Abstract
Influenza vaccines used today are strain specific and need to be adapted every year to try and match the antigenicity of the virus strains that are predicted to cause the next epidemic. The strain specificity of the next pandemic is unpredictable. An attractive alternative approach would be to use a vaccine that matches multiple influenza virus strains, including multiple subtypes. In this review, we focus on the development and clinical potential of a vaccine that is based on the conserved ectodomain of matrix protein 2 (M2) of influenza A virus. Since 1999, a number of studies have demonstrated protection against influenza A virus challenge in animal models using chemical or genetic M2 external domain (M2e) fusion constructs. More recently, Phase I clinical studies have been conducted with M2e vaccine candidates, demonstrating their safety and immunogenicity in humans. Ultimately, and possibly in the near future, efficacy studies in humans should provide proof that this novel vaccine concept can mitigate epidemic and even pandemic influenza A virus infections.
Keywords: clinical trial, influenza A, M2e, matrix protein 2, pandemic, universal vaccine
Influenza is a vaccine-preventable disease. Currently, licensed seasonal influenza vaccines provide considerable benefit to children, healthy young adults, the elderly and people with a condition that increases the risk of complications following influenza virus infection [1,2]. Influenza vaccination is especially recommended for the elderly and certain risk groups, including immunocompromised individuals and people with a cardiovascular or pulmonary disorder. Human influenza is caused by influenza A and B viruses, which are classified as Orthomyxoviridae, a virus family in which the members are characterized by the presence of an envelope and a segmented negative-stranded RNA genome. Hemagglutinin (HA) and neuraminidase (NA), the two membrane-anchored glycoproteins of influenza A and B virions, constitute the receptor-binding and receptor-destroying viral molecules, respectively, as well as the major antigenic determinants (Figure 1). The presence of serum antibodies that inhibit the hemagglutination activity of influenza A and B virus particles in an in vitro assay correlate well with protection against seasonal influenza [3]. Nearly all currently licensed vaccines are derived from virus propagated on chick embryos, a production system that has been in place for over 60 years. Inactivated whole virion can be used as a vaccine, but most manufacturers produce a split-virus or subunit vaccine, which have a weaker reactogenicity than whole-virion vaccines [4]. Live-attenuated influenza vaccines are also used in Russia and have been licensed by the US FDA since 2003, initially for use in healthy individuals aged 5-49 years, and with a recent extension to children of 2-5 years of age [5,6]. Although there is some evidence that live-attenuated influenza vaccines can induce slightly cross-reactive anti-HA responses in children [7], the principle aim of both available human influenza vaccine types is to induce an antibody response that best matches the HA of the virus strain likely to cause the next epidemic.
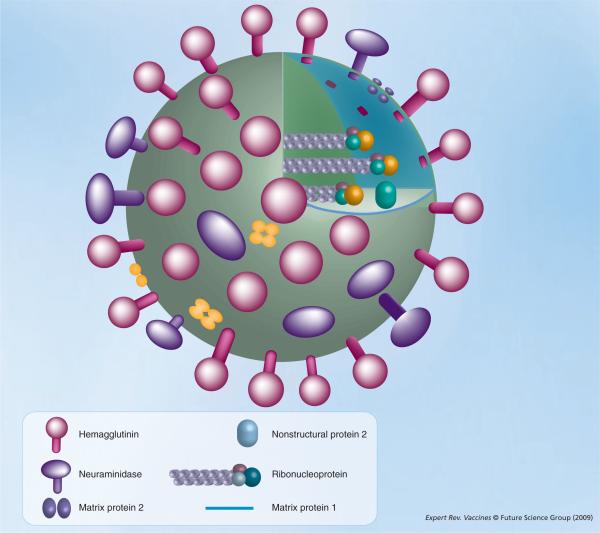
Figure 1. Influenza A virion.
There are three proteins embedded in the influenza A virion envelope: hemagglutinin, neuraminidase and matrix protein 2. Below the envelope is the matrix, composed of matrix protein 1, which surrounds the segmented negative-stranded RNA genome. The genome consists of eight segments that are packed into ribonucleoprotein complexes, with the three polymerase subunits (turquoise, teal and orange) assembled at the genomic RNA termini. Nonstructural protein 2 is also incorporated in the virion.
Is there room for improving current influenza vaccines, especially by overcoming the rigorous strain specificity? If so, where and how can a vaccine based on the third influenza A virus membrane protein, matrix protein 2 (M2), the focus of this review, offer an alternative? Most efforts exploiting M2 for vaccine development are directed towards M2e, the external domain of the M2 protein, because of its accessibility to antibodies.
A first answer to these questions concerns vaccine-production capacity. Today, the global capacity of seasonal influenza vaccines for human use is approximately 400 million doses, or less than 7% of the world population [8]. Two main factors that limit production capacity are the high turnover rate of strain-specific vaccines and the short production time imposed by the seasonality of influenza epidemics in temperate climate zones. Production starts when the WHO proposes the vaccine strain composition for the Northern hemisphere in February, and the vaccine is delivered to the market starting in October. By focusing current influenza vaccines on a protective immune response directed against HA, their composition has to keep pace with the continuously changing immunogenicity of the HA expressed by circulating influenza A and B viruses. This gradual antigenic change, termed antigenic `drift', results from the selection of escape virus strains with altered amino acid residues in two or more of the five major antigenic regions of HA [9]. The selection of these escape mutants is driven by the prevailing herd immunity in the human host, directed against the major antigenic sites of HA. Notably, antigenic drift also occurs in NA [10], but current influenza vaccines are only standardized on the basis of their HA immunogenicity. Based on global surveillance of influenza viruses by reference laboratories, a WHO expert panel predicts, twice a year, which virus strains will most likely cause the next epidemic. Since 1977, two A subtype strains (one H3N2 [HA subtype 3 and NA sub-type 2] and one H1N1) and one B strain have been circulating in the human population. Therefore, each recommendation for the vaccine composition has to be made for the three virus types. For example, for the winter season of 2008-2009, all three vaccine strains are different from the strains that were represented in the 2007-2008 vaccine [101]. Although the strains selected for inclusion in seasonal influenza vaccines usually agree well with the prevailing epidemic strain in the ensuing winter, occasionally the antigenic match is poor, resulting in partial or complete vaccine failure. For example, an antigenic mismatch occurred during the winter season of 2005-2006 in the Northern hemisphere, where both circulating H3N2 and B viruses poorly matched the vaccine [11]. Focusing on a conserved protective influenza viral antigen, such as M2, would not require annual revision of the influenza A vaccine content, and would even allow stockpiling of the vaccine. Since there are no time constraints, not one but two or more immunizations can be administered, thus considerably consolidating the immune response.
A future pandemic influenza outbreak is feared, although no one can predict the subtype, let alone the strain, that will cause it. Current pandemic vaccine preparedness efforts largely focus on the production and clinical evaluation of vaccines based on the HA antigen derived from strains with presumed pandemic potential. These strains include avian H5N1 viruses that have sporadically infected humans in Asia, Africa and the Middle East, with a more than 60% fatality rate [12]. Since the first human H5N1 cases were reported, the pandemic alertness has increased considerably and this has definitely promoted research on and clinical evaluation of new vaccine concepts for influenza [13]. However, an inactivated H5N1 vaccine that is produced and available now will most likely have a negligible impact in controlling disease following a pandemic outbreak caused by a different influenza subtype, such as H2N2 or H7N7. Even if the next pandemic is caused by H5N1, it is unlikely that a currently available H5N1 vaccine would antigenically match the H5N1 strain sweeping the globe, since the virus would drift and diversify at an alarming rate [14]. Nevertheless, the use of an oil-in-water emulsion-based adjuvant system together with a recombinant H5N1 split-virion vaccine can induce cross-clade humoral immunity, determined in an in vitro assay, in humans [15]. On the other hand, the typical sequence of avian M2e differs from that of human M2e by four amino acid residues (Figure 2). This sequence variation occurs at positions 11, 14, 16 and 20 of M2e [16].
Figure 2. The M2e consensus sequences of human (top) and avian (bottom) influenza A viruses.
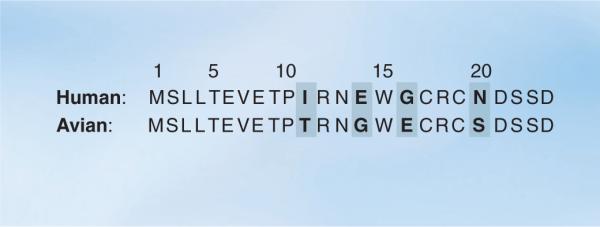
Amino acid differences between the two sequences are highlighted in bold. The consensus human and avian M2e sequences were obtained by aligning 3913 human and 2994 avian M2 amino acid sequences, respectively, as present in the Influenza Virus Resource database (National Center for Biotechnology Information) on 3 February 2009, using MUSCLE version 3.6 [76].
Immunity against M2e in response to natural infection or induced by conventional influenza vaccines is weak to non-existent [17-22]. Therefore, the concept of using M2(e) as a vaccine is radically different from the conventional approach, which aims to induce an adaptive immune response resembling that of a host that has controlled the infection, which correlates with the presence of humoral anti-HA and anti-NA immunity. Influenza viruses have successfully evolved to cope with humoral anti-HA and anti-NA immunity by drifting away from the prevalent herd immunity and occasionally even by completely shifting HA (and NA) subtype. By contrast, an immunoprotective M2 vaccine, even if it were used on a global scale, would confront the virus with a new hurdle that cannot be overcome by the known shift or drift mechanisms. M2e-escape virus mutants have been isolated from challenged severe combined immunodeficiency mice that had been injected with passively administered anti-M2e monoclonal antibodies [23]. These escape viruses had an altered M2e mutations at a single amino acid position (P10). However, even after 11 consecutive passages in immunocompetent mice that had been actively vaccinated with M2e, no escape viruses could be isolated, suggesting that the likelihood of emergence of fit M2e escape viruses is low [24].
Influenza control by anti-M2 immunity
The M2 protein was first described by Lamb and Choppin [25] and characterized in more detail in 1985 by the same group [26]. This tetrameric membrane protein is translated from a splice variant of the mRNA coding for the matrix protein M1. M2 is a so-called `viroporin' with pH-controlled, passive proton-selective channel activity [27,28]. Following endocytosis, this activity of M2 promotes acidification of the virion interior, a process that unties the interactions between the viral ribonucleoprotein complexes and the matrix and allows the entry of the viral core constituents into the cytosol. Following genome replication and progeny virus maturation, the proton-transfer function of M2 slightly increases the pH of the Golgi lumen, thereby, preventing HA from adopting its low pH-induced membrane fusion conformation, at least in the case of particular H5 and H7 viruses [29-31]. Finally, M2e is involved in the incorporation of M2 in newly formed virions [32,33], and the 54-amino acid residue cytoplasmic tail of M2 is needed for proper particle formation, presumably by interacting with M1 [34,35].
The first evidence that anti-M2 immunity has antiviral activity was demonstrated in vitro using a mouse monoclonal antibody directed against M2e [36]. This antibody, 14C2, was isolated following immunization of mice with M2 protein derived from cells infected with influenza A virus; it was extracted from a denaturing polyacrylamide gel and emulsified with Freund's adjuvant. This monoclonal antibody could reduce the plaque size of many but not all influenza A virus strains expressing M2 that were recognized by 14C2. Passive administration of this 14C2 monoclonal antibody also reduces virus titers in the lungs of mice infected with influenza A virus [37]. Recently, human anti-M2e monoclonal antibodies were isolated and evaluated prophylactically as well as therapeutically in adoptive transfer experiments in mice for protection against influenza A virus challenge [38]. Interestingly, intraperitoneal injection of anti-M2e antibodies in cotton rats protects these animals from influenza-induced tachypnea (increased respiratory rate), suggesting that these antibodies could indeed provide clinical benefit against influenza when used for passive immunoprophylaxis [39].
While antibody-based therapies are an attractive strategy for treating severe human influenza disease, for example following zoonotic infection by highly pathogenic avian influenza strains [12,40], inducing adaptive anti-M2 immunity with a prophylactic vaccine would be far more cost effective and practical for controlling an influenza epidemic or pandemic.
Following the original report of a M2e-based influenza A vaccine [41], several groups have reported that immune responses induced by recombinant M2-based vaccines can protect against influenza virus challenge. Most of these studies focused on M2e fusion constructs, using mouse models and occasionally ferrets. Protection was measured by reduced host morbidity, host mortality and virus replication following challenge with human or avian influenza A viruses (Table 1). In 1995, Slepushkin and coworkers reported vaccination with complete M2 protein as antigen [42]. These authors partially purified an M2-containing membrane fraction derived from a recombinant insect-cell expression system. They demonstrated that vaccination reduced morbidity and virus replication in challenged mice. Although this M2 vaccine preparation induced M2e-specific antibodies, including M2e-directed antibodies, attempts to transfer immunity by serum to naive mice were not successful. This result is at odds with the passive immunization studies using anti-M2e monoclonal antibodies reported previously [36]. More recent vaccination studies using full-length M2-protein where either based on gene vaccination or viral vectors encoding M2 and, thereby, not only circumvent the difficult task of purifying membrane-anchored M2 for use as a vaccine antigen, but also induce cellular immune responses [16,43,44]. M2 contains cytotoxic T-cell epitopes against which (cytotoxic) cellular immune responses can be elicited, for example by a DNA vaccine alone or by priming with a DNA vaccine and boosting with an adenoviral vector coding for the same influenza genes; these cells contribute to protection against experimental infection in mice and ferrets [16,43-45]. However, the clinical impact of M2-specific cellular responses during a natural infection is probably negligible in view of the immunodominant cellular responses that are directed mainly against nucleoprotein [46].
Table 1.
Overview of M2-based vaccine studies.
| Antigen | Vaccine type | Carrier | Type of antigen | Ref. |
|---|---|---|---|---|
| M2 | Protein | Human | [42] | |
| DNA vaccine | Human, avian | [16,43,44,71] | ||
| Adenoviral vector | Human, avian | [16,44] | ||
| M2e | Peptide | [47] | ||
| M2e | Virus-like particle | HBVc | [41,52,54,69,72,73] | |
| PaMV | [49] | |||
| HPV | [50] | |||
| Qβ | [48] | |||
| M2e | Protein | -/GST | Human | [55,57] |
| CTA1-DD | Human | [60] | ||
| KLH | Human, avian | [16,54] | ||
| tGCN4 | Human | [65] | ||
| M2e | Synthetic | Synthetic multiantigenic carrier | Human | [22] |
| Liposome | Human, avian | [53] | ||
| M2e | DNA | NP | Human, avian | [74,75] |
HBVc: Hepatitis B virus core; HPV: Human papillomavirus; KLH: Keyhole limpet hemocyanin; M2: Matrix protein 2; M2e: M2 external domain; PaMV: Petunia asteroid mosaic tombusvirus; tGCN4: Transgenic general control nondepressible-4.
M2e, on the other hand, is conserved across influenza A viruses and is soluble; however, its drawback is that it is only 23 amino acids long. Although one study reported protection of mice against lethal challenge after immunization with adjuvanted M2e peptide [47], most efforts to produce an M2e vaccine have made use of chemical or genetic M2e fusion constructs to a variety of carriers (Table 1). Our group was the first to report protection against influenza A virus challenge after vaccination with an M2e fusion construct [41]. Protection was demonstrated against challenge with H1N1 and H3N2 viruses, it could be conferred by intranasal immunization, and it was dependent on anti-M2e IgG antibodies, which were induced at high levels using recombinant M2e-hepatitis B virus core (HBVc) virus like-particles (VLPs). Other M2e VLPs, including M2e displayed on papaya mosaic virus, human papillomavirus and phage Qβ-derived VLPs, have since been used successfully to protect mice against influenza A virus challenge [48-50]. VLPs as antigen carriers allow the presentation of foreign epitopes in an ordered array, which promotes a strong immune response [51]. In addition, recombinant VLPs may be tuned to increase their stability and the immunogenicity of the presented antigen of interest [52].
Other M2e fusion constructs that were shown to provide protection from influenza A virus challenge in mouse models include M2e-linkage to keyhole limpet hemocyanin, Neisseria meningitides outer membrane complex (OMPC), bovine serum albumin, gluthation S-transferase, a synthetic multiple antigen peptide, flagellin and CTA1-DD [16,21,22,53-58]. CTA1-DD is a powerful and safe recombinant mucosal adjuvant that targets the enzymatically active A1 part of cholera toxin to immunoglobulin-expressing cells, such as B cells [59]. Some M2e fusion constructs reportedly induce a M2e-specific CD4+ T-cell response [58,60], which may contribute to protection by stimulating antibody class switching and affinity maturation of M2e-specific antibodies.
Mode of action & clinical application of M2e vaccines
Mice are excellent models for identifying the mechanisms of action of vaccines because many protocols and mutant strains are available for studying in vivo vaccine efficacy when specific immune functions are altered. Such in vivo studies are crucial for defining genuine correlates of protection. For example, a study by Huber et al. revealed that anti-influenza humoral immunity induced by an influenza subunit vaccine was dependent on opsonophagocytosis of influenza virions by macrophages and was not the result of virus neutralization by vaccine-induced antibodies [61]. Protection induced by M2e-directed vaccines depends largely on anti-M2e antibodies, but contradictory findings have been reported on the role of natural killer (NK)/NK T-cell host cells versus complement [16,38,62]. It was recently reported that FcγRIII, an IgG-binding receptor present on many immune cells, such as NK cells, macrophages and neutrophils, plays an important role in protection against a lethal respiratory tract influenza challenge by anti-M2e human monoclonal antibodies [38]. Most likely, the effector mechanism involves the binding of anti-M2e IgG to infected cells, which abundantly express M2 at their cell surface [26], followed by elimination of the infected cell by killing (e.g., complement mediated or NK cell mediated) or phagocytosis (e.g., by macrophages) (Figure 3). Eliminating the infected cells suppresses new virus production and spreading. However, the route of vaccine administration and the mode of challenge that is used to evaluate protection play important roles in determining correlates of protection. Mozdzanowska and coworkers used a mouse challenge model in which influenza A virus infection is initiated in the nasal tissues and then proceeds further into the lower respiratory tract. They reported that serum anti-M2e IgG levels following immunization with M2e synthetic conjugate vaccine did not necessarily correlate with protection, defined as the decrease in virus titers in the respiratory tract compared with untreated, infected controls [63]. However, following subcutaneous immunization with their M2e vaccine, protection did correlate with the level of a subset (~15%) of the anti-M2e serum IgGs that were specific for native, tetrameric M2. By contrast, intranasal immunization of mice with the same construct provided better protection despite the induction of lower levels of serum IgG specific for native M2. Most likely, local airway-associated immunity, comprising mucosal anti-M2e IgA and activated M2e-specific B and T cells, was induced more efficiently after intranasal vaccination compared with parenteral immunization. Furthermore, mucosal M2e-specific immunity in the upper respiratory tract presumably operates more effectively against nasal challenge. Clearly, further research is needed to identify the mechanism of action of anti-M2e immunity, since this information will be essential to identify true correlates of protection and to set up assays for measuring them in vitro. Such assays will be very helpful for further clinical development of M2e vaccines and will ultimately provide more reliable correlates of protection.
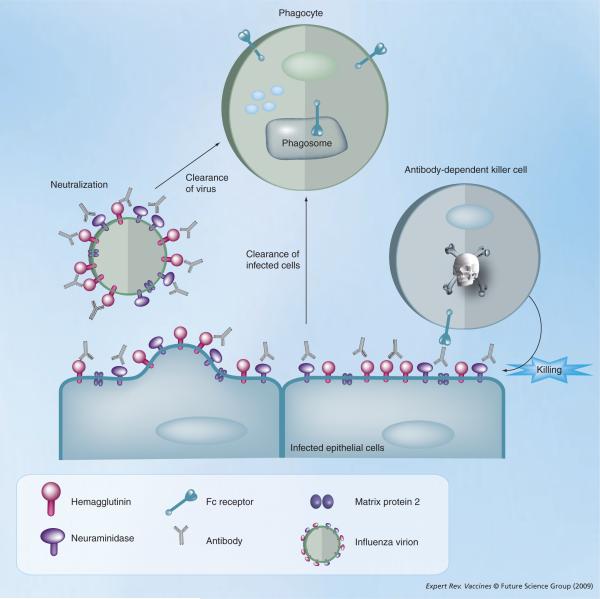
Figure 3. Proposed in vivo immune mechanism of action of anti-M2e and anti-HA antibodies.
Infected epithelial cells display hemagglutinin, neuraminidase and matrix protein 2 (M2) on the cell membrane in the course of infection. Anti-M2 external domain (M2e) antibodies can bind to M2 present on infected cells and the resulting immune complexes can be recognized by Fc-receptor bearing effecter cells, such as natural killer cells, leading to killing of the infected cell by antibody-dependent cellular cytotoxicity (right). Alternatively, anti-M2e opsonized infected cells can be taken up by phagocytes in a Fc-receptor-dependent way. The canonical mode of action of anti-hemagglutinin (HA) antibodies is through the direct neutralization of virions (left). In addition, anti-HA antibodies can promote uptake of virions by phagocytes and bind to HA on infected cells followed by removal of infected cells by phagocytes.
There has been a time gap of nearly 10 years between the publication of the initial proof-of-concept findings [41] and the first Phase I clinical studies, which were initiated in 2007. What has happened during that time? First, to take a novel vaccine from the bench into the clinic takes 10-12 years [64]. Second, the entirely new concept of raising immune protection by using a viral peptide (M2e) that is naturally not or hardly immunogenic differs fundamentally from the conventional vaccine approach, where the aim of the vaccine is to mimic (part of) the immunity that is present in the blood of convalescent patients. In the mean time, different independent studies confirmed that the M2e vaccine approach is indeed protective in animal models involving influenza A virus challenge. Third, anti-M2e antibodies do not neutralize influenza virus in vitro, whereas, anti-HA antibodies raised by conventional vaccines do. As mentioned previously, the level of these hemagglutination-inhibitory antibodies in serum correlates with protection against disease caused by an antigenically matching influenza virus infection; although, whether the in vitro test truly reflects the in vivo mechanism of protection remains an open question. Correlates of protection are not yet known for a M2e-based vaccine and defining these correlates in humans requires, in the first place, efficacy studies that demonstrate clinical benefit by M2(e) vaccines.
Four companies have reported preliminary results from Phase I clinical studies with M2e-based vaccines: Acambis Inc. (Cambridge, UK), Cytos Biotechnology (Schlieren, Switzerland), Merck & Co Inc. (NJ, USA), and VaxInnate Corp. (NJ, USA) [102]. Two genetic (M2e-HBVc and M2e-flagellin) and two chemical (M2e-Qβ phage and M2e-OMPC) M2e fusions were evaluated for safety, immunogenicity and tolerance with or without adjuvant. Although the results of these trials have not been reported yet in the scientific literature, it appears that humans have negligible anti-M2e serum titers before vaccination and that the tested M2e vaccine formulations are immunogenic and well tolerated.
Summary & conclusion
During the last three decades, many approaches have been pursued to develop a vaccine that can induce cross-protective immunity against influenza A. Such a vaccine could overcome the recurrent uncertainty regarding the antigenic match between the proposed vaccine seed strains and the subsequent epidemic strain(s) that cause(s) disease 6-9 months later. In addition, if a cross-protective vaccine could also induce hetero subtypic immunity, it could mitigate a pandemic influenza outbreak. Based on all studies reported to date, M2e remains a very promising candidate for a universal influenza A vaccine. M2e is highly conserved across all human influenza A viruses, it is an extracellular target and, therefore, accessible to specific antibodies, and M2e-based vaccines can be easily produced in efficient recombinant prokaryotic-expression systems that require minimal containment. M2e fusions to VLPs are intrinsically very immunogenic and generally very stable. For these reasons, they are promising candidates for further clinical evaluation. Mucosal delivery may be the preferred way for future clinical evaluation, especially when the goal is to develop a pandemic vaccine. Furthermore, efforts to design M2e vaccine antigens for induction of antibodies that preferentially react with the native, tetrameric membrane-anchored form of M2 may improve efficacy [65]. The mechanism of action of M2e-specific immunity is still poorly understood. Anti-M2e antibodies play an essential role, presumably in concert with NK cells and phagocytes. However, the route of administration and the vaccine formulation also contribute to the type and site of anti-M2e immune induction, which, in turn, determines the mechanism(s) of protection that becomes operational upon challenge. Ultimately, the effectiveness of an M2e-vaccine has to be determined by clinical trials in humans, an endeavor that will require substantial resources but may answer the key question: is it possible to make an effective universal influenza vaccine for humans?
Expert commentary
The M2e vaccine approach has now entered the early clinical test phase. However, in the wake of efficacy studies in human volunteers, two critical questions have been raised regarding the prospect of introducing M2e vaccination into the general population. There may be a concern that introducing immune pressure on M2e will promote the evolution of antigenic escape viruses. Such escape viruses were actually reported in a severe combined immunodeficiency mouse model in which a panel of anti-M2e monoclonal antibodies was used to suppress virus replication [23]. However, M2e mutations were confined to a single residue (P10) and were not detected following multiple passages of virus under anti-M2e immune pressure in normal, immunocompetent mice [24]. The possibility of the emergence of M2e-escape mutant viruses after extensive vaccination with an M2e vaccine can not be excluded. However, there is no experimental evidence to support this scenario and, furthermore, unlike the case of emergent amantadine-resistant strains, M2e vaccination induces a polyclonal anti-M2e response, meaning it would require two or more amino acid residue changes to escape the immune protection. In addition, such M2e mutant viruses would have to gain a selective advantage over wild-type M2e-coding viruses in order to become dominant and, unless M2e vaccination covers almost 100% of the population, such a vaccine-induced selection environment remains unrealistic. Nevertheless, monitoring the genetic diversity of influenza virus should be continued regardless of the use of M2e vaccines.
Should a M2e vaccine be used as a stand-alone vaccine or should it be added to seasonal vaccines? In our opinion, clinical effectiveness of this universal influenza A vaccine candidate should be tested using clinical efficacy as a readout, which may require approximately 2000 volunteers [66]. Based on early in vitro studies and on the results of experiments performed in mouse models, M2e immunity is infection permissive. It is unknown how this will translate into clinical benefit in humans under natural challenge and transmission conditions. There are several, equally important considerations as to why the M2e vaccine should be developed as a stand alone. First, the M2e vaccine is produced in a highly efficient microbial expression system and, thus, can be made very cheaply. This advantage would be lost if it has to be admixed with an expensive conventional vaccine derived from virus growth. Furthermore, owing to supply and time constraints, only a single administration of seasonal vaccine is administered, while the M2e vaccine needs at least one booster immunization. Perhaps most important of all, especially with regard to pandemic preparedness, the M2e vaccine, unlike all commercially available vaccines, is not strain specific. Therefore, there is no time constraint. One does not have to wait until the strain characteristics of the forthcoming epidemic can be guessed with some degree of confidence. With respect to pandemic preparedness, it is estimated that the time delay between the certified emergence of a new pandemic strain, characterized by human transmissibility and possibly high pathogenicity, and availability of a vaccine seed strain of lower pathogenicity and high virus production yields in embryonated chicken eggs, is approximately 4-6 months. This should be compared with a recombinant M2e vaccine that can be made, distributed and administered long before any epidemic or pandemic is on the horizon.
Five-year view
What are the important milestones, apart from the necessity to continue clinical testing, that lie ahead and that could bring us closer to a universal human influenza vaccine? First, it would be valuable if a universal influenza B vaccine could be developed as well. Although only one subtype of this virus is known, its HA and NA are equally subject to antigenic drift and two distinct antigenic lineages are discerned [67]. The Merck group has reported on a cross-reactive universal influenza B vaccine that is based on the conserved fusion peptide of the viral HA [68]. A M2e-like approach, based on the conserved ectodomain of NB or BM2, might also provide protection against multiple influenza B strains, and so it should be explored further. The influenza A M2e and the conserved influenza B target epitope could be integrated into the same fusion construct or could be delivered as separate vaccine antigens.
Based on animal studies, vaccination with M2e vaccines by the intranasal route improves protection against influenza virus challenge [41,48,52,63,69]. This is likely due to stimulation of local immunity and may involve cellular responses as well, particularly when mucosal vaccination is performed in the presence of potent mucosal adjuvants [70]. A safe mucosal vaccine would also improve vaccine take and compliance because no needles are required. Most important, especially for mass-vaccination campaigns, is the elimination of accidental transmission of blood-borne diseases through contaminated needles. Finally, more attention will probably be paid to the design and testing of improved M2e vaccines that mimic tetrameric M2e as present on the surface of infected cells. Determining the structure of the M2 ectodomain would be an important step forward and may help the design of such tetrameric M2e vaccines.
Key issues.
It is unclear to what extent the efficacy studies of matrix protein 2 (M2) and M2 external domain (M2e) vaccines in animal models, usually involving a lethal challenge dose, can predict clinical outcome. Efficacy studies under natural transmission conditions and in other species will be necessary to evaluate this.
A stand-alone M2e vaccine should be evaluated further in humans, using clinical benefit as readout.
The performance of M2e-based vaccines can be modulated by different adjuvant formulations. This should be further explored in the clinical setting and/or in monkeys.
The intranasal route of M2e-vaccine administration improves protection in animal challenge models. Intranasal vaccination of humans should be explored further.
The mechanism of action of M2e vaccines requires further investigation. Protection following parenteral M2e-vaccine administration mainly induces anti-M2e antibodies that operate in concert with immune cells; these should be further characterized in vivo and in vitro.
Correlates of protection need to be developed in order to identify reliable parameters for predicting the success rate of vaccination programs.
It will be important to discriminate between anti-M2e antibodies that preferentially bind to monomeric M2e peptide versus tetrameric M2e as present in native M2. Do both contribute to protection?
Acknowledgments
Financial & competing interests disclosure Influenza vaccine research in the group of Xavier Saelens and Walter Fiers is supported by Acambis Inc. (now Sanofi Pasteur), NIH grant 1 R01 AI055632-01A1, Ghent University BOF B/05930/01 and IOF Stepstone IOF08/STEP/001 grants, and fellowships from FWO-Vlaanderen and IWT. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
No writing assistance was utilized in the production of this manuscript.
References
- 1.Jefferson TO, Rivetti D, Di Pietrantonj C, Rivetti A, Demicheli V. Vaccines for preventing influenza in healthy adults. Cochrane Database Syst. Rev. 2. 2007 doi: 10.1002/14651858.CD001269.pub3. CD001269. [DOI] [PubMed] [Google Scholar]
- 2.Nichol KL, Nordin JD, Nelson DB, Mullooly JP, Hak E. Effectiveness of influenz vaccine in the community-dwelling elderly. N. Engl. J. Med. 2007;357(14):1373, 1381. doi: 10.1056/NEJMoa070844. [DOI] [PubMed] [Google Scholar]
- 3.de Jong JC, Palache AM, Beyer WE, et al. Haemagglutination-inhibiting antibody to influenza virus. Dev. Biol. (Basel) 2003;115:63–73. [PubMed] [Google Scholar]
- 4.al-Mazrou A, Scheifele DW, Soong T, Bjornson G. Comparison of adverse reactions to whole-virion and split-virion influenza vaccines in hospital personnel. CMAJ. 1991;145(3):213–218. [PMC free article] [PubMed] [Google Scholar]
- 5.Belshe R, Lee MS, Walker RE, Stoddard J, Mendelman PM. Safety, immunogenicity and efficacy of intranasal, live attenuated influenza vaccine. Expert Rev. Vaccines. 2004;3(6):643–654. doi: 10.1586/14760584.3.6.643. [DOI] [PubMed] [Google Scholar]
- 6.Rudenko LG, Lonskaya NI, Klimov AI, Vasilieva RI, Ramirez A. Clinical and epidemiological evaluation of a live, cold-adapted influenza vaccine for 3-14-year-olds. Bull. WHO. 1996;74(1):77–74. [PMC free article] [PubMed] [Google Scholar]
- 7.Mendelman PM, Rappaport R, Cho I, et al. Live attenuated influenza vaccine induces cross-reactive antibody responses in children against an A/Fujian/411/2002-like H3N2 antigenic variant strain. Pediatr. Infect. Dis. J. 2004;23(11):1053–1055. doi: 10.1097/01.inf.0000143643.44463.b1. [DOI] [PubMed] [Google Scholar]
- 8.The MIV Study Group The macro-epidemiology of influenza vaccination in 56 countries, 1997-2003. Vaccine. 2005;23(44):5133–5143. doi: 10.1016/j.vaccine.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl Acad. Sci. USA. 2003;100(12):7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilbourne ED, Johansson BE, Grajower B. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc. Natl Acad. Sci. USA. 1990;87(2):786–790. doi: 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skowronski DM, Masaro C, Kwindt TL, et al. Estimating vaccine effectiveness against laboratory-confirmed influenza using a sentinel physician network: results from the 2005-2006 season of dual A and B vaccine mismatch in Canada. Vaccine. 2007;25(15):2842–2851. doi: 10.1016/j.vaccine.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, et al. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 2008;358(3):261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 13.Claas EC, Osterhaus AD, vanBeek R, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351(9101):472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Holmes EC. Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Biol. Evol. 2006;23(12):2336–2341. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- 15.Leroux-Roels I, Bernhard R, Gerard P, et al. Broad clade 2 cross-reactive immunity induced by an adjuvanted clade 1 rH5N1 pandemic influenza vaccine. PLoS ONE. 2008;3(2):e1665. doi: 10.1371/journal.pone.0001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tompkins SM, Zhao ZS, Lo CY, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg. Infect. Dis. 2007;13(3):426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Black RA, Rota PA, Gorodkova N, Klenk HD, Kendal AP. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J. Gen. Virol. 1993;74(Pt 1):143–146. doi: 10.1099/0022-1317-74-1-143. [DOI] [PubMed] [Google Scholar]
- 18.Drummond JE, Shaw EE, Antonello JM, et al. Design and optimization of a multiplex anti-influenza peptide immunoassay. J. Immunol. Methods. 2008;334(12):11–20. doi: 10.1016/j.jim.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Feng J, Zhang M, Mozdzanowska K, et al. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006;3:102. doi: 10.1186/1743-422X-3-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitikoon P, Strait EL, Thacker EL. The antibody responses to swine influenza virus (SIV) recombinant matrix 1 (rM1), matrix 2 (M2), and hemagglutinin (HA) proteins in pigs with different SIV exposure. Vet. Microbiol. 2008;126(13):51–62. doi: 10.1016/j.vetmic.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Li H, Chen YH. N-terminus of M2 protein could induce antibodies with inhibitory activity against influenza virus replication. FEMS Immunol. Med. Microbiol. 2003;35(2):141–146. doi: 10.1016/S0928-8244(03)00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mozdzanowska K, Feng J, Eid M, et al. Induction of influenza type A virus-specific resistance by immunization of mice with a synthetic multiple antigenic peptide vaccine that contains ectodomains of matrix protein 2. Vaccine. 2003;21(1920):2616–2626. doi: 10.1016/s0264-410x(03)00040-9. [DOI] [PubMed] [Google Scholar]
- 23.Zharikova D, Mozdzanowska K, Feng J, Zhang M, Gerhard W. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. J. Virol. 2005;79(11):6644–6654. doi: 10.1128/JVI.79.11.6644-6654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerg. Infect. Dis. 2006;12(4):569–574. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb RA, Choppin PW. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology. 1981;112(2):729–737. doi: 10.1016/0042-6822(81)90317-2. [DOI] [PubMed] [Google Scholar]
- 26.Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40(3):627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 27.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451(7178):591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stouffer AL, Acharya R, Salom D, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451(7178):596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciampor F, Thompson CA, Grambas S, Hay AJ. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22(3):247–258. doi: 10.1016/0168-1702(92)90056-f. [DOI] [PubMed] [Google Scholar]
- 30.Sakaguchi T, Leser GP, Lamb RA. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell. Biol. 1996;133(4):733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi K, Lamb RA. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 1994;68(2):911–919. doi: 10.1128/jvi.68.2.911-919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwatsuki-Horimoto K, Horimoto T, Noda T, et al. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 2006;80(11):5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 2005;79(6):3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 2008;82(20):10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 2006;80(16):8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988;62(8):2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treanor JJ, Tierney EL, Zebedee SL, Lamb RA, Murphy BR. Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J. Virol. 1990;64(3):1375–1377. doi: 10.1128/jvi.64.3.1375-1377.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang R, Song A, Levin J, et al. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res. 2008;80(2):168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Straight TM, Ottolini MG, Prince GA, Eichelberger MC. Antibody contributes to heterosubtypic protection against influenza A-induced tachypnea in cotton rats. Virol. J. 2008;5:44. doi: 10.1186/1743-422X-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N. Engl. J. Med. 2007;357(14):1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- 41.Neirynck S, Deroo T, Saelens X, et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999;5(10):1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 42.Slepushkin VA, Katz JM, Black RA, et al. Protection of mice against influenza A virus challenge by vaccination with baculovirus-expressed M2 protein. Vaccine. 1995;13(15):1399–1402. doi: 10.1016/0264-410x(95)92777-y. [DOI] [PubMed] [Google Scholar]
- 43.Lalor PA, Webby RJ, Morrow J, et al. Plasmid DNA-based vaccines protect mice and ferrets against lethal challenge with A/Vietnam/1203/04 (H5N1) influenza virus. J. Infect. Dis. 2008;197(12):1643–1652. doi: 10.1086/588431. [DOI] [PubMed] [Google Scholar]
- 44.Lo CY, Wu Z, Misplon JA, et al. Comparison of vaccines for induction of heterosubtypic immunity to influenza A virus: cold-adapted vaccine versus DNA prime-adenovirus boost strategies. Vaccine. 2008;26(17):2062–2072. doi: 10.1016/j.vaccine.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 45.Jameson J, Cruz J, Ennis FA. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 1998;72(11):8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rimmelzwaan GF, Boon AC, Voeten JT, et al. Sequence variation in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. Virus Res. 2004;103(12):97–100. doi: 10.1016/j.virusres.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Wu F, Huang JH, Yuan XY, Huang WS, Chen YH. Characterization of immunity induced by M2e of influenza virus. Vaccine. 2007;25(52):8868–8873. doi: 10.1016/j.vaccine.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 48.Bessa J, Schmitz N, Hinton HJ, et al. Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design. Eur. J. Immunol. 2008;38(1):114–126. doi: 10.1002/eji.200636959. [DOI] [PubMed] [Google Scholar]
- 49.Denis J, Acosta-Ramirez E, Zhao Y, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26(2728):3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 50.Ionescu RM, Przysiecki CT, Liang X, et al. Pharmaceutical and immunological evaluation of human papillomavirus viruslike particle as an antigen carrier. J. Pharm. Sci. 2006;95(1):70–79. doi: 10.1002/jps.20493. [DOI] [PubMed] [Google Scholar]
- 51.Bachmann MF, Rohrer UH, Kundig TM, et al. The influence of antigen organization on B cell responsiveness. Science. 1993;262(5138):1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 52.De Filette M, Min Jou W, Birkett A, et al. Universal influenza A vaccine: optimization of M2-based constructs. Virology. 2005;337(1):149–161. doi: 10.1016/j.virol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Ernst WA, Kim HJ, Tumpey TM, et al. Protection against H1, H5, H6 and H9 influenza A infection with liposomal matrix 2 epitope vaccines. Vaccine. 2006;24(24):5158–5168. doi: 10.1016/j.vaccine.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Fan J, Liang X, Horton MS, et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine. 2004;22(2324):2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 55.Frace AM, Klimov AI, Rowe T, Black RA, Katz JM. Modified M2 proteins produce heterotypic immunity against influenza A virus. Vaccine. 1999;17(18):2237–2244. doi: 10.1016/s0264-410x(99)00005-5. [DOI] [PubMed] [Google Scholar]
- 56.Huleatt JW, Nakaar V, Desai P, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008;26(2):201–214. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 57.Liu W, Peng Z, Liu Z, et al. High epitope density in a single recombinant protein molecule of the extracellular domain of influenza A virus M2 protein significantly enhances protective immunity. Vaccine. 2004;23(3):366–371. doi: 10.1016/j.vaccine.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 58.Mozdzanowska K, Furchner M, Zharikova D, Feng J, Gerhard W. Roles of CD4+ T-cell-independent and -dependent antibody responses in the control of influenza virus infection: evidence for noncognate CD4+ T-cell activities that enhance the therapeutic activity of antiviral antibodies. J. Virol. 2005;79(10):5943–5951. doi: 10.1128/JVI.79.10.5943-5951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eriksson AM, Schon KM, Lycke NY. The cholera toxin-derived CTA1-DD vaccine adjuvant administered intranasally does not cause inflammation or accumulate in the nervous tissues. J. Immunol. 2004;173(5):3310–3319. doi: 10.4049/jimmunol.173.5.3310. [DOI] [PubMed] [Google Scholar]
- 60.Eliasson DG, El Bakkouri K, Schon K, et al. CTA1-M2e-DD: a novel mucosal adjuvant targeted influenza vaccine. Vaccine. 2008;26(9):1243–1252. doi: 10.1016/j.vaccine.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 61.Huber VC, Lynch JM, Bucher DJ, Le J, Metzger DW. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001;166(12):7381–7388. doi: 10.4049/jimmunol.166.12.7381. [DOI] [PubMed] [Google Scholar]
- 62.Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004;172(9):5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 63.Mozdzanowska K, Zharikova D, Cudic M, Otvos L, Gerhard W. Roles of adjuvant and route of vaccination in antibody response and protection engendered by a synthetic matrix protein 2-based influenza A virus vaccine in the mouse. Virol. J. 2007;4:118. doi: 10.1186/1743-422X-4-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buckland BC. The process development challenge for a new vaccine. Nature Med. 2005;11(4 Suppl):S16–S19. doi: 10.1038/nm1218. [DOI] [PubMed] [Google Scholar]
- 65.De Filette M, Martens W, Roose K, et al. An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J. Biol. Chem. 2008;283(17):11382–11387. doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Govaert TM, Thijs CT, Masurel N, et al. The efficacy of influenza vaccination in elderly individuals. A randomized double-blind placebo-controlled trial. JAMA. 1994;272(21):1661–1665. [PubMed] [Google Scholar]
- 67.Lin YP, Gregory V, Bennett M, Hay A. Recent changes among human influenza viruses. Virus Res. 2004;103(12):47–52. doi: 10.1016/j.virusres.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 68.Bianchi E, Liang X, Ingallinella P, et al. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J. Virol. 2005;79(12):7380–7388. doi: 10.1128/JVI.79.12.7380-7388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Filette M, Ramne A, Birkett A, et al. The universal influenza vaccine M2e-HBc administered intranasally in combination with the adjuvant CTA1-DD provides complete protection. Vaccine. 2006;24(5):544–551. doi: 10.1016/j.vaccine.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 70.Stephenson I, Zambon MC, Rudin A, et al. Phase I evaluation of intranasal trivalent inactivated influenza vaccine with nontoxigenic Escherichia coli enterotoxin and novel biovector as mucosal adjuvants, using adult volunteers. J. Virol. 2006;80(10):4962–4970. doi: 10.1128/JVI.80.10.4962-4970.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jimenez GS, Planchon R, Wei Q, et al. Vaxfectin-formulated influenza DNA vaccines encoding NP and M2 viral proteins protect mice against lethal viral challenge. Hum. Vaccin. 2007;3(5):157–164. doi: 10.4161/hv.3.5.4175. [DOI] [PubMed] [Google Scholar]
- 72.De Filette M, Fiers W, Martens W, et al. Improved design and intranasal delivery of an M2e-based human influenza A vaccine. Vaccine. 2006;24(4446):6597–6601. doi: 10.1016/j.vaccine.2006.05.082. [DOI] [PubMed] [Google Scholar]
- 73.De Filette M, Martens W, Smet A, et al. Universal influenza A M2e-HBc vaccine protects against disease even in the presence of pre-existing anti-HBc antibodies. Vaccine. 2008;26(51):6503–6507. doi: 10.1016/j.vaccine.2008.09.038. [DOI] [PubMed] [Google Scholar]
- 74.Heinen PP, Rijsewijk FA, de Boer-Luijtze EA, Bianchi AT. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J. Gen. Virol. 2002;83(Pt 8):1851–1859. doi: 10.1099/0022-1317-83-8-1851. [DOI] [PubMed] [Google Scholar]
- 75.Laddy DJ, Yan J, Kutzler M, et al. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS ONE. 2008;3(6):e2517. doi: 10.1371/journal.pone.0002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.US CDC www.cdc.gov.
- 102.WHO tables on the clinical trials of pandemic influenza prototype vaccines www.who.int/vaccine_research/diseases/influenza/flu_trials_tables/en/index.html.