Abstract
Tracking of single particles in optical microscopy has been employed in studies ranging from material sciences to biophysics down to the level of single molecules. The technique intrinsically circumvents ensemble averaging and may therefore reveal directly mechanistic details of the involved dynamic processes. Such processes range from translational and rotational motion to spectral dynamics. We distinguish between conventional a posteriori tracking of objects (e.g. from the sequences of images) and the experimentally more refined ‘on-the-fly’ tracking technique. In this technique, the observation volume of the microscope is kept centred with respect to the moving object via a feedback algorithm. This approach brings a series of advantages in comparison with the tracking from images, ranging from a superior spatio-temporal resolution (2–50 nm and 1–32 ms) to the capability of inferring additional data (e.g. fluorescence lifetime, emission spectrum, polarization, intensity dynamics) from an object as it moves over several microns in three dimensions. In this contribution, we describe the principle of the tracking technique as implemented on a two-photon laser scanning microscope and illustrate its capabilities with experimental data, from particles labelled with different dyes moving in a liquid to the characterization of small fluorescently labelled protein assemblies in living cells.
Keywords: three-dimensional particle tracking, high-resolution microscopy, spectroscopy, biophysics
1. Introduction
The two main strengths of optical microscopy lie in its essentially non-invasive character, and in the possibility of gathering information from a small sub-volume in a three-dimensional specimen, as long as it is effectively transparent. Optical microscopy is, for this reason, among the most suitable techniques to examine biological samples, in which many phenomena take place in several microns inside the aqueous environment of a living cell or tissue. The spatial resolution of an optical microscope is characterized by the size of the focal spot (the aforementioned small sub-volume), which is described mathematically by the point spread function (PSF) of the instrument. The PSF depends essentially on the optical illumination and detection geometries and on the wavelength of the light used. In practice the optical resolution is of the order of a few 100 nm. This stands in contrast to the spatial resolution achievable with other forms of microscopy, such as electron, force and tunnelling microscopy, where nanometre resolution and below is commonplace—albeit with the typical restriction to surfaces (or layers not thicker than 100 nm) of specially prepared samples (e.g. conducting materials, solids, proteins adsorbed to surfaces, etc.). Any improvement or possibility of circumventing the resolution limit in optical microscopy is, of course, highly desirable.
Improvements based solely on optical concepts, for example the increase of the aperture in the 4-π microscope (Hell 1994) or orthogonal detection geometry in theta-microscopy (Stelzer & Lindek 1994), reduce the PSF size to approximately 100 nm in all three dimensions. The size of specimens for these studies is limited because they have to be placed in the field of view of two (or more) microscope objectives. Other approaches to increase the resolution make use of luminescent labels, which range from individual organic fluorophores (Tamarat et al. 2000), carbon nanotubes (Lefebvre et al. 2004) and fluorescent proteins (Campbell et al. 2002) to noble-metal nanoparticles (Huang et al. 2007) and quantum dots (Cai et al. 2007; Hammer et al. 2007). These labels can be attached physically or chemically to specific proteins, chromatin sequences, lipid membranes, organelles, etc. By engineering the luminescence response of fluorescent dye molecules in the focal spot via stimulated emission depletion (Klar et al. 2000), it is possible to reduce the volume of the conventional PSF by an order of magnitude and to achieve a spatial resolution of the order of 20–30 nm (Donnert et al. 2006). Another, perhaps more subtle, way of obtaining information optically on the length scale of nanometres exploits the additional information contained in the label's emission, such as its luminescence lifetime, photon quantum yield, spectrum, polarization, etc. which are influenced by the emitter's immediate environment. Förster resonant energy transfer, for example, may take place between pairs of dyes separated by a few nanometres with a sharp (1/r6) distance dependence (as well as orientation and spectral dependencies—omitted for simplicity). This distance dependence can be used to gauge distances between pairs of dye molecules, for example, when they are attached specifically to different parts of a macromolecule. Pairs of fluorescent dyes can be used to monitor dynamic structural changes in the substrate they are attached to, revealing mechanistic details on the millisecond and nanometre scales (Ha 2001).
Another general strategy to reach higher resolution is to detect the labels or labelled objects on an individual basis. In this case, it is possible to resolve the distributions of properties directly in space and time, as long as the individual components of the distribution are well separated. If the object or label is small enough to be considered a point-like emitter, its intensity distribution on the image plane of the microscope is defined via the instrument's PSF. While the object itself may remain unresolved, the centre of this distribution can be determined with arbitrary accuracy, depending ultimately on the signal-to-noise ratio (SNR; Bobroff 1986). One implication of this is the possibility of completely abolishing the diffraction limit restrictions by measuring the position of one particle at a time. One particular technique that exploits this principle is known as photo-activated localization microscopy (PALM; Betzig et al. 2006) or stochastic optical reconstruction microscopy (STORM; Rust et al. 2006), in which a collection of labels is turned on and off on an individual basis. Since each emitter's position can be located with an arbitrary accuracy (typically of the order of 20 nm), the resulting image of the superimposed positions yields a map of the studied specimen with a resolution corresponding to the localization accuracy. The major limitation, presently, in this particularly new technique is that, in order to obtain a representative image of the specimen, the acquisition time is of the order of several minutes. Three-dimensional images, being reconstructions from a stack of images acquired at different focal planes, are thus typically obtained from fixed or immobilized specimens. In two dimensions, however, it is possible to use the technique in unfixed samples and to analyse the frame-by-frame movement of individual photoactivated molecules (Manley et al. 2008), as in conventional low-concentration single-molecule tracking experiments (Sonnleitner et al. 1999; Hellriegel et al. 2004).
The approach presented in this article produces high-resolution maps in a similar way, using single-particle tracking (SPT). While, traditionally, SPT is used to draw conclusions on the modes of motion of the particle itself (directed transport versus diffusion, the influence of local viscosity, etc.; Saxton & Jacobson 1997), the trajectories also allow for the visualization of substrates along which the tracked object moves. It is possible to infer the size and shape of diffusion barriers, extent and tortuosity of motion paths, presence of specific structural elements, etc.—as demonstrated in materials science (e.g. Jung et al. 2007; Kirstein et al. 2007). In summary, the experimenter obtains a map of the regions that are accessible to the tracked object—much as the tip of a tunnelling microscope is used to map the surface of a specimen. Conventional SPT methods, however, are—as PALM and STORM—based on image analysis, in which the individual particle is located on an image via fitting its PSF and subsequently tracked in a sequence of frames. This form of SPT is usually limited to the instrument's focal plane, and is, thus, essentially a two-dimensional technique. Expansion to three dimensions, via the acquisition of image stacks consisting of slices along the optical axis, has the typical drawback of being time intensive to acquire, failing to detect dynamics on the sub-second time scale. Another typical disadvantage of acquiring temporal sequences of three-dimensional stacks lies in the comparatively high photodegradation of the sample during the acquisition. Table 1 summarizes the typical resolution limits of the various techniques mentioned in this section.
Table 1.
Comparison between different types of optical microscopy methods. (The scale of length in the focal plane (r) and along the optical axis (z) denotes the typical (as opposed to best-possible) values achieved with the method. Some methods do not strictly require labelling, but are typically used with labels. In the time-scale column, we distinguish between imaging and point detection times. Point detecting methods have almost arbitrary temporal resolution. However, this point has to be raster scanned in order to obtain images. In the case of PALM/STORM, a sufficient number of individual emitters have to be acquired to produce images. In the case of three-dimensional SPT, the time needed to obtain one point is given by the orbit time; larger maps depend on the trajectory length and point density.)
| microscopy method | scale r, z (nm) | limit | label | restrictions | time scale | e.g. references |
|---|---|---|---|---|---|---|
| conventional | 300, 3000 | yes: diffr. | no | transparency | camera (ms) | Pawley (2006) |
| confocal | 300, 900 | yes | transparency | image: scanner (s) point detector | Pawley (2006) | |
| 4π | 100, 190 | yes | thin specimen | Hell (1994) | ||
| θ | 100, 100 | yes | thin specimen | Stelzer & Lindek (1994) | ||
| STED | 30, 30 | yes | depletable dye | Klar et al. (2000) | ||
| PALM/STORM | 20, 50–100 | no: SNR | yes | switchable dye | pop. (10 min) | Betzig et al. (2006) and Rust et al. (2006) |
| two-dimensional SPT image | 30, – | yes | focal plane two dimensions | camera (ms) | Hellriegel et al. (2004) | |
| three-dimensional SPT | 2–20, 50–100 | yes | select particles | orbit (0.5–32 ms) trajectory length | Levi et al. (2005a) | |
| FRET | 0–10 | yes | not >10 nm | point detector | Ha (2001) |
A different approach to track particles in three dimensions based on a two-photon microscopy set-up has been developed recently in our laboratory (Kis-Petikova & Gratton 2004; Levi et al. 2005a). The probe volume (described by the PSF) is kept ‘on top’ of the tracked particle in three dimensions using mirror and focus controls via a feedback algorithm (details are given in §2). The photodegradation of the entire specimen is effectively suppressed because only a small sub-volume of the whole specimen (i.e. the region around the particle of interest) is being exposed to light during measurement. The trajectories of the particles may span several microns in all the three dimensions, while still maintaining a high spatio-temporal resolution. At SNRs of 100–10, the accuracy typically ranges from ±2 to ±50 nm in the focal plane and from ±50 to ±500 nm along the optical axis. The temporal resolution, limited mainly by the acquisition hard- and software, is typically in the range of 1–32 ms. Moreover, the circumstance in which the focus is kept on top of the tracked particle can be exploited to further analyse the light coming from the moving object to gather (i) its emission intensity, (ii) luminescence spectrum, (iii) emission lifetime, and (iv) polarization; all ‘in sync’ with the acquisition of the trajectory. In the following sections, we review the implementations of the three-dimensional SPT technique and its expansion to simultaneously acquired multiple parameters, we demonstrate its capability of producing high-accuracy multi-parameter three-dimensional maps of different substrates and finally we show the possibility of achieving a high level of detail in biophysical studies in vivo.
2. Experimental section
2.1 Three-dimensional tracking
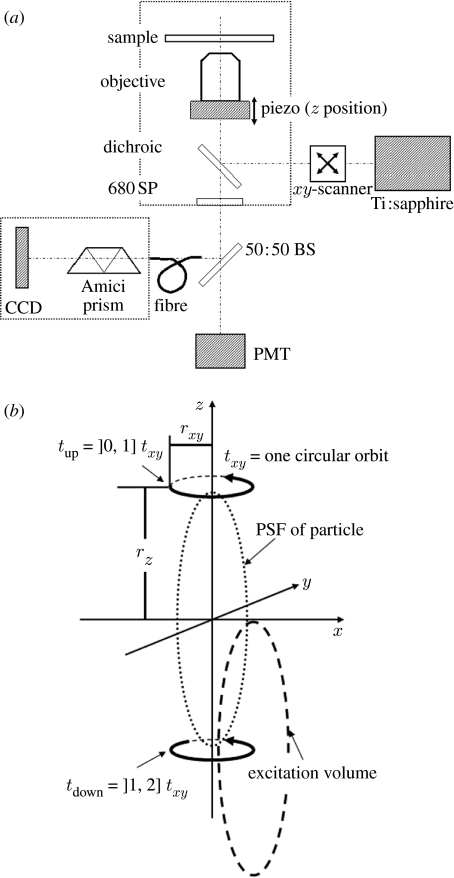
The three-dimensional spatial position of a particle can be determined by scanning the excitation volume of a two-photon microscope in a three-dimensional orbit around the particle's PSF (figure 1) and by subsequently analysing the fluorescence intensity profile along the orbit. While the principle has been discussed in detail in Kis-Petikova & Gratton (2004) and Levi et al. (2005a) for the case of two-photon microscopy, it is easily transferable, e.g. to laser scanning confocal microscopy. Some of the theoretical background will be summarized here.
Figure 1.
Schematic of experimental setup. (a) The two-photon microscopy setup used is shown as a simple schematic diagram. (b) Representation of the orbital scan for three-dimensional tracking (drawn approx. to scale—the width of the PSF is typically of the order of 300 nm).
The intensity profile is a function of the relative position of the particle's PSF centre (xp, yp and zp) with respect to the excitation PSF in one orbit centred at xc, yc and zc,
| (2.1) |
where
| (2.2) |
are the positions of the excitation PSF in the orbit at any given time t as a function of the orbit frequency fo and radius r (the indices xy and z denote the focal plane and the optical axis, respectively). Using a Gaussian–Lorentzian approximation for the two-photon PSF, we obtain
| (2.3) |
for the intensity profile, where Further, Ib is the background intensity, and w0 the PSF's waist. Within the constants are I0 (the normalized intensity at rxy, rz, Δx, Δy, Δz=0) as well as λ the wavelength of the excitation light.
Starting from the case in which the particle's PSF is centred with the orbit, that is, with xc, yc, zc=xp, yp, zp, equation (2.3) becomes
| (2.4) |
Because the whole expression becomes time invariant. Upon displacing the particle with respect to the orbit's centre, equation (2.3) will be modulated with time. The centre of the scanned orbit, as the frame of reference, can be assigned to 0, so that the particle positions (xp, yp and zp) with respect to the (known) centre of the orbit can be obtained via fitting using equation (2.3). When the particle is far away from the centre of the orbit, the exponential factor in equation (2.3) becomes small, eventually bringing the signal below the noise level. In this limit, I(Δx, Δy, Δz)∼Ib is also time invariant.
One of the most important results outlined in Kis-Petikova & Gratton (2004) deals with the accuracy with which the particle's position can be determined from fitting equation (2.3) to an experimentally acquired intensity profile. Ultimately, it is limited by the number of acquired photons, but also depends on the matching between the PSF and the orbit's radii. From the mathematical perspective, the changes in the intensity trace along the orbit are most pronounced where the PSF has the largest slope, i.e. in the focal plane the orbit's radius corresponds to 0.5w0. When this is the case, the positioning accuracy for a particle that is centred with the orbit is maximal. For a given number of acquired photons, the accuracy even exceeds that of the equivalent positioning from a raster scanned image, because all the photons come from the most sensitive part of the ‘image’. In practice, however, because the particle is moving, it will always move out of this ‘perfect’ condition. For that reason, it proves to be more practical to choose a radius slightly larger than 0.5w0. When this is the case, the particle moves from a less sensitive configuration into a more sensitive one. The trade-off between a smaller number of acquired photons and the improved positioning accuracy has an optimum at a radius of 0.8w0 in the focal plane and ca 1.8w0 along the optical axis (cf. figure 1b and discussion in Kis-Petikova & Gratton (2004)—w0 is the beam waist in the focal plane).
The main step in a feedback algorithm for particle tracking is finding the particle position after it has moved off the centre, and the rapid re-centring of the orbit with respect to the particle at the new position. Fitting the intensity traces using equation (2.3), while most accurate, is a computationally intensive procedure. The automatic feedback is realized using an algorithm based on the FFT of the intensity transient using the first two Fourier coefficients. This operation is computationally fast but less accurate because the higher order coefficients are discarded. Still, because all intensity traces are stored, it is possible to obtain the most precise position estimations of the particle via fitting after the data have been recorded—see discussion in Kis-Petikova & Gratton (2004).
The first two Fourier coefficients—DC and AC1 (or simply AC)—resulting from the FFT of one orbit's intensity trace are used for tracking. AC is a complex number and its modulo |AC| and phase φ are defined as
| (2.5) |
| (2.6) |
The modulation, defined as the quotient MOD=|AC|/DC, serves as an indicator for the distance of the particle to the orbit centre, whereas the in-plane angle of the particle with respect to the orbit centre is easily obtained from the phase φ of the AC term, with respect to the angle at t=0 (which is set to 0° after calibration).
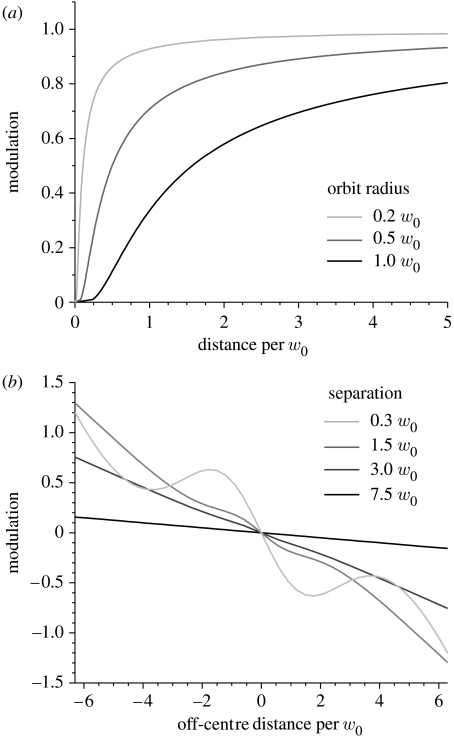
Starting from the case of the particle resting exactly at the orbit's centre, that is, xc, yc, zc=xp, yp, zp, gives a constant value for I(Δx, Δy, Δz), thus rendering the value for |AC|=0. The value for MOD in this case is 0, and the phase undefined. As we slightly move the particle off the orbit's centre, the |AC| value increases and the phase becomes defined. In this region, the variation of DC is small compared with that of |AC|. As a consequence, we see that the slope of MOD versus distance is large for small distances. As the distance between the particle and the orbit becomes large, both |AC| and DC level off to 0 and MOD levels off to unity (figure 2a).
Figure 2.
Modelled modulation versus distance lookup tables. All units are given as fractions of the in-focus beam waist (w0). (a) The lookup table for the estimation of distance in the plane of the orbits. The extreme curves are for too small and too large orbit radii, respectively. Each curve in (b) represents the lookup table for the estimation of distance along the optical axis, using different separations between the upper and lower orbits. Too large separations (above 3.0w0) lead to ambiguous results.
The behaviour of MOD versus particle distance depends on the relationship between the PSF size and orbit radius. The estimation accuracy for the distance is directly related to the slope of this curve. If the orbit radius is small in comparison with the PSF (e.g. 0.2w0 in figure 2a), we see that the MOD curve is essentially delineating the PSF—it is initially shallow at small distances, becomes steeper and levels off at long distances from the centre. The slope of the lookup table increases with orbit radius, improving the estimation accuracy for a given MOD value. Above a certain radius (e.g. 1.0w0 in figure 2a), the curve already levels off at small distances, meaning that the error is large even when the particle is still within the orbit boundaries. The same reasoning is true also for distance estimation in the z-direction. However, owing to the way the orbits are scanned, we do not have access to a continuously oscillating function but rather two intensities integrated over the entire upper and entire lower orbits. DC is thus the simple average and the equivalent to AC is the difference between the signals. The functional form of the MOD versus distance plot in the z direction is shown in figure 2b. The slope of this curve, again, determines the accuracy of the estimation. For orbit-to-centre distances between 0.3w0 and 3w0, the plot is almost linear, giving a simple relationship between MOD and distance. Above 3w0 the nonlinearity of the curve starts to become more pronounced—the presence of plateaus and minima gives rise to ambiguous distance estimations. Also at such separations, the signal becomes impracticably low. The ideal value is found to be approximately 1.8w0.
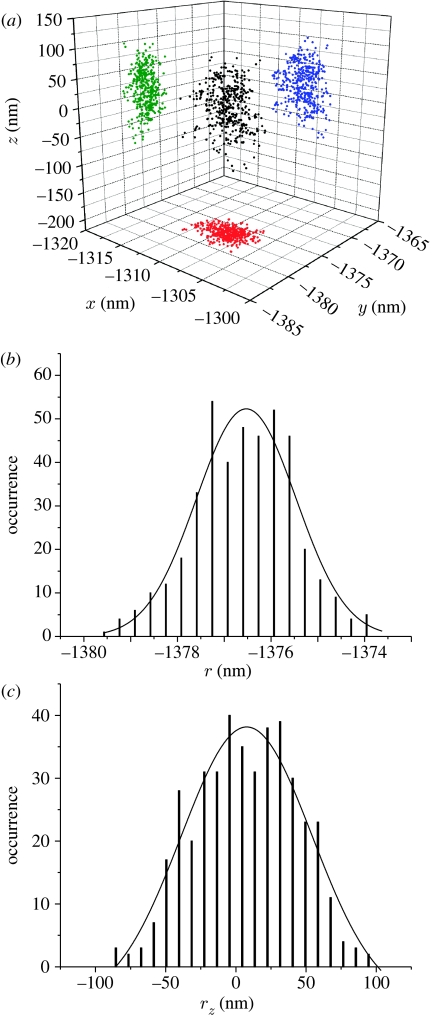
All lookup tables and the initial angle are determined by imaging beads and gratings at known distances and angles from the orbit centre. Shown in figure 3 are the retrieved positions of an immobile luminescent, 5 nm-sized particle in a sol-gel glass matrix. The particle is detected with SNR∼100 (10 000 detected photons per second versus 100 photons per second background counts). The standard deviation of the retrieved positions is a measure of the accuracy with which the tracking program finds the positions of the particle. In the focal plane, the error is ±1.25 nm; along the optical axis, the error is ±46.7 nm.
Figure 3.
(a–c) Retrieved positions from an immobile nanoparticle embedded in a sol-gel glass. The widths of the position's histograms describe the positioning error, which is laterally ±1.25 nm and axially ±46.7 nm. (A total of 470 points were acquired at 32 ms per position, the intensity remained constant during the whole acquisition period and the SNR was approx. 100.) (b) Χ2/d.f.=35.46; R2=0.92. Y0=−0.04±3.70. Xc=−1376.53±0.07. w=2.13±0.25. A=140±22. (c) Χ2/d.f.=16.06; R2=0.93. Y0=−5.98±4.77. Xc=7.72±1.99. w=93.39±11.69. A=5166±1095.
2.2 Microscopy set-up for multi-parameter acquisition
The two-photon microscope set-up (figure 1) is similar to the one used in Levi et al. (2005a). It is based on an inverted microscope stand (IX70, Olympus, Tokyo, Japan) using a 60× water immersion objective (1.2 NA) and appropriate dichroic and rejection filters (700dcspxr, et680sp-2p8, respectively, from Chroma Technology Corp, Rockingham, VT, USA). The excitation source is a tunable Ti:sapphire laser (tunable from 690 to 1040 nm, with 120 fs pulse lengths; Chameleon Ultra, Coherent, Palo Alto, CA, USA). The laser beam can be variably attenuated with a neutral density filter revolver shortly before entering the dichroic filter casing in the microscope. Typical wavelength and power (measured at objective back aperture) for the particle studies are 800 nm and 1 mW. For the in vivo study, we use 940 nm excitation and ca 4 mW. The three-dimensional scanning is achieved by combining galvano motor-driven mirrors (Cambridge Technologies, Watertown, MA, USA) for in-plane (or x, y−) beam scanning with a piezo-objective positioner (Physik Instrumente GmbH & Co. KG, Karlsruhe, Germany) to scan along the optical axis (or in z). The microscope set-up is mounted on an actively vibration-damped optical table (Smart Table UT, Newport Corporation, Irvine, CA, USA).
In the detection path, the emission can be split into two channels using either 50 : 50, polarization or dichroic beam splitters. In the first channel, the luminescence is detected with a photomultiplier tube (H7422P-40, Hamamatsu, Tokyo, Japan). Its output signal is processed by a constant fraction discriminator and the resulting pulse train subsequently counted and recorded using a data acquisition card (ISS, Champaign, IL, USA). In the second channel, the emission can be detected with a second PMT-discriminator set-up, as in the first channel. Alternatively, the luminescence can be analysed with a custom-made prism-CCD spectrometer (which uses a Princeton Instruments CCD, Trenton, NJ, USA), in a similar way to that described in Jung et al. (2007). The experiments presented in this article were all acquired with the 50 : 50 beam-splitter in the detection path.
2.3 Samples
2.3.1 Labelled particles dried on the surface and in glycerine
Latex beads (100 nm size) with different chromophores (Fluospheres, green–yellow, red and crimson, from Molecular Probes, Eugene, OR, USA) were sonicated and filtered (size exclusion) immediately before being dispersed on a cleaned microscopy coverslip. The coverslips were kept in a Hellmanex II (Hellma GmbH & Co. KG, Müllheim, Germany) bath for 2 days and rinsed with ethanol immediately before use. We note here that the cleaning step is essential to achieve an efficient wetting of the coverslip. The thin film of the aqueous bead suspension evaporates quickly, so that the beads remain well separated (as opposed to bead agglomerates that form on non-treated coverslips). The beads remain immobilized on the coverslip surface after evaporation of the solvent. For the studies in glycerine, we added 1 part of the bead suspension to 10 parts of glycerine. The nominal glycerine concentration in this mixture is 91 per cent by volume or 93 per cent by weight (using a density of 1.23 g ml−1 for glycerine).
2.3.2 In vivo studies
HEK293 cells, modified to produce green-fluorescent protein (GFP) and RFP chimeras of the urokinase plasminogen activator receptor (uPAR), were available from the laboratory of Caiolfa et al. (2007). The cells were kept in an incubator at 37°C during measurements.
3. Results and discussion
In this section, we highlight the results from a series of studies that were made using the tracking technique and set-up described above. In the first set of results, we demonstrate some capabilities of the method, namely the possibility of acquiring high-resolution three-dimensional maps and the capability of simultaneously obtaining spectra using labelled latex beads. In this set, we first track immobilized beads along a predefined three-dimensional trajectory, which itself is smaller than the PSF, and confirm the typical error of the apparatus, this time for a moving object. We then acquire the trajectories of beads in a glycerine water mixture and confirm that the retrieved diffusion behaviour is consistent with theory. Finally, we test the capability of recording simultaneously the emission spectra of the particles using a mixture of beads with different chromophores in glycerine. The second set of results show how the simultaneous acquisition of multiple parameters, in synchronization with the tracking of protein assemblies in living cells, opens the door to a new level of detail in the characterization of biophysical processes, also illustrating the method's high-resolution three-dimensional mapping capabilities.
3.1 High-resolution three-dimensional maps and multiple-parameter acquisition
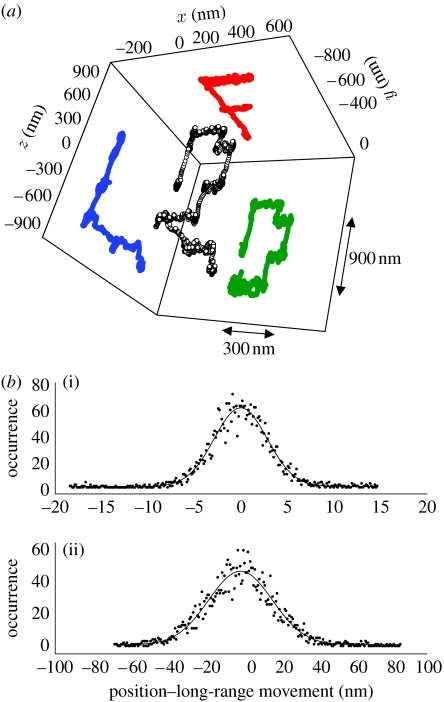
A coverslip with immobilized, spherical and fluorescently labelled latex beads of 100 nm diameter was fixed on a piezo stage (Physik Instrumente GmbH & Co. KG, Karlsruhe, Germany). This assembly was then mounted on the tracking microscope set-up. In this way, it is possible to move the beads along an arbitrary, known path in the focal plane of the microscope (x, y). The entire stage can be moved in z with the microscope's controls, giving complete control over the three-dimensional motion path of the sample. A bead is selected for tracking and the stage is moved manually in a three-dimensional trajectory (the stage speeds range from 0.2 to 1 μm s−1). The trajectory was chosen to be comparable in size to the dimensions of the PSF, and to have sub-structures of the order of 10–50 nm. The recovered trajectory shown in figure 4a was acquired with 32 ms per retrieved position; the total time for the entire movement of the particle was of the order of 30 s. Features of the path, with a size of 20–50 nm, become easily visible (for example the ‘step’ in the letter D, or the overshoot in the horizontal dash of the letter F).
Figure 4.
An immobile nanometre-sized luminescent bead has been moved along a specified path in three dimensions. The lateral movement (x, y) was controlled with a piezo stage and axial movement (z) with the microscope stand control. (a) The size of the complete trajectory is comparable to that of the PSF itself (noted by the arrows). (b(i),(ii)) The histograms shown have been computed from the difference between the long-range movement and each position. The widths of these histograms, of 5.71 nm and 34.3 nm (lateral and axial, respectively), represent a measure for the instantaneous apparent jitter about the predefined path and are an indicator of the positioning accuracy. (b(i)) Lateral Gaussian fit, w=5.71±0.09; (b(ii)) axial Gaussian fit w=34.3±0.71.
We can distinguish between the real jitter of the stage and the apparent jitter due to the limited positioning accuracy of the set-up. The long-range movement can be subtracted from the individual positions, resulting in the histograms shown in figure 4b). The result is that the fast fluctuations, which characterize the accuracy with which each position is determined, have a standard deviation of 5.8 nm in the focal plane and 34 nm along the optical axis (SNR∼50). This also means that the larger fluctuations or overshoots of the order of 6–20 nm (again, for example, visible in the letter D) probably result from real oscillations of the stage. We note also that these oscillations are more pronounced in the z-direction. These could be a consequence of the comparatively massive piezo mount ‘ringing’ on the comparatively lighter microscope stage and/or electronic noise in the control of the stage. While, on the one hand, this poses a limiting factor for the resolution of the apparatus, on the other hand, this also shows that, at the level of resolution of a few nanometres, very faint ‘external’ movements start to interfere with the measurement and have to be considered both in the set-up design as well as in the data analysis. We expect to overcome these limitations and to explore the full resolution of the apparatus in further studies, after revising the mechanical instrument damping and electronics.
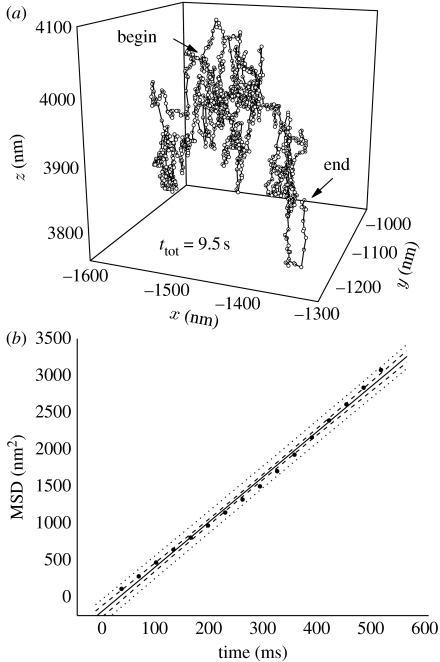
To further test the apparatus, we turned to a different sample, in which the particle does not move in any predefined way. We expected to verify that the retrieved trajectories and the analysis thereof are consistent with physical diffusion models. To this end, we tracked 100 nm labelled beads diffusing in a mixture of water and glycerine. A typical three-dimensional trajectory of a 100 nm bead diffusing in glycerine is shown in figure 5a. The total time of this trajectory is 9.5 s and the time interval between individual positions is 32 ms. The trajectory has a chaotic appearance, as expected for a random walk, and covers a distance of approximately 300 nm in all directions.
Figure 5.
Latex beads (100 nm) in glycerine. (a) This three-dimensional trajectory of one bead covers a total time of 9.5 s and a region of approximately 300 nm in all directions. (b) Evaluation of the diffusion coefficient using the linear relationship between mean-square displacement and observation time. The recovered diffusion coefficient is D=1.02×10−4 μm2 s−1. Circle, raw data; solid line, linear fit (slope=6.12±0.11); dashed line, confidence bands (5–95%); dotted line, prediction bands (5–95%).
We computed the mean square displacements 〈r2〉 in dependence of the temporal separation between all positions (i.e. for steps separated by 32, 64, 96, 128, … ms) and obtained 〈r2(t)〉, which obeys the linear relationship
in the case of a random walk in three dimensions. The experimental data and the linear regression are shown in the graph in figure 5b. We note here that only the first portion (approx. 10%) of the data has sufficient statistical significance to obtain D from a linear regression (see argumentation in Saxton & Jacobson 1997). The value for the diffusion coefficient of this trajectory is D=(1.02±0.05)×10−4 μm2 s−1. We use this value for D in the Stokes–Einstein equation
with T=293 K and r=5×10−8 m and obtain a viscosity of η=42.1 kg m−1 s or 421 cP. We consider an error of 5 K for the temperature and 20 per cent in bead radius and retrieve a glycerine concentration range from 92 to 96 wt% (in water) from lookup tables (Dow Chemical Company 2008). The diffusion coefficients of five further beads have been evaluated in the same way as described above and range between 1.01×10−4 and 1.07×10−4 μm2 s−1. We note that, within the assumed errors, the data stand in good agreement with the conventional diffusion model.
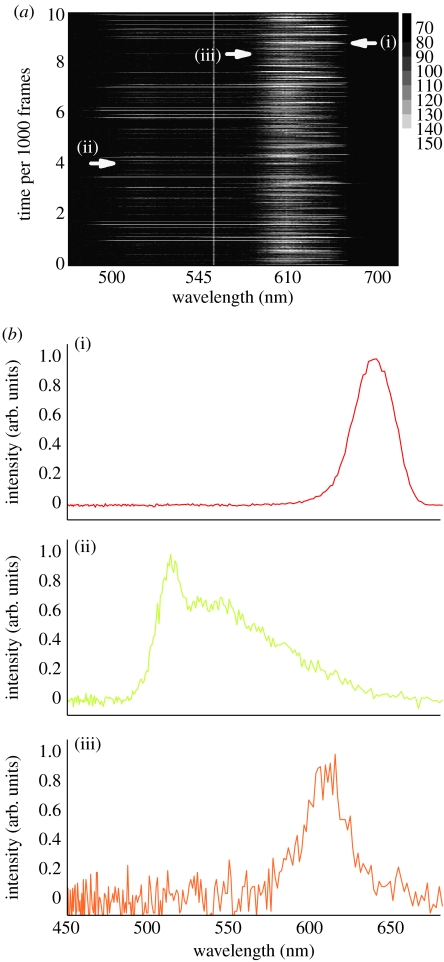
With the set-up described above, it is possible to obtain the emission spectra of the particles under investigation in synchronization with tracking. We tested this capability using a mixture of beads labelled with three different dyes. In the first case, we tracked different immobilized particles (that can be moved with the stage as described above) and recorded their emission spectra. The trajectory and the sequence of spectra were recorded by two independent detectors and computers. The spectral integration time can be varied typically from 50 ms to 2 s. In figure 6, we show a sequence of 10 000 spectra taken consecutively with 50 ms integration time. This sequence was obtained from a fixed point in a liquid glycerine drop containing a mixture of latex beads. As the individual beads move through the observation volume, their emission spectrum becomes visible. The exemplary spectra shown in figure 6b show a pronounced difference in the spectrum quality going on from the red and green to the orange species. This is so because the excitation wavelength is optimized for the two-photon excitation of the red species.
Figure 6.
Sequence of spectra from a mixture of labelled beads passing through the observation volume. (a) Complete raw sequence of 10 000 spectra. The integration time per spectrum is 50 ms. (b) The individual stripes taken from the sequence depicted in (a) were corrected for background and normalized. (i) Red, (ii) yellow–green and (iii) orange. They show the distinguishable spectra of the three different beads.
The spectral resolution of this prism-CCD spectrometer is of the order of 10 nm. Superior spectral resolution (1 nm or so) and a linear dispersion can be achieved by using gratings instead of the prism, however, at the expense of signal intensity (and transportability). The quantum yield of the prism-CCD spectrometer is comparatively high (approx. 80%) and can be used to analyse the fluorescence of individual molecules (e.g. as demonstrated in Jung et al. (2007), if the emission signal is routed directly into the spectrometer. At present, however, we introduce a major loss element by coupling the spectrometer to the detection path of the microscope via a glass fibre.
3.2 Multiple-parameter three-dimensional single-particle tracking in vivo
In this second example, we turn to an in vivo study and demonstrate how multi-parameter three-dimensional SPT reveals details that are not readily obtained by other microscopy techniques. The biological problem at hand is related to protein dynamics in the cellular membrane (their transport, oligomerization, etc.). Studies of membrane proteins in vivo are difficult to realize or limited on the experimental level, owing to the geometrical, three-dimensional shape of a cell. For this reason, most studies are limited either to a small cross section of the membrane or, when available, to the flatter region at the bottom of the cell, the basal membrane or lamellipodia. In the latter case, the fact that the cell is in contact with the cover slide is often a major point of criticism. Another source of experimental difficulty lies in the fact that often the signal of interest (e.g. coming from a labelled protein) is overwhelmed or distorted by the interference of autofluorescent species. This is frequently counteracted by increasing the concentration of the species of interest (e.g. by boosting the expression levels of a labelled protein) to levels that are often of questionable physiological significance. With three-dimensional SPT, both the problems can be bypassed, as long as particles are identifiable on an individual basis. First, there is no need to remain limited to a two-dimensional section or to the basal membrane. Second, because it is possible to select individual particles, the experimenter does not need artificially high levels of expression. Finally, with the capability of obtaining the particle's emission spectrum, the experimenter obtains immediately a confirmation if the particle under investigation is displaying the correct label (or, in the negative case, if the signal is coming from unspecific autofluorescence). However, the necessity to single out individual particles also poses a limit to the three-dimensional SPT technique: the probability that different particles will cross the scanned orbits is proportional to the concentration of particles and their mobility. When the concentration is high (or when the particles move fast), it is not possible to assign one trajectory to one particular particle. This reasoning also applies to the influence of autofluorescence on the signal, for example if the autofluorescence cannot be spectrally filtered and if it is high in intensity and not spatially confined to a distinguishable region in the cell.
The HEK293 cells used in this study produce a receptor, uPAR, that has been modified with the insertion of a GFP as a fluorescent label—care has been taken to ensure that this modification does not introduce any artefact in the functioning of the protein, or in the biology of the specimen (Caiolfa et al. 2007). In conventional two-photon microscopy image sequences (figure 7a), we can readily see particles moving in close proximity to the cellular membrane. The particles remain in the field of view for a few frames and vanish, most probably due to motion along the optical axis. This major problem of conventional tracking in microscopy, as mentioned above, the vanishing of particles (vesicles, proteins, small agglomerates, etc.) from the field of view, is avoided in three-dimensional SPT. The individual particles remain observable for a longer period of time, limited only by the extent of photobleaching.
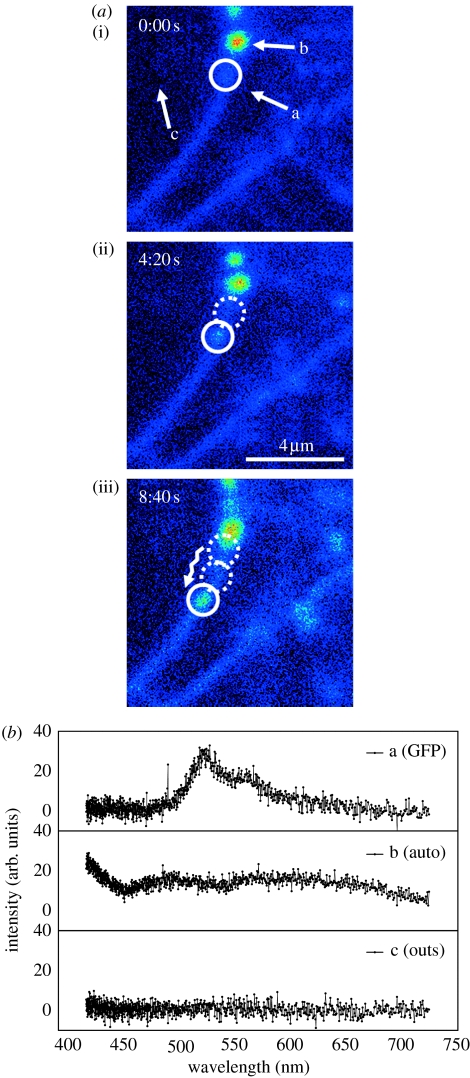
Figure 7.
(a,b) Image sequence showing a cross section of a cellular membrane. Circles indicate the particle of interest being transported along the membrane. The spectra (1 s acquisition time) were taken from the marked particles a and b after the acquisition of the image sequence, and from region c (without tracking).
An important experimental detail can be highlighted in this particular example. In the case shown in figure 7, the experimenter would be easily tempted to choose the bright, well-defined spot (marked b) on the upper left part of the image for tracking. We draw attention to the (barely visible) particle (marked a) in the circle. We started tracking both the particles immediately after the last frame in figure 7a. The difference between these two particles becomes apparent in the emission spectra shown in figure 7b. The spectrum of particle a can be assigned to the spectrum of GFP, indicating that the protein of interest is contained in the diffusing spot. On the other hand, the spectrum of particle b is unspecific (note that the spectra are corrected for background, acquired in region c). By using multi-parameter three-dimensional SPT, the experimenter can ascertain that the particle being tracked contains the correct species.
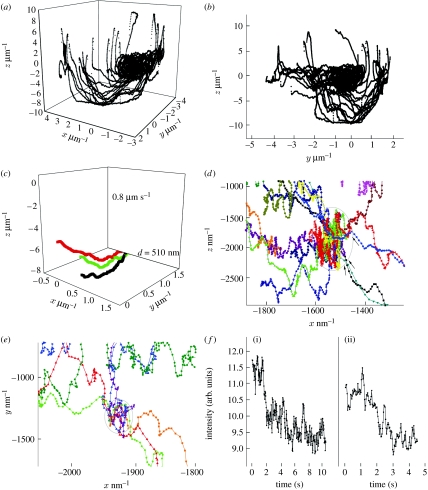
A series of trajectories from approximately 50 different particles are recorded successively over a period of approximately 10 min—all of them having been identified via their spectrum. We record details of the receptor transport that have, up to now, never been observed in vivo with such a level of spatio-temporal accuracy. In the example shown here, we overlay the 50 trajectories of individual, small GFP–uPAR aggregates. The trajectories clearly delineate the three-dimensional structure of the cell membrane (figure 8a,b). The complete set of trajectories is acquired over a period of several minutes, sequentially. We have to consider that on this time-scale the movement of the whole cell and its structure are not negligible. For this reason, we do not expect all the trajectories to overlap, forming a perfect layer delineating the cell membrane. Despite this comparatively slow movement, we recognize, firstly, that some trajectories follow the same path (figure 8c) and, secondly, that there are specific regions in the membrane to which different particles get transported (figure 8d,e).
Figure 8.
Trajectories of uPAR aggregates near the cell membrane. (a) A total of 51 trajectories (taken sequentially over a period of approx. 10 min) delineate the cell membrane in three dimensions, the y,z-projection is shown in (b). (c) These three trajectories from different particles span about 7 s in time; they have been acquired minutes apart. The particles move in the same direction, along the same region in space, with approximately the same velocity. (d,e) Zoom-ins from (b) (note the scale now in nm); both show a distinct region (marked by circle) in which trajectories end. (f) These two exemplary intensity versus time traces show abrupt fluctuations.
Furthermore, we observe that the intensity versus time behaviour of the particles shows abrupt bleaching and blinking (two examples shown in figure 8f). The photobleaching takes place on a time scale of a few seconds. The typical signal in the beginning of a trajectory ranges from 500 to 1000 counts per second. The background signal near the cell membrane is spatially heterogeneous ranging from 50 to 150 counts per second, affecting the SNR, which varies largely from 3 to 20. The signal at the end of a trajectory drops to 200 counts per second owing to photobleaching, and is barely above the background. At that point, the tracking algorithm will lose the particle. While the few (5–10) seconds are sufficient to recognize different mechanisms responsible for particle trafficking, on the other hand this also imposes a time limit per trajectory. However, because the bleaching is not continuous, as would be observed for a large (more than 20) ensemble of dyes, we can assert that the particles tracked are agglomerates of a few (4–10) individual molecules. The abrupt fluctuations observed in the recorded emission intensity versus time traces are a signature of the presence of a few individual emitters. All the traces recorded show this behaviour.
The overall velocity of the aggregates in the most rectilinear regions of their respective trajectories is of the order of 400–800 nm s−1 (comparable to the velocities observed in myosin/actin transport). Some trajectories were found to end in a specific region of the cell. The size of these regions, given the error in positioning, cannot be determined with arbitrary accuracy, but is below 200 nm. The zoomed-in sections in figure 8d,e show two such regions. We do not observe a fluorescence intensity build-up in these regions, meaning that the fluorescent species are being removed from this region. At present it is not clear whether the aggregates followed in this study have, or not, been exposed to the extracellular matrix. In the first case, the aggregates would be in the process of being transported along the cell membrane for deployment. In the second case, the aggregates would be in the process of being internalized, e.g. for degradation or recycling. Experiments in which we monitor the binding of the receptor to extracellular species labelled with a different fluorophore are currently underway.
In summary, this study shows that, with three-dimensional SPT, it is possible to follow and characterize details of the transport of small protein aggregates in a living cell. Furthermore, owing to the capability of recording emission spectra, it is possible to verify whether the particle under observation does indeed contain the (labelled) protein of interest. The recorded intensity behaviour can, in addition, be used to estimate the number of fluorophores in the tracked object.
4. Summary and Concluding remarks
In this article, we show that the feedback implementation of SPT offers substantial advantages with respect to conventional camera-based methods. First, there is a substantial gain in spatio-temporal resolution of the order of nanometres and milliseconds—over a large volume of several microns in (we emphasize) all three dimensions. Second, it is possible to obtain additional information such as spectrum or other fluorescence parameters during the tracking of the particle, because the feedback algorithm ensures that the focus is kept centred on the particle of interest. Another important feature of our tracking method is the capability of tracking more than one particle simultaneously. This is achieved by multiplexing the determination of the position of several particles with time. If a particle remains within the range of a scanned orbit during the acquisition time of the other particles, then the scanning algorithm will ‘pick it up’ again and re-centre when the orbit returns to that specific position. We demonstrated this feature in a previous publication (Levi et al. 2005b). The application to biological samples shows that bleaching does not hinder the capability to follow a particle composed of a few individual fluorophores, it merely limits the observation time, which for the cases shown here is long enough to recognize a specific path in the cell and therefore allows us to infer biological mechanisms responsible for the transport of protein assemblies in the cell. On the other hand, the observation of the bleaching behaviour, e.g. the observation of distinct bleaching events, is a valuable additional parameter, which allows us to discern, for example, between large and small agglomerates of fluorophores.
Overall, the position feedback method presented here allows new molecular parameters to be measured during the tracking. These parameters could be crucial to determine molecular events that otherwise cannot be observed using conventional camera-based methods.
Acknowledgments
We acknowledge NIH grants P41 RR03155 that provided funding for this work. We would like also to thank Dr Valeria Caiolfa and Dr Moreno Zamai for providing us with the HEK293 cells and for the discussion of the cell experiments.
Footnotes
One contribution of 9 to a Theme Supplement ‘Quantitative fluorescence microscopy: The 1st international Theodor Förster lecture series’.
References
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. doi:10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Bobroff N. Position measurement with a resolution and noise limited detector. Rev. Sci. Instrum. 1986;57:1152–1157. doi:10.1063/1.1138619 [Google Scholar]
- Cai W.B., Hsu A.R., Li Z.B., Chen X.Y. Are quantum dots ready for in vivo imaging in human subjects? Nanoscale Res. Lett. 2007;2:265–281. doi: 10.1007/s11671-007-9061-9. doi:10.1007/s11671-007-9061-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiolfa V.R., et al. Monomer–dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies. J. Cell Biol. 2007;179:1067–1082. doi: 10.1083/jcb.200702151. doi:10.1083/jcb.200702151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. doi:10.1073/pnas.082243699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnert G., Keller J., Medda R., Andrei M.A., Rizzoli S.O., Lurmann R., Jahn R., Eggeling C., Hell S.W. Macromolecular-scale resolution in biological fluorescence microscopy. Proc. Natl Acad. Sci. USA. 2006;103:11440–11445. doi: 10.1073/pnas.0604965103. doi:10.1073/pnas.0604965103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow Chemical Company. 2008. Viscosity of aqueous glycerine solutions. See http://www.dow.com/glycerine/resources/table18.htm. [Google Scholar]
- Ha T. Single molecule fluorescence resonance transfer. Methods. 2001;25:78. doi: 10.1006/meth.2001.1217. doi:10.1006/meth.2001.1217 [DOI] [PubMed] [Google Scholar]
- Hammer N.I., Emrick T., Barnes M.D. The cutoff below 500 nm in the OPV photoluminescence is due to the choice of filters. Nanoscale Res. Lett. 2007;2:282–290. doi:10.1007/s11671-007-9062-8 [Google Scholar]
- Hell S.W. Improvement of lateral resolution in far-field light microscopy using two-photon excitation with offset beams. Opt. Commun. 1994;106:19–24. doi:10.1016/0030-4018(94)90050-7 [Google Scholar]
- Hellriegel C., et al. Diffusion of single streptocyanine molecules in the. nanoporous network of sol–gel glasses. J. Phys. Chem. B. 2004;108:14 699–14 709. doi:10.1021/jp049412a [Google Scholar]
- Huang X., Jain P.K., El-Sayed I.H., El-Sayed M.A. Gold nanoparticles: interesting opticals properties and recent applications in cancer diagnostics and therapy. Nanomedicine. 2007;2:681–693. doi: 10.2217/17435889.2.5.681. doi:10.2217/17435889.2.5.681 [DOI] [PubMed] [Google Scholar]
- Jung C., Hellriegel C., Michaelis J., Bräuchle C. Single molecule traffic in mesoporous materials: translational, orientational and spectral dynamics. Adv. Mater. 2007;19:956. doi:10.1002/adma.200602173 [Google Scholar]
- Kirstein J., Platschek B., Jung C., Brown R., Bein T., Bräuchle C. Exploration of nanostructured channel systems with single-molecule probes. Nat. Mater. 2007;6:303–310. doi: 10.1038/nmat1861. doi:10.1038/nmat1861 [DOI] [PubMed] [Google Scholar]
- Kis-Petikova K., Gratton E. Distance measurement by circular scanning of the excitation beam in a two-photon microscope. Microsc. Res. Tech. 2004;63:34–49. doi: 10.1002/jemt.10417. doi:10.1002/jemt.10417 [DOI] [PubMed] [Google Scholar]
- Klar T.A., Jakobs S., Dyba M., Egner A., Hell S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl Acad. Sci. USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. doi:10.1073/pnas.97.15.8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre J., Fraser J.M., Finnie P., Homma Y. Photoluminescence from an individual single-walled carbon nanotube. Phys. Rev. B. 2004;69:075 403. doi:10.1103/PhysRevB.69.075403 [Google Scholar]
- Levi V., Ruan Q., Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys. J. 2005a;88:2919–2928. doi: 10.1529/biophysj.104.044230. doi:10.1529/biophysj.104.044230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi V., Ruan Q., Plutz M., Belmont A.S., Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005b;89:4275–4285. doi: 10.1529/biophysj.105.066670. doi:10.1529/biophysj.105.066670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S., Gillette J.M., Patterson G.H., Shroff H., Hess H.F., Betzig E., Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. doi:10.1038/nmeth.1176 [DOI] [PubMed] [Google Scholar]
- Pawley J.B. 3rd edn. Springer Science and Business Media; New York, NY: 2006. Handbook of biological confocal microscopy. [Google Scholar]
- Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–796. doi: 10.1038/nmeth929. doi:10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M.J., Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. doi:10.1146/annurev.biophys.26.1.373 [DOI] [PubMed] [Google Scholar]
- Sonnleitner A., Schütz G.J., Schmidt T. Free Brownian motion of individual lipid molecules in biomembranes. J. Biophys. 1999;77:2638–2642. doi: 10.1016/S0006-3495(99)77097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer E.H.K., Lindek S. Fundamental reduction of the observation volume in far-field light microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Opt. Commun. 1994;111:536–547. doi:10.1016/0030-4018(94)90533-9 [Google Scholar]
- Tamarat P., Maali A., Lounis B.L., Orrit M. Ten years of single-molecule spectroscopy. J. Phys. Chem. A. 2000;104:1–16. doi: 10.1021/jp992505l. doi:10.1021/jp992505l [DOI] [PubMed] [Google Scholar]