Abstract
How genomic expression differs as a function of life history variation is largely unknown. Atlantic salmon exhibits extreme alternative life histories. We defined the gene-expression signatures of wild-caught salmon at two different life stages by comparing the brain expression profiles of mature sneaker males and immature males, and early migrants and late migrants. In addition to life-stage-specific signatures, we discovered a surprisingly large gene set that was differentially regulated - at similar magnitudes, yet in opposite direction - in both life history transitions. We suggest that this co-variation is not a consequence of many independent cellular and molecular switches in the same direction but rather represents the molecular equivalent of a physiological shift orchestrated by one or very few master regulators.
Keywords: Life history, alternative reproductive tactics, migration, microarrays, gene expression, gene-expression signature, ecological genomics, module
INTRODUCTION
Major life history transitions are common and are thus of central interest in evolutionary and behavioural biology. Age at maturation and age at migration are two life history traits involved in such transitions with direct effects on fitness. Age at maturation can vary widely among individuals of the same population. For example, sneaker males do not fight to gain access to females like resource monopolizing bourgeois males, but rather sneak into a reproductive act by imitating females. Sneaker males are almost always smaller, hold a peripheral locations and are usually younger than bourgeois males (Taborsky, 2001). Another striking life history transition is large scale migration found in insects (monarch butterflies, (Drake and Gatehouse, 1995)), reptiles (turtles, (Morreale et al., 1996; Polovina et al., 2004)), amphibians (Semlitsch, 2008), fishes (e.g., salmon, eels, (Dodson, 1997)), as well as birds and mammals ((Hobson, 1999)), where the same individual is found in completely different habitats at various stages of its life, owing to a journey that can span thousands of kilometres. In many species, age at migration can vary extensively among individuals of the same population (Metcalfe and Thorpe, 1990; Theriault and Dodson, 2003).
These forms of life history plasticity have been hypothesized to have their molecular roots in the organization and the differential expression of the genome (Stearns and Magwene, 2003; West-Eberhard, 2003). Many studies have been conducted on the ultimate evolutionary and ecological factors leading to important life history transitions (Stearns, 1992), but little is known about the regulation of individual genes or sets of genes associated with these different life history stages (but see for example (Aubin-Horth et al., 2005a; Aubin-Horth et al., 2005b; Roff, 2007; Whitfield et al., 2003). Further, we know even less, for any vertebrate species, about genome-scale changes in brain gene expression, as an individual transition through different stages of its life history. Are the gene-expression signatures (Nevins and Potti, 2007) representative of particular life history stages composed of specific genes that are differentially expressed only at a particular life stage? Or can the same set of genes be “reused” in contrasting ways and in different combinations, giving rise to the life history variations observed?
The Atlantic salmon (Salmo salar) is an ideal model system for studying the gene-expression signatures of life history transitions. Salmon have a complex life cycle composed of a phase of juvenile birth and growth in freshwater, followed by a migration to a saltwater habitat where substantial body growth is achieved, before homing to natal waters for reproduction in freshwater (reviewed in (Verspoor et al., 2007)). Age at migration to the sea varies among individuals within the same population, which can be referred to as “early” and “late” migrants (reviewed in (Garcia de Leaniz et al., 2007)). Furthermore, salmon exhibit alternative reproductive tactics. Migratory males (upon their return to the native stream) adopt a bourgeois male reproductive tactic, fight over territories and access to females, while early-maturing sneaker males reproduce at a younger age and smaller size (without first migrating to sea) by stealing fertilizations from the territorial males (Fleming, 1996; Hutchings and Myers, 1994). If they survive reproduction, these sneaker males can also eventually migrate to sea and return as large migratory males.
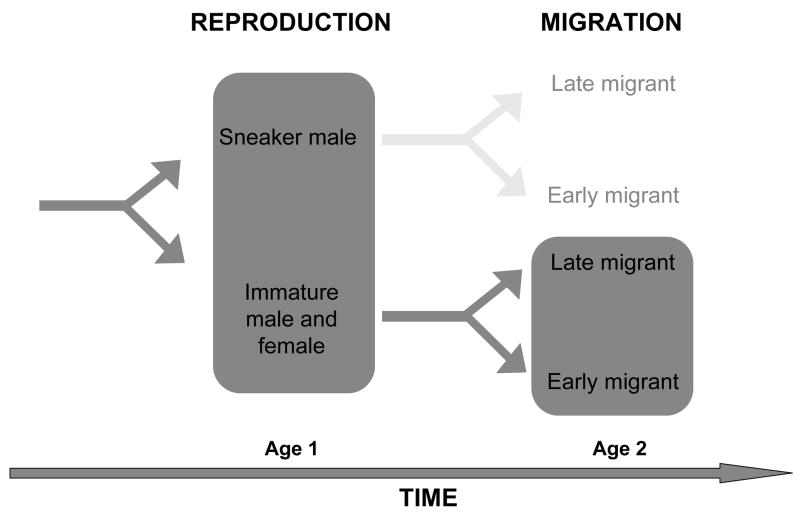
We have studied the changes in brain gene expression in two key life history transitions: early sexual maturation transition in males, as well as seaward migration transition, in wild-caught Atlantic salmon from tributaries of the Connecticut River in Western Massachusetts, USA, using a cDNA microarray (n=1827 genes). We sampled separately at each stage of the life history (figure 1). We did so at time points when the decisions for these life history transitions are already apparent, either in the fall or in the spring, respectively. We then compared the gene sets identified in each experiment and asked whether there was any overlap between those sets. Studies of genomic changes associated with physiological shifts have shown that it is possible to identify genes that are related to life cycle transitions and differences in life histories. Such studies include gene expression variation in brown adipose tissues during hibernation in ground squirrel (Hittel and Storey, 2001) and differences in transcription in liver between populations of brown trout that complete their life cycle in freshwater compared to populations that migrate ((Giger et al., 2008), reviewed in (St-Cyr and Aubin-Horth, 2009)). We chose to study gene expression in the brain, as it is the site of integration of external and internal signals and of regulation of numerous biological processes through neural and endocrine outputs.
Figure 1.
Life history transitions in Atlantic salmon are related to alternative reproductive tactics in males and alternative age at sea migration. We studied variation in expression profiles (highlighted in dark grey) separately between 1 year-old mature sneaker males and immature males at the reproduction stage in the fall, and between 2 year-old early migrants (ready to leave for saltwater) and late migrants (that will stay in freshwater for another year) at the migration stage in the spring. We studied the spring migration transition only in individuals (males and females) that were not previously mature the preceding fall.
METHODS
Field sampling
We sampled individuals in tributaries of the Connecticut River (Sawmill River and West brook) in Western Massachusetts, USA. We used electrofishing to collect fish. We sampled and processed individual fish sequentially. We immediately euthanized each fish in buffered MS-222 (MS-222 100 mg l−1 and sodium bicarbonate 100 mg l−1, pH 7). We measured fork length and mass and determined sex and sexual maturity by dissection. We then dissected the whole brain and transferred it to RNAlater storage solution (Ambion) in less than 15 minutes. We ensured that only one person conducted the brain dissections, in a consistent manner.
Sampling mature and immature males
We first collected 40 1-year-old individuals during the reproduction period in the fall ((October 10th, 2002; see (Aubin-Horth et al., 2005a), at which time a male can be sexually immature or a mature sneaker. Females at that age can only be immature and we therefore focused in the present study only on the mature and immature males, although immature females were included in the original sampling and microarray analysis found in (Aubin-Horth et al., 2005a) (see dark-shaded box in figure 1). Four sneaker males and four immature males were used in the microarray analysis (supplementary figure 1) presented here (the original analysis used twelve individuals as it included four immature females, see (Aubin-Horth et al., 2005a) for details).
Sampling early and late migrants
We performed the second study in the spring prior to sea migration when fish were aged 2-year-old. At this time of life, an individual (male or female) can become ready for saltwater challenge, a process called smolting, and migrate to sea (which we term “early migrant”). Alternatively, that individual can stay in freshwater for at least another year, before transitioning to sea (which we term “late migrant”). We sampled 17 immature individuals in the pre-migration season of spring (March 21st, 2006). Six of them were used in the microarray analysis: three early migrants (1 male and 2 females) and three late migrants (1 male and 2 females) (supplementary figure 2). The pre-migration sampling season was 42 days before a sharp increase of average daily water temperature above 9 degrees Celsius in that tributary and 63 days before reaching 640 degree-days since January 1st. These two thermal indicators have been related to downstream migration in salmon (Zydlewski et al., 2005). In accordance with these indicators, the fish we sampled were all still in freshwater, but late migrants and early migrants could already be differentiated by morphology, colouration and body size (Pearlstein et al., 2007). The early migrants had an average final fork length of 127 mm and a weight of 22.2 g, while non-migrating fish had an average final fork length of 108.3 mm and weight of 13.06 g. Furthermore, growth and maturity history since the first summer of life were known, as each fish in that tributary is individually marked (PIT tag) and that repeated recapture had been performed over their lifetime in that stream (Letcher and Gries, 2003). With that individual information, we found that fish that we identified as early migrants had a higher relative growth in fork length in the six months that preceded spring sampling (13% increase in length) than non-migrating fish (2.5% increase in length) (t-test, p = 0.0018). Higher growth during this life period is a characteristic of future smolts, which supports our classification based on color and overall morphology at capture. Information on each fish captured using the pit tag system allowed us to sample only individuals that were not mature males during the preceding fall, to limit the number of possible life histories (see dark-shaded box in figure 1). However, we are aware that including early and late migrants that were previously early maturing sneaker males would have greatly enlightened our comprehension of Atlantic salmon life cycle variation.
Microarray experiments
We determined in two separate microarray experiments which genes change in expression in the brain between 1) early maturing sneaker males and immature males (biological replicates: four mature males compared to four immature males) and 2) between early and late sea migrants (biological replicates: one early migrating male and two early migrating females compared to one late migrating male and two late migrating females). We homogenized brain tissue (Tissue Tearor, Biospec Products) and extracted total RNA according to protocol (Trizol, Invitrogen). To control for technical variation, we performed several independent reverse transcriptions (RT) on total RNA extracted from each brain sample, instead of one large reverse transcription reaction that is then split for use on different microarray slides. We followed a 3DNA array 350 protocol (Genisphere, as in (Aubin-Horth et al., 2005a)). We performed four RT per fish for the reproduction experiment and three RT per fish for the migration experiment. The cDNA obtained from each fish is used on four different microarrays in the reproduction experiment and on three microarrays in the migration experiment. We thus have technical replicates for each fish. Each fish of a given group (sneaker male, immature male, early migrant, late migrant) is a biological replicate for that phenotype (see supplementary figure 1, where one microarray hybridization is represented by an arrow). We used a loop design comparison with biological and technical replicates and dye-reversals, such that an individual of a given group was directly compared to individuals of the other group (sneaker male with immature male; early migrant with late migrant), without the need for a reference sample ((Churchill, 2002) (Townsend, 2003), see supplementary figure 1). On the supplementary figure, an individual is linked to other individuals to whom it was directly compared by an arrow. An arrowhead indicates that the sample was labelled with Cy5 dye, while an arrowtail indicates that the sample was labelled with Cy3 dye. We used a total of 24 microarrays in the first experiment (including control females), and 9 microarrays in the second experiment (represented by the number of arrows in each loop in supp. Figure 1). The microarray used is described in (Rise et al., 2004) (NCBI GEO GPL966), and is referred to as the “3.7K cDNA array” by GRASP, as used in (Aubin-Horth et al., 2005a). Clone sequences, library of origin, and annotation are available from GRASP (http://web.uvic.ca/cbr/grasp/) and from the DFCI gene index database for Atlantic salmon release 3.0 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=salmon). Arrays were scanned with an Axon 4000B scanner using Genepix 4.0 software (Molecular devices).
Statistical and bioinformatics analyses of differential gene expression
We analysed each experiment separately. We imported the data into R software (v1.9 (R Development Core Team, 2004)) and normalized using the Linear Models for Microarray Data package (LIMMA v2.9.8 (Smyth et al., 2003)). We normalised background-corrected mean intensities using a robust spline. The comparison of interest was the average difference in gene expression between early maturing sneaker males and immature males in experiment 1 and early and late migrants in experiment 2. We fitted a model with a coefficient for each fish. In each experiment, one of the individuals was treated as a baseline reference, in order to extract the contrast between groups of interest (Smyth, 2004). We obtained fold changes, standard errors, t-statistics and p-values in LIMMA. The comparison between mature sneaker males and immature males (and that included immature females) we reported previously relied on different normalisation and statistical methods (Aubin-Horth et al., 2005a). Because we analysed the two datasets in a consistent manner, we could then conduct a meta-analysis for comparison of the two life cycle transitions. We considered genes to be implicated in a life stage if they varied between two phenotypes with an adjusted p-value of 0.05, corrected for multiple tests. An adjusted p-value is corrected for false discovery rate, e.g. false positives, which are inherent to hypothesis testing in general. A lower (adjusted) threshold p-value means that less false positives are called (saying that a gene changes in expression when it doesn’t) compared to a p-value of 0.05, but it also raises the false negative rate (omitting genes that do vary in expression). A balance between false positives and false negatives is important and several methods of setting the adjusted p-value have been proposed. We used the Benjamini-Hochberg false discovery rate correction (Benjamini and Hochberg, 1995; Smyth, 2004). Results from the first microarray experiment comparing early maturing sneaker males and immature males were corroborated using quantitative real time PCR (see Aubin-Horth et al. 2005 for details). We obtained data for 4298 array features in both experiments, representing 1827 unique genes, based on the DFCI gene indices annotation. For the purpose of the analysis, we consider being immature and living in freshwater to be the original developmental state of a juvenile salmon. Moving to a new life history stage, (becoming sexually mature, or preparing to leave for saltwater), is the modified state. Thus, genes up-regulated in the original state (immature males and late migrants) are up only relative to the new modified state; in other words, at least some of these genes can also be considered as down-regulated in the modified state. It should be clear that some genes may also be up-regulated in the original state to actively control the developmental trajectory to prevent transitioning to the modified state (as discussed in (Aubin-Horth et al., 2005a).
Gene-expression signature
We defined a gene-expression signature as the set of genes that vary significantly in expression between two phenotypes. We then compared the gene-expression signature of the reproduction and the migration life history transitions, obtained from the separate microarray experiments, to determine whether the molecular substrates underlying the reproduction and migration life history transitions are distinct or shared. We determined how many and which genes were independently classified as significantly differentially expressed in the two experiments. Because we used a fairly stringent statistical cut-off, we consider this number to be a conservative estimate of the extent of overlap in gene expression signatures (FDR-adjusted p-value equivalent to 10−4). We then used Spearman rank correlation to determine if expression levels of the genes contained in this shared set were correlated between the two experiments. In cases where more than one array feature represented a gene of interest, we calculated the mean expression from the different features and used the resulting value for the correlation analysis
RESULTS AND DISCUSSION
Gene-expression signatures for each life history stage
Early reproduction stage
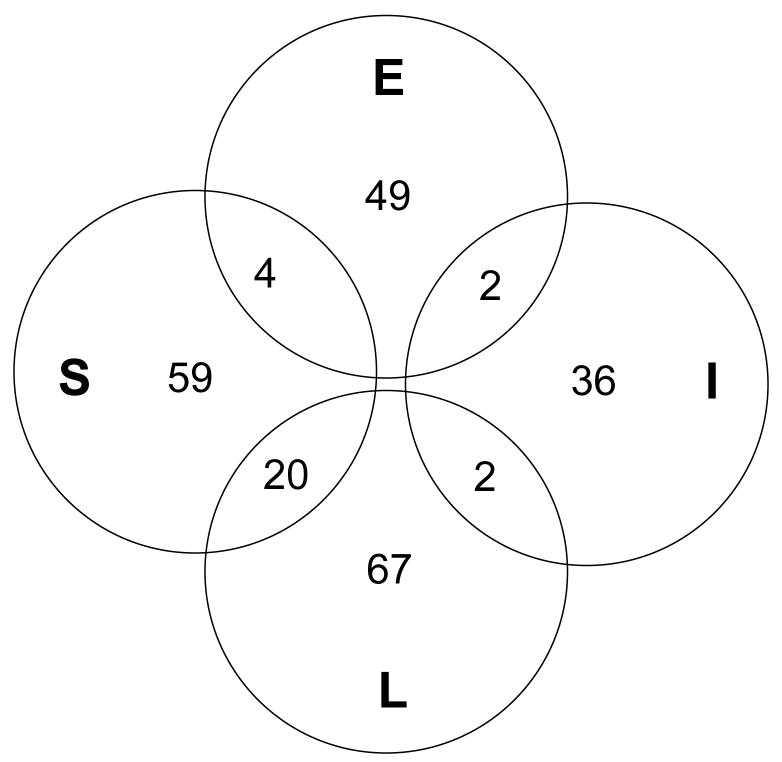
Ninety-five of the 123 genes differentially regulated at the reproduction life stage were specific to this life history transition (supp. table 1). The remaining 28 genes were regulated at this stage as well as at the migration stage (see below). Of these 95 specific genes, 59 showed increased expression in the brains of early maturing sneaker males, while 36 genes were more expressed in immature males (figure 2, “S” and “I”). Among genes with increased expression specifically in sneaker males, we found genes involved in synaptic function and plasticity (activity-dependent synaptic remodelling (Huh et al., 2000)), neurogenesis (Delta-like protein B precursor, (Haddon et al., 1998)) and genes encoding enzymes involved in the synthesis of the gaseous neurotransmitter nitric oxide that has neuropeptide regulation functions (argininosuccinate lyase and iNOS/NOS2 gene for inducible nitric oxide synthase (Wiesinger, 2001) (Cioni et al., 2002)). All these suggest that neural plasticity, including learning and memory formation, is activated in sneaker males at time of reproduction compared to males that do not mature. Also, soxP1 was up-regulated in sneakers, which may indicate maturation of pituitary for reproduction (Ito et al., 1995). These findings point to higher neuroendocrine activation in these males, which makes sense in the light of the reproductive axis activation. Glutamine synthetase, which was more expressed in immature males, controls intra-cerebral glutamine synthesis from glutamate. Its activity is inhibited by NMDA receptors and NO (Kosenko et al., 2003), which is in accordance with our finding of increased NO synthesis in sneaker males compared to immature males. These results are consistent with the gene expression data we reported in (Aubin-Horth et al., 2005a), and this new meta-analysis shows that many of these genes shown to vary in expression between mature sneaker males and immature males are specific to this stage of the life history.
Figure 2.
Genes forming the unique gene-expression signatures found for mature sneaker males (S), immature males (I), early migrants (E) and late migrants (L) and the 28 genes that are found in more than one gene-expression signature.
Twenty-eight genes that differed in expression between mature sneaker males and immature males also varied significantly between early and late migrants (table 1, supp. table 1, figure 2). Among them, gonadotropin alpha and beta were up-regulated in sneaker males (table 1). These adenohypophysal hormones regulate the reproductive axis by stimulating the gonads to produce gametes and sex steroids, which is in accordance with the early sexual maturation observed in sneaker males. The rod-like opsin gene expression is up-regulated in sneaker males and down-regulated in early migrants (or up-regulated in late migrants). The rod-like opsin found in the pineal gland of salmon described by (Philp et al., 2000) is closest in sequence to the exo-rhodopsin found in the pineal gland of the zebrafish, according to FCI gene index (Atlantic salmon v4.0). In the zebrafish, this exo-rhodopsin has been shown to be related to melatonin biosynthesis by the zebrafish pineal gland and thus to the regulation of circadian and seasonal rhythms (Pierce et al., 2008). It has been suggested that the pineal gland is involved in sexual maturation and change in growth patterns in fish, but conflicting results have been reported (Falcon et al., 2007) although to which extent these processes affect the life history transitions discussed here remains to be seen.
Table 1.
Overlap is found between the gene-expression signature of the reproduction and migration life stages. Brain expression profiles of mature sneaker males were compared to immature males in one experiment and early migrants were compared to late migrants in another experiment. Genes significantly differentially expressed were then compared between the two experiments. Annotations are based on consensus between the GRASP (http://web.uvic.ca/cbr/grasp/) and the DFCI gene index database for Atlantic salmon release 3.0 annotations. Complete information is found in supplementary table 2.
|
Up-regulated in mature sneaker males and early migrants |
| Globin alpha |
| Globin beta |
| Retinol binding protein |
| Unknown_TC48219 |
|
Up-regulated in mature sneaker males and down-regulated in early migrants |
| Apolipoprotein |
| cGMP-phosphodiesterase |
| Class III alcohol dehydrogenase |
| Ependymin |
| Glycerol-3-phosphate dehydrogenase |
| Gonadotropin alpha 1 |
| Gonadotropin alpha 2 |
| Gonadotropin beta (FSH) |
| Na/K ATPase alpha |
| Prolactin |
| Proopiomelanocortin |
| Rod-like opsin |
| Selenoprotein |
| Somatolactin |
| Somatotropine |
| Thr3 |
| Transposase |
| Unknown_TC39918 |
| Unknown_TC43698 |
| Unknown_TC50727 |
|
Down-regulated in mature sneaker males and up-regulated in early migrants |
| Unknown_singleton_srkc^8^61 |
| Unknown_TC34256 |
|
Down-regulated in mature sneaker males and in early migrants |
| Unknown_TC35270 |
| Unknown_TC36017 |
Migration stage
Among the 144 genes differentially regulated between early and late migrants, 116 were specific to this life history transition, with 49 that were up-regulated specifically in the brain of early migrants and 67 in the brains of late migrants/residents (supp. table 1). (See discussion below for the remaining 28 genes that we also found to be differentially regulated in the reproduction stage). We can thus evaluate genomic-scale transcriptional changes in the brain in a vertebrate that prepares to migrate. Activation of transcription of three odorant and vomeronasal receptor candidate genes in the olfactory epithelium of immature salmon has been shown in a time-series experiment following salmon during their development into migrating individuals, before the transition to saltwater (Dukes et al., 2004). That study and ours show that it is possible to detect differences at the transcription level between late migrants and early migrants of the same age, before complete metamorphosis and migration to saltwater.
We found that a type II iodothyronine deiodinase-encoding gene was up-regulated specifically in the brains of early migrant individuals. This enzyme is the principal player in the activation and inactivation of thyroxine (T4), the major secretory product of the thyroid. Regulation of this enzyme is important in the context of saltwater migration in salmon, as circulating levels of T4 rise during this period. Both thyroid hormones, T3 and T4, have been proposed to be directly involved in downstream swimming behaviour (reviewed in (Iwata, 1995)). Additionally, T3 and T4 are both important in neural development, learning and memory (Rivas and Naranjo, 2007). T4 is activated in peripheral tissues by outer ring deiodination to its more potent form, triiodothyronine (T3) (Bianco et al., 2002). In salmonids, T3 has a higher affinity for thyroid hormone receptors (TRs) than T4 (Bres and Eales, 1986). These two hormones can in turn be inactivated by inner ring deiodination (Bianco et al., 2002). In Atlantic salmon, a large rise in the ratio of T4 outer ring deiodination activity over T3 inner ring deiodination activity in the brain has been shown during the late phase of transformation from the juvenile (parr) to the (pre-migratory) smolt, indicating the potential for a positive T3 balance in the brain (Specker et al., 2000). T3 has been shown to activate receptors mainly locally when formed in the brain. Our gene expression results thus suggest that the individuals classified as “migrants” were most likely preparing for imminent behavioural and physiological changes and downstream migration to saltwater. Interestingly, the neuropeptides isotocin and arginine vasotocin showed increased expression uniquely in resident individuals in the spring. This result may seem surprising given the involvement of these hormones in the regulation of reproductive behaviour, stress response, blood pressure and osmoregulation ((Aubin-Horth et al., 2007; Bond et al., 2007; Goodson and Bass, 2001; Hyodo and Urano, 1991). However, as shown by these studies and others, the functions of these hormones are complex and can vary widely across species and physiological contexts.
We found that some of the genes that differed in expression between early and late migrants were also found to vary significantly between mature sneaker males and immature males (supp. Table 1, figure 2). This gene set includes ependymin, pro-opiomelanocortin and members of the growth hormone family (prolactin, somatolactin, somatotropin). The regulation of members of the growth hormone family (prolactin, somatolactin, somatotropin) is overall consistent with their role in smolt transformation (Agustsson et al., 2003). The down-regulation of somatotropin (growth hormone) in early migrants may seem unexpected, given its well-known role in saltwater adaptation (Prunet et al., 1989), though it may have a different function in the brain and the timing of our sampling (see methods) may have affected expression as well. The hormonally induced glycoprotein ependymin has been shown to be involved in learning and memory formation in fishes (Schmidt, 1995). The fact that all these genes are regulated in both life history transitions raises the questions whether they are part of a larger functional unit.
A Gene Module Underlying Life History Transitions
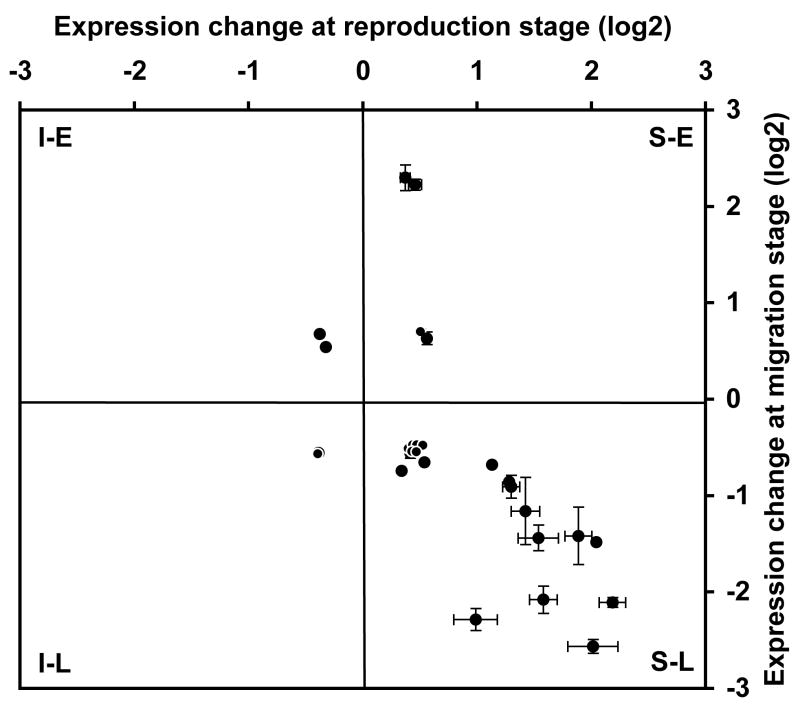
The strength of genomics lies in the capacity to extract unexpected patterns and sets of co-regulated genes, or modules, from the comparison of large datasets. In molecular systems biology, a module can simply be defined as a set of co-regulated genes or proteins (Segal et al., 2004). Such modules can serve as building blocks for the assembly of complex processes and phenotypes. For example, a conserved sex-related module was found when comparing brain gene expression in males and females of different primate species (human, macaque and marmoset, (Reinius et al., 2008)). Similarly, (Renn et al., 2008) uncovered gene modules associated with reproduction or social dominance. These insights could only be obtained by a higher order analysis of large-scale transcription data (Landry and Aubin-Horth, 2007; Nevins and Potti, 2007). In our study, we found a considerable overlap between the gene-expression signatures of the two life stages, with 28 genes whose expression differentiated a mature sneaker male from an immature male and an early from a late migrant (table 1 and supp. table 2). This number represents about 20% of the genes implicated in the reproduction and the migration life stages. For these genes associated with very different life history transitions, four possible expression pattern categories exist in principle, as indicated by each quadrant in figure 3: up-regulated in 1) sneaker males and late migrants (S-L), 2) sneaker males and early migrants (S-E), 3) immature males and late migrants (I-L) or 4) immature males and early migrants (I-L). Our null hypothesis was that the 28 genes in the life history transition gene set would fall into these categories at random, such that the same number of genes will be found in each category. Surprisingly, we found that most (20/28 = 71%) of these common genes that were up-regulated in mature sneaker males during the first life history transition were also up-regulated in late migrants during the second transition (figure 3, lower right quadrant). This is significantly different from the expected random distribution, Chi-square test, χ2 = 32.571, p< 0.0001). Even more remarkable, we also found that genes that showed large expression differences in the sneaker/immature males comparison also exhibited large differences in the early/late migrant comparison (figure 3). In fact, the magnitude of fold-differences in expression levels covaried significantly between the two experiments (Spearman rank correlation ρ= 0.70, p = 0.0002).
Figure 3.
Twenty out of the 28 genes differentially regulated in both the reproduction and the migration life history transitions are up-regulated in sneaker males in the fall (compared with immature males) and up-regulated in late migrant individuals in the spring (compared with early migrants) (lower right quadrant). Genes in this life history transition module also covary in magnitude of fold change.
Our study thus makes the striking finding that the gene networks associated with two very different life history decisions share a common set of >20 key genes. We propose that these genes represent a putative life history transition module. Interestingly, many members of this module are involved in neuroendocrine processes and their individual function is well documented. However, they have rarely been studied together or within a genomic context. We hypothesize that the extent of co-variation present in this life history transition module is not simply a consequence of many independent cellular and molecular switches in the same direction, but rather represents the molecular equivalent of a physiological (homeostatic) shift orchestrated by one or very few master regulators. Future studies should therefore focus on experiments that would identify these putative master regulators, and the 28 genes regulated in both transitions should provide a promising substrate. For example, one could ask whether the promoters of at least a good fraction of these genes share certain regulatory motifs, which would point to novel candidate transcription factors as the master regulators.
Twenty of the 28 genes are regulated in an opposing manner between the reproduction and the migration life stages (compared to the original developmental state represented by immature male and late migrant): these genes are up-regulated in sneakers in the fall (compared to immature males) and down-regulated in early migrants in the following spring (compared to immature late migrants that stay in freshwater for another year). For example, while gonadotropin genes are up-regulated in a sneaker male’s brain in the fall in preparation for reproduction (Amano et al., 1995; Antonopoulou et al., 1999; Waring et al., 1996), our results suggest that these same genes must be down-regulated in individuals preparing for a thousand kilometre-long saltwater migration. The genes that comprise the life history transition module are likely important in creating and maintaining two vastly different phenotypes (Aubin-Horth and Dodson, 2004; Moore, 1996; Rowe and Thorpe, 1990; Saunders et al., 1982; Saunders et al., 1985; Waring et al., 1996). A similar conclusion was advanced recently by (Renn et al., 2008) in a study comparing brain transcription profiles of males that transition between a socially dominant, reproductive, phenotype and a subordinate (reproductively inactive) stage.
We thus find a striking opposition in the direction of gene expression between reproduction and the migration life stage, as 71% of genes that are differentially expressed both at the reproduction and the migration life stages are in opposite directions between sneaker males and migrating individuals. Eight of the 28 genes (29%) regulated at both life history stages do not follow this pattern of covariation. For the most part these genes could not be annotated (table 1), making it difficult to interpret their expression pattern. The exception is provided by genes that are up-regulated in mature sneaker males and early migrants: Alpha and beta globins are important in oxygen transport, suggesting higher metabolic demands in sneaker males and individuals preparing for a seaward migration. Retinol binding protein is involved in transport of retinol (vitamin A), which in turn affects several traits in vertebrates, including brain development, vision, growth and learning and memory (Lane and Bailey, 2005), which could be related to transitions to new life stages faced by sneaker males and migrating individuals. All these hypotheses will need to be further tested in future studies.
Our results provide for the first time a molecular representation of a developmental conflict that has so far been demonstrated only at the endocrine level. For example, in masou salmon (Oncorhynchus masou), the same steroid hormones circulating at high levels in mature sneaker males compared with immature individuals (T, 11-KT and E(2)) inhibit downstream migration in pre-migrating individuals (Munakata et al., 2001). The activity pattern of the α1, α2 and β gonadotropins are entirely consistent with this finding. The same applies to pro-opiomelanocortin, a versatile regulator of feeding, energy balance, reproduction, stress response and pigmentation (Raffin-Sanson et al., 2003), even though the complexity of this system has yet to be unravelled in salmonids. It is possible that it could be implicated, among other things, in the striking coloration change seen in the silvery and uniform migrant smolts compared to the dark marks observed in parr (Verspoor et al., 2007).
Understanding the molecular basis underlying dramatically different life history stages and uncovering gene function in a natural context has become a major focus in ecological genomics, as it allows an integration of proximate with evolutionary insights (Hofmann, 2003; Landry and Aubin-Horth, 2007). Our results provide an important step forward in this effort. Each life history stage appears to be associated with a specific gene network, which acts in concert with a relatively small gene module that is important in the regulation of life history transitions in general. In other words, life history and behavioural variation in Atlantic salmon are orchestrated by distinct, yet overlapping gene networks. Life history transition modules such as the one presented here may well be at the basis of the life history variations observed in many animal species. With the increasing availability of genome-scale profiling techniques in model systems in ecology, evolution and behaviour we can now begin to test the generality of our findings.
Supplementary Material
Genes forming the unique gene-expression signatures found for mature sneaker males, immature males, early migrants and late migrants and the 28 genes that are found in more than one gene-expression signature. Physical location on microarray, Genbank and/or GRASP ID, TIGR contig ID, GRASP and TIGR annotation and GO terms, fold change coefficient and FDR corrected p-value in each comparison from LIMMA and whether the gene is up-regulated in mature sneaker males, immature males, early migrants and/or late migrants are presented.
Experimental hybridization design used in the two separate microarray experiments. A) Four sexually mature sneaker males (S) were compared to four immature males (I) and four immature females (F) of the same age. Individuals that are compared are linked by an arrow. The arrowhead indicates labelling with Cy5 dye and the arrow tail indicates that the sample was labelled with Cy3 fluorescent dye. Only results comparing males of the two types are used in the present meta-analysis, original results were reported in Aubin-Horth et al (2005). B) Three early migrants (1 male and 2 females; EM1, EF1, EF2) were compared to three late migrants (1 male and 2 females; LM1, LF1, LF2).
Acknowledgments
Thanks to Keith Nislow, Kitty Griswold, Todd Dubreuil, Matthew O’Donnell and Sarah Annis for help with sampling in the field and Yehoda Martei for technical assistance in the laboratory. Christian Landry helped with bioinformatics and with the gene annotation by merging data from GRASP and DFCI databases. Lin Huffman, Christian Landry, Dagan Lavee and Susan Renn provided helpful comments on previous versions of this manuscript. Funding was provided by FQRNT (Post doctoral fellowship), NSERC (Post doctoral fellowship and Discovery grant) and an Olin fellowship from the Atlantic Salmon Federation (NAH), the US Forest Service, Northern Research Station (BHL) and the Bauer Center for Genomics Research and the Institute for Molecular & Cellular Biology at UT Austin (HAH).
Footnotes
AUTHORS’ CONTRIBUTIONS
NAH, BL and HAH conceived and designed the experiments and did field sampling. NAH performed the gene expression laboratory experiments and statistical analysis. NAH wrote the manuscript together with HAH. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agustsson T, et al. Pituitary gene expression of somatolactin, prolactin, and growth hormone during Atlantic salmon parr-smolt transformation. Aquaculture. 2003;222:229–238. [Google Scholar]
- Amano M, et al. Short photoperiod accelerates preoptic and ventral telencephalic salmon GnRH synthesis and precocious maturation in underyearling male masu salmon. Gen Comp Endocrinol. 1995;99:22–27. doi: 10.1006/gcen.1995.1080. [DOI] [PubMed] [Google Scholar]
- Antonopoulou E, et al. Feedback control of gonadotropins in Atlantic salmon, Salmo salar, male parr. I. Castration effects in rematuring and nonrematuring fish. Gen Comp Endocrinol. 1999;114:132–41. doi: 10.1006/gcen.1998.7249. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, et al. Masculinized dominant females in a cooperatively breeding species. Mol Ecol. 2007;16:1349–58. doi: 10.1111/j.1365-294X.2007.03249.x. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, Dodson JJ. Influence of individual body size and variable thresholds on the incidence of a sneaker male reproductive tactic in Atlantic salmon. Evolution Int J Org Evolution. 2004;58:136–44. doi: 10.1111/j.0014-3820.2004.tb01580.x. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, et al. Alternative life histories shape brain gene expression profiles in males of the same population. Proc Biol Sci. 2005a;272:1655–62. doi: 10.1098/rspb.2005.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N, et al. Interaction of Rearing Environment and Reproductive Tactic on Gene Expression Profiles in Atlantic Salmon. J Heredity. 2005b;96:261–78. doi: 10.1093/jhered/esi030. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bianco A, et al. Biochemistry, Cellular and Molecular Biology, and Physiological Roles of the Iodothyronine Selenodeiodinases. Endocrine Reviews. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- Bond H, et al. Effect of acute restraint on hypothalamic provasotocin mRNA expression in flounder, Platichthys flesus. Gen Comp Endocrinol. 2007;153:221–227. doi: 10.1016/j.ygcen.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Bres O, Eales J. Thyroid hormone binding to isolated trout (Salmo gairdneri) liver nuclei in vitro; binding affinity, capacity and chemical specificity. Gen Comp Endocrinol. 1986;61:29–39. doi: 10.1016/0016-6480(86)90246-7. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32(Suppl):490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Cioni C, et al. Nitric oxide and neuromodulation in the caudal neurosecretory system of teleosts. Comp Biochem Physiol B Biochem Mol Biol. 2002;132:57–68. doi: 10.1016/s1096-4959(01)00532-2. [DOI] [PubMed] [Google Scholar]
- Dodson J. Fish migration: an evolutionary perspective. In: Godin JJ, editor. Behavioural ecology of teleost fishes. Oxford university press; Oxford: 1997. pp. 10–36. [Google Scholar]
- Drake V, Gatehouse A. Insect Migration: Tracking resources through space and time. Cambridge University Press; Cambridge: 1995. [Google Scholar]
- Dukes J, et al. Odorant receptor gene expression changes during the parr–smolt transformation in Atlantic salmon. Molecular Ecology. 2004;13:2851–2857. doi: 10.1111/j.1365-294X.2004.02252.x. [DOI] [PubMed] [Google Scholar]
- Falcon J, et al. Melatonin effects on the hypothalamo-pituitary axis in fish. Trends Endocrinol Metab. 2007;18:81–8. doi: 10.1016/j.tem.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Fleming I. Reproductive strategies of Atlantic salmon: ecology and evolution. Rev Fish Biol Fish. 1996;6:379–416. [Google Scholar]
- Garcia de Leaniz C, et al. A critical review of inherited adaptive variation in Atlantic salmon. Biological reviews. 2007;82:173–211. doi: 10.1111/j.1469-185X.2006.00004.x. [DOI] [PubMed] [Google Scholar]
- Giger T, et al. Population transcriptomics of life-history variation in the genus Salmo. Mol Ecol. 2008;17:3095–108. doi: 10.1111/j.1365-294X.2008.03820.x. [DOI] [PubMed] [Google Scholar]
- Goodson J, Bass A. Social behavior functions and related anatomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res Brain Res Rev. 2001;35:246–265. doi: 10.1016/s0165-0173(01)00043-1. [DOI] [PubMed] [Google Scholar]
- Haddon C, et al. Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 1998;125:359–370. doi: 10.1242/dev.125.3.359. [DOI] [PubMed] [Google Scholar]
- Hittel D, Storey K. Differential expression of adipose- and heart-type fatty acid binding proteins in hibernating ground squirrels. Biochimica et Biophysica Acta. 2001;1522:238–243. doi: 10.1016/s0167-4781(01)00338-4. [DOI] [PubMed] [Google Scholar]
- Hobson K. Tracing origins and migration of wildlife using stable isotopes: a review. Oecologia. 1999;120:314–326. doi: 10.1007/s004420050865. [DOI] [PubMed] [Google Scholar]
- Hofmann HA. Functional genomics of neural and behavioral plasticity. J Neurobiol. 2003;54:272–82. doi: 10.1002/neu.10172. [DOI] [PubMed] [Google Scholar]
- Huh GS, et al. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–9. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings JA, Myers RA. The evolution of alternative mating strategies in variable environments. Evolutionary Ecology. 1994;8:256–268. [Google Scholar]
- Hyodo S, Urano A. Changes in expression of provasotocin and proisotocin genes during adaptation to hyper- and hypo-osmotic environments in rainbow trout. J Comp Physiol B. 1991;161:549–556. doi: 10.1007/BF00260744. [DOI] [PubMed] [Google Scholar]
- Ito M, et al. A rainbow trout SRY-type gene expressed in pituitary glands. FEBS Lett. 1995;377:37–40. doi: 10.1016/0014-5793(95)01307-5. [DOI] [PubMed] [Google Scholar]
- Iwata M. Downstream migratory behavior of salmonids and its relationship with cortisol and thyroid hormones: A review. Aquaculture. 1995;135:131–139. [Google Scholar]
- Kosenko E, et al. Glutamine synthetase activity and glutamine content in brain: modulation by NMDA receptors and nitric oxide. Neurochem Int. 2003;43:493–499. doi: 10.1016/s0197-0186(03)00039-1. [DOI] [PubMed] [Google Scholar]
- Landry CR, Aubin-Horth N. Ecological annotation of genes and genomes through ecological genomics. Mol Ecol. 2007;16:4419–21. doi: 10.1111/j.1365-294X.2007.03504.x. [DOI] [PubMed] [Google Scholar]
- Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–93. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Letcher BH, Gries G. Effects of life history variation on size and growth in stream-dwelling Atlantic salmon. J Fish Biol. 2003;62:97–114. [Google Scholar]
- Metcalfe N, Thorpe J. Determinants of geographical variation in the age of seaward-migrating salmon, Salmo salar. Journal of Animal Ecology. 1990;59:135–145. [Google Scholar]
- Moore A. Electrophysiological and endocrinological evidence that F-series prostaglandins function as priming pheromones in mature male Atlantic salmon (Salmo salar) parr. J Exp Biol. 1996;199:2307–16. doi: 10.1242/jeb.199.10.2307. [DOI] [PubMed] [Google Scholar]
- Morreale SJ, et al. Migration corridor for sea turtles. Nature. 1996;384:319–320. [Google Scholar]
- Munakata A, et al. The involvement of sex steroid hormones in downstream and upstream migratory behavior of masu salmon. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2001;129:661–669. doi: 10.1016/s1096-4959(01)00365-7. [DOI] [PubMed] [Google Scholar]
- Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–9. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- Pearlstein JH, et al. Early discrimination of Atlantic salmon smolt age: Time course of the relative effectiveness of body size and shape. Trans Am Fish Soc. 2007;136:1622–1632. [Google Scholar]
- Philp AR, et al. A novel rod-like opsin isolated from the extra-retinal photoreceptors of teleost fish. FEBS Lett. 2000;468:181–8. doi: 10.1016/s0014-5793(00)01217-5. [DOI] [PubMed] [Google Scholar]
- Pierce LX, et al. Novel functions for Period 3 and Exo-rhodopsin in rhythmic transcription and melatonin biosynthesis within the zebrafish pineal organ. Brain Res. 2008;1223:11–24. doi: 10.1016/j.brainres.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polovina JJ, et al. Forage and migration habitat of loggerhead (Caretta caretta) and olive ridley (Lepidochelys olivacea) sea turtles in the central North Pacific Ocean. Fisheries Oceanography. 2004;13:36–51. [Google Scholar]
- Prunet P, et al. Smoltification and seawater adaptation in Atlantic salmon (Salmo salar): Plasma prolactin, growth hormone, and thyroid hormones. General and Comparative Endocrinology. 1989;74:355–364. doi: 10.1016/s0016-6480(89)80031-0. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing. Vienna, Austria: 2004. URL http://www.R-project.org. [Google Scholar]
- Raffin-Sanson M, et al. Proopiomelanocortin, a polypeptide precursor with multiple functions: from physiology to pathological conditions. Eur J Endocrinol. 2003;149:79–90. doi: 10.1530/eje.0.1490079. [DOI] [PubMed] [Google Scholar]
- Reinius B, et al. An evolutionarily conserved sexual signature in the primate brain. PLoS Genet. 2008;4:e1000100. doi: 10.1371/journal.pgen.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S, et al. Fish & Chips: Functional Genomics of Social Plasticity in an African Cichlid Fish. Journal of Experimental Biology. 2008;211 doi: 10.1242/jeb.018242. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rise ML, et al. Development and application of a salmonid EST database and cDNA microarray: data mining and interspecific hybridization characteristics. Genome Res. 2004;14:478–90. doi: 10.1101/gr.1687304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M, Naranjo J. Thyroid hormones, learning and memory. Genes Brain Behav. 2007;6:40–44. doi: 10.1111/j.1601-183X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- Roff DA. Contributions of genomics to life-history theory. Nat Rev Genet. 2007;8:116–125. doi: 10.1038/nrg2040. [DOI] [PubMed] [Google Scholar]
- Rowe DK, Thorpe JE. Differences in growth between maturing and non-maturing male Atlantic salmon, Salmo salar L., parr. J Fish Biol. 1990;36:643–658. [Google Scholar]
- Saunders RL, et al. Precocious sexual maturation and smoltification in male Atlantic salmon (Salmo salar) Aquaculture. 1982;28:211–229. [Google Scholar]
- Saunders RL, et al. Effects of photoperiod on juvenile growth and smolting of Atlantic salmon and subsequent survival and growth in sea cages. Aquaculture. 1985;45:55–66. [Google Scholar]
- Schmidt R. Cell-adhesion molecules in memory formation. Behav Brain Res. 1995;66:65–72. doi: 10.1016/0166-4328(94)00126-z. [DOI] [PubMed] [Google Scholar]
- Segal E, et al. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–8. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Semlitsch RD. Differentiating Migration and Dispersal Processes for Pond-Breeding Amphibians. Journal of Wildlife Management. 2008;72:260–267. [Google Scholar]
- Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, et al. Statistical issues in microarray data analysis. In: Brownstein MJ, Khodursky AB, editors. Functional Genomics: Methods and Protocols. Humana Press; Totowa: 2003. pp. 111–136. [Google Scholar]
- Specker J, et al. Parr–smolt transformation in Atlantic salmon: thyroid hormone deiodination in liver and brain and endocrine correlates of change in rheotactic behavior. Canadian Journal of Zoology. 2000;78:696–705. [Google Scholar]
- St-Cyr S, Aubin-Horth N. Integrative and genomics approaches to uncover the mechanistic bases of fish behavior and its diversity. Comp Biochem Physiol A Mol Integr Physiol. 2009;152:9–21. doi: 10.1016/j.cbpa.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Stearns SC. The Evolution of Life Histories. Oxford University Press; Oxford: 1992. [Google Scholar]
- Stearns SC, Magwene P. The naturalist in a world of genomics. Amer Nat. 2003;161:171–180. doi: 10.1086/367983. [DOI] [PubMed] [Google Scholar]
- Taborsky M. The evolution of bourgeois, parasitic, and cooperative reproductive behaviors in fishes. J Hered. 2001;92:100–10. doi: 10.1093/jhered/92.2.100. [DOI] [PubMed] [Google Scholar]
- Theriault V, Dodson JJ. Body size and the adoption of a migratory tactic in brook charr. Journal of Fish Biology. 2003;63:1144–1159. [Google Scholar]
- Townsend JP. Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics. 2003;4:41. doi: 10.1186/1471-2164-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspoor E, et al. The Atlantic Salmon: Genetics, Conservation and Management. Blackwell; Oxford: 2007. [Google Scholar]
- Waring CP, et al. Milt and endocrine responses of mature male Atlantic salmon (Salmo salar L.) parr to water-borne testosterone, 17,20 beta-dihydroxy-4-pregnen-3-one 20-sulfate, and the urines from adult female and male salmon. Gen Comp Endocrinol. 1996;103:142–9. doi: 10.1006/gcen.1996.0105. [DOI] [PubMed] [Google Scholar]
- West-Eberhard M-J. Developmental Plasticity and Evolution. Oxford University press; Oxford: 2003. [Google Scholar]
- Whitfield CW, et al. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–9. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Wiesinger H. Arginine metabolism and the synthesis of nitric oxide in the nervous system. Progress in Neurobiology. 2001;64:365–391. doi: 10.1016/s0301-0082(00)00056-3. [DOI] [PubMed] [Google Scholar]
- Zydlewski G, et al. Evidence for cumulative temperature as an initiating and terminating factor in downstream migratory behavior of Atlantic salmon (Salmo salar) smolts. Can J Fish Aquat Sci. 2005;62:68–78. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes forming the unique gene-expression signatures found for mature sneaker males, immature males, early migrants and late migrants and the 28 genes that are found in more than one gene-expression signature. Physical location on microarray, Genbank and/or GRASP ID, TIGR contig ID, GRASP and TIGR annotation and GO terms, fold change coefficient and FDR corrected p-value in each comparison from LIMMA and whether the gene is up-regulated in mature sneaker males, immature males, early migrants and/or late migrants are presented.
Experimental hybridization design used in the two separate microarray experiments. A) Four sexually mature sneaker males (S) were compared to four immature males (I) and four immature females (F) of the same age. Individuals that are compared are linked by an arrow. The arrowhead indicates labelling with Cy5 dye and the arrow tail indicates that the sample was labelled with Cy3 fluorescent dye. Only results comparing males of the two types are used in the present meta-analysis, original results were reported in Aubin-Horth et al (2005). B) Three early migrants (1 male and 2 females; EM1, EF1, EF2) were compared to three late migrants (1 male and 2 females; LM1, LF1, LF2).