Abstract
The endoribonuclease L (RNase L) is the effector of the 2–5A system, a major enzymatic pathway involved in the molecular mechanism of interferons (IFN). RNase L is a very unusual nuclease with a complex mechanism of regulation. It is a latent enzyme, expressed in nearly every mammalian cell type. Its activation requires its binding to a small oligonucleotide, 2–5A. 2–5A is a series of unique 5′-triphosphorylated oligoadenylates with 2′–5′ phosphodiester bonds. By regulating viral and cellular RNA expression, RNase L plays an important role in the antiviral and antiproliferative activities of IFN and contributes to innate immunity and cell metabolism. The 2–5A/RNase L pathway is implicated in mediating apoptosis in response to viral infections and to several types of external stimuli. Several recent studies have suggested that RNase L could have a role in cancer biology and evidence of a tumor suppressor function of RNase L has emerged from studies on the genetics of hereditary prostate cancer.
Keywords: Interferon, RNase L, RNA expression, apoptosis, prostate, virus, cancer
I) Introduction
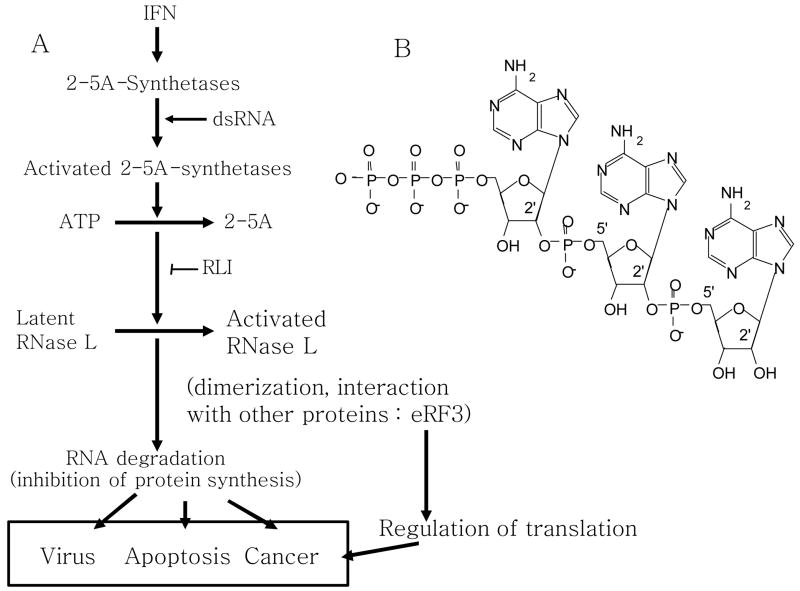
The endoribonuclease L (RNase L) is the effector of the 2–5A system, a major enzymatic pathway regulated by interferons (IFN) [1] (Figure 1). The 2–5A system is one of the two antiviral pathways induced by IFN and activated by double stranded RNA (dsRNA), the other is mediated by the dsRNA dependent protein kinase (PKR) [2]. RNase L is a very unusual nuclease. It is a latent enzyme, expressed in nearly every mammalian cell type. Its activation requires its binding to a small oligonucleotide, 2–5A (Figure 1). 2–5A itself is very unusual, consisting of a series of 5′-triphosphorylated oligoadenylates with 2′–5′ phosphodiester bonds in contrast to the 3′–5′ linkages found in RNA and DNA. The initial and essential observation was made by Ian Kerr’s group in 1974 reporting an IFN-induced increase in the sensitivity of protein synthesis to inhibition by dsRNA [3]. Peter Lengyel’s group observed increased nuclease activity in extracts of interferon treated cells incubated with dsRNA [4, 5]. The identification by Ian Kerr’s group of the activators of this nuclease, 2–5A [6] and of the enzyme responsible for their synthesis, the 2–5A-synthetase [7–9], led to the discovery of the 2–5A pathway. Clemens and Williams directly demonstrated a nuclease, now recognized as RNase L, that was activated by 2–5A [10]. In brief, IFN induces the transcription of several 2–5A synthetase genes whose protein products are in turn activated by dsRNA and produce 5′-triphosphorylated, 2′,5′-oligoadenylates (2–5A) from ATP [11]. 2–5A bind specifically to RNase L, which is activated to cleave single stranded RNAs at UpN (mainly UpU and UpA) sequences in single-stranded regions of RNA [12, 13]. The only known function of 2–5A is to bind and activate RNase L. 2–5A is very unstable in cells and in serum. It is dephosphorylated by general phosphatases at its 5′ end, leaving the so-called core oligoadenylate that is unable to efficiently activate RNase L [14]. 2–5A can also be degraded from the 2′,3′-termini by a 2′-phosphodiesterase [15].
Figure 1.
A) The 2–5A/RNase L pathway
B) Structure of the 2–5A4: 2′–5′ oligoadenylates tetramer.
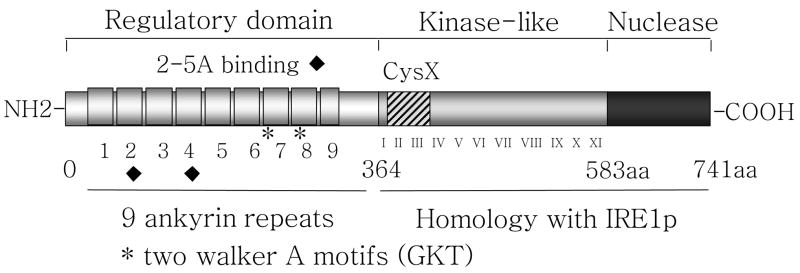
The chemical and/or enzymatic synthesis of several analogues of 2–5A with increased metabolic stability has allowed the determination of the regions of the oligomer which interact with RNase L and that are absolutely necessary for the activation of RNase L [16]. Only oligomers with more than two adenylates are able to activate RNase L. The 5′-monophosphate, 2′, 5′-phosphodiester bonds and the 3′-hydroxyl groups of the second adenosine (from the 5′ terminus) are critical for its biological activity. The 5′ terminal adenine base of 2–5A is vital for binding with RNase L, the adenine of the 2′ terminal adenosine is absolutely critical for activation of the nuclease function of the enzyme but not for its binding. The cloning of RNase L in 1993, allowed the subsequent elucidation of its remarkable properties [1] (Figure 2). The recent crystal structure of the 2–5A binding domain of RNase L in a complex with 2–5A provides a detailed view of these interactions at the atomic level [17].
Figure 2.
Structure of the RNase L.
II) RNase L structure
RNase L was first described as a 185 kDa complex by gel filtration experiments [18] then as a 78–80 kDa protein in gel denaturing conditions [19, 20]. These two forms seem to coexist in cells, their ratio depending on the cell type and experimental procedures use to detect RNase L [21–23]. The two forms of RNase L bind 2–5A and have 2–5A-dependent nuclease activity. Another small protein (40–46 kDa) which binds 2–5A and cleaves poly(U) has been observed in mouse spleen and liver and in EAT (Ehrlich ascites tumor) cells [19, 21, 24]. This low molecular weight protein seems to be due to a proteolytic cleavage of RNase L [25, 26].
Cloning of RNase L demonstrates that human RNase L is a 741-amino-acid protein with a molecular mass of 83,539 kDa [1]. RNase L is widely expressed in different cell types of mammals. Recently the expression of an alternative spliced RNase L variant, lacking the third exon, was shown in peripheral blood leukocytes [27]. RNase L consists of three domains: an N-terminal ankyrin repeat domain, a protein kinase homology domain and a C-terminal ribonuclease domain (Figure 2). The N-terminal domain could be considered as the regulatory domain of RNase L. It is composed of eight complete and one partial ankyrin motifs (R1–R9). Two walker A motifs (ATP or GTP fixation) are located within R7 and R8 [28]. Ankyrin motifs function in mediating many different protein-protein interactions [29]. Surprisingly, the ankyrin motifs in RNase L interact with an oligonucleotide, 2–5A. Several studies with N-terminal truncations of RNase L in ankyrin motifs or mutation in the two walker A motifs have shown that R1, 7, 8, 9 are essential for 2–5A binding [1, 30, 31]. The crystal structure of the N-terminal ankyrin repeat domain of RNase L complexed with 2–5A shows that the bound 2–5A directly interacts with R2–R4 [17]. That study indicates that R2–R4 constitute the 2–5A binding pocket and that R7–R9 may be necessary for structural integrity of RNase L rather than directly binding 2–5A. To determine more precisely the role of the amino acids residues surrounding the 2–5A binding site, several mutants were constructed based on the crystal structure [32].
Binding with 2–5A induces a conformational change in the ankyrin domain of RNase L believed to unmask the C-terminal ribonuclease domain allowing homodimerization of RNase L and activation of its nuclease activity [30, 33–35]. The dimerization and activation of RNase L requires a molar ratio of 1:1 between RNase L and 2–5A [33, 36, 37]. The C-terminal half of RNase L is homologous with the kinase/endoribonuclease, IRE1p, which functions in the unfolded protein response (UPR) in Saccharomyces cerevisiae, Caenorhabditis elegans and vertebrates including Homo sapiens [34, 38, 39]. RNase L and IRE1 have homology in their nuclease domains but also in their kinase or kinase-like domains. However, whereas the kinase function of IRE1p is well established, RNase L has not been shown to have kinase activity. However, mutation of the lysine 392 in the protein kinase II domain leads to a greatly reduced activity of RNase L which was correlated to a defect in the ability of RNase L to dimerize [40]. Several amino acids in C-terminal domain of RNase L are required for catalysis, including R667 and H672 [34]. In addition, Tyr712 and Phe716 are important for both binding and cleavage of RNA [41]. As mentioned above, RNase L is an endoribonuclease with minimal sequence specificity. Its first RNA targets to be identified were viral mRNA and rRNA [42–45]. But more recently, several cellular mRNAs regulated by RNase L were identified. This is an indication that RNase L, and the 2–5A pathway, could have a wide biological role in cell physiology.
III) Biological activities of RNase L
1) Antiviral activity
The type I IFN response is the host’s frontline defence against viral infections and occurs prior to the onset of the adaptive immune response. IFN activity is critical to limit virus propagation before the development of a full immune response. Type I IFN regulates transcription of a number of genes which inhibit or block viral replication through diverse mechanisms. Since the 1980’s, many studies have established that the 2–5A/RNase L pathway plays a central role in the antiviral activity of IFN. During viral infections, many viruses produce dsRNA structures that can activate 2–5A-synthetases. The presence of 2–5A has been demonstrated in cells infected with encephalomyocarditis (EMC) virus [46], vaccinia virus [47] or reovirus [48]. The role played by RNase L in IFN-induced antiviral activity has clearly been demonstrated by transfection of 2–5A or stabilized 2–5A analogues in intact cells [49–54]. The cloning of 2–5A-synthetase [55, 56] and RNase L [1] allowed the definitive demonstration of the antiviral activity of RNase L. The constitutive expression of the 40 kDa 2–5A-synthetase confers resistance to EMCV and mengovirus [57, 58] and over expression of the 69 kDa form of 2–5A-synthetase protein in human cells inhibits the replication of EMCV [59]. Similarly, the antiviral activity of IFN against EMCV was inhibited by the expression of a dominant negative mutant of RNase L [28] or treatment with a 2–5A antagonist [49, 53, 54]. In vivo evidence for the antiviral role of the 2–5A system was provided by studies with RNase L−/− mice, which have enhanced susceptibility to infections with EMCV, Coxsackievirus B4,West Nile virus and herpes simplex virus 1 [60–63]. In addition, it has been shown that RNase L is activated in West Nile virus-infected cells and plays a role in the cellular antiviral response to flaviviruses [64]. The RNase L pathway has also been implicated in the response to IFN-beta during acute ocular herpes simplex virus type 1 infection (HSV-1) of mouse trigeminal-ganglia (TG) cells [65].
a) Mechanism of action
A localized model of activation of RNase L could lead to preferential degradation of viral vs. cellular RNA. Viral replicative intermediates or viral mRNA possessing double-stranded structures can activate 2–5A-synthetase resulting in localized production of 2–5A and activation of RNase L [66–68]. 2–5A synthetase may bind to replicative intermediates of RNA viruses and be activated by these RNA structures [69]. Moreover, complexes between EMCV dsRNA and 2–5A-synthetase that produce 2–5A from ATP have been isolated from virus-infected cells [70]. Several viral RNAs can activate 2–5A-synthetase including adenoviral VAI RNA [71], TAR RNA of human immunodeficiency virus type 1 (HIV-1) [72], REX-RE RNA of Human T-cell Leukemia virus type I (HTLV-1) [73], and EBER-1 RNA of Epstein-Barr virus [74]. Cleavage of viral mRNA by RNase L in intact cells has been demonstrated during treatment of vesicular stomatitis virus-infected cells with 2–5A [75], HIV-1 and EMCV infection [44, 45]. Interestingly, limited activation of RNase L depends on the levels of 2–5A. Low levels of 2–5A are sufficient to observe a biological response: a reduction in EMCV RNA. In presence of high concentrations of 2–5A an extensive rRNA cleavage is observed, indicating of a widespread degradation of host cell RNA [44].
b) Inhibition of RNase L activity by viruses
Several viruses have developed different strategies to counteract the antiviral activity of 2–5A/RNase L pathway. In some virus infected cells, despite the presence of viral dsRNA structures able to activate 2–5A-synthetase and the presence of high concentrations of 2–5A, no RNase L activity could be detected. During herpes simplex virus type 1 and 2 infections, natural 2–5A analogs are synthesized in parallel with authentic 2–5A. These 2–5A derivatives are weak activator of RNase L and able to inhibit RNase L activation by authentic 2–5A [76]. Such 2–5A related compounds, unable to activate RNase L, are also observed in simian virus 40 (SV40) infected cells [77]. Hepatitis C virus (HCV) may escape RNase L activity through selection of sequence variants [78, 79]. As expected, RNase L cleaves HCV mRNA predominantly after UA and UU dinucleotides in single-stranded (loop) regions leading to fragments of 200 to 500 bases in length. HCV mRNAs from relatively IFN-resistant genotypes (HCV 1a and 1b) have fewer UA and UU dinucleotides than HCV mRNAs from more IFN-sensitive genotypes (HCV 2a, 2b, 3a and 3b). During IFN therapy, HCV 1b mRNA accumulates silent mutations preferentially at UA and UU dinucleotides, possibly to escape RNase L activity. Perhaps as a consequence, patients infected with HCV 1b viruses are cured less frequently than patients infected with HCV genotype 2 or 3 [78]. To synthesize 2–5A, 2–5A-synthetase needs to be activated by dsRNA. Some viral proteins which possess the capacity of binding dsRNA directly compete with 2–5A synthetase. In fact, during vaccinia virus infection, 2–5A-synthetase is inhibited and 2–5A is degraded [80]. Inhibition of 2–5A synthetase during vaccinia virus infection is mediated by the E3L, viral encoded dsRNA binding proteins [81–83]. During influenza A virus infection, the viral protein NS1A sequesters dsRNA and inhibits 2–5A-synthetase activation [84]. Some viruses recruit a host protein to inhibit RNase L activity. This is the case for EMCV and HIV-1 which lead to increase expression of the Ribonuclease L Inhibitor (RLI) [45, 85, 86]. RLI is an ATP binding cassette (ABC) protein now classified as ABCE1 protein. RLI can form a heterodimer with RNase L and inhibit the binding of 2–5A by RNase L in a noncompetitive manner [85]. During HIV-1 infection, RLI also interacts with the nucleocapsid domain of Gag and play a role in HIV-1 capsid assembly [87].
2) Regulation of mRNA translation
RLI is not the only cellular protein which can form heterodimer with RNase L. One of us (C.B.) has previously identified an RNA binding protein (RNABP) which associates with RNase L. This complex has been observed in different cell extracts by co-immunoprecipitation, gel filtration and pull-down assays in a 2–5A dependent and single-stranded RNA independent manner [23, 88]. Recently RNABP was identified (by C.B. and collaborators) as the translation termination release factor eRF3/GSPT1 [89]. After activation by 2–5A, RNase L can interact with eRF3. This association can help to localize RNase L to its mRNA target, but it is also a way to modulate eRF3 activity. Importantly the regulation of eRF3 activity depends of the 2–5A oligomer size activating RNase L. Binding of 2–5A3 or 2–5A4 induces a conformational change in RNase L that promotes its interaction with eRF3. In one conformational change, the eRF3-RNase L interaction brings RNase L into close association with the mRNA, where it can act as an endoribonuclease. But binding with 2–5A3 can induce another conformational change leading to an RNase L-eRF3 complex that can modulate translation termination and promote ribosomal readthrough of a termination codon. Moreover, RNase L regulates the +1 frameshifting of the antizyme 1 mRNA in IFN treated cells. This was the first report implicating a nuclease, RNase L, in the translation regulation of a cellular mRNA independently of its nuclease activity.
3) Regulation of cellular mRNA stability
2–5A-synthetase levels vary with growth and hormonal status in a variety of cell types and tissues [90, 91]. These results suggested that 2–5A levels, and consequently RNase L activity, may play a role in metabolism of uninfected cells and in the antiproliferative effects of IFN. It has been known for more than 30 years that IFN-treated cells show enhanced sensitivity to inhibition of protein synthesis by dsRNA [3], this is also true for cells treated by 2–5A [92], mediated by RNase L that degrades mRNA and rRNA [69, 93–95]. 2–5A-synthetases are activated by double stranded region of cellular RNA which leads to 2–5A synthesis [96, 97]. But the identification of specific mRNAs regulated by RNase L was difficult to detect in intact cells until the advent of DNA microarray technology [98]. Activation of RNase L by exogenous 2–5A leads to general degradation (70%) of cellular RNA [99]. Studies of subcellular localization of RNase L showed that RNase L could be associated with polysomes [24], mitochondria [100], cytoskeleton [101] and nuclei [102], suggesting that RNase L could regulates the stability of different cellular RNA species in different cellular compartments. Conditional expression of RNase L showed that RNase L regulates MyoD mRNA stability and muscle cell differentiation [103]. RNase L in mitochondria regulates mitochondrial mRNA stability and possibly contributing to the antiproliferative activity of IFNs [100]. Interestingly, RNase L regulates the stability of PKR mRNA and of two interferon-stimulated genes: ISG43 and ISG15 [104, 105] suggesting one function of RNase L is to limit the IFN response.
4) Apoptosis
The 2–5A/RNase L pathway is implicated in mediating apoptosis in response to viral infections and to several types of external stimuli [60, 106–111]. How might RNase L activation lead to apoptosis? The degradation of 28S and 18S rRNA by RNase L in intact ribosomes has been long known as a hallmark of IFN and viral infections [42, 43]. Cleavage of 28S rRNA by RNase L maps to the L1 protuberance implicated in formation of the exit or E site of the ribosome, possibly interfering with release of deacylated tRNA [112]. By comparison, the fungal ribonuclease, α-sarcin, and the RNA modifying enzyme, ricin A chain, and uv light each lead to ribotoxic stress responses involving damage to the 3′-end of the large ribosomal RNA. These treatments activate the stress-activate protein kinases, c-jun NH2-terminal kinases (JNKs) [113, 114]. JNK activation in response to uv irradiation has been linked to apoptosis through Bax [115]. Furthermore, JNK activation by the topoisomerase I inhibitor, camptothecin, or by the protein synthesis inhibitor, anisomycin, sensitizes prostate cancer cells to anti-fas mediated apoptosis [116]. Interestingly, phosphorylation of JNK in response to dsRNA is deficient in RNase L-null cells [112]. Furthermore, it has been shown using JNK1−/−JNK2−/− mouse embryonic fibroblast cells that the JNKs are essential for apoptosis mediated by RNase L [117]. Apoptosis initiated by RNase L requires caspase 3 activity and is characterized by the appearance of cytosolic cytochrome c indicating involvement of mitochondria [110]. In fact, IFNα treatment induces down regulation of mitochondrial mRNAs, which leads to a decrease in cellular ATP levels and suppression of mitochondrial functions and in particular a decrease in mitochondrial protein levels [118, 119]. The consequence of such a degradation of mitochondrial mRNA and inhibition of protein synthesis is a loss of mitochondrial membrane potential which results in osmotic swelling and cytochrome c release [120]. Yanase and co-workers have shown that during IFNα induced apoptosis there is a release of cytochrome c from mitochondria, a decline in mitochondrial membrane potential and caspase 3 activation [121]. The different partners of the 2–5A pathway are localized in mitochondria: RNase L, RLI and p69 form of 2–5A-synthetase [100, 122, 123]. This pathway is induced by IFNα treatment and RNase L participates in mitochondrial mRNA degradation. Mitochondrial mRNA remained stable during IFNα treatment in cells where RNase L activity was decreased by transfection of RNase L antisense or RLI sense cDNAs [100]. A similar stabilization of mitochondrial mRNA is observed in RNASEL −/− mouse embryonic fibroblast cells [124]. If mitochondrial mRNAs are stabilized by inhibiting RNase L activity, no cytochrome c release or caspase 3 activation are observed during IFNα treatment (CB, unpublished observation).
Ultimately, sustained activation of RNase L triggers a mitochondrial pathway of apoptosis that eliminates virus-infected cells [106, 111]. Both the antiviral and tumor suppressor activities of RNase L in vivo could be due to its pro-apoptotic activity, limiting viral spread or tumor growth. In fact, Aimin Zhou and co-workers demonstrated recently that RNase L can potently inhibit fibrosarcoma growth in vivo [125].
IV) Implications in pathology
1) Role of RNase L in the biology of prostate cancer
Because genetic lesions in RNase L impair apoptotic responses, such mutations could possibly contribute to malignancy [60, 107]. Recently, evidence of a tumor suppressor function of RNase L has emerged from studies on the genetics of hereditary prostate cancer. Prostate cancer has a complex etiology and is influenced by genetics, but also by androgens, diet, and other environmental factors [126]. Sporadic prostate cancer displays an age-related increase in prevalence, while familial prostate cancer often displays earlier-onset disease. Hereditary prostate cancer (HPC) stricken families, with three or more first degree relatives per family, account for 43% of early onset cases (<55 years old) and 9% of all cases [127]. HPC genetics is complex with many genes proposed as susceptibility factors in this syndrome. Interestingly, the prototype of these genes, HPC1, maps to the RNase L gene, RNASEL [128, 129]. Several germline mutations or variants in HPC1/RNASEL have been observed among hereditary prostate cancer cases (reviewed in ref. [130]). For instance, a common (35% allelic frequency) missense variant of RNase L, in which a G to A transition at nucleotide position 1385 (G1385A) results in a glutamine instead of arginine at amino acid position 462 (R462Q). A controlled sib-pair study implicated the RNase L “Q” variant in up to 13% of unselected prostate cancer cases [129]. One and two copies of the mutated gene increased the risk of prostate cancer by about 150% and 200%, respectively. The RNase L “Q” variant at residue 462 in the kinase-like region had a 3-fold decrease in catalytic activity compared to the wild-type enzyme, due to an impairment in enzyme dimerization [129, 131]. However, while several case-controlled genetic and epidemiologic studies support the involvement of RNASEL (and notably the R462Q variant) in prostate cancer etiology [128, 129, 132, 133], others do not [134–136], suggesting that either population differences or environmental factors, such as viral infection, may modulate the impact of RNASEL on prostatic carcinogenesis. Therefore, there is a possibility that the linkage of RNase L alterations to HPC might reflect enhanced susceptibility to a viral agent. To test this hypothesis, RNA derived from wild-type and RNase L variant (R462Q) prostate tumors was examined for evidence of viral sequences, by hybridization to a DNA microarray composed of the most conserved sequences of all known human, animal, plant and bacterial viruses [137, 138]. Because the array contains highly conserved sequences within viral nucleic acids, it can detect viruses not explicitly represented. These studies identified the novel retrovirus, xenotropic murine leukemia related virus (XMRV) in 8 (40%) of 20 R462Q homozygous prostate cancer tissues, and in 1 (1.5%) of 66 tissues that harbored at least one copy of the wild-type allele. Three complete genomes were sequenced, sharing >98% nucleotide and >99% protein sequence identity. The virus encodes four major proteins, gag, pro, pol, and env. XMRV is more closely related to the xenotropic and polytropic than to the ecotropic murine retroviruses. Complete viral genome sequences were obtained from three strains and partial sequences were obtained for another six XMRV strains. XMRV is a canonical gammaretrovirus, with gag, pro-pol and env genes, and is not closely related to any endogenous human retroviral (HERV) elements. In addition, XMRV sequences are not present in any human genome sequences that have been reported to date [139]. Recently, the group of one of us (R.H.S.) constructed a complete infectious clone for XMRV strain VP62 [140]. In the same study, XMRV provirus integration sites were mapped in DNA isolated from human prostate tumor tissue. These findings represent the first detection of xenotropic MuLV-like agents in humans, and reveal a strong association between infection with the virus and defects in RNase L activity. In addition, these studies provide evidence that RNase L functions as an antiviral enzyme in humans.
2) Pancreatic cancer and colorectal cancer
As prostate cancer occurs in some familial pancreatic families, Bartsch and co-workers evaluated the role of two variants of RNASEL gene: E265X and R462Q in the etiology of pancreatic cancer [141]. That study showed that the RNASEL R462Q variant might be associated with an increased risk for sporadic pancreas cancer and with more aggressive tumors in familial pancreatic cancer. Additional, large scale efforts will be important to validate these results.
Also, the R462Q variant of RNase L correlated with earlier age of onset of hereditary non-polyposis colorectal cancer [142].
3) Chronique fatigue syndrome (CFS)
Chronic fatigue syndrome (CFS) is an illness characterized by long-lasting fatigue accompanied by non specific symptoms [143, 144]. Several reports indicated the up-regulation of components of the 2–5A/RNase L pathway in extracts of peripheral blood mononuclear cells (PBMCs) from CFS patients [145, 146] as well as the accumulation of a low molecular weight 2–5A-binding protein of 37 kDa [147]. It has been proposed that this polypeptide could be a biochemical marker for CFS [148, 149]. However, levels of the 37 kDa polypeptide appeared to vary with time within individual patients [150]. The polypeptide is an apparent degradation product of the native RNase L due to an increased proteolytic activity in CFS PBMC extracts. In vitro, an equivalent degradation of RNase L could be observed when recombinant RNase L was incubated with human leucocyte elastase [151]. In CFS patients, the majority of 2–5A oligomers produced are 2–5A dimers which fail to bind or activate RNase L [33]. 2–5A trimer and tetramer binding appeared to stabilize RNase L in PBMC cell extracts from CFS patients. These observations suggest that in CFS there is increased proteolytic activity in PBMC causing accumulation of the 37 kDa polypeptide [152].
V) Conc lusion
RNase L is an unusual and fascinating enzyme unique among know nucleases in its complex mechanism of action. Considerable progress has been made since the discovery of the 2–5A pathway in 1978 and its cloning in 1993. Nevertheless, much work remains to be done in understanding how RNase L contributes to innate immunity and cell metabolism. We anticipate many additional findings over the coming years of the roles of RNase L in human pathology, in particular cancer and viral diseases.
Acknowledgments
Investigations described here in from CB’s laboratory were supported by grants from Centre National de la Recherche Scientifique (CNRS), Association pour la Recherche sur le Cancer (ARC) and Ligue Contre le Cancer and grant CA044059 from the U.S. National Institutes of Health (National Cancer Institute) to R.H.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2–5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 2.Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA- activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 3.Kerr IM, Brown RE, Ball LA. Increased sensitivity of cell-free protein synthesis to double-stranded RNA after interferon treatment. Nature. 1974;250:57–59. doi: 10.1038/250057a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown GE, Lebleu B, Kawakita M, Shaila S, Sen GC, Lengyel P. Increased endonuclease activity in an extract from mouse Ehrlich ascites tumor cells which had been treated with a partially purified interferon preparation: dependence of double-stranded RNA. Biochem Biophys Res Commun. 1976;69:114–122. doi: 10.1016/s0006-291x(76)80280-x. [DOI] [PubMed] [Google Scholar]
- 5.Sen GC, Lebleu B, Brown GE, Kawakita M, Slattery E, Lengyel P. Interferon, double-stranded RNA and mRNA degradation. Nature. 1976;264:370–373. doi: 10.1038/264370a0. [DOI] [PubMed] [Google Scholar]
- 6.Kerr IM, Brown RE. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci U S A. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts WK, Hovanessian A, Brown RE, Clemens MJ, Kerr IM. Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature. 1976;264:477–480. doi: 10.1038/264477a0. [DOI] [PubMed] [Google Scholar]
- 8.Hovanessian AG, Brown RE, Kerr IM. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- 9.Kerr IM, Brown RE, Hovanessian AG. Nature of inhibitor of cell-free protein synthesis formed in response to interferon and double-stranded RNA. Nature. 1977;268:540–542. doi: 10.1038/268540a0. [DOI] [PubMed] [Google Scholar]
- 10.Clemens MJ, Vaquero CM. Inhibition of protein synthesis by double-stranded RNA in reticulocyte lysates: evidence for activation of an endoribonuclease. Biochem Biophys Res Commun. 1978;83:59–68. doi: 10.1016/0006-291x(78)90397-2. [DOI] [PubMed] [Google Scholar]
- 11.Rebouillat D, Hovanessian AG. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J Interferon Cytokine Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- 12.Floyd SG, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′–5′)oligoadenylate-- dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 13.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action--sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 14.Silverman RH. Functional analysis of 2–5A-dependent RNase and 2–5a using 2′,5′-oligoadenylate-cellulose. Anal Biochem. 1985;144:450–460. doi: 10.1016/0003-2697(85)90141-1. [DOI] [PubMed] [Google Scholar]
- 15.Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, Haruyama H, Ichikawa K, Yamashita M, Matsuoka T, Iijima Y. Identification of 2′-phosphodiesterase, which plays a role in the 2–5A system regulated by interferon. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 16.Torrence PF, Xiao W, Li G, Khamnei S. Development of 2′,5′-oligonucleotides as potential therapeutic agents. Curr Medic Chem. 1994;1:176–191. [Google Scholar]
- 17.Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT. Structural basis for recognition of 2′,5′-linked oligoadenylates by human ribonuclease L. Embo J. 2004;23:3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slattery E, Ghosh N, Samanta H, Lengyel P. Interferon, double-stranded RNA, and RNA degradation: activation of an endonuclease by (2′–5′)An. Proc Natl Acad Sci U S A. 1979;76:4778–4782. doi: 10.1073/pnas.76.10.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floyd SG, Yoshie O, Lengyel P. Interferon action. Covalent linkage of (2′–5′)pppApApA(32P)pCp to (2′– 5′)(A)n-dependent ribonucleases in cell extracts by ultraviolet irradiation. J Biol Chem. 1982;257:8584–8587. [PubMed] [Google Scholar]
- 20.Silverman RH, Jung DD, Nolan SN, Dieffenbach CW, Kedar VP, SenGupta DN. Purification and analysis of murine 2–5A-dependent RNase. J Biol Chem. 1988;263:7336–7341. [PubMed] [Google Scholar]
- 21.Bisbal C, Salehzada T, Lebleu B, Bayard B. Characterization of two murine (2′–5′)(A)n-dependent endonucleases of different molecular mass. Eur J Biochem. 1989;179:595–602. doi: 10.1111/j.1432-1033.1989.tb14588.x. [DOI] [PubMed] [Google Scholar]
- 22.Bayard B, Bette BP, Aliau S. Affinity purification and characterization of (2′–5′)oligo(adenylate)- dependent RNase from mouse spleen. Eur J Biochem. 1994;223:403–410. doi: 10.1111/j.1432-1033.1994.tb19007.x. [DOI] [PubMed] [Google Scholar]
- 23.Le Roy F, Laskowska A, Silhol M, Salehzada T, Bisbal C. Characterization of RNABP, an RNA binding protein that associates with RNase L. J Interferon Cytokine Res. 2000;20:635–644. doi: 10.1089/107999000414817. [DOI] [PubMed] [Google Scholar]
- 24.Salehzada T, Silhol M, Lebleu B, Bisbal C. Polyclonal antibodies against RNase L. Subcellular localization of this enzyme in mouse cells. J Biol Chem. 1991;266:5808–5813. [PubMed] [Google Scholar]
- 25.Krause D, Silverman RH, Jacobsen H, Leisy SA, Dieffenbach CW, Friedman RM. Regulation of ppp(A2′p)nA-dependent RNase levels during interferon treatment and cell differentiation. Eur J Biochem. 1985;146:611–618. doi: 10.1111/j.1432-1033.1985.tb08695.x. [DOI] [PubMed] [Google Scholar]
- 26.Bayard B, Zhou A. Affinity blotting assay for 2–5A-dependent RNase. Anal Biochem. 1992;200:108–114. doi: 10.1016/0003-2697(92)90284-e. [DOI] [PubMed] [Google Scholar]
- 27.Rennert H, Sadowl C, Edwards J, Bantly D, Molinaro RJ, Orr-Urtreger A, Bagg A, Moore JS, Silverman RH. An Alternative Spliced RNASEL Variant in Peripheral Blood Leukocytes. J Interferon Cytokine Res. 2006;26:820–826. doi: 10.1089/jir.2006.26.820. [DOI] [PubMed] [Google Scholar]
- 28.Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH. A dominant negative mutant of 2–5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. Embo J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong B, Silverman RH. A bipartite model of 2–5A-dependent RNase L. J Biol Chem. 1997;272:22236–22242. doi: 10.1074/jbc.272.35.22236. [DOI] [PubMed] [Google Scholar]
- 31.Diaz-Guerra M, Rivas C, Esteban M. Full activation of RNaseL in animal cells requires binding of 2–5A within ankyrin repeats 6 to 9 of this interferon-inducible enzyme. J Interferon Cytokine Res. 1999;19:113–119. doi: 10.1089/107999099314252. [DOI] [PubMed] [Google Scholar]
- 32.Nakanishi M, Tanaka N, Mizutani Y, Mochizuki M, Ueno Y, Nakamura KT, Kitade Y. Functional characterization of 2′,5′-linked oligoadenylate binding determinant of human RNase L. J Biol Chem. 2005;280:41694–41699. doi: 10.1074/jbc.M507424200. [DOI] [PubMed] [Google Scholar]
- 33.Dong B, Silverman RH. 2–5A-dependent RNase molecules dimerize during activation by 2–5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 34.Dong B, Niwa M, Walter P, Silverman RH. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. Rna. 2001;7:361–373. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakanishi M, Goto Y, Kitade Y. 2–5A induces a conformational change in the ankyrin-repeat domain of RNase L. Proteins. 2005;60:131–138. doi: 10.1002/prot.20474. [DOI] [PubMed] [Google Scholar]
- 36.Cole JL, Carroll SS, Kuo LC. Stoichiometry of 2′,5′-oligoadenylate-induced dimerization of ribonuclease L. A sedimentation equilibrium study. J Biol Chem. 1996;271:3979–3981. doi: 10.1074/jbc.271.8.3979. [DOI] [PubMed] [Google Scholar]
- 37.Cole JL, Carroll SS, Blue ES, Viscount T, Kuo LC. Activation of RNase L by 2′,5′-oligoadenylates. Biophysical characterization. J Biol Chem. 1997;272:19187–19192. doi: 10.1074/jbc.272.31.19187. [DOI] [PubMed] [Google Scholar]
- 38.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 39.Urano F, Bertolotti A, Ron D. IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci. 2000;113(Pt 21):3697–3702. doi: 10.1242/jcs.113.21.3697. [DOI] [PubMed] [Google Scholar]
- 40.Dong B, Silverman RH. Alternative function of a protein kinase homology domain in 2′, 5′-oligoadenylate dependent RNase L. Nucleic Acids Res. 1999;27:439–445. doi: 10.1093/nar/27.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakanishi M, Yoshimura A, Ishida N, Ueno Y, Kitade Y. Contribution of Tyr712 and Phe716 to the activity of human RNase L. Eur J Biochem. 2004;271:2737–2744. doi: 10.1111/j.1432-1033.2004.04202.x. [DOI] [PubMed] [Google Scholar]
- 42.Wreschner DH, James TC, Silverman RH, Kerr IM. Ribosomal RNA cleavage, nuclease activation and 2–5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li XL, Blackford JA, Hassel BA. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinand C, Montavon C, Salehzada T, Silhol M, Lebleu B, Bisbal C. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2–5A/RNase L pathway in human T cells. J Virol. 1999;73:290–296. doi: 10.1128/jvi.73.1.290-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams BR, Golgher RR, Brown RE, Gilbert CS, Kerr IM. Natural occurrence of 2–5A in interferon-treated EMC virus-infected L cells. Nature. 1979;282:582–586. doi: 10.1038/282582a0. [DOI] [PubMed] [Google Scholar]
- 47.Rice AP, Roberts WK, Kerr IM. 2–5A accumulates to high levels in interferon-treated, vaccinia virus- infected cells in the absence of any inhibition of virus replication. J Virol. 1984;50:220–228. doi: 10.1128/jvi.50.1.220-228.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsen TW, Maroney PA, Baglioni C. Synthesis of (2′–5′)oligoadenylate and activation of an endoribonuclease in interferon-treated HeLa cells infected with reovirus. J Virol. 1982;42:1039–1045. doi: 10.1128/jvi.42.3.1039-1045.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watling D, Serafinowska HT, Reese CB, Kerr IM. Analogue inhibitor of 2–5A action: effect on the interferon-mediated inhibition of encephalomyocarditis virus replication. Embo J. 1985;4:431–436. doi: 10.1002/j.1460-2075.1985.tb03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Defilippi P, Huez G, Verhaegen LM, De CE, Torrence P, Content J. Antiviral activity towards VSV and Mengo virus of a chemically stabilized 2–5A analog upon microinjection into HeLa cells. Prog Clin Biol Res. 1985;202:141–146. [PubMed] [Google Scholar]
- 51.Defilippi P, Huez G, Verhaegen LM, De CE, Imai J, Torrence P, Content J. Antiviral activity of a chemically stabilized 2–5A analog upon microinjection into HeLa cells. Febs Lett. 1986;198:326–332. doi: 10.1016/0014-5793(86)80430-6. [DOI] [PubMed] [Google Scholar]
- 52.Bisbal C, Bayard B, Silhol M, Leserman L, Lebleu B. 2′5′ Oligoadenylate analogues:synthesis, biological activities and intracellular delivery. Prog Clin Biol Res. 1985;202:89–95. [PubMed] [Google Scholar]
- 53.Bisbal C, Silhol M, Lemaitre M, Bayard B, Salehzada T, Lebleu B, Perree TD, Blackburn MG. 5′-modified agonist and antagonist (2′–5′)(A)n analogues: synthesis and biological activity. Biochemistry. 1987;26:5172–5178. doi: 10.1021/bi00390a041. [DOI] [PubMed] [Google Scholar]
- 54.Charachon G, Sobol RW, Bisbal C, Salehzada T, Silhol M, Charubala R, Pfleiderer W, Lebleu B, Suhadolnik RJ. Phosphorothioate analogues of (2′–5′)(A)4: agonist and antagonist activities in intact cells. Biochemistry. 1990;29:2550–2556. doi: 10.1021/bi00462a017. [DOI] [PubMed] [Google Scholar]
- 55.Merlin G, Chebath J, Benech P, Metz R, Revel M. Molecular cloning and sequence of partial cDNA for interferon-induced (2′–5′)oligo(A) synthetase mRNA from human cells. Proc Natl Acad Sci U S A. 1983;80:4904–4908. doi: 10.1073/pnas.80.16.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marie I, Hovanessian AG. The 69-kDa 2–5A synthetase is composed of two homologous and adjacent functional domains. J Biol Chem. 1992;267:9933–9939. [PubMed] [Google Scholar]
- 57.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 58.Rysiecki G, Gewert DR, Williams BR. Constitutive expression of a 2′,5′-oligoadenylate synthetase cDNA results in increased antiviral activity and growth suppression. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh A, Sarkar SN, Sen GC. Cell growth regulatory and antiviral effects of the P69 isozyme of 2–5 (A) synthetase. Virology. 2000;266:319–328. doi: 10.1006/viro.1999.0085. [DOI] [PubMed] [Google Scholar]
- 60.Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. Embo J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng X, Silverman RH, Zhou A, Goto T, Kwon BS, Kaufman HE, Hill JM. Increased severity of HSV-1 keratitis and mortality in mice lacking the 2–5A-dependent RNase L gene. Invest Ophthalmol Vis Sci. 2001;42:120–126. [PubMed] [Google Scholar]
- 62.Flodstrom-Tullberg M, Hultcrantz M, Stotland A, Maday A, Tsai D, Fine C, Williams B, Silverman R, Sarvetnick N. RNase L and double-stranded RNA-dependent protein kinase exert complementary roles in islet cell defense during coxsackievirus infection. J Immunol. 2005;174:1171–1177. doi: 10.4049/jimmunol.174.3.1171. [DOI] [PubMed] [Google Scholar]
- 63.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale M, Jr, Diamond MS. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. J Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Austin BA, James C, Silverman RH, Carr DJ. Critical role for the oligoadenylate synthetase/RNase L pathway in response to IFN-beta during acute ocular herpes simplex virus type 1 infection. J Immunol. 2005;175:1100–1106. doi: 10.4049/jimmunol.175.2.1100. [DOI] [PubMed] [Google Scholar]
- 66.Nilsen TW, Baglioni C. Mechanism for discrimination between viral and host mRNA in interferon- treated cells. Proc Natl Acad Sci U S A. 1979;76:2600–2604. doi: 10.1073/pnas.76.6.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nilsen TW, Weissman SG, Baglioni C. Role of 2′,5′-oligo(adenylic acid) polymerase in the degradation of ribonucleic acid linked to double-stranded ribonucleic acid by extracts of interferon-treated cells. Biochemistry. 1980;19:5574–5579. doi: 10.1021/bi00565a018. [DOI] [PubMed] [Google Scholar]
- 68.Baglioni C, De BA, Williams GJ. Cleavage of nascent reovirus mRNA by localized activation of the 2′–5′-oligoadenylate-dependent endoribonuclease. J Virol. 1984;52:865–871. doi: 10.1128/jvi.52.3.865-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baglioni C, Minks MA, Maroney PA. Interferon action may be mediated by activation of a nuclease by pppA2′p5′A2′p5′A. Nature. 1978;273:684–687. doi: 10.1038/273684a0. [DOI] [PubMed] [Google Scholar]
- 70.Gribaudo G, Lembo D, Cavallo G, Landolfo S, Lengyel P. Interferon action: binding of viral RNA to the 40-kilodalton 2′–5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desai SY, Patel RC, Sen GC, Malhotra P, Ghadge GD, Thimmapaya B. Activation of interferon-inducible 2′–5′ oligoadenylate synthetase by adenoviral VAI RNA. J Biol Chem. 1995;270:3454–3461. doi: 10.1074/jbc.270.7.3454. [DOI] [PubMed] [Google Scholar]
- 72.Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 73.Mordechai E, Kon N, Henderson EE, Suhadolnik RJ. Activation of the interferon-inducible enzymes, 2′,5′-oligoadenylate synthetase and PKR by human T-cell leukemia virus type I Rex-response element. Virology. 1995;206:913–922. doi: 10.1006/viro.1995.1014. [DOI] [PubMed] [Google Scholar]
- 74.Sharp TV, Raine DA, Gewert DR, Joshi B, Jagus R, Clemens MJ. Activation of the interferon-inducible (2′–5′) oligoadenylate synthetase by the Epstein-Barr virus RNA, EBER-1. Virology. 1999;257:303–313. doi: 10.1006/viro.1999.9689. [DOI] [PubMed] [Google Scholar]
- 75.Hovanessian AG, Wood JN. Anticellular and antiviral effects of pppA(2′p5′A)n. Virology. 1980;101:81–90. doi: 10.1016/0042-6822(80)90485-7. [DOI] [PubMed] [Google Scholar]
- 76.Cayley PJ, Davies JA, McCullagh KG, Kerr IM. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur J Biochem. 1984;143:165–174. doi: 10.1111/j.1432-1033.1984.tb08355.x. [DOI] [PubMed] [Google Scholar]
- 77.Hersh CL, Brown RE, Roberts WK, Swyryd EA, Kerr IM, Stark GR. Simian virus 40-infected, interferon-treated cells contain 2′,5′-oligoadenylates which do not activate cleavage of RNA. J Biol Chem. 1984;259:1731–1737. [PubMed] [Google Scholar]
- 78.Han JQ, Barton DJ. Activation and evasion of the antiviral 2′–5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. Rna. 2002;8:512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han JQ, Wroblewski G, Xu Z, Silverman RH, Barton DJ. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J Interferon Cytokine Res. 2004;24:664–676. doi: 10.1089/jir.2004.24.664. [DOI] [PubMed] [Google Scholar]
- 80.Paez E, Esteban M. Resistance of vaccinia virus to interferon is related to an interference phenomenon between the virus and the interferon system. Virology. 1984;134:12–28. doi: 10.1016/0042-6822(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 81.Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rivas C, Gil J, Melkova Z, Esteban M, Diaz GM. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)- induced 2–5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 83.Xiang Y, Condit RC, Vijaysri S, Jacobs B, Williams BR, Silverman RH. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J Virol. 2002;76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′–5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 86.Martinand C, Salehzada T, Silhol M, Lebleu B, Bisbal C. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2–5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur J Biochem. 1998;254:238–247. doi: 10.1046/j.1432-1327.1998.2540248.x. [DOI] [PubMed] [Google Scholar]
- 87.Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem. 2006;281:3773–3784. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- 88.Salehzada T, Silhol M, Steff AM, Lebleu B, Bisbal C. 2′,5′-Oligoadenylate-dependent RNase L is a dimer of regulatory and catalytic subunits. J Biol Chem. 1993;268:7733–7740. [PubMed] [Google Scholar]
- 89.Le Roy F, Salehzada T, Bisbal C, Dougherty JP, Peltz SW. A newly discovered function for RNase L in regulating translation termination. Nat Struct Mol Biol. 2005;12:505–512. doi: 10.1038/nsmb944. [DOI] [PubMed] [Google Scholar]
- 90.Stark GR, Dower WJ, Schimke RT, Brown RE, Kerr IM. 2–5A synthetase: assay, distribution and variation with growth or hormone status. Nature. 1979;278:471–473. doi: 10.1038/278471a0. [DOI] [PubMed] [Google Scholar]
- 91.Krishnan I, Baglioni C. Regulation of 2′5′-oligo(A) polymerase activity in quiescent human fibroblasts treated with interferon. Virology. 1981;111:666–670. doi: 10.1016/0042-6822(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 92.Williams BR, Kerr IM. Inhibition of protein synthesis by 2′–5′ linked adenine oligonucleotides in intact cells. Nature. 1978;276:88–90. doi: 10.1038/276088a0. [DOI] [PubMed] [Google Scholar]
- 93.Clemens MJ, Williams BR. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- 94.Schmidt A, Zilberstein A, Shulman L, Federman P, Berissi H, Revel M. Interferon action: isolation of nuclease F, a translation inhibitor activated by interferon-induced (2′–5′) oligo-isoadenylate. Febs Lett. 1978;95:257–264. doi: 10.1016/0014-5793(78)81006-0. [DOI] [PubMed] [Google Scholar]
- 95.Nilsen TW, Maroney PA, Baglioni C. Double-stranded RNA causes synthesis of 2′,5′-oligo(A) and degradation of messenger RNA in interferon-treated cells. J Biol Chem. 1981;256:7806–7811. [PubMed] [Google Scholar]
- 96.Nilsen TW, Maroney PA, Robertson HD, Baglioni C. Heterogeneous nuclear RNA promotes synthesis of (2′,5′)oligoadenylate and is cleaved by the (2′,5′)oligoadenylate-activated endoribonuclease. Mol Cell Biol. 1982;2:154–160. doi: 10.1128/mcb.2.2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Molinaro RJ, Jha BK, Malathi K, Varambally S, Chinnaiyan AM, Silverman RH. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Malathi K, Paranjape JM, Bulanova E, Shim M, Guenther-Johnson JM, Faber PW, Eling TE, Williams BR, Silverman RH. A transcriptional signaling pathway in the IFN system mediated by 2′–5′-oligoadenylate activation of RNase L. Proc Natl Acad Sci U S A. 2005;102:14533–14538. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hovanessian AG, Wood J, Meurs E, Montagnier L. Increased nuclease activity in cells treated with pppA2′p5′A2′p5′ A. Proc Natl Acad Sci U S A. 1979;76:3261–3265. doi: 10.1073/pnas.76.7.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Le Roy F, Bisbal C, Silhol M, Martinand C, Lebleu B, Salehzada T. The 2–5A/RNase L/RNase L Inhibitor (RNI) Pathway Regulates Mitochondrial mRNAs Stability in Interferon alpha -treated H9 Cells. J Biol Chem. 2001;276:48473–48482. doi: 10.1074/jbc.M107482200. [DOI] [PubMed] [Google Scholar]
- 101.Tnani M, Aliau S, Bayard B. Localization of a molecular form of interferon-regulated RNase L in the cytoskeleton. J Interferon Cytokine Res. 1998;18:361–368. doi: 10.1089/jir.1998.18.361. [DOI] [PubMed] [Google Scholar]
- 102.Bayard BA, Gabrion JB. 2′,5′-Oligoadenylate-dependent RNAse located in nuclei: biochemical characterization and subcellular distribution of the nuclease in human and murine cells. Biochem J. 1993;296:155–160. doi: 10.1042/bj2960155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bisbal C, Silhol M, Laubenthal H, Kaluza T, Carnac G, Milligan L, Le RF, Salehzada T. The 2′–5′ oligoadenylate/RNase L/RNase L inhibitor pathway regulates both MyoD mRNA stability and muscle cell differentiation. Mol Cell Biol. 2000;20:4959–4969. doi: 10.1128/mcb.20.14.4959-4969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Khabar KS, Siddiqui YM, al-Zoghaibi F, al-Haj L, Dhalla M, Zhou A, Dong B, Whitmore M, Paranjape J, Al-Ahdal MN, Al-Mohanna F, Williams BR, Silverman RH. RNase L mediates transient control of the interferon response through modulation of the double-stranded RNA-dependent protein kinase PKR. J Biol Chem. 2003;278:20124–20132. doi: 10.1074/jbc.M208766200. [DOI] [PubMed] [Google Scholar]
- 105.Li XL, Blackford JA, Judge CS, Liu M, Xiao W, Kalvakolanu DV, Hassel BA. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2–5A system in attenuation of the interferon response. J Biol Chem. 2000;275:8880–8888. doi: 10.1074/jbc.275.12.8880. [DOI] [PubMed] [Google Scholar]
- 106.Castelli JC, Hassel BA, Wood KA, Li XL, Amemiya K, Dalakas MC, Torrence PF, Youle RJ. A study of the interferon antiviral mechanism: apoptosis activation by the 2–5A system. J Exp Med. 1997;186:967–972. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Castelli JC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li XL, Hsu YT, Silverman RH, Youle RJ. The role of 2′–5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 108.Diaz GM, Rivas C, Esteban M. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology. 1997;236:354–363. doi: 10.1006/viro.1997.8719. [DOI] [PubMed] [Google Scholar]
- 109.Maitra RK, Silverman RH. Regulation of human immunodeficiency virus replication by 2′,5′-oligoadenylate-dependent RNase L. J Virol. 1998;72:1146–1152. doi: 10.1128/jvi.72.2.1146-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rusch L, Zhou A, Silverman RH. Caspase-dependent apoptosis by 2′,5′-oligoadenylate activation of RNase L is enhanced by IFN-beta. J Interferon Cytokine Res. 2000;20:1091–1100. doi: 10.1089/107999000750053762. [DOI] [PubMed] [Google Scholar]
- 111.Zhou A, Paranjape JM, Hassel BA, Nie H, Shah S, Galinski B, Silverman RH. Impact of RNase L overexpression on viral and cellular growth and death. J Interferon Cytokine Res. 1998;18:953–961. doi: 10.1089/jir.1998.18.953. [DOI] [PubMed] [Google Scholar]
- 112.Iordanov MS, Paranjape JM, Zhou A, Wong J, Williams BR, Meurs EF, Silverman RH, Magun BE. Activation of p38 mitogen-activated protein kinase and c-Jun NH(2)-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Chen SL, Magun BE. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol. 1997;17:3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Magun BE. Ultraviolet radiation triggers the ribotoxic stress response in mammalian cells. J Biol Chem. 1998;273:15794–15803. doi: 10.1074/jbc.273.25.15794. [DOI] [PubMed] [Google Scholar]
- 115.Lei K, Nimnual A, Zong WX, Kennedy NJ, Flavell RA, Thompson CB, Bar-Sagi D, Davis RJ. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol Cell Biol. 2002;22:4929–4942. doi: 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Costa-Pereira AP, McKenna SL, Cotter TG. Activation of SAPK/JNK by camptothecin sensitizes androgen-independent prostate cancer cells to Fas-induced apoptosis. Br J Cancer. 2000;82:1827–1834. doi: 10.1054/bjoc.2000.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li G, Xiang Y, Sabapathy K, Silverman RH. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J Biol Chem. 2004;279:1123–1131. doi: 10.1074/jbc.M305893200. [DOI] [PubMed] [Google Scholar]
- 118.Lou J, Anderson SL, Xing L, Rubin BY. Suppression of mitochondrial mRNA levels and mitochondrial function in cells responding to the anticellular action of interferon. J Interferon Res. 1994;14:33–40. doi: 10.1089/jir.1994.14.33. [DOI] [PubMed] [Google Scholar]
- 119.Lewis JA, Huq A, Najarro P. Inhibition of mitochondrial function by interferon. J Biol Chem. 1996;271:13184–13190. doi: 10.1074/jbc.271.22.13184. [DOI] [PubMed] [Google Scholar]
- 120.Bernardi P, Scorrano L, Colonna R, Petronilli V, Di LF. Mitochondria and cell death. Mechanistic aspects and methodological issues. Eur J Biochem. 1999;264:687–701. doi: 10.1046/j.1432-1327.1999.00725.x. [DOI] [PubMed] [Google Scholar]
- 121.Yanase N, Ohshima K, Ikegami H, Mizuguchi J. Cytochrome c release, mitochondrial membrane depolarization, caspase-3 activation, and Bax-alpha cleavage during IFN-alpha-induced apoptosis in Daudi B lymphoma cells. J Interferon Cytokine Res. 2000;20:1121–1129. doi: 10.1089/107999000750053799. [DOI] [PubMed] [Google Scholar]
- 122.Hovanessian AG, Laurent AG, Chebath J, Galabru J, Robert N, Svab J. Identification of 69-kd and 100-kd forms of 2–5A synthetase in interferon-treated human cells by specific monoclonal antibodies. Embo J. 1987;6:1273–1280. doi: 10.1002/j.1460-2075.1987.tb02364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Besse S, Rebouillat D, Marie I, Puvion DF, Hovanessian AG. Ultrastructural localization of interferon-inducible double-stranded RNA-activated enzymes in human cells. Exp Cell Res. 1998;239:379–392. doi: 10.1006/excr.1997.3908. [DOI] [PubMed] [Google Scholar]
- 124.Chandrasekaran K, Mehrabian Z, Li XL, Hassel B. RNase-L regulates the stability of mitochondrial DNA-encoded mRNAs in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2004;325:18–23. doi: 10.1016/j.bbrc.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 125.Liu W, Liang SL, Liu H, Silverman R, Zhou A. Tumour suppressor function of RNase L in a mouse model. Eur J Cancer. 2006 doi: 10.1016/j.ejca.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 126.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 127.Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, Walsh PC. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993;150:797–802. doi: 10.1016/s0022-5347(17)35617-3. [DOI] [PubMed] [Google Scholar]
- 128.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, Moses T, Ewing C, Gillanders E, Hu P, Bujnovszky P, Makalowska I, Baffoe-Bonnie A, Faith D, Smith J, Stephan D, Wiley K, Brownstein M, Gildea D, Kelly B, Jenkins R, Hostetter G, Matikainen M, Schleutker J, Klinger K, Connors T, Xiang Y, Wang Z, De Marzo A, Papadopoulos N, Kallioniemi OP, Burk R, Meyers D, Gronberg H, Meltzer P, Silverman R, Bailey-Wilson J, Walsh P, Isaacs W, Trent J. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 129.Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, Klein EA, Catalona WJ, Nupponen N, Carpten JD, Trent JM, Silverman RH, Witte JS. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32:581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 130.Silverman RH. Implications for RNase L in prostate cancer biology. Biochemistry. 2003;42:1805–1812. doi: 10.1021/bi027147i. [DOI] [PubMed] [Google Scholar]
- 131.Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, Trent JM, Isaacs WB, Casey G, Silverman RH. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 2003;63:6795–6801. [PubMed] [Google Scholar]
- 132.Rennert H, Bercovich D, Hubert A, Abeliovich D, Rozovsky U, Bar-Shira A, Soloviov S, Schreiber L, Matzkin H, Rennert G, Kadouri L, Peretz T, Yaron Y, Orr-Urtreger A. A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am J Hum Genet. 2002;71:981–984. doi: 10.1086/342775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rokman A, Ikonen T, Seppala E, Nupponen N, Autio V, Mononen N, Bailey-Wilson J, Trent J, Carpten J, Matikainen M, Koivisto PA, Tammela TL, Kallioniemi OP, Schleutker J. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70:1299–12304. doi: 10.1086/340450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Downing SR, Hennessy KT, Abe M, Manola J, George DJ, Kantoff PW. Mutations in ribonuclease L gene do not occur at a greater frequency in patients with familial prostate cancer compared with patients with sporadic prostate cancer. Clin Prostate Cancer. 2003;2:177–180. doi: 10.3816/cgc.2003.n.027. [DOI] [PubMed] [Google Scholar]
- 135.Wiklund F, Jonsson BA, Brookes AJ, Stromqvist L, Adolfsson J, Emanuelsson M, Adami HO, Augustsson-Balter K, Gronberg H. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin Cancer Res. 2004;10:7150–7156. doi: 10.1158/1078-0432.CCR-04-0982. [DOI] [PubMed] [Google Scholar]
- 136.Maier C, Haeusler J, Herkommer K, Vesovic Z, Hoegel J, Vogel W, Paiss T. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br J Cancer. 2005;92:1159–1164. doi: 10.1038/sj.bjc.6602401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, Latreille JP, Wilson RK, Ganem D, DeRisi JL. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, Ganem D, DeRisi J, Chow SA, Silverman RH. An infectious retrovirus susceptible to an interferon antiviral pathway from human prostate tumors. Proc Natl Acad Sci. 2007;104:1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bartsch DK, Fendrich V, Slater EP, Sina-Frey M, Rieder H, Greenhalf W, Chaloupka B, Hahn SA, Neoptolemos JP, Kress R. RNASEL germline variants are associated with pancreatic cancer. Int J Cancer. 2005;117:718–722. doi: 10.1002/ijc.21254. [DOI] [PubMed] [Google Scholar]
- 142.Kruger S, Silber AS, Engel C, Gorgens H, Mangold E, Pagenstecher C, Holinski-Feder E, von Knebel Doeberitz M, Moeslein G, Dietmaier W, Stemmler S, Friedl W, Ruschoff J, Schackert HK. Arg462Gln sequence variation in the prostate-cancer-susceptibility gene RNASEL and age of onset of hereditary non-polyposis colorectal cancer: a case-control study. Lancet Oncol. 2005;6:566–572. doi: 10.1016/S1470-2045(05)70253-9. [DOI] [PubMed] [Google Scholar]
- 143.Behan PO, Behan WM, Bell EJ. The postviral fatigue syndrome--an analysis of the findings in 50 cases. J Infect. 1985;10:211–222. doi: 10.1016/s0163-4453(85)92488-0. [DOI] [PubMed] [Google Scholar]
- 144.Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group [see comments] Ann Intern Med. 1994;121:953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 145.Suhadolnik RJ, Reichenbach NL, Hitzges P, Sobol RW, Peterson DL, Henry B, Ablashi DV, Muller WE, Schroder HC, Carter WA, et al. Upregulation of the 2–5A synthetase/RNase L antiviral pathway associated with chronic fatigue syndrome. Clin Infect Dis. 1994:S96–104. doi: 10.1093/clinids/18.supplement_1.s96. [DOI] [PubMed] [Google Scholar]
- 146.Suhadolnik RJ, Reichenbach NL, Hitzges P, Adelson ME, Peterson DL, Cheney P, Salvato P, Thompson C, Loveless M, Muller WE, et al. Changes in the 2–5A synthetase/RNase L antiviral pathway in a controlled clinical trial with poly(I)-poly(C12U) in chronic fatigue syndrome. In Vivo. 1994;8:599–604. [PubMed] [Google Scholar]
- 147.Suhadolnik RJ, Peterson DL, O’Brien K, Cheney PR, Herst CV, Reichenbach NL, Kon N, Horvath SE, Iacono KT, Adelson ME, De MK, De BP, Charubala R, Pfleiderer W. Biochemical evidence for a novel low molecular weight 2–5A-dependent RNase L in chronic fatigue syndrome. J Interferon Cytokine Res. 1997;17:377–385. doi: 10.1089/jir.1997.17.377. [DOI] [PubMed] [Google Scholar]
- 148.De Meirleir K, Bisbal C, Campine I, De Becker P, Salehzada T, Demettre E, Lebleu B. A 37 kDa 2–5A binding protein as a potential biochemical marker for chronic fatigue syndrome. Am J Med. 2000;108:99–105. doi: 10.1016/s0002-9343(99)00300-9. [DOI] [PubMed] [Google Scholar]
- 149.Tiev KP, Demettre E, Ercolano P, Bastide L, Lebleu B, Cabane J. RNase L levels in peripheral blood mononuclear cells: 37-kilodalton/83-kilodalton isoform ratio is a potential test for chronic fatigue syndrome. Clin Diagn Lab Immunol. 2003;10:315–316. doi: 10.1128/CDLI.10.2.315-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tiev KP, Briant M, Ziani M, Cabane J, Demettre E, Lebleu B. Variability of the RNase L isoform ratio (37 kiloDaltons/83 kiloDaltons) in diagnosis of chronic fatigue syndrome. Clin Diagn Lab Immunol. 2005;12:366. doi: 10.1128/CDLI.12.2.366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Demettre E, Bastide L, D’Haese A, De Smet K, De Meirleir K, Tiev KP, Englebienne P, Lebleu B. Ribonuclease L proteolysis in peripheral blood mononuclear cells of chronic fatigue syndrome patients. J Biol Chem. 2002;277:35746–35751. doi: 10.1074/jbc.M201263200. [DOI] [PubMed] [Google Scholar]
- 152.Fremont M, El Bakkouri K, Vaeyens F, Herst CV, De Meirleir K, Englebienne P. 2′,5′-Oligoadenylate size is critical to protect RNase L against proteolytic cleavage in chronic fatigue syndrome. Exp Mol Pathol. 2005;78:239–246. doi: 10.1016/j.yexmp.2005.01.003. [DOI] [PubMed] [Google Scholar]