Abstract
Systems theoretic tools (i.e. mathematical modelling, control, and feedback design) advance the understanding of robust performance in complex biological networks. We highlight phase entrainment as a key performance measure used to investigate dynamics of a single deterministic circadian oscillator for the purpose of generating insight into the behaviour of a population of (synchronized) oscillators. More specifically, the analysis of phase characteristics may facilitate the identification of appropriate coupling mechanisms for the ensemble of noisy (stochastic) circadian clocks. Phase also serves as a critical control objective to correct mismatch between the biological clock and its environment. Thus, we introduce methods of investigating synchrony and entrainment in both stochastic and deterministic frameworks, and as a property of a single oscillator or population of coupled oscillators.
Keywords: circadian clocks, phase response curves, coupled oscillators, control, stochastic simulations
1. Introduction
Undergirding a biological system are networks of interacting components. To elucidate the mechanisms employed by these networks, biological experimentation and intuition are by themselves insufficient (Kitano 2002). In the field of systems biology, investigators characterize dynamics via mathematical models and apply systems theory with the goal of guiding further experimentation to better understand the biological network that gives rise to robust performance (Fall et al. 2005). An ideal example of biological complexity is the circadian clock, which coordinates daily physiological behaviours of most organisms. The robust timekeeping of the circadian clock may be correlated to its internal structure (a synchronized network of coupled oscillators) and its ability to be entrained by the environment (most notably Sun cycles).
To better understand, characterize and control the mechanisms that give rise to robust circadian performance, we study its phase dynamics. Given that the circadian clock architecture is hierarchical and depending upon where in the hierarchy we analyse phase, we apply different methods and strategies of investigation. Several mathematical models have been developed which aim to characterize the network underlying circadian rhythmicity in a variety of organisms including mammals, flies, plants and algae (Leloup & Goldbeter 1998, 2003; Tyson et al. 1999; Ueda et al. 2001; Forger & Peskin 2003; Smolen et al. 2004). Of these models, we consider those specific to Mus musculus (house mouse) and Drosophila melanogaster (fruit fly).
Left in constant conditions, the clock will free-run with a period of only approximately 24 hours such that its internal time, or phase, drifts away from that of its environment. Thus, vital to a circadian clock is its ability to entrain to external time through environmental factors (Daan & Pittendrigh 1976; Boulos et al. 2002; Dunlap et al. 2004). To study the timekeeping of circadian clocks, we employ systems theory to analyse two complementary cases: one involving the network of coupled oscillators and the other involving a single coherent oscillator. To study synchronization and entrainment of circadian oscillators at different levels of the hierarchy, we use ordinary differential equations (ODEs), stochastic differential equations (SDEs) and a discrete stochastic model in both the network and single-cell setting. In §2, we outline the biological questions we are investigating and describe the techniques best suited to answer each. In §3, we simulate a population of mammalian (Mus) neurons using a discrete stochastic model. The challenges associated with achieving synchronization are addressed via the study of a single oscillator. In §4, we simulate an SDE model of the mammalian network and analyse the period of the synchronized neurons by studying the phase response behaviour of a single deterministic cell. Section 5 describes a strategy that makes use of the light-induced circadian phase response to correct phase mismatch that arises when there is a difference between internal and external time.
2. Background
For most species, including M. musculus and D. melanogaster, the behaviour of a single clock cell is controlled by a gene regulatory network. Some genes, such as the period and clock genes, are shared by multiple species whereas others are particular to the organism. The gene/protein components form a complex transcriptional regulatory structure made up of both positive and negative feedback loops that describe the competitive activation and inhibition of circadian mRNA transcription. Many of the mathematical models of the circadian clock in the literature consist of ODEs that exhibit limit cycle behaviour. This is the class of models addressed in this work, with extensions to discrete stochastic behaviour as well.
Three genes key to the D. melanogaster clock are period (per), timeless (tim) and Drosophila clock (dClk). Here, we study the mathematical model of the clock as developed by Ueda et al. (2001). In Ueda et al. (2001), there is a negative feedback loop in which per and tim mRNA cycle in phase translate to PER and TIM proteins and form PER:TIM heterodimers that downregulate per and tim mRNA transcription. An additional negative feedback loop involves dClk mRNA, which cycles anti-phase to per and tim mRNA. After translation, dCLK protein forms a heterodimer with the protein Cycle (CYC). dCLK:CYC then downregulates dClk mRNA transcription. The loops are interlocked via additional regulation—dCLK:CYC upregulates per and tim mRNA transcription and PER:TIM upregulates dClk mRNA transcription (see fig. 1 in Ueda et al. (2001)).
The network structure in a M. musculus cell involves three forms of period (per1, per2 and per3), two forms of cryptochrome (cry1 and cry2), Brain and muscle Arnt-like protein-1 (Bmal1), clock (clk), and rev-erb α. The per and cry genes and proteins form a negative feedback loop similar to the PER:TIM negative feedback loop in Drosophila—a form of PER becomes a heterodimer with a form of CRY, which then enters the nucleus and indirectly downregulates transcription of PER and CRY. This indirect regulation is modulated by the BMAL1 and CLK proteins. Additionally, REV-ERB α indirectly upregulates PER and CRY. In this work, we study two detailed models of the mammalian clock. One is the 16-state version of Leloup & Goldbeter (2003), while the other is the 71-state model by Forger & Peskin (2003). Both models take into account the regulatory networks outlined above, using different assumptions about the kinetics necessary to capture certain subprocesses. Leloup & Goldbeter lump the various forms of per and cry into a single form of per and cry, respectively. Forger & Peskin model per1, per2, cry1 and cry2, but not per3.1 Light is the dominant external input used to coordinate the cellular clock with the environment. The presence of regular sunlight entrains the clock to acquire 24-hour periodicity in addition to an appropriate phase relationship with respect to the light/dark cycle. The action of light on the clock causes it to shift its phase to align the clock's internal (subjective) time with the external (environmental) time. The precise mechanism through which light acts on circadian clock cells is not yet known, but evidence suggests that it acts by rapid induction of per expression (Reppert & Weaver 2002). Thus, in both mammalian models, light is modelled as an increase in the rate of per transcription.
In both the M. musculus and D. melanogaster systems, there is stochastic behaviour at the single-cell level. Through the coupling of these cells, the population of stochastic oscillators becomes coherent. We aim to investigate how the communication of single cells confers robustness in the mammalian network,2 and how the stable, coherent master clock is entrained by an external signal. Thus, the mathematical model used to investigate coupling and entrainment relies on the nature of the investigation, the method, and the computational constraints associated with the investigation.
In §3, we seek to determine whether the coupling mechanism is capable of reproducing data at many different levels. We aim to capture the temporal variability of the data through the use of a discrete stochastic model, a quality that was not assessed in a previous study (To et al. 2007). Thus, we make use of the most detailed, realistic version of the system that is currently available. In this manner, we analyse the type of noise that results from complex biophysical behaviour, as opposed to idealized noise added to a simple model. In §4, we introduce stochasticity via multiplicative white noise and simulate the model as a system of stochastic differential equations (SDEs). By moving from a discrete stochastic to a continuous stochastic framework, the noise we consider is idealized (rather than variability arising ‘naturally’) and we do not capture all effects due to low molecule counts. However, the results reflect biological data (Ueda et al. 2002). The SDE simulation is less computationally complex than a discrete stochastic simulation, which is important because we simulate many different coupling models. To better understand the results of the SDE simulations, we make use of the noiseless, ODE version of the model. The combination of these methods allows for the assessment of more complicated systems. Finally, in §5, we investigate the interaction between an external cue (most notably, light) and the coherent oscillator. Assuming the coherent oscillator defines a synchronized population of single circadian cells and knowing that the optimal control algorithm is computationally expensive, phase resetting properties of the clock are examined via a set of ODEs that characterize the dynamics of a single cell. Table 1 provides a summary of the objectives, tools and resources used to investigate the synchrony and entrainment problems outlined for each section.
Table 1.
A summary of relevant studies concerning the synchronization and entrainment of fly and mammalian circadian clocks as they relate to the content of this paper.
| section 3 | section 4 | section 5 | |
|---|---|---|---|
| objective | to determine whether the VIP coupling mechanism reproduces mammalian SCN data | to determine how the coupling mechanism may identify circadian periodicity | to investigate light as a control target to optimize the re-entrainment of biological oscillators |
| problem | tuning | prediction | control |
| tools | bifurcation analysis. The study of deterministic/stochastic models of single cells | pIPRC analysis. The study of deterministic single-cell models | the application of model predictive control (MPC) |
| model type | discrete stochastic models of coupled oscillators | SDE models of coupled oscillators | ODE models of the clock as a single oscillator |
| references | Leloup & Goldbeter (2003) and To et al. (2007) | Ueda et al. (2002) | Forger & Peskin (2003) |
3. Modelling coupled stochastic mammalian neurons
In mammals, the circadian master clock resides in the suprachiasmatic nucleus (SCN), located in the hypothalamus (Reppert & Weaver 2002). It is a network of multiple autonomous noisy oscillators, which communicate via neuropeptides to synchronize and form a coherent oscillator (Herzog et al. 2004; Liu et al. 2007). This coherent oscillator then coordinates the timing of daily behaviours, such as the sleep/wake cycle. Biological experiments, however, demonstrate that uncoupled neurons in the SCN are either damped or sloppy oscillators (Aton et al. 2005). Thus, coupling in the SCN causes a collection of stochastic, unreliable oscillators to form a robust oscillator that can be reliably reset. To unravel the design principles behind this remarkable behaviour, mathematical models must incorporate the stochastic properties of the single cell, while coupling the population of cells through biophysical components. A putative coupling agent is the vasoactive intestinal (neuro)peptide (VIP; Herzog et al. 2004), whose intercellular concentration levels peak during the subjective day. Preliminary results demonstrate that controlled VIP pulses cause phase shifts similar to those resulting from light pulses, suggesting that the VIP signal and target are similar to those of light and may be modelled correspondingly. The target of VIP signalling is therefore assumed to be per transcription (Piggins et al. 1995).
Experimental data demonstrate that isolated (uncoupled) neurons exhibit both a broad distribution of periods and temporal (cycle-to-cycle) variability (Herzog et al. 2004). Hao et al. (2006) and To et al. (2007) postulated mechanisms through which VIP signals are received by a cell via signal cascades culminating in the modulation of the parameter associated with per transcription. Using an ODE model, To et al. incorporate this coupling mechanism into a population of non-identical cells, each of which is based on the gene regulatory network model of Leloup & Goldbeter (2003). They simulate scenarios with no coupling (the cells drift out of phase) and with coupling (the cells form a coherent oscillator), demonstrating that their mechanism is capable of creating the spontaneous synchronization seen in experimental data. Likewise, their simulations show a broad distribution of periods across cells. However, because they use a deterministic model, they do not reproduce cycle-to-cycle variation. We develop a discrete stochastic model based on that in To et al. (2007) incorporating intrinsic noise, and consequently temporal variability.3 We simulate this model in a two-dimensional grid of 25 SCN neurons using a software package (StochKit, http://www.cs.ucsb.edu/∼cse/StochKit) based on the stochastic simulation algorithm (SSA; Gillespie 1976, 1977). Successful synchronization of 25 coupled cells validates the mechanism in the presence of noise.
3.1 Simulating a single cell
The introduction of noise alters the behaviour of the single cells such that additional tuning is required to achieve synchrony. In particular, because VIP signalling ultimately manifests as modulation of the rate of per transcription, special attention must be paid to the levels of per mRNA and its rate of transcription, νsP(t). The basal rate νsP0 characterizes the behaviour of an isolated cell: per mRNA oscillations are damped when νsP0<1.2 and sustained when νsP0≥1.2 (figure 1). Fig. 6 in Leloup & Goldbeter (2004) shows the period versus νsP0, which peaks at νsP0=1.5 with a period of 23.8 hours. The depletion or accumulation of per mRNA that occurs when νsP0 is below 1.2 indicates that the balance is upset between the transcription rate and the combination of the transport (from nucleus to cytoplasm) and degradation. For the coupled population to exhibit synchrony, we have observed that the median value of νsP(t) must stay within the range that produces oscillations in an individual cell. Thus to achieve synchrony, the basal transcription rate νsP0 has been set to 1.5 for the coupling topology and volume used in this work. At this basal transcription rate, all isolated cells are oscillators.
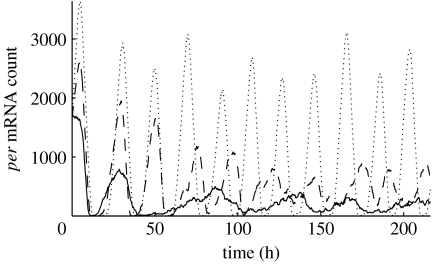
Figure 1.
Per mRNA population as a function of basal transcription rate νsP0 in uncoupled cells. The solid line represents a simulation where νsP0=1, the dashed line νsP0=1.5 and the dotted line νsP0=2. For νsP0 below 1.2, per mRNA concentrations exhibit damped oscillations for the 10 day period simulated.
3.2 Simulating a population
Simulation of a 5×5 grid of cells shows that the VIP coupling mechanism is capable of achieving synchrony (figure 2) between stochastic cells exhibiting temporal variability in period and amplitude. To measure the phase coherence of the cells in a simulation, we use the radius r(t) of the complex order parameter (Strogatz 2000), computed according to
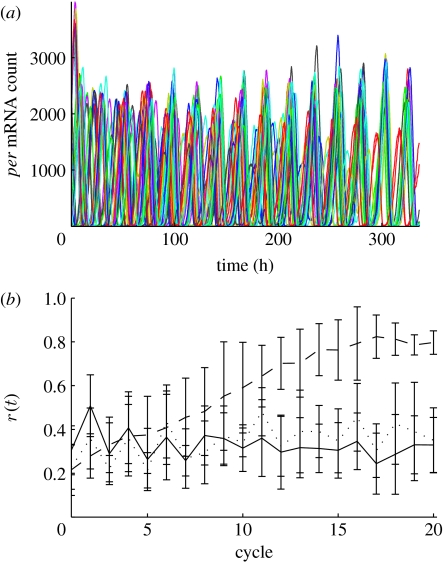
where N is the number of cells; θj is the phase of the jth oscillator; and Ψ(t) is the average phase. If the oscillators are in phase, r(t)≈1. Here the mean r(t) across 10 simulations begins at 0.2–0.3 with uniform random initial phase and increases to 0.8 in 14 cycles when coupled. These data demonstrate the effective response of intercellular signalling that gives rise to phase synchrony.
Figure 2.
(a) The time series of per mRNA concentration is shown for a single SSA simulation of a 5×5 grid of cells. (b) For 10 SSA simulations of the grid, we show the mean degree of phase coherence with error bars indicating the standard deviation; the solid line represents simulations where νsP0=1, the dashed line νsP0=1.5 and the dotted line νsP0=2.
As in Gonze et al. (2004), proximity to the bifurcation point νsP0=1.06 (Leloup & Goldbeter 2004) in the deterministic ODE model predicts oscillatory behaviour in the stochastic simulation based on the same cell model. Unlike the results in Gonze et al. (2005), synchronization of the 5×5 grid of cells does not occur when the coupling parameter is below the bifurcation point where individual cells are damped oscillators. This is due to both the nature of the individual cell model and the properties of the coupling signal, which are dependent on the grid connectivity, size and coupling strength. The range of νsP0, which permits synchronization using mean-field coupling, appears to be bounded on the low end by the bifurcation point. Further investigation will be needed to deduce how the upper bound depends on the coupling network; but for this choice of connectivity and coupling strength νsP0 has a fairly narrow range that will allow synchronization. Presumably this is due to the independent νsP0 signals overpowering the common coupling signals (Jensen 2002). In Bernard et al. (2007) this self-feedback is modelled as an autocrine signal that is scaled with the same coupling strength as signals from other cells. As the coupling strength is increased, the autocrine signal is also increased. This is analogous to increasing νsP0 in this model to produce rhythmic cells. Maywood et al. (2006) observed that in an intact SCN the loss of VIP coupling had the effect of suppressing rhythmicity in many cells and loss of synchrony in the cells that retained rhythmicity. Whether the difference in behaviours can be attributed to heterogeneity in the cells themselves or how they are coupled within the SCN remains an open question (Yamaguchi & Shimizu 1984).
4. Analysing coupled D. melanogaster neurons
As demonstrated in §3, single-cell simulation can be used to tune the simulation of a population, bringing about the desired synchrony. In this section, we present single-cell analysis as a complement to simulation and demonstrate that it yields a better understanding of the behaviour of a coupled population. We analyse the phase behaviour of an SDE model of 100 coupled neurons of the D. melanogaster circadian pacemaker. In an investigation of potential coupling mechanisms, Ueda et al. (2002) developed a framework with 960 potential coupling mechanisms, each of which includes a component (source) that generates a ‘synchronizing factor’ sent to neighbouring cells in a hexagonal lattice. This synchronizing factor then acts as a signal by modulating a given target parameter within each cell.
The authors show that a subset of the potential coupling mechanisms produces spontaneous (phase) synchronization among the cells. We expand upon their analyses by simulating the population for each of the 960 coupling mechanisms, 84 of which produce (phase) synchrony. However, not all synchronized systems display the same behaviour. Notably, the period of oscillation is different for each coupling mechanism and ranges between 20 and 37 hours. Because we are interested in rhythms that are circadian, we study the 82 coupling mechanisms that produce periods within 20–28 hours. The data describe what will happen, but leave unanswered questions such as why some source/target pairs are speeding up the oscillations and some are slowing them down, and how an adjustment of the relative timing between the signal and the cell's phase would change the timing.
4.1 Theoretical background
There is a rich literature concerning the mathematical analysis of coupled oscillators (Kuramoto 1984; Hoppensteadt & Izhikevich 1997; Winfree 2001; Brown et al. 2004). The most heavily studied systems, such as the Kuramoto model, assume that interactions among oscillators are sinusoidal (Kuramoto 1984; Strogatz 2000) or that they perturb state velocities directly (Brown et al. 2004). In circadian clock models, a signal sent to an oscillator ultimately manifests as the manipulation of a single parameter. Thus, to study the effects of signalling on the phase behaviour, we must examine the effects of parametric manipulation on phase behaviour. We use the parametric impulse phase response curve (pIPRC; Taylor et al. 2008), which predicts the oscillator's velocity change in response to parametric perturbation. To use the pIPRC, we study a single neuron, modelled as a set of ODEs with a stable attracting limit cycle . The solution along the limit cycle is periodic with period τ, and we describe its progress along the cycle by its phase ϕ. When the clock is unperturbed, the phase progresses at the same rate as time, dϕ(x(t, p))/dt≡1. When the clock is perturbed (via a change in the jth parameter), the velocity response is predicted by the pIPRC
Computation is relatively straightforward, requiring only the solution of the adjoint linear variational equation and the differentiation of f with respect to pj (Taylor et al. 2008). This requires the system Jacobian (which can be estimated numerically or generated exactly via automatic differentiation) and the state trajectories over time. Interpretation is also straightforward. Consider a signal Δpj(t, ϕ) which is a function of either time or phase. This signal will change the oscillator's velocity according to Δϕ/Δt≈pIPRCj(ϕ)Δpj(t, ϕ). For example, figure 3a shows the pIPRC for the parameter associated with the maximal rate of degradation for clock component tim mRNA. From hours 0 to 3, there is a so-called dead zone—a region in which a signal will cause no change in the phase velocity. It is followed by a delay zone and then by an advance zone. Another interpretation of the pIPRC is that, for a pulse of duration Δt, a phase shift is caused according to . Using this interpretation, the pIPRC predicts phase response curves (PRCs), which have been used by experimentalists in the circadian community for over 40 years (Johnson 1999). The PRC characterizes the clock's time-dependent sensitivity to a given stimulus (in this work, a pulse of light or a neuropeptide) by mapping the stimulating signal's arrival time to the resultant phase shift (Daan & Pittendrigh 1976). The pIPRC can be said not only to predict the PRC but also to characterize the timing behaviour of an oscillator alone.
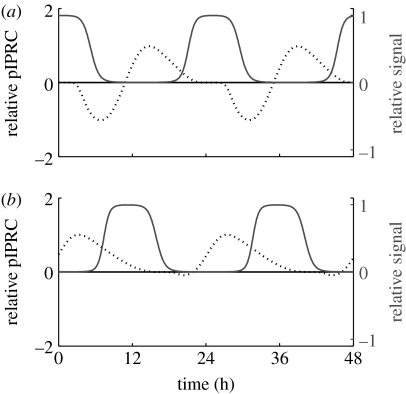
Figure 3.
The pIPRC (black dotted line) and signal trace (grey solid line) for coupling mechanisms that cause the coupled SDE system to synchronize with (a) long periods (slow oscillations; observed τ=25.2119) and (b) short periods (fast oscillations; observed τ=22.6764). All curves are relative to the target parameter's nominal value.
4.2 Methodology
Each candidate coupling mechanism is described in Ueda et al. (2002) by the ODEs4 that govern the ith cell. Each cell contains the components outlined in §2, e.g. PER, TIM, and dCLK mRNA and proteins. A candidate coupling mechanism involves a source Yi (1 of the 10 components of the cell model) and a target reaction (associated with 1 of the 24 rate parameters of the cell model) in addition to rules about their interactions. Yi up- or downregulates the synchronizing factor Xi. The concentration of Xi is dependent upon its dynamics in both cell i and its neighbouring cells according to eqn. B.1 of Ueda et al. (2002), i.e.
| (4.1) |
where the cells indexed by m are the neighbours of cell i and SX, AX, aX, DX, LX, D0 and Dc are constant parameters (see appendix B in Ueda et al. 2002). The values of aX, SX and AX are set depending upon the regulation of Xk by Yk (see table B1 in Ueda et al. 2002). The levels of Xi in turn up- or downregulate a target parameter pj in the ith cell according to
| (4.2) |
where M, k and Xh are constant parameters. The modulated parameter is then used in place of . The signal is
| (4.3) |
To understand the period of the synchronized cells of the Drosophila clock for a given coupling mechanism, we study the relationship between the signal (Δpj) and the pIPRC for the target. To acquire the signal, we capitalize on the stable synchrony and make the additional assumption that it is reasonable to ignore any effects due to stochasticity—in a noiseless synchronized system, each neuron contains the same concentration of X at the same time. This enables us to use a deterministic single-cell simulation. Equation (4.1) is altered to incorporate the assumption that Xi=Xg for all cells g, leading to
| (4.4) |
In the simulation, equations (4.2) and (4.3) provide the signal at each time step, but we prevent the cell from ‘signalling itself’ by disregarding and using . The signal and pIPRCj are computed with the same single-cell ODEs, share the same period and can be plotted together for the ease of comparison. Two example signals along with their associated pIPRCs are shown in figure 3.
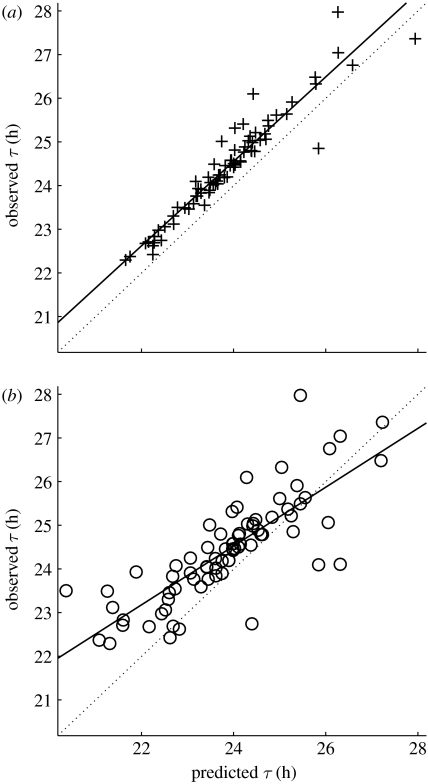
Before we begin our analysis, we evaluate the predictive power of the single-cell deterministic model and of the pIPRC for each potential coupling mechanism. First, we predict the period of the synchronized SDE population by simulating a single-cell ODE model, allowing it to signal itself. As above, we replace equation (4.1) with equation (4.4). Unlike above, we use . After several cycles, it converges to a new limit cycle with a new period—a prediction of the period of the synchronized system. In figure 4a we plot our prediction for the new period against the observed period of the SDE model of the full population. The square of the Pearson correlation coefficient, R2, is 0.91 and the data are located within an hour of the observed values.
Figure 4.
Observed versus predicted periods. For each coupling mechanism that produces synchrony there is a plus or a circle. (a) The x-axis position indicates the period predicted using a single cell signalling itself. (b) The x-axis position indicates the period predicted using the pIPRC. For both subplots the y-axis position indicates the period observed once collection of noisy cells becomes (and remains) synchronized. Perfect predictions fall on the dotted line. The solid lines represent the best fit by linear regression analysis, with (a) R2=0.91 (plus) for the single-cell data and (b) R2=0.65 (open circle) for the pIPRC-predicted data.
Next, we use the pIPRC directly to make similar predictions. Under the assumptions that the oscillators are identical and that the coupling is sufficiently weak (and thus ϕ does not deviate significantly from time), we use the method of averaging to predict the change in period. This is analogous to methodology used in the coupled oscillator literature (Kuramoto 1984),5 but is a novel application of the pIPRC. By replacing time with ϕ as the independent variable, we predict the change in period by assessing the effect of the signal on the oscillator over a single cycle. Integrating over the cycle
we find that the predictions are qualitatively accurate. Figure 4b shows the predicted period change of the synchronized system versus the observed period change of the synchronized system. The R2 value between the observed and predicted periods is 0.65. The data are more scattered than those from the full cell simulation, though the majority are within 1 hour of perfect prediction. We conclude that although neither of these methods is a perfect predictor, their qualitative correctness supports our approach.
4.3 Phase response behaviour
Figure 3a shows the signal and pIPRC for a coupling mechanism that causes the system to slow down. In this mechanism, nuclear PER:TIM upregulates the synchronizing factor, which upregulates the maximal rate of degradation for clock component tim mRNA. Figure 3b shows the signal and pIPRC related to a coupling mechanism in which cytoplasmic dCLK upregulates the synchronizing factor, which upregulates the maximal rate of dClk mRNA degradation. The pIPRCs by themselves provide insight into the oscillator. First we observe that the pIPRCs associated with dClk mRNA degradation and tim mRNA degradation peak in anti-phase with each other. This is an intuitive result, as the traces of dClk mRNA and tim mRNA peak in anti-phase with each other (see Ueda et al. 2002; figure 1). Less intuitive is the absence of an appreciable delay zone in the dClk mRNA degradation pIPRC. This is in direct contrast with the tim mRNA degradation pIPRC, which shows a large delay region. We might expect dClk and tim mRNA degradation to have similar but anti-phase effects, because the dynamics concerning dClk and tim mRNA have a similar construction (both are regulated by the same proteins, but with opposite effects). However, intuition alone fails to capture the full effects of the complex intracellular dynamics.
The relationship between the signal and pIPRC allows us to understand how the period of the SDE population is determined. In figure 3a, the signal arrives at the tail end of the advance zone and is active during the dead zone and the first half of the delay zone, leading to a cycle that is slower than nominal. Other synchronization-causing coupling mechanisms that upregulate tim mRNA degradation use different components to regulate the synchronizing factor. This results in different phase relationships between the signal and pIPRC, and ultimately in periods that are both fast and slow (see electronic supplementary material, figure 1). In figure 3b, the pIPRC shows nearly negligible delay regions. It follows that, regardless of the phase relationship between the signal and target, the oscillator will respond by speeding up. In fact, there exist two additional coupling mechanisms that upregulate dClk mRNA degradation, and they produce populations with short periods as well (see electronic supplementary material, figure 2).
In both of these mechanisms (and in all mechanisms that produce synchrony), the relationship between the signal and target meets the criteria for stable entrainment (data not shown). If the signal arrives early (because the phase of the system is a little behind), the system is sped up more (or slowed down less) than usual and vice versa. The study of the pIPRC and signal is consistent with the observed behaviour—the mutual entrainment is stable and the system remains synchronized. However, meeting the conditions for mutual entrainment is merely a necessary factor; there exist pairs that meet the requirements for stable entrainment but do not yield a transition to synchrony (data not shown). This is due to the relatively large perturbations caused by the signal in conjunction with the stochasticity in the simulations.
5. Phase entrainment
Just as signalling among cells serves to synchronize the phase of the network, signals from the environment serve to entrain, or reset, the phase of emergent coherent oscillators. More specifically, environmental factors such as light induce phase shifts that calibrate an organism's internal phase to external time.
Exploring phase resetting through controlled light pulses is not a recent interest. In the early 1970s, Daan & Pittendrigh investigated light-induced phase shifts in free-running organisms through the development of PRCs. Watanabe et al. (2001) build upon Daan & Pittendrigh's investigation of light-induced phase shifts in free-running organisms by proving that entrainment in mammals involves both advance and delay components of the PRC. Boulos et al. (2002) extend the application of PRCs by establishing bright light treatment as a means to accelerate circadian re-synchronization rates. Similarly, Forger & Peskin (2005) invoke calculus of variations and an analysis of phase space to find the optimum stimuli that start, stop and reset the phase of a simplified biological oscillator. In a previous study, we optimized the re-synchrony of a detailed mammalian circadian oscillator to the environment through the systematic application of light. We showed that a closed-loop model predictive control (MPC) algorithm is an effective (and realistic) means of resetting the organisms' phase (Bagheri et al. 2007). Through MPC, we were able to eliminate an induced phase difference between an organism (whose biological clock is modelled as a single deterministic oscillator) and the natural 24 hour light/dark environment. In other words, we synchronized the timing of the internal circadian clock with that of the Sun cycle. Here, we investigate phase resetting dynamics as a function of the MPC tuning parameters (described in §5.2) as well as the attributes of the driving force (light).
5.1 A general nonlinear mammalian model
The circadian dynamics of a single deterministic M. musculus limit cycle oscillator (Forger & Peskin 2003) serves as the example system. The model is generalized as a set of nonlinear ODEs with time t, n-length state vector x(t), environmental light input L(t), controlled light input u(t) and system dynamics f(x(t), L(t), u(t))
In this paper, the nominal model (a version of the model that has converged to the natural light/dark environment where u(t)=0 and L(t) oscillates as a square wave between values 3.39×10−2 and 0) is used to define the reference r(t). A circadian time of 0 reflects dawn while a circadian time of 12 reflects dusk, assuming regular 24 hour day/night cycles.
5.2 Optimizing the manipulated control profile
MPC (Morari & Lee 1999) describes a strategy that relies on a (most often linear) mathematical representation of the system (in this case, circadian rhythms) to predict the output, or response, of the system with respect to a given control input. With phase entrainment as the objective, we define the output as a measure of phase synchrony with respect to the control variable, light. By simulating the future behaviour of the system, we are able to optimize the control input based on the cost, or quantified fitness, of the predicted response. Hence, MPC is used to increase the resynchronization rate of circadian oscillators through the systematic addition of light. The algorithm samples the system output (protein concentrations) every ts=2 hours, updating the mathematical model and simulating a discrete time system with index . The manipulated light profile u(k) optimizes the performance objective on a time interval extending from the current time to the current time plus a prediction horizon (the time interval over which the simulation of the predictive mathematical model is run) P=54 hours, where . This horizon allows the algorithm to take control action at the current time in response to a forecasted error. The move horizon M limits the number of controlled light pulses within the prediction horizon such that u(k) spans a time interval ; by definition, the move horizon must be less than or equal to the prediction horizon. Beyond M hours of simulation, the control input defaults to u(k)=0. Future behaviours for a variety of control inputs are computed according to the model of the plant.
Given umin=−3.39×10−2, umax=3.39×10−2 and L(k)+u(k)≥0, the cost function penalizes the normalized predicted error between the reference and controlled trajectories e(k) and its corresponding control sequence u(k). To avoid penalizing transient effects, the state error is weighted uniformly over the move horizon (reflected in the first m diagonal values of the p×p matrix Q) and with increasing weight of slope 2 over the prediction horizon (reflected in the p−m to p diagonal values of Q).6 The cost of applying a light input is weighted uniformly with a magnitude of 1 as reflected in the diagonal values of the m×m matrix R.
The performance of an M-length control input is measured by
Only the first move of the lowest cost control sequence u* is implemented. Therefore, the sequence of actually implemented control moves may differ significantly from the sequence of control moves calculated at a particular time step. This discrepancy disappears as the prediction and move horizons near infinity. Feedback is incorporated by using the next measurement to update the optimization problem. Once the controlled state trajectories converge to within 15% of the reference state trajectories, the system is considered to have recovered its phase in hours. For further details concerning the development and implementation of MPC as applied to circadian networks, the reader is referred to Bagheri et al. (2007).
5.3 Tuning the MPC parameters
The best fit control sequence is determined by enumerating the solutions over a grid in the solution space (light magnitude as a function of time). The algorithm approaches a globally optimal solution as the total possible quantization steps of the control input increase. We test the efficacy of the algorithm with respect to a quantization of 2, 4, 8 and 16 steps. Results suggest that the shorter recovery time may not outweigh the increase in computation time (figure 5a). Therefore, we investigate solutions for 2 and 8 possible control values.
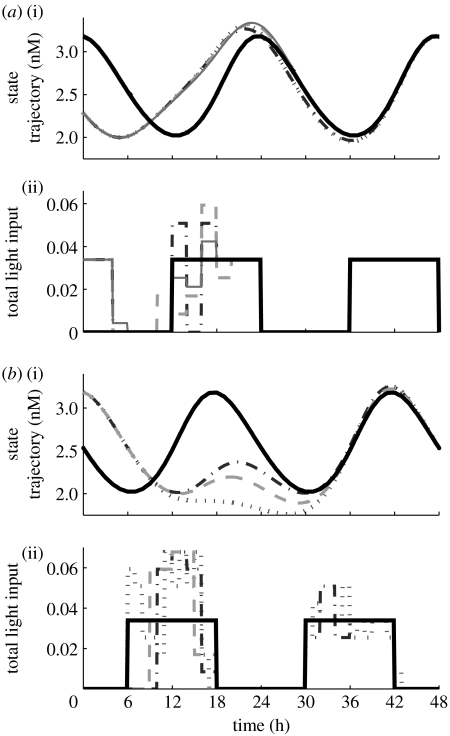
Figure 5.
(a(i),b(i)) State dynamics as they converge to the reference trajectory, the bold solid lines. (a(ii),b(ii)) The light profiles used to reset the phase differences; these sequences also converge to nominal light/dark cycles, the bold solid lines. Control action begins at (a) 12 hours and (b) 18 hours, resetting the phase differences (IP) of (a) 8 and (b) 6 hours. (a) The recovery time associated with a control input that allows 2 (dotted lines), 4 (dot-dashed lines), 8 (dashed lines) and 16 (solid lines) possible values is 36.8, 35.6, 34.7 and 34.6 hours, respectively. (b) The recovery time associated with a 1 (dotted lines), 2 (dot-dashed lines) and 3 (dashed lines) hour control input is 36.7, 34.7 and 35.2 hours, respectively.
Similarly, we evaluate the algorithm with respect to a control input of duration of 1, 2 and 3 hours (reflecting a move horizon of 3, 6 and 9 hours, respectively). Although shorter light pulses offer a more dynamic manipulated variable profile, it shortens the move horizon and may reduce the usefulness of MPC (figure 5b). Conversely, a longer pulse may reduce the possible control profiles since extended exposure to light leads to arrhythmic behaviour (Ohta et al. 2005). We set the duration of control to 2 hours.
5.4 Phase recovery dynamics
The nonlinear properties of biological oscillators often cause different phase resetting times with respect to the initial condition (IC, the circadian time at which the control begins) and initial phase difference (IP, the amount of circadian time that needs to be recovered). We generate phase recovery dynamics for the open-loop algorithm7 where environmental light/dark cycles entrain the system (figure 6), and the closed-loop MPC algorithm where the manipulated control variable (light) has 2 and 8 possible values (figure 7). Since nonlinearity also challenges the uniqueness of optimal light sequences, we choose the control profile that minimizes the magnitude of total admitted light, hence penalizing the weighted sum of u(·) (refer to §5.2).
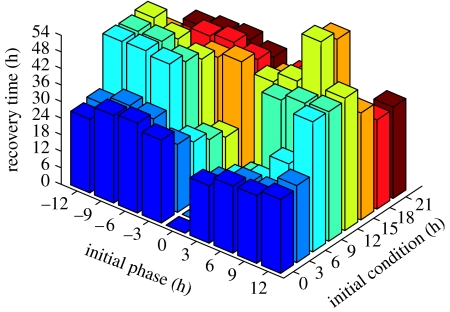
Figure 6.
The open-loop (natural) phase recovery dynamics as a function of the initial condition (IC) and initial phase difference. The shading of recovery times (shown on the vertical axis) is consistent along the IC and among figure 7a and figure 7b.
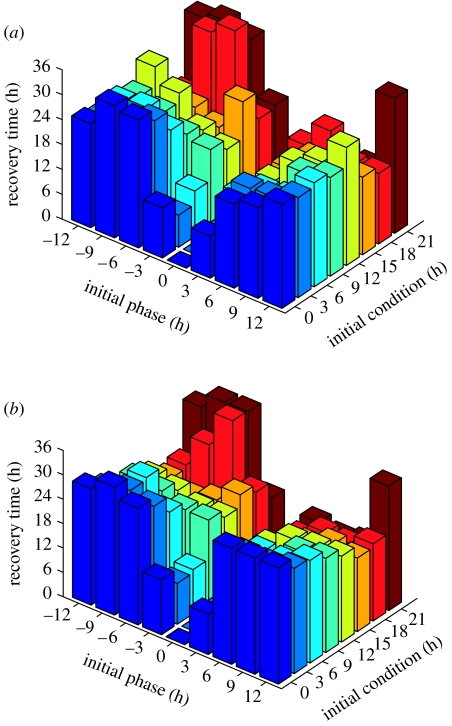
Figure 7.
(a) Two-step and (b) eight-step closed-loop controls. The shading of closed-loop MPC recovery times (shown on the vertical axis) as a function of the IC and IP is consistent along the IC. The vertical axes, however, are not the same; refer to table 2 for a direct comparison of phase resetting dynamics among the presented algorithms. An IC=12 hours is one of the few data sets whose recovery time decreases with greater control flexibility.
Recovery times are described as a function of both the IC and initial phase difference to better visualize the nonlinear dynamic behaviour of phase resetting. The open-loop environmental control strategy (figure 6) requires (at most) 64.7 hours to synchronize a ±12 hour initial phase difference beginning at an IC of 12 hours (table 2; Bagheri et al. 2007). The closed-loop two-step algorithm (figure 6) improves upon the maximum recovery time as it requires only 36.8 hours to recover a 6 hour initial phase difference (at IC=18 hours). Surprisingly, the eight-step algorithm (figure 6) does not significantly improve phase resetting as it requires 34.7 hours to recover the same conditions. For this reason, a two-step algorithm (which requires approx. 0.17 hours of computation time per simulation) is more efficient for real-world applications—such as light therapy—than the eight-step algorithm (which requires approx. 3.5 hours of computation time per simulation).
Table 2.
Maximum recovery times (in hours) with respect to ICs and initial phase differences (also in hours). (The italic text reflects the maximum recovery time over the entire set.)
| open loop | two-step algorithm | eight-step algorithm | ||||
|---|---|---|---|---|---|---|
| IC | recovery | IP | recovery | IP | recovery | IP |
| 0 | 32.50 | −6 | 31.80 | −9 | 31.40 | −9 |
| 3 | 31.40 | −9 | 28.00 | −6 | 28.10 | −9 |
| 6 | 47.50 | −9 | 26.00 | −9 | 28.60 | −9 |
| 9 | 46.90 | ±12 | 23.80 | ±12 | 23.20 | ±12 |
| 12 | 64.70 | ±12 | 28.40 | ±12 | 21.10 | ±12 |
| 15 | 61.70 | ±12 | 24.70 | −3 | 21.50 | −3 |
| 18 | 40.80 | 6 | 36.80 | −6 | 34.70 | −6 |
| 21 | 35.50 | −6 | 34.00 | −9 | 34.30 | −6 |
Although the closed-loop algorithm significantly increases re-synchronization rates, using two possible control values is often just as effective as using eight possible control values; the flexibility of control has little effect on the MPC algorithm. Such results support the hypothesis that, in general, natural light/dark cycles are not optimized to reset large phase differences (Bagheri et al. 2007). Instead, organisms may have evolved to efficiently reset small phase differences since rapid transit across multiple time zones is a recent innovation. Although the medical community has explored the benefits of light therapy, there are very few algorithmic approaches to optimal light pulsing and none that employs the MPC algorithm presented here. The admission of morning light, for instance, has already been considered as an antidepressant by realigning the internal clock with the environment (Lewy et al. 2006). Studies suggest that the human circadian clock mechanism functions similarly to those of other organisms (Boivin et al. 1996). This similarity may be attributed to shape/amplitude characteristics of their respective PRCs. This parallel motivates the experimental application of controlled light pulses for phase resetting in mammals, despite the lack of quantitatively predictive mammalian models. Furthermore, melatonin has proven to be a key circadian phase resetting agent for totally blind people who cannot synchronize to environmental day/night cycles (or do so at an abnormal time; Lewy et al. 2006). Therefore, melatonin may be used individually (in cases to treat the totally blind), or in combination with light to provide more effective phase resetting. Although the applications are plenty, we acknowledge that the means through which we quantify phase in this study (namely, by collecting data at the molecular level) may not be readily feasible. However, behavioural and/or physiological parameters that are controlled by and correlated with the circadian clock's dynamics (such as activity or body temperature) are easily accessible. Assuming we link such physiological markers with circadian phase, one can implement this phase re-entrainment algorithm in practice.
Further studies may include the investigation of phase resetting dynamics as a function of environmental day length. This application would address circadian-related disorders common among people who live near either pole. As the day length decreases, people are exposed to less light and may be at a higher risk of de-synchronizing their internal clocks with that of their environment (Lamberg 1994; Moore 1997).
6. Conclusion
We employ systems theoretic tools to investigate the circadian phase properties of single deterministic models, and populations of stochastic models. Our investigations have illustrated that computational techniques applied to single-cell data are fundamental for tuning and predicting the behaviour of oscillatory phenomena at the population level, since the results of such investigations point to the coupling mechanisms that give rise to spontaneously synchronized networks of stochastic biophysical nodes. Without such insight, we would not have been able to reproduce the synchrony observed in the SCN. As a result, it is important for experimental biologists to adopt the tools necessary to analyse the structure of both in vitro and in vivo systems.
Biologists may further benefit from the analysis of mathematical models, since the circadian phase response may target areas of further experimental interest. Our investigation of the Drosophila clock model, for instance, suggested that only 82 (out of 960) cellular coupling mechanisms would provide synchrony, significantly decreasing the number of candidates an experimentalist may consider. Even further, circadian phase analysis may point to non-photic control targets that may be used in place of, or in combination with, light as a means of optimizing phase resetting.
In summary, the study of synchronization supports the reverse engineering of the clock in the SCN while providing a foundation to engineer other communication networks. Analysis of the pIPRC provides separation of the timing characteristics of the oscillator and signal. Altering the signal (duration, magnitude or shape) can have profound effects prescribed by the pIPRC. For example, it is possible to speed up an oscillator when (and only when) there is an advance area in the target pIPRC. To synchronize, the signal must meet the conditions for stable entrainment. Through MPC, we solve for a sequence of light pulses that resets the circadian clock in a fraction of the time required by natural open-loop entrainment. The study of circadian phase entrainment provides a forum to address the re-synchronization properties of the clock, at both the single-cell and population levels.
Acknowledgments
This project was supported in part by the ICB grant DAAD19-03-D-0004; NIH grant GM078993; NSF IGERT grant DGE02-21715; NIH grant EB007511; Army Research Office grant W911NF-07-1-0279; and the Research Participation Programme between the US DOE and AFRL/HEP.
Footnotes
A previous version of this paper was presented at the 2nd conference on Foundations of Systems Biology in Engineering, Stuttgart, Germany, September 2007.
One contribution of 10 to a Theme Supplement ‘Biological switches and clocks’.
per3 is not taken into account because biological evidence shows that its disruption does not have a strong effect on clock performance (Forger & Peskin 2003).
The study of phase synchrony via the coupling of a population of circadian oscillators is better understood in mammals than in flies. In fact, it remains an open question as to whether or not fly clock cells are indeed coupled (Hardin 2005).
Gillespie (2000) offers an explanation of the conditions under which Langevin and deterministic chemical kinetics approximations are valid. This is usually the case when populations of all the reactant species are sufficiently large. This may not be true in the biological system being modelled, and is not the case in the predictions of the Leloup & Goldbeter deterministic model (some species concentrations approach zero during low points in the oscillatory cycle); therefore a deterministic approximation using ODEs may not be completely adequate for our purposes.
This model is simulated as an SDE with multiplicative noise added to each of the rate parameters, but is expressed as a collection of ODEs.
Kuramoto and others use the method of averaging to compute the so-called interaction function between oscillators. The interaction function for the synchronized system provides the period change.
m and p reflect the number of discrete time points that make up the move and prediction horizons, respectively: m=M/ts and p=P/ts.
The open-loop algorithm is one in which there is no feedback; the light input does not depend on the current phase of the system. Instead nominal/environmental 24 hour light cycles are used without a priori state information.
Supplementary Material
Model description
References
- Aton S.J., Colwell C.S., Harmar A.J., Waschek J., Herzog E.D. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. doi:10.1038/nn1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri N., Stelling J., Doyle III F.J. Circadian phase entrainment via nonlinear model predictive control. Int. J. Robust Nonlinear Control. 2007;17:1555–1571. doi:10.1002/rnc.1209 [Google Scholar]
- Bernard S., Gonze D., Cajavec B., Herzel H., Kramer A. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput. Biol. 2007;3:e68. doi: 10.1371/journal.pcbi.0030068. doi:10.1371/journal.pcbi.0030068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin D.B., Duffy J.F., Kronauer R.E., Czeisler C.A. Dose–response relationships for resetting of human circadian clock by light. Nature. 1996;379:540–542. doi: 10.1038/379540a0. doi:10.1038/379540a0 [DOI] [PubMed] [Google Scholar]
- Boulos Z., Macchi M., Stürchler M., Stewart K., Brainard G., Suhner A., Wallace G., Steffen R. Light visor treatment for jet lag after westward travel across six time zones. Aviat. Space Environ. Med. 2002;73:953–963. [PubMed] [Google Scholar]
- Brown E., Moehlis J., Holmes P. On the phase reduction and response dynamics of neural oscillator populations. Neural Comput. 2004;16:673–715. doi: 10.1162/089976604322860668. doi:10.1162/089976604322860668 [DOI] [PubMed] [Google Scholar]
- Daan S., Pittendrigh C. A functional analysis of circadian pacemakers in nocturnal rodents. II. The variability of phase response curves. J. Comp. Physiol. 1976;106:253–266. doi:10.1007/BF01417857 [Google Scholar]
- Dunlap J., Loros J., DeCoursey P., editors. Chronobiology: biological timekeeping. Sinauer Associates, Inc; Sunderland, MA: 2004. [Google Scholar]
- Fall C., Marland E., Wagner J., Tyson J. Springer; New York, NY: 2005. Computational cell biology. [Google Scholar]
- Forger D., Peskin C. A detailed predictive model of the mammalian circadian clock. Proc. Natl Acad. Sci. USA. 2003;100:14 806–14 811. doi: 10.1073/pnas.2036281100. doi:10.1073/pnas.2036281100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger D., Peskin C. Stochastic simulation of the mammalian circadian clock. Proc. Natl Acad. Sci. USA. 2005;102:321–324. doi: 10.1073/pnas.0408465102. doi:10.1073/pnas.0408465102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 1976;22:403–434. doi:10.1016/0021-9991(76)90041-3 [Google Scholar]
- Gillespie D. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. doi:10.1021/j100540a008 [Google Scholar]
- Gillespie D. The chemical langevin equation. J. Chem. Phys. 2000;113:297–306. doi:10.1063/1.481811 [Google Scholar]
- Gonze D., Halloy J., Goldbeter A. Emergence of coherent oscillations in stochastic models for circadian rhythms. Phys. A-Stat. Mech. Appl. 2004;342:221–233. doi:10.1016/j.physa.2004.04.082 [Google Scholar]
- Gonze D., Bernard S., Waltermann C., Kramer A., Herzel H. Spontaneous synchronization of coupled circadian oscillators. Biophys. J. 2005;89:120–129. doi: 10.1529/biophysj.104.058388. doi:10.1529/biophysj.104.058388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Zak D., Sauter T., Schwaber J., Ogunnaike B. Modeling the VPAC2-activated cAMP/PKA signaling pathway: from receptor to circadian clock gene induction. Biophys. J. 2006;90:1560–1571. doi: 10.1529/biophysj.105.065250. doi:10.1529/biophysj.105.065250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E. The circadian timekeeping system of drosophila. Curr. Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. doi:10.1016/j.cub.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Herzog E.D., Aton S.J., Numano R., Sakaki Y., Tei H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythm. 2004;19:35–46. doi: 10.1177/0748730403260776. doi:10.1177/0748730403260776 [DOI] [PubMed] [Google Scholar]
- Hoppensteadt F., Izhikevich E. Applied mathematical sciences. vol. 126. Springer; New York, NY: 1997. Weakly connected neural networks. [Google Scholar]
- Jensen R.V. Synchronization of driven nonlinear oscillators. Am. J. Phys. 2002;70:607–619. http:link.aip.orglink?AJP706071 [Google Scholar]
- Johnson C. Forty years of PRCs: what have we learned? Chronobiol. Int. 1999;16:711–743. doi: 10.3109/07420529909016940. doi:10.3109/07420529909016940 [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. doi:10.1126/science.1069492 [DOI] [PubMed] [Google Scholar]
- Kuramoto Y. Springer; Berlin, Germany: 1984. Chemical oscillations, waves, and turbulence. [Google Scholar]
- Lamberg L. William Morrow and Company, Inc; New York, NY: 1994. Bodyrhythms: chronobiology and peak performance. [Google Scholar]
- Leloup J.-C., Goldbeter A. A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J. Biol. Rhythms. 1998;13:70–87. doi: 10.1177/074873098128999934. doi:10.1177/074873098128999934 [DOI] [PubMed] [Google Scholar]
- Leloup J.-C., Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc. Natl Acad. Sci. USA. 2003;100:7051–7056. doi: 10.1073/pnas.1132112100. doi:10.1073/pnas.1132112100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup J.-C., Goldbeter A. Modeling the mammalian circadian clock: sensitivity analysis and multiplicity of oscillatory mechanisms. J. Theor. Biol. 2004;230:541–562. doi: 10.1016/j.jtbi.2004.04.040. doi:10.1016/j.jtbi.2004.04.040 [DOI] [PubMed] [Google Scholar]
- Lewy A.J., Lefler B.J., Emens J.S., Bauer V.K. The circadian basis of winter depression. Proc. Natl Acad. Sci. USA. 2006;103:7414–7419. doi: 10.1073/pnas.0602425103. doi:10.1073/pnas.0602425103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. doi:10.1016/j.cell.2007.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood E.S., Reddy A.B., Wong G.K.Y., O'Neill J.S., O'Brien J.A., McMahon D.G., Harmar A.J., Okamura H., Hastings M.H. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. doi:10.1016/j.cub.2006.02.023 [DOI] [PubMed] [Google Scholar]
- Moore R. Circadian rhythms: basic neurobiology and clinical applications. Annu. Rev. Med. 1997;48:253–266. doi: 10.1146/annurev.med.48.1.253. doi:10.1146/annurev.med.48.1.253 [DOI] [PubMed] [Google Scholar]
- Morari M., Lee J. Model predictive control: past, present and future. Comput. Chem. Eng. 1999;23:667–682. doi:10.1016/S0098-1354(98)00301-9 [Google Scholar]
- Ohta H., Yamazaki S., McMahon D.G. Constant light desynchronizes mammalian clock neurons. Nat. Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. doi:10.1038/nn1395 [DOI] [PubMed] [Google Scholar]
- Piggins H., Antle M., Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J. Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S., Weaver D. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. doi:10.1038/nature00965 [DOI] [PubMed] [Google Scholar]
- Smolen P., Hardin P.E., Lo B.S., Baxter D.A., Byrne J.H. Simulation of Drosophila circadian oscillations, mutations, and light responses by a model with VRI, PDP-1, and CLK. Biophys. J. 2004;86:2786–2802. doi: 10.1016/S0006-3495(04)74332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogatz S. From Kuramoto to Crawford: exploring the onset of synchronization in populations of coupled oscillators. Physica D. 2000;143:1–20. doi:10.1016/S0167-2789(00)00094-4 [Google Scholar]
- Taylor S.R., Gunawan R., Petzold L.R., Doyle F.J., III Sensitivity measures for oscillating systems: application to mammalian circadian gene network. IEEE Trans. Automat. Contr. 2008;153:177–188. doi: 10.1109/TAC.2007.911364. doi:10.1109/TAC.2007.911364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T.-L., Henson M.A., Herzog E.D., Doyle F.J., III A molecular model for intercellular synchronization in the mammalian circadian clock. Biophys. J. 2007;92:3792–3803. doi: 10.1529/biophysj.106.094086. doi:10.1529/biophysj.106.094086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.J., Hong C.I., Thron C.D., Novak B. A simple model of circadian rhythms based on dimerization and proteolysis of PER and TIM. Biophys. J. 1999;77:2411–2417. doi: 10.1016/S0006-3495(99)77078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H.R., Hagiwara M., Kitano H. Robust oscillations within the interlocked feedback model of drosophila circadian rhythm. J. Theor. Biol. 2001;210:401–406. doi: 10.1006/jtbi.2000.2226. doi:10.1006/jtbi.2000.2226 [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Hirose K., Iino M. Intercellular coupling mechanism for synchronized and noise-resistant circadian oscillators. J. Theor. Biol. 2002;216:501–512. doi: 10.1006/jtbi.2002.3000. doi:10.1006/jtbi.2002.3000 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Deboer T., Meijer J. Light-induced resetting of the circadian pacemaker: quantitative analysis of transient versus steady-state phase shifts. J. Biol. Rhythm. 2001;16:564–573. doi: 10.1177/074873001129002259. doi:10.1177/074873001129002259 [DOI] [PubMed] [Google Scholar]
- Winfree A. Springer; New York, NY: 2001. The geometry of biological time. [Google Scholar]
- Yamaguchi Y., Shimizu H. Theory of self-synchronization in the presence of native frequency distribution and external noises. Phys. D: Nonlin. Phenomena. 1984;11:212–226. doi:10.1016/0167-2789(84)90444-5 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Model description