permutation procedure in the framework of the multivariate regression model (See Appendix II for details) to maintain the relationship between the pair-wise similarity and X1 in the permutations.
Obtain Q = (In-HX1)CSC(In-HX1), and the observed pseudo F statistic Fobs according to (2).
- Randomly permute the rows and columns of Q simultaneously B times. For each resultant matrix , b = 1, …, B, obtain the corresponding pseudo F statistic
- Calculate the empirical p value as
Comment 1
When calculating in Step 2, we only need to calculate the diagonal terms of the matrix for the evaluation of the trace. The same is true for the evaluation of . This can reduce computing time substantially when the number of subjects n is large (n > 2,000 in a typical GWAS application)
Comment 2
When there are no covariates to be adjusted for (i.e., X1 is a column of 1’s), we have Q = CSC. Then, the above permutation procedure becomes the one suggested by McArdle and Anderson [2001].
USING DISTRANCE BASED REGRESSION FOR THE EVALUATION AND CORRECTION OF PS
PS occurs when cases and controls have different population genetic backgrounds. Thus, under PS, it is expected that genetic similarity between two subjects depends on their case and control status. The pseudo F statistic derived under the distance-based regression model can be used to test for the existence of PS formally, that is, to assess the genetic background difference between the case and control groups. The general pseudo F statistic that allows for adjustment by other covariates can be useful in situations where we intend to evaluate whether PS still exists after certain potential ancestral confounding factors, such as self-identified ethnicity or selected principal components [Price et al., 2006; Patterson et al., 2006] have already been adjusted for. In the following sections, we describe a procedure for the selection of PCs for the correction of PS.
The definition of the similarity matrix
Suppose that in a GWAS with a total of n subjects, including r cases and s controls, we use a panel of M structural inference SNPs for characterizing the genetic background of individuals. The genotype at a marker locus is coded as 0, 1 or 2, corresponding to the copy number of a chosen allelic type. Let gim be the genotype measured at SNP m for the ith individual. We follow Price et al. [2006] in measuring the genetic background similarity between two subjects. We first standardize each genotype coding as , with being the allele frequency for the mth marker. The similarity between the ith and jth individuals is defined as
| (3) |
Then we have a similarity matrix S = (sij)n×n. Other types of genetic similarity metrics can be used.
PS evaluation based on the pseudo F statistic
Using the similarity matrix defined above, we can evaluate the relationship between the genetic similarity level and the case/control status using the pseudo F statistic given by (1). In this setting, X2 = d, where d is a column of case/control status indictors, with 1 for cases, and 0 for controls; X1 is the matrix of adjusted covariates, which could include covariates, such as a set of selected PCs [Price et al., 2006; Patterson et al., 2006], for the correction of PS.
A procedure to select PCs for the correction of PS
First, the pseudo F statistic without adjusting for PCs is applied to assess the extent of PS. If the associated p value is larger than a given threshold α (e.g., 0.05), we do not do any PC adjustment. Otherwise, we seek to identify a minimal number of PCs whose adjustment in the pseudo F statistic can result in a non-significant p value, i.e., a p value larger than α. We start with the L PCs, y1, y2, ⋯, yL, having the largest eigenvalues, with L large enough (say 50) so that the pseudo F statistic, which evaluates the relationship between case/control status and the level of genetic similarity after the adjustment for L PCs, has its p value larger than α. That is, there is little evidence of PS after the adjustment of the top L of PCs.
To keep the number of adjusted for PCs minimal while allowing for an adequate adjustment, we use a backward elimination procedure to remove PCs that do not appear to have “a significant effect” in reducing the level of PS. To restrict the searching space, we rank those L PCs in an increasing order according to their Wilcoxon rank-sum test statistics comparing the distributions of cases and controls along individual PCs, and we define them in that order as y(1), y(2), ⋯, y(L), with y(j) being the PC with the jth smallest Wilcoxon rank-sum test statistic among {y1, y2, ⋯, yL}. This order is to be followed when searching for PCs to be removed, that is, PCs on which cases and controls have most comparable distributions will be checked for elimination first. This makes sense intuitively, as those PCs with lower ranks (small Wilcoxon rank-sum statistics) are less likely to be helpful in the correction of PS, compared with PCs with higher ranks. Below is a summary of the backward elimination procedure, called PC-Finder:
- Define E to be the current set of selected PCs, starting with
- Iterate the following two steps for f = 1 to L.
- Obtain the pseudo F statistic with the adjustment for PCs within the set E\{y(j)}.
- If the associated p value is larger than α, remove y(j) from E, otherwise leave the set E unchanged.
The set E at the end of L iterations is the final set of PCs to be used for adjustment in the association analysis.
In the above procedure, we exclude the PC y(j) whenever the adjustment using the remaining PCs can correct for PS adequately (i.e., the p value from the pseudo F test is larger than α).
APPLICATIONS
APPLICATIONS TO CANCER GENETIC MARKERS OF SUSCEPTIBILITY GWAS
The Cancer Genetic Markers of Susceptibility (CGEMS) project has conducted two multi-stage GWAS of breast cancer and prostate cancer [Hunter et al., 2007; Yeager et al., 2007; Thomas et al., 2008]. For the initial stage of the GWAS of breast cancer, cases and controls were drawn from the Nurses’ Health Study (NHS) cohort. The prostate cancer initial scan used cases and controls collected from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Both studies used a nested case-control design for the sample collection. For illustration purpose, the test set for the PLCO prostate cancer study consisted of 1,171 prostate cancer cases and 1,094 controls, while the test set for the NHS breast cancer study included 1,140 breast cancer cases and 1,138 controls. By exchanging the control groups of the two studies, we created two additional GWAS that mimic a non-standard but potentially cost-effective design, which draws cases and controls independently from two studies. Thus, we have the following four GWAS: PLCO cases vs. PLCO controls (PLCOca-PLCOco), NHS cases vs. NHS controls (NHSca-NHSco), PLCO cases vs. NHS controls (PLCOca-NHSco), and NHS cases vs. PLCO controls (NHSca-PLCOco).
A panel of 12,898 previously identified structure inference autosomal SNPs was used to assess the genetic background of each subject [Yu et al., 2008]. We used formula (3) for the calculation of the similarity matrix for each considered GWAS based on genotypes measured on this panel of structural inference SNPs. To evaluate the type I error inflation, as measured by the over-dispersion factor, for a chosen association test statistic before and after the correction of PS, a set of 241,238 genomic control SNPs that were in low linkage disequilibrium with any nearby structure inference SNPs were identified [Yu et al., 2008]. For each genomic-control SNP, we applied 1-df likelihood ratio test derived from the standard logistic regression, with or without adjusting for chosen PCs while assuming an additive genetic risk model. Following Devlin and Roeder [1999], the over-dispersion factor can be estimated as the ratio of the median of likelihood ratio test statistics over the set of genomic control SNPs and the expected median of the chosen test statistics under the null hypothesis (i.e., in the present case, 0.455, the median of Chi-square distribution with one degree freedom).
When we applied the pseudo F statistic without the adjustment of PCs to each of the two original designed GWAS, we found no significant difference in genetic background between cases and controls (p values of 0.123 for PLCOca-PLCOco; and 0.104 for NHSca-NHSco; see Table 1). Thus, we do not expect a strong confounding effect by PS in either study. In fact, the estimated over-dispersion factor λ is 1.025 in PLCOca-PLCOco and 1.005 in NHSca-NHSco.
TABLE I.
P values of the pseudo F-test and Over-dispersion factors (λ) for CGEMS data set.
| Unadjusteda |
PC-Finderb |
EIGENSTRATc |
||||
|---|---|---|---|---|---|---|
| Data set | p value | λ | p value | λ | p value | λ |
| NHSca vs NHSco | 0.104 | 1.005 | -- | -- | -- | -- |
| PLCOca vs PLCOco | 0.123 | 1.025 | -- | -- | -- | -- |
| NHSca vs PLCOco | <10-3 | 1.062 | 0.052 | 1.007 | 0.052 | 1.010 |
| PLCOca vs NHSco | <10-3 | 1.090 | 0.054 | 1.020 | 0.029 | 1.036 |
P-values for the pseudo F test and over-dispersion factors (λ) for the association test, both tests are not adjusted for PS.
P-values for the pseudo F test and over-dispersion factors (λ) for the association test, both tests are adjusted by PCs selected by the PC-Finder.
P-values for the pseudo F test and over-dispersion factors (λ) for the association test, both tests are adjusted by the top 10 ranked PCs
By contrast, when we applied the pseudo F statistic to either of the two reconstructed GWAS, PLCOca-NHSco and NHSca-PLCOco, we found a significant difference in genetic background between the case and control groups (p value < 0.001 for both PLCOca-NHSco and NHSca-PLCOco, Table 1). As expected, the estimated over-dispersion factor λ in either of two scans is much higher than those observed in the two original scans (λ = 1.090 in PLCOca-NHSco, and λ = 1.062 in NHSca-PLCOco). The demonstrated genetic background difference between samples from PLCO and samples from NHS could be due in part to the difference in geographic locations of the source populations which were sampled, as people from distinct regions tend to have different genetic structure [Yu et al., 2008].
To correct for PS in PLCOca-NHSco, we applied the proposed PC selection procedure and identified the 1st, 2nd, 4th, 35th, and 55th PCs (ranked according to corresponding eigenvalues in the decreasing order) for the adjustment. After adjustment with those PCs, the p value for the resultant pseudo F statistic was 0.054. The dispersion factor λ for the 1-df likelihood ratio test with adjustment for the 5 chosen PCs was reduced to 1.020 from its original level of 1.090.
For NHSca-PLCOco, using the PC-Finder we identified the 1st, 2nd, 3rd, 7th, and 10th PCs for the correction of PS, with the p value for the resultant pseudo F statistic being 0.052. Again, the dispersion factor λ after the adjustment of the chosen PCs was 1.007, which was much reduced from its original level of 1.062.
Some of the selected PCs in both the PLCOca-NHSco and NHSca-PLOCco examples are not the top ranked ones, reflecting PC-Finder’s requirement that the distribution of identified PCs be different between cases and controls.
SIMULATION
Simulation design for the evaluation of pseudo F statistic
To evaluate the type I error rate for using the pseudo F statistic as a test for the existence of PS, we generated 1,000 datasets consisting of r cases and the same number of controls from a common population, with r = 500 and 1000. For each simulated dataset, we considered 10,000 structural inference SNPs, with their minor allele frequencies (MAFs) being independently drawn from the uniform distribution U(0.1,0.5). Given the MAF for a SNP, its genotypes were randomly assigned to cases and controls according to the genotype frequencies under an assumption of fitness for Hardy-Weinberg proportion. We obtained the similarity matrix using genotypes measured on the 10,000 structural inference SNPs, and applied the unadjusted pseudo F statistic to evaluate the relationship between the similarity level and case/control status.
To assess the power of the pseudo F statistic, we generated datasets with various level of PS based on the two GWAS from the CGEMS projects. As shown in the Application section and in Yu et al. [2008] the samples from PLCO and NHS apparently have different genetic backgrounds. Thus, we expect a certain degree of PS to exist when we compare a group of prostate cancer cases, which were all sampled from PLCOca, with a mixed group of controls sampled from the two original control groups (PLCOco and NHSco). Letting the mixture proportion ρ be the proportion of controls sampled from NHSco, we considered various levels for ρ in the simulation. For a given mixture proportion ρ and the sample size r, we generated 1,000 datasets with r cases from PLCOca, r × ρ controls from NHSco, and r × (1-ρ) controls from PLCOco. Similar to the application described in Section 3.1, we used genotypes measured on the set of 12,898 structural inference SNPs for the calculation of the similarity matrix.
Simulation design for the evaluation of PC-Finder
We conducted simulation studies to evaluate the type I error rate of the association test, which adjusts for the selected PCs by the PC-Finder for the correction of PS. The association test is the 1-df likelihood ratio test derived from the standard logistic regression. For comparison, we also considered adjustment for the top 10 PCs as suggested by Price et al. (2006), as well as no adjustment.
To evaluate the type I error rate, we generated 1,000 datasets with each data set consisting of 1,000 cases and 1,000 controls sampled from two subpopulations. Within each dataset, 60% and 40% of the cases, as well as 40% and 60% of the controls, were from subpopulations 1 and 2, respectively. We assumed that Hardy-Weinberg equilibrium (HWE) holds within each subpopulation. To generate genotypes, we used the same algorithm as the one in Price et al. [2006]. The minor allele frequency (MAF) for each SNP in the two subpopulations were generated independently from the Beta distribution with two parameters, p(1-FST)/FST and (1-p)(1-FST)/FST, with the inbreeding coefficient FST = 0.01, and p, the ancestry population allele frequency, being drawn from the uniform distribution over [0.1, 0.5]. For each simulated dataset, we generated 10,000 structural inference SNPs and used genotypes measured on them for the construction of the similarity matrix. We also generated a separate set of 1000 genomic control SNPs for the evaluation of the over-dispersion factor and empirical type I error.
In addition, we carried out simulation using the CGEMS data set. We generated 100 datasets, with each consisting 800 cases sampled from the PLCO prostate cancer case group (PLCOca) and 800 controls sampled from the NHS control group (NHSco). Similar to the application described in Section 3.1, we used the set of 12,898 structural inference SNPs for the calculation of the similarity matrix, and the set of 241,238 genomic control SNPs for the evaluation of over-dispersion factor and empirical type I error.
Simulation design for the power comparison
Price et al. [2006] suggested adjustment for the top 10 PCs for the correction of PS. Although they interpreted their simulation results to show that the adjustment of a few more unnecessary PCs did not have any noticeable negative impact on the power of the association test, Yu et al. [2008] demonstrated substantial loss in power in an example that the unnecessary adjustment of population substructure (even one PC). Here, following the design of Yu et al. [2008], we conducted a simulation study to compare the power of the following three association tests, the test without the adjustment of PS, the test with the adjustment of PCs selected by the PC-Finder, and test with the adjustment of top 10 PCs.
Here is the brief summary of the simulation design. More details were given in Yu et al. [2008]. The considered source population consists of two equal sized subpopulations, and the disease risk model for both subpopulations is
| (4) |
where E is an exposure measure with 0 for unexposed subjects and 1 for exposed ones, and G is the genotype score at the disease locus with 0 for having none of the disease risk allele and 1 for having at least one copy of disease risk allele. In either subpopulation, we assume that E is independent of G, and the odds ratios for E and G are both 1.3, that is, exp(γ) = exp(β) = 1.3. We also assume that, in the first subpopulation, Pr1(E = 1) = ξ and Pr1(G = 1) = η, and in the second subpopulation, Pr2(E = 1) = η and Pr2(G = 1) = ξ. Given (ξ,η,γ,β), the value for μ is chosen in such a way that the resultant disease prevalence in each subpopulation is 5%. Under this setting, there is no PS for the association test that adjusts for the effect of E, since the two subpopulations have the same disease prevalence.
Each simulated dataset consists of 1,000 cases and 1,000 controls that are randomly collected form the source population. We generate 1,001 independent SNPs including 1 disease-associated SNPs for the power evaluation and 1,000 independent disease-unrelated SNPs for the PS correction. The genotypes at the disease-associated SNPs were simulated according to the method given by Price et al. [2006]. Genotypes at 1,000 disease-unrelated SNPs for subjects sampled from either subpopulation were simulated using the same algorithm described under the design for the evaluation of PC-Finder. To save the computing time, we chose a relatively large inbreeding coefficient value (FST = 0.2) to ensure that 1,000 independent disease-unrelated SNPs were enough to recognize the population structure. For each of the three considered tests, the effect of E was always adjusted. For the test using the PC-Finder for the PS adjustment, we chose α = 0.05 as the P-value threshold.
Simulation results for the evaluation of pseudo F statistic
At the significance level of 0.05, based on 1,000 simulated datasets the empirical type I error rate for the pseudo F statistic was 0.055 when the sample size was 500, and was 0.052 when the sample size was 1,000. Both values are close to the nominal level.
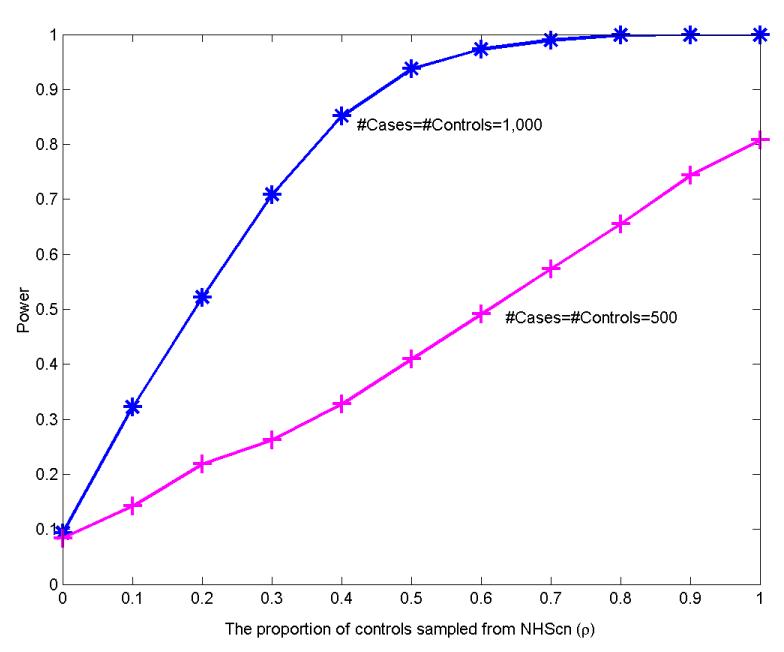
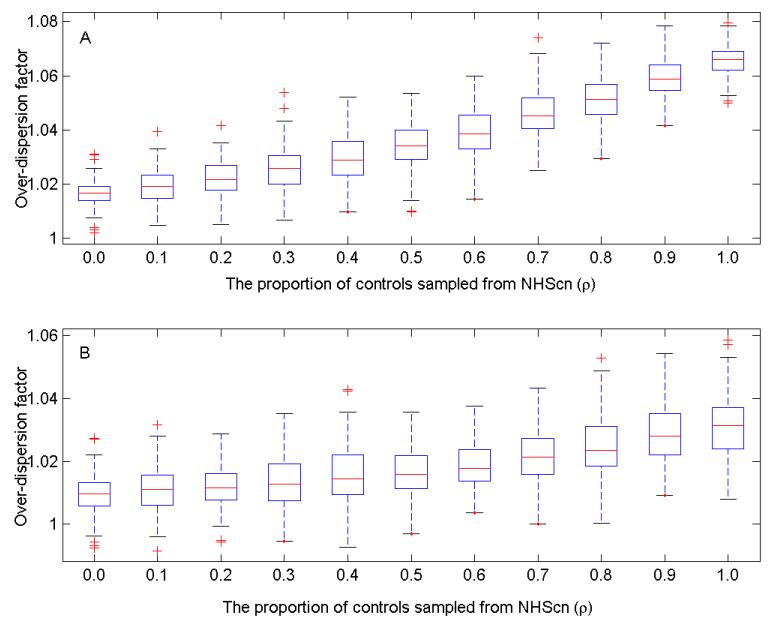
Fig. 1 summarizes the power of the pseudo F statistic to detect PS at a significance level of 0.05. We can see from Fig. 1 that the power increases as the control mixture proportion ρ increases. This is consistent with the pattern shown in Fig. 2, which suggests that the genomic inflation factor tends to increase as more controls are sampled from NHSco. In Fig. 2, we provide the box-plot for the estimated over-dispersion factor based on 100 datasets (randomly chosen from 1,000 generated ones for the sake of saving computing time) for a given mixture proportion ρ. The over-dispersion factor was for the association test (1-df likelihood ratio test) without adjustment for any PC; it was estimated based on testing results on the set of 241,238 genomic control SNPs described in the Section 3.1. Based on a comparison of the power under two sample sizes (500 and 1,000) with the same control mixture proportion level ρ, it is evident that the power to detect PS increases with the sample size.
Fig. 1.
Power of the pseudo F test for detecting population stratification. Results are based on 1,000 replicates at the significance level 0.05. Within each dataset, the case group consists of a random sample of prostate cancer cases from the GWAS of prostate cancer; the control group is a mixture of two samples, one from the GWAS of prostate cancesr, the other from the GWAS of breast cancer cases.
Fig. 2.
Boxplots of over-dispersion factor based on 100 replicates. Within each dataset, the case group consists of a random sample of prostate cancer cases from the GWAS of prostate cancer; the control group is a mixture of two samples, one from controls in GWAS of prostate cancer, the other from controls in GWAS of breast cancer. (A) Results under the sample size of 1,000 cases and 1,000 controls. (B) Results under the sample size of 500 cases and 500 controls.
Simulation results for the evaluation of PC-Finder
Fig. 3 shows simulation results based on 1,000 generated datasets, with each consisting of cases and controls unevenly sampled from two distinct subpopulations. Ideally, we need just one PC that separates the two subpopulations for the correction of PS. In 953 out of 1,000 replications, the PC-Finder identified one single PC. For the association test without any PC adjustment, we observed inflated type I error and an over-dispersion factor. For the association test adjusting for the top 10 PCs (ranked by their corresponding eigenvalues) or for PCs identified by the PC-Finder, the PS was corrected for adequately. The number of PCs chosen by the PC-Finder tended to be much smaller than 10.
Fig. 3.
Performance comparisons among three PC selection strategies in simulations where the study population consists of two subpopulations. Results are summarized based on 1,000 simulated datasets, with each consisting of 1,000 cases and 1,000 controls. (A) Boxplots of number of chosen PCs; (B) Boxplots of empirical type I errors under the significance level of 0.05 and (C) Boxplots of over-dispersion factors.
Fig. 4 shows simulation results based on 100 replicated datasets, with each consisting of 800 cases from PLCOca and 800 controls from NHSco. We found either of the two strategies---choosing the top 10 ranked PCs or using PC-Finder---worked well for the correction of PS. But PC-Finder can keep the number of PCs adjusted for to a minimum by excluding the “unnecessary” ones.
Fig. 4.
Performance comparisons among three PC selection strategies in simulations where replicated datasets are assembled from the two GWAS in the CGEMS. Results are summarized based on 100 generated datasets, with each consisting of 800 cases and 800 controls. (A) Boxplots of number of chosen PCs; (B) Boxplots of empirical type I errors under the significance level of 0.05 and (C) Boxplots of over-dispersion factors.
In summary, depending on the extent of PS, PC-Finder can adaptively pick up PCs whose use in adjustment can reduce the level of PS.
Simulation results for the power comparison
Table 2 shows the power for the three considered association tests based on 1,000 replicated datasets under a given (ξ,η) configuration. It is not surprising to see from the table that the test without the adjustment of PS has the highest power, since there is no PS under the considered scenario where two subpopulations have the same level of disease risk. The power of the test using PC-Finder is slightly lower than the one without the adjustment of PS, reflecting the 5% chance that the pseudo F test was significant at the P-value threshold of 0.05. The test consistently adjusting for top 10 PCs has the worst performance. Results shown in Table 2 clearly demonstrate that the adjustment of unnecessary PCs could lead to a substantial loss in power, but that the PC-Finder performs nearly as well as no adjustment.
Table 2.
Power comparisons for the three PC selection strategies.
| (ξ,η)a | Unadjustedb | PC-Finderc | EIGENSTRATd |
|---|---|---|---|
| (0.1,0.9) | 0.62 | 0.61 | 0.44 |
| (0.2,0.8) | 0.79 | 0.78 | 0.67 |
| (0.3,0.7) | 0.83 | 0.82 | 0.76 |
| (0.4,0.6) | 0.84 | 0.83 | 0.82 |
Values for ξ, the probability of being exposed to the environment risk, and for η, the probability of having at least one copy of the disease risk allele, for a subject from subpopulation I. These values are reversed for subpopulation II.
Results based on the association test derived from the logistic regression model adjusting for only the exposure factor.
Results based on the association test derived from the logistic regression model adjusting for both the exposure factor and PCs selected by PC-Finder.
Results based on the association test derived from the logistic regression model adjusting for both the exposure factor and the top 10 ranked PCs.
DISCUSSION
We have proposed a formal PC selection procedure, called PC-Finder, to identify an optimal set of PCs whose adjustment can adequately mitigate the confounding effect of PS caused by differences in the genetic background between cases and controls. We have demonstrated its robust application using two GWAS from the CGEMS projects. Simulation studies based on empirical data from the CGEMS projects, as well as simulated populations, showed that PC-Finder can control for PS with greater power than currently used methods.
Our previously proposed PC selection procedure [Yu et al., 2008] requires the estimation of the genomic control inflation factor in each permutation step, and is only computationally feasible for choosing PCs among a limited number of candidates. PC-finder, on the other hand, uses only the genetic similarity matrix, and can search through a large number of PCs reasonably fast because the permutation procedure used for estimating the p value associated with the pseudo F statistic is computationally efficient. For example, it takes about 150 seconds on a Pentium 4, 3.0 GHz personal computer to run 1,000 permutations on a sample of 500 cases and 500 controls when trying to estimate the p value for the pseudo F statistic with adjustment for 5 covariates when usingour freely available R code software
The pseudo F test provides a sensible measure for the extent of PS. We used a P-value threshold of 0.05 to decide whether or not to include PCs for the correction of PS. In practice, it is always helpful to apply this test in conjunction with other PS diagnostic measures, such as the Q-Q plot and the over-dispersion factor, in order to have a more thorough evaluation of PS.
PS assessment and correction have become an integral part of GWAS analysis, particularly in multi-stage GWAS designs. The proposed pseudo F statistic and the PC-Finder procedure can control for the effects of PS in GWAS with greater power than other PCA methods. Finally, we have implemented the PC-Finder procedure using R code and the software is freely available.
ACKNOWLEDGEMENTS
We thank B.J. Stone for her helpful comments. This research utilized the high-performance computational capabilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, Maryland, USA (http://biowulf.nih.gov). Q Li is supported in part by the Knowledge Innovation Program of the Chinese Academy of Sciences, Nos. 30465W0 and 30475V0.
APPENDIX I: DERIVATION OF THE PSEUDO F STATISTIC
Here we provide the motivation for the derivation of the general pseudo F statistic that allows for the adjustment of X1. We first use multidimensional scaling to transform the problem into a multivariate regression analysis of principal coordinates, then re-express the obtained test statistic in terms of the similarity matrix.
We first assume the similarity matrix S is semi-positive (the assumption will be dropped later). We define the corresponding distance matrix D = (dij)n×n, with . According to Theorem 14.22 in Mardia et al. [2003], we can represent each subject i as a point with the coordinates (yi1, yi2,⋯, yiL), called its principal coordinates, in L dimensional Euclidean space, such that , that is, the Euclidean distance between the two points corresponding to subjects i and j is given by dij. The following algorithm can be used to find the principal coordinates for each subject:
Let C = In-n-1JJT be the centering matrix, with J being a column of 1’s of length n.
Obtain CSC, the centered matrix of S.
For CSC, find its L non-zero eigenvalues, η1≥η2≥⋯≥ηL> 0 and their corresponding scaled eigenvectors, y1, y2, ⋯, yL, with yj = (y1j, y2j, ⋯, ynj)T and , 1≤j≤L.
Form the n × L matrix Y = (y1:y2:⋯ :yL). The ith row of Y provides the principal coordinates for subject i.
According to the above algorithm, CSC =YYT, which specifies the relationship between the similarity matrix and the principal coordinates.
To test whether X2 influences the pairwise similarity given by S, we can evaluate its effect on the principal coordinates through the following multivariate linear regression model,
| (A1) |
where β and γ are the regression coefficient matrices, with their jth column representing the vector of coefficients corresponding to eigenvector yj. We call model (A1) the full model. The null hypothesis we want to test is H0 : γ = 0. Under the null, the full model (A1) becomes the following reduced model,
| (A2) |
The pseudo F statistic for testing H0 : γ = 0, derived by comparing the residuals of models (A1) and (A2), can be written as
| (A3) |
where HX = X(XTX)-1XT and are the projection matrices onto the subspace spanned by X and by X1, respectively. This pseudo F statistic becomes the standard F statistic with proper scaling when the number of columns in Y is one.
Since CSC = YYT, the pseudo F statistic (A3) can be expressed in terms of the original similarity matrix S as follows,
| (A4) |
Thus, through multidimensional scaling, we obtain the pseudo F statistic for evaluating the effect of X2 on the pair-wise similarity given by S, while adjusting for the effect of X1. In the above derivation, we assume for motivation purposes that S is semi-positive. But the same statistic given by (A4) can be used for any given similarity matrix.
Since (In-HX)(In-HX1) = (In-HX), and (HX-HX1)(In-HX1)=HX-HX1, the pseudo F statistic given by (A4) can also be written as
APPENDIX II: BASIS OF THE PERMUTATION PROCEDURE
To motivate the permutation procedure, we start with the matrix of principal coordinates Y under the model given by (A1). We want to generate datasets under the null hypothesis H0 : γ = 0 for the evaluation of the p value associated with the F statistic given by (A3). The residual permutation procedure in the framework of multivariate regression model can be used for this purpose.
We represent the observed outcome (the matrix of principal coordinates) as
Within each residual permutation step, we regenerate each subject’s outcome by randomly shuffling the rows in the residuals matrix (In-HX1)Y derived under the reduced model (A2) and add them back to the original fitted values. Thus, the newly generated outcome Y* can be written as Y* = HX1Y+P(In-HX1)Y, where P is the permutation matrix corresponding to the permutation step. Following (A3), the pseudo F-statistic corresponding to the newly generated outcome Y* can be written as
Notice that since CSC = YYT, we have
According to the properties of orthogonal projections, we can represent F* as
| (A5) |
It is clear from (A5) that the residual permutation is equivalent to the simultaneous permutation of rows and columns of the matrix Q = (In-HX1)CSC(In-HX1). The summary of the permutation procedure is given in the main text.
REFERENCES
- Cox DR, McCullagh P. Some aspects of analysis of covariance. Biometrics. 1982;38:541–561. [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager N, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yu K. Improved correction for population stratification in genome-wide association studies by identifying hidden population structures. Genet Epidemiol. 2008;32:215–226. doi: 10.1002/gepi.20296. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Academic Press; New York: 2003. [Google Scholar]
- McArdle BH, Anderson JM. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- Nievergelt CM, Libiger O, Schork NJ. Generalized analysis of molecular variance. PLoS Genet. 2007;3:467–478. doi: 10.1371/journal.pgen.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:2074–2093. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott J, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Wessel J, Schork NJ. Generalized genomic distance-based regression methodology for multilocus association analysis. Am J Hum Genet. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Yu K, Chatterjee N, Wheeler W, Li Q, Wang S, et al. Flexible design for following up positive findings. Am J Hum Genet. 2007;81:540–551. doi: 10.1086/520678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Wang Z, Li Q, Wacholder S, Hunter D, et al. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapala MA, Schork NJ. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. PNAS. 2006;103:19430–19435. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhang S, Zhao H, Cooper RS. Association mapping, using a mixture model for complex traits. Genet Epidemiol. 2002;23:184–196. doi: 10.1002/gepi.210. [DOI] [PubMed] [Google Scholar]