Abstract
We report on the rational design and synthesis of a new type of bioactive poly(ethylene glycol) diacrylate (PEGDA) macromers, cyclic Arg-Gly-Asp (cRGD)-PEGDA, to mimic the cell-adhesive properties of extracellular matrix (ECM), aiming to create biomimetic scaffolds with controlled spatial organization of ligands and enhanced cell binding affinity for tissue engineering. To attach the cRGD peptide in the middle of PEGDA chain, a tailed cRGD peptide, c[RGDfE(SSSKK-NH2)] (1) was synthesized with c(RGDfE) linked to a tail of SSSKK. The tail consists of a spacer with three serine residues, and a linker with two lysine residues for conjugating with acryloyl-PEG-NHS (5) to create cRGD-PEGDA (6). cRGD-PEGDA possesses good ability of photopolymerization to fabricate hydrogel scaffolds under UV radiation. Surface morphology and composition analysis demonstrates that cRGD-PEGDA hydrogels were well-constructed with porous three-dimensional (3D) structures and uniform distribution of cRGD ligands. Our results show that cRGD-PEGDA hydrogels facilitate endothelial cell (EC) adhesion and spreading on the hydrogel surfaces, and exhibit significantly higher EC population in comparison with linear RGD-modified hydrogels at low peptide incorporation. Since ligand presentation in biomimetc scaffolds plays an important role in controlling cell behaviors, cRGD-PEGDA has great advantages of controlling hydrogel properties and ligand spatial organization in the resulting scaffolds. Furthermore, cRGD-PEGDA is an attractive candidate for the future development of tissue engineering scaffolds with optimum cell adhesive strength and ligand density.
Keywords: hydrogel, cyclic RGD peptide, biomimetic scaffold, tissue engineering, endothelial cell
INTRODUCTION
The shortage of available organ donors has driven the growth and expansion of the field of tissue engineering as promising alternative to organ transplantation (1–6). Scaffolds play crucial roles in tissue engineering including providing support and space for cells, encouraging cell adhesion and growth, and guiding cell function (7–15). Hydrogels are excellent scaffolding materials for repairing and regenerating a variety of tissues, such as cartilage, bone and vasculature (16–22). They are attractive because they can provide a highly swollen three dimensional (3D) environment similar to soft tissues and allow diffusion of nutrients and cellular waste through the elastic network. Synthetic poly(ethylene glycol) (PEG)-based hydrogels have advantages over natural hydrogels, such as the ability of photopolymerization into any shape, injectable capacity for tissue repair, and adjustable mechanical properties to fit various applications (23). However, most synthetic hydrogels typically exhibit minimal or no intrinsic biological activity (16,17), and do not provide an ideal environment for culturing anchorage-dependent cells, such as endothelial cells (ECs) and smooth muscle cells (SMCs). To facilitate cell adhesion, small peptides derived from extracellular matrix (ECM) proteins, have been important targets for the covalent coupling type of biospecific modification (24–30). For example, Arg-Gly-Asp (RGD)-modified PEG monoacrylate was copolymerized with PEG diacrylate (PEGDA) to create cell-adhesive PEG hydrogels (31–33), but the extent of incorporation of RGD-modified PEG monoacrylate into PEGDA hydrogels is limited because increasing incorporation of monoacrylated monomers affects the mechanical properties and the swelling ratio of the resultant hydrogels (34–36).
To address these limitations, we recently reported on a linear RGD-modified PEGDA macromer, RGD-PEGDA with a GRGDSP peptide attached in the middle of PEG chain, which stimulates cell adhesion, and good control over the hydrogel mechanical properties (37). However, a further consideration is that the RGD sequence is exposed at the tip of a loop in the cell-binding domain of fibronectin, which presents the ligand with a spatial constraint that results in increased specificity for cell binding (38,39). It is a challenge to incorporate cyclic RGD (cRGD) peptides into PEGDA hydrogels with controlled spatial organization to mimic the native RGD loop structure. Previous studies have demonstrated that cRGD peptides have the advantage of increasing the specificity to integrin αvβ3 and enhance biological activity up to 240 times, in comparison with linear analogues (40–43). Research in our laboratory has shown that cyclo(Arg-Gly-Asp-D-Phe-Lys) [c(RGDfK)] and cyclo(Arg-Gly-Asp-D-Phe-Glu) [c(RGDfE)] more efficiently compete with fibronectin for specific binding to integrin αvβ3 on human pulmonary artery ECs, compared with the linear GRGDSP peptide (44). Therefore, we hypothesize that the incorporation of cRGD peptides into the PEGDA hydrogels will better mimic the native fibronectin cell-binding domain and benefit the cell specific adhesion.
Here, we report on the design and synthesis of a new type of bioactive PEGDA macromers, cRGD-PEGDA (6), to mimic the cell-adhesive properties of ECM, aiming to create biomimetic scaffolds with controlled spatial organization and enhanced cell binding affinity for tissue engineering. To attach the cRGD peptide in the middle of PEGDA chain, a tailed cRGD peptide, c[RGDfE(SSSKK-NH2)] (1) was synthesized with c(RGDfE) linked to a tail of SSSKK, which consists of a spacer with three serine residues, and a linker with two lysine residues for conjugating with acryloyl-PEG-NHS (5) to create cRGD-PEGDA (6). Since ligand presentation in biomimetc scaffolds plays an important role in controlling cell behaviors, cRGD-PEGDA is an attractive candidate for the development of well-defined tissue engineering scaffolds with spatial control of ligand organization, and optimization of adhesive strength through controlling ligand density.
EXPERIMENTAL
Materials
9-Fluoromethoxycarbonyl (Fmoc)-protected amino acids, O-(benzotriazol-1-yl)-N,N,N’N’-tetramethyluronium hexafluorophosphate (HBTU) and 1-hydroxybenzotrilazole (HOBt) were purchased from AnaSpec (San Jose, CA). Amino acids had various side chain protect groups, such as Otert-butyl (tBu) for Asp and Ser, O-tert-butoxycarbonyl (Boc) for Lys, and 2,2,4,6,7-pentamethyldihydrobenzofurn-5-sulfonyl (Pbf) for Arg, while allyl (All) was used to protect α-COOH of Glu (Fmoc-Glu-OAll). Fmoc-aminomethyl-3,5-dimethoxyphenoxy-valeric acid (PAL) resin (with a loading of 0.6 mmol/g), tetrakis-(triphenyl phosphine) palladium (0) [Pd(PPh3)4], piperidine, N,N-dimethylformamide (DMF),and N,N-diisopropylethylamine (DIEA) were purchased from Sigma-Aldrich. 1-Hydroxy-1-azabenzotriazoleuronium (HATU) was purchased from GenScript (Piscataway, NJ). PEGDA (Mw 6,000) and Linear RGD-PEGDA with a sequence of GRGDSP were synthesized according to our previous method (37). Acryloyl-PEG-NHS (5, Mw, 3,400) was purchased from Nektar (Huntsville, AL). The cleavage cocktail of Reagent K containing 85% (v/v) TFA was prepared prior to use according to the method described previously (45). Irgacure 2959 (I2959, 2-hydroxy-1-[4-(hydroxyethoxy)phenyl]-2-methyl-1-propane) was used as water-soluble photoiniators, which was provided by Ciba (Tarrytown, NY).
Instrumentation
Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry was performed on a Bruker Biflex III MALDI-TOF mass spectrometer by dissolving the sample and the matrix of 2,5-dihydroxybenzoic acid in 1:1 (v/v) ethanol and water, and mass spectra were typically accumulated from 200 laser shots. The surface morphology of freeze-dried hydrogels was examined by scanning electron microscopy (SEM) on a Hitachi S-4500 SEM instrument. Samples were coated 50 Å of platinum prior to SEM imaging. Attenuated total reflectance-Fourier transform infrared (ATR-FTIR) spectra were obtained on a Bio-Rad FTS-575C FTIR spectrometer equipped with a UMA-500 microscope. A germanium crystal was used in the micro-ATR attachment to achieve a sampling depth of 0.1–0.2 µm. X-ray photoelectron spectroscopy (XPS) analysis was carried out on a PHI 5000 VersaProbe X-ray photoelectron spectrometer with the incident radiation consisting of Mg Kα X-ray and the take-off angle fixed at 45°. Phase contrast photomicrographs of cells were obtained on a Nikon Diaphot 200 Inverted Phase Contrast Microscope with a CCD camera using Metamorph software package.
Synthesis of tailed cyclic RGD peptide, c[RGDfE(SSSKK-NH2)] (1)
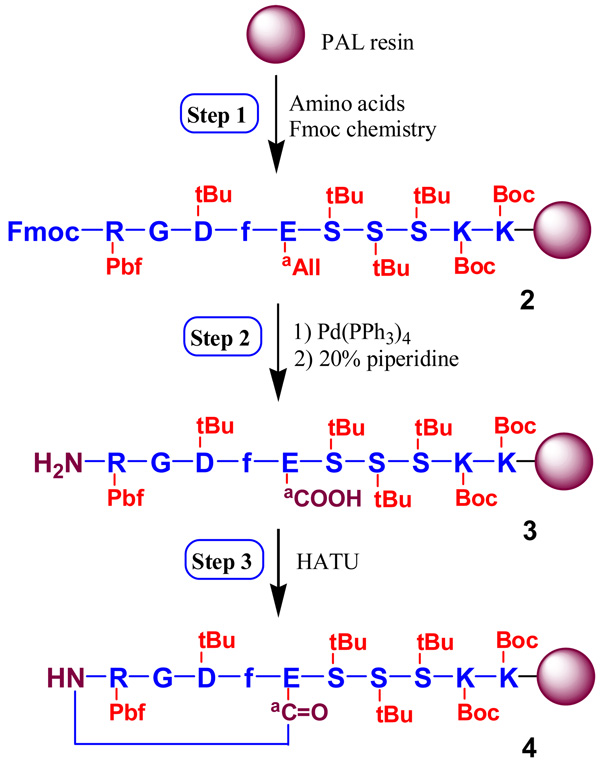
Peptides were synthesized by a three-step synthesis method in a 0.25 mmol scale with PAL resin to produce an amide C-terminus on a 433A Automatic Peptide Synthesizer (Applied Biosystems). In general, couplings were performed with a 4-fold excess of Fmoc-amino acid in the presence of HBTU/DIEA/HOBt (1:1:0.5, v/v/v) for 30 min. Fmoc groups were cleaved with 20% (v/v) piperidine in DMF for 10 min. Briefly, (1) protected linear RGD peptides on resin (2) with a sequence of RGDfE(SSSKK) on resin and an α-allyl protective group on Glu was synthesized using standard Fmoc strategy; (2) selectively deprotected peptides on resin (3) were obtained by the removal of α-allyl group on Glu manually on Peptide Synthesizer by three equivalents of Pd(PPh3)4, followed by the removal of Fmoc on Arg at the N-terminus by 20% (v/v) piperidine in DMF; (3) protected cyclic RGD peptides on resin (4) were formed by cyclization between the amine group (NH2) on Arg and the α-carboxylic group (COOH) on Glu using HATU. The final cyclized peptides (1) were cleaved from the resins using Reagent K, and were further purified by reverse phase-HPLC on a Waters 2690 Alliance system (62% yield). MALDI-TOF MS: m/z = 1,121.9 for [M+H]+ (calculated mass 1,122.1); m/z = 1,143.9 for [M+Na]+ (calculated mass 1,144.1).
Synthesis of cRGD-PEGDA (6)
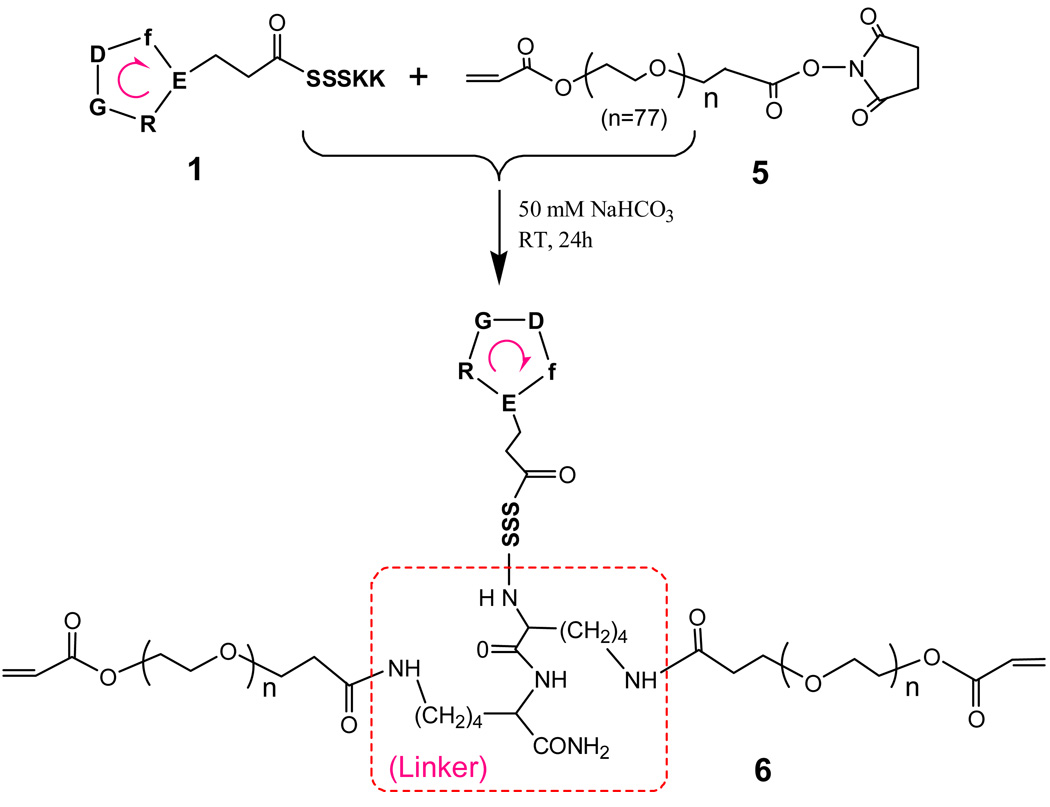
c[RGDfE(SSSKK-NH2)] 1 was dissolved in 50 mM of sodium bicarbonate buffer (pH 8.2). Acryloyl-PEG-NHS (5) with two-fold molar amount as the cRGD peptide, was dissolved in 50 mM of sodium bicarbonate buffer (pH 8.2), and added dropwise to the aqueous peptide solution. The mixture was stirred at room temperature for 24 h, and then dialyzed against water with membranes of Mw Cutoff 5,000 for 48 h. The final power product was obtained by lyophilization for 48 h (65% yield). MALDI-TOF MS: a maximum peak at 7,658 and a unique distribution with a repeating Mw difference of 44 (corresponding to one PEG repeating unit).
Fabrication of hydrogels
Hydrogel solution containing 20% (w/v) of cRGD-PEGDA or PEGDA and 0.1% (w/v) of Irgacure 2959 (I2959) photoinitiator in PBS was sterilized by filtering through a 0.2-µm nylon syringe filter, and was added to each well of a sterilized 6-well stainless steel mold. The plate was placed under a UV lamp (365 nm, 5–10 mW/cm2) for 10 min irradiation. Hydrogel disks (8 mm in diameter, 1 mm in thickness) were obtained by lifting the mold plate and transferred to a 24-well culture plate. The swelling ratio (Q) of hydrogels was determined gravimetrically. The resulting hydrogel was placed in a vial containing 4 mL of PBS and incubated under gentle shaking (80 rpm) at 37° for 48 hours. Excess fluid was removed to obtain the weight of the wet hydrogel (Wwet). The hydrogel was then freeze-dried to obtain the weight of the dried hydrogel (Wdry). The swelling ratio (Q) was calculated by using the equation: Q = Wwet/Wdry.
Cell culturing on hydrogels
Human pulmonary artery endothelial cells (ECs) (passage 7, Cambrex) were grown to 80–90% confluence using complete growth medium (CGM) in 25-cm2 tissue culture flasks (Costar) at 37°C and 5% CO2. Confluent cells were trypsinized and resuspended in Opti-MEM® I (Gibco). EC suspension (1 mL, ~105 cells) was added to each well of the 24-well tissue culture plate containing hydrogel disks. Cells were allowed to attach for 2 h, and then the EC-seeded hydrogel disks were transferred to another 24-well culture plate with CGM and placed in the incubator at 37°C and 5% CO2. Culture media was changed every 2 days. Cells were imaged using a Nikon Diaphot 200 Inverted Phase Contrast Microscope. EC growth and viability were quantified 96 h after seeding by MTS assays, using CellTiter cell proliferation assay kit (Promega). The absorbance at 490 nm was measured. ANOVA with Tukey’s comparison of means (a = 0.05, Minitab 15) was used to assess statistic differences.
RESULTS AND DISCUSSION
Rational design of well-defined hydrogel scaffolds
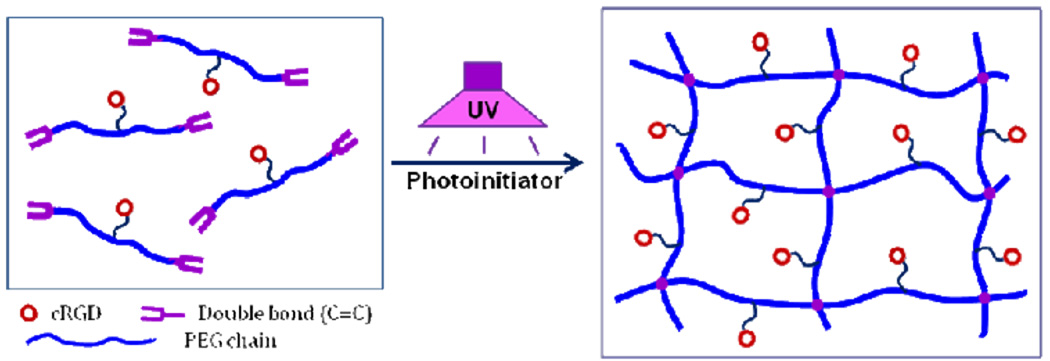
The presentation of peptide ligands in scaffolds plays important roles in controlling cell adhesion and motility behaviours, and mechanical properties of tissue engineered constructs. Our design principle of hydrogel scaffolds is that the preconstruction of a bioactive PEGDA macromer with cRGD peptide attachment in the middle of its chain, will lead to controlled ligand spatial organization and ligand distribution in the resulting photopolymerized hydrogel scaffolds, as shown in Figure 1. Since this designed novel macromer has two C=C double bonds on both ends with a similar PEGDA backbone structure, high cRGD peptide density can be incorporated into the hydrogels without affecting the hydrogel physical properties. Furthermore, this cRGD-modified macromer can also be able to copolymerize with other monomers to form cell-adhesive hydrogels with less incorporation due to the higher binding ability of cRGD ligands, compared with linear RGD peptides.
Figure 1.
Rational design of cyclic RGD (cRGD)-modified PEGDA macromer for fabrication of biomimetic hydrogel scaffolds with controlled cRGD organization. A novel type of bioactive PEGDA macromers was designed with a cRGD peptide attached in the middle of PEGDA chain, which can be photopolymerized to form 3D hydrogel networks with uniform cRGD distribution.
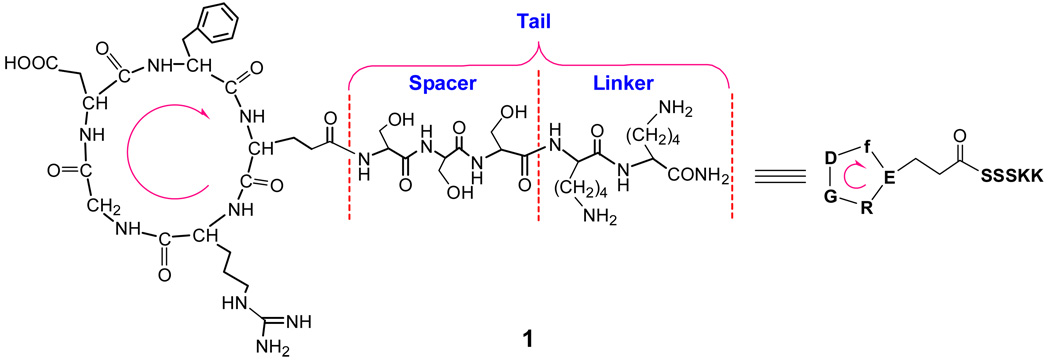
Design and synthesis of tailed cyclic RGD peptide
To attach the cRGD peptide in the middle of PEGDA chain, a tailed cRGD peptide, c[RGDfE(SSSKK-NH2)] (1) was designed with c(RGDfE) linked to a tail of SSSKK through the Glu (E) residue, as shown in Figure 2. The tail consists of a spacer with three serine residues and two lysine residues. The hydrophilic spacer of SSS is used to increase the mobility of the c(RGDfE) ligand, which may lead to enhanced binding ability. Another consideration is that a SSS sequence next to Glu can prevent the unexpected glutarimide formation, which took place with a GGG spacer next to Glu in our previous studies (45). The KK acts as a linker with two free amine groups that provide two active sites for conjugating with other molecules. Compared with c(RGDfK) (which has limited spacer length from the side chain of lysine and only one free amino group on Lys for conjugation), the design of tailed cyclic RGD peptide is more flexible (due to the spacer) and affords two or more conjugating sites through the linker, provided two or more lysine residues are attached.
Figure 2.
Chemical structure of designed cyclic RGD peptide, c[RGDfE(SSSKK-NH2)] (1) with a tail of SSSKK, which consists of a spacer with three serine residues and a linker with two lysine residues for further conjugation with other molecules. The arrow shows the direction of the amino acid sequence from N-terminus to C-terminus prior to the cyclization of the linear peptide, RGDfE(SSSKK-NH2).
The tailed cyclic peptide 1 was synthesized by orthogonal solid phase peptide synthesis (SPPS), as shown in Scheme 1, using Fmoc-protected glutamic acid α-allyl ester (Fmoc-Glu-OAll) on a PAL resin. The allyl group on Glu was used as the orthogonal protecting group for on-resin cyclization with Fmoc strategy. It can be selectively removed to create a free carboxylic group and enable the construction of a lactam bridge between carboxylic and N-terminal amino groups while the peptide is still attached to the resin. This synthesis includes three major steps. First, a protected linear peptide on PAL resin (2) was synthesized. Then, allyl and Fmoc groups were removed sequentially by Pd(PPh3) and piperidine to produce a selectively deprotected linear peptide on the resin (3). Finally, cyclization of 3 was carried out using HATU to generate the protected cyclic peptide of c[RGDfE(SSSKK-NH2)] on the resin (4). The final deprotected cyclic peptide 1 was obtained by the cleavage of peptides from 4 using Reagent K containing 85% TFA, and purified by reverse-phase HPLC. Its structure was confirmed by MALDI-TOF MS analysis.
Scheme 1.
Orthogonal solid phase synthesis of c[RGDfE(SSSKK-NH2)] 1 by a three-step method
Synthesis of cRGD-PEGDA macromer
A monoacrylated PEG macromer with a carboxylic end group activated by N-hydroxysuccinimide (NHS), acryloyl-PEG-NHS (5, Mw 3,400), was used to react with the two free amine groups from the linker (KK) of the tailed cyclic peptide 1 to produce cRGD-PEGDA (6), as shown in Scheme 2. The formation of the resulting cRGD-PEGDA macromer 1 was verified by MALDI-TOF mass analysis. The maximum peak at 7,658 corresponds to the structure of 6, which resulted from the conjugation of 2 molar 5 (Mw 3,400) and one molar 1 (Mw 1,121.1) with elimination of 2 molar NHS (Mw 115.1). This novel bioactive PEGDA macromer 1 combines the photo-polymerization ability of PEGDA and the bioactivity of cRGD peptides, and possesses the ability to form cell-adhesive hydrogels with uniform distribution of cRGD peptides in the middle of PEG chains through the whole hydrogel.
Scheme 2.
Synthesis of cRGD-modified PEGDA macromer, cRGD-PEGDA 6.
Hydrogel fabrication and characterization
Hydrogels with 20% (w/v) of macromers were fabricated in the form of thin disks (8 mm in diameter, 1 mm in thickness) by photopolymerization. The swelling ratio (Q) and mesh size (ξ) results are listed in Table 1. The number-average molecular weight between cross-links (Mc) and average mesh size were calculated according to the Peppas and Merrill model (46,47). The presence of ionic moieties in hydrogels makes the theoretical treatment of swelling much more complex (48). Given that the macromer of cRGD-PEGDA has only two charge groups on each long PEG chain and the resulting hydrogels are not highly charged, we still use the Peppas and Merrill model in order to simply the calculations (49). The hydrogel swelling properties and cross-linking density are particularly important from a tissue-engineering perspective, since they impact transport and overall cell viability, and influence cell behavior. Under the same processing conditions, hydrogels made from cRGD-PEGDA exhibited higher swelling ratio with Q =17.6 than the control hydrogels from PEGDA with Q = 13.0, as listed in Table 1. As a result, cRGD-PEGDA hydrogels have lower cross-linking density with a higher Mc (2,326 g/mol) and greater mesh size (ξ = 85.4 Å) than the PEGDA hydrogels (Mc =1,791 g/mol, ξ = 64.7 Å). These results are consistent with the theoretical analysis that cRGD-PEGDA possesses a longer PEG chain (Mw ~ 6,800) and much higher molecular weight (7,658) after cRGD attachment, compared with PEGDA (Mw = 6,303).
Table 1.
Properties of hydrogels from 20% (w/v) macromers
| Hydrogel | Mw (g/mol) a | Q | Mc (g/mol) | ξ (Å) |
|---|---|---|---|---|
| PEGDA | 6,303 | 13.0 | 1,791 | 64.7 |
| cRGD-PEGDA | 7,658 | 17.6 | 2,326 | 85.4 |
Initial molecular weight of macromers determined by MALDI MS
Hydrogel surface morphology
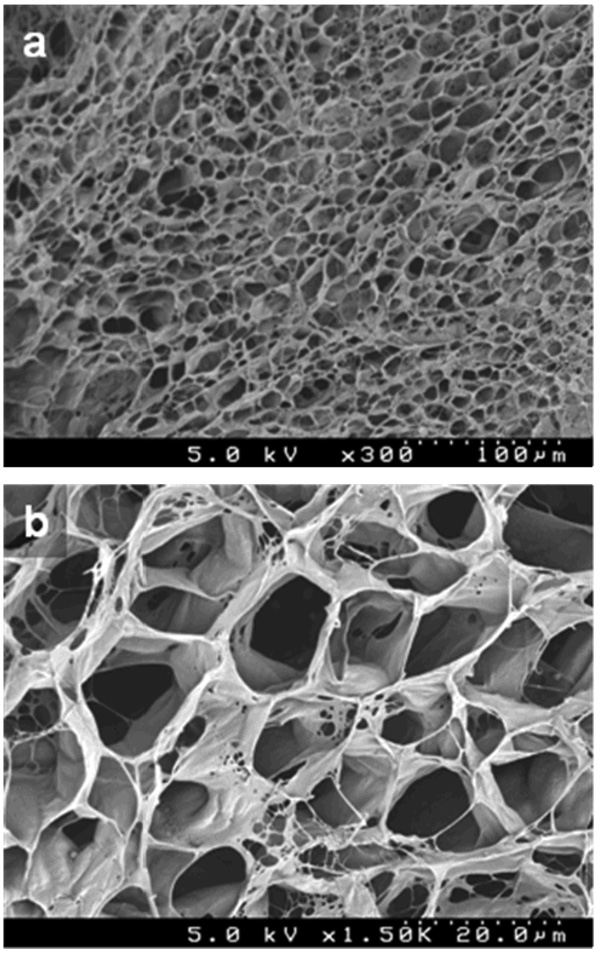
To obtain insight into the three-dimensional (3D) structure of the cRGD-PEGDA hydrogel, we used SEM to characterize the surface morphology, as shown in Figure 3. The samples were prepared by a freeze-drying method and followed by coating a thin layer of platinum (50 Å) prior to SEM imaging. There are some reports on SEM morphology of dried PEGDA hydrogels showing microporous structures (50,51), but much care must be taken when making the dried samples for SEM analysis, since the removal of water held within the porous hydrogel scaffolds may disrupt the structural stability and result in collapse and some artifacts upon drying (52). Our results revealed that dried cRGD-PEGDA hydrogels possess a highly porous structure (Figure 3a). The pore sizes range from 10 to 50 µm. More features were observed in the SEM image with higher amplification (Figure 3b). There are some bigger pores containing small pores, and some fibrillar networks on the pore walls. In addition, most of the pores are interconnected. It demonstrates that the cRGD-PEGDA hydrogel has the microscopic porous structure with a complex 3D construction in the dried state.
Figure 3.
Surface morphology of 20% (w/v) cRGD-PEGDA hydrogel after freeze-drying. (a) SEM photograph with lower magnification (300×); (b) SEM photograph with higher magnification (1,500×).
Hydrogel surface composition analysis
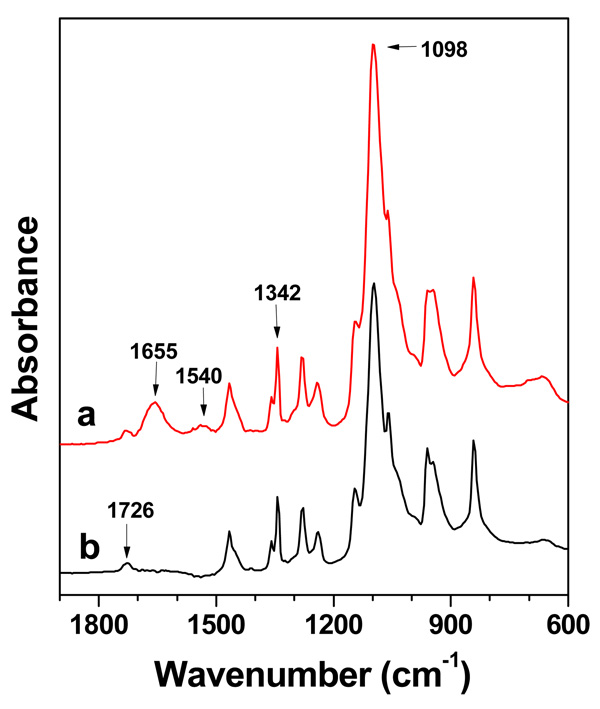
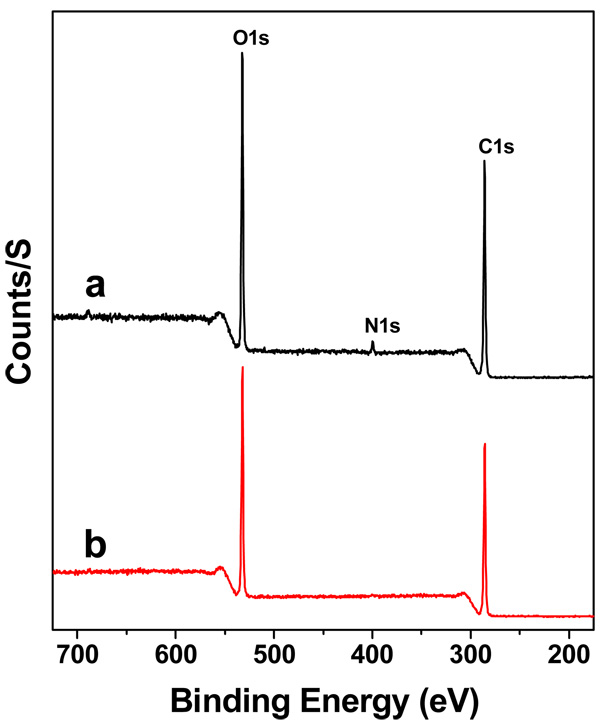
The peptide ligand presentation in scaffolds plays an important role in controlling cell motility behaviours for tissue engineering (54–56). To verify the presence and distribution of cRGD peptide ligands on the cRGD-PEGDA hydrogel surface, we used ATR-FTIR (Figure 4) and XPS (Figure 5) to characterize the surface functional groups and surface composition, respectively. ATR-FTIR analysis revealed that PEGDA hydrogels (Figure 4b) show characteristic peaks at 1,098, 1,342 and 1,726 cm−1, which correspond to -C-O-symmetric stretching, -C-H2 bending and -C=O stretching from ester bonds, respectively. The ART-FTIR spectrum of cRGD-PEGDA (Figure 4a) shows all these three characteristic peaks, which demonstrates cRGD-PEGDA has the similar backbone structure as PEGDA. However, cRGD-PEGDA shows two distinct peaks at 1,655 and 1,540 cm−1, which correspond to the vibrations of amide (-CO-NH) I and II bands, respectively. These amide groups are from the cRGD peptides, indicating that cRGD ligands are present on the hydrogel surface. XPS analysis reveals that both surfaces of cRGD-PEGDA (Figure 5a) and PEGDA (Figure 5b) hydrogels have similar peaks at binding energy of 531 and 285 eV, which corresponds to O1s and C1s, respectively. However, cRGD-PEGDA hydrogels has a new peak at binding energy of 398 eV, which is attributed to N1s resulted from cRGD peptides. Quantitative analysis shows that there is 2.9% of nitrogen presented on the cRGD-PEGDA hydrogel surface, which is very close to the theoretical nitrogen content of 2.8%. This indicates that the hydrogel has well-defined structure with cRGD peptides uniformly presented on the surface and distributed through the material.
Figure 4.
Surface analysis by ATR-FTIR spectroscopy. (a) ATR-FTIR spectrum of cRGD-PEGDA hydrogels after freeze-drying; (b) ATR-FTIR spectrum of PEGDA hydrogels after freeze-drying. cRGD-PEGDA hydrogels show two distinct peaks of amide I and II bands at 1,655 and 1,540 cm−1, respectively, indicating that cRGD ligands are present on the hydrogel surface.
Figure 5.
Surface composition analysis by XPS. (a) XPS spectrum of cRGD-PEGDA hydrogels after freeze-drying; (b) XPS spectrum of PEGDA hydrogels after freeze-drying. XPS analysis reveals that cRGD-PEGDA hydrogels has a new peak at binding energy of 398 eV, which is attributed to N1s resulted from cRGD peptides. Quantitative analysis shows that there is 2.9% of nitrogen presented on the cRGD-PEGDA hydrogel surface.
Two-dimensional (2D) EC seeding and culturing on hydrogel surfaces
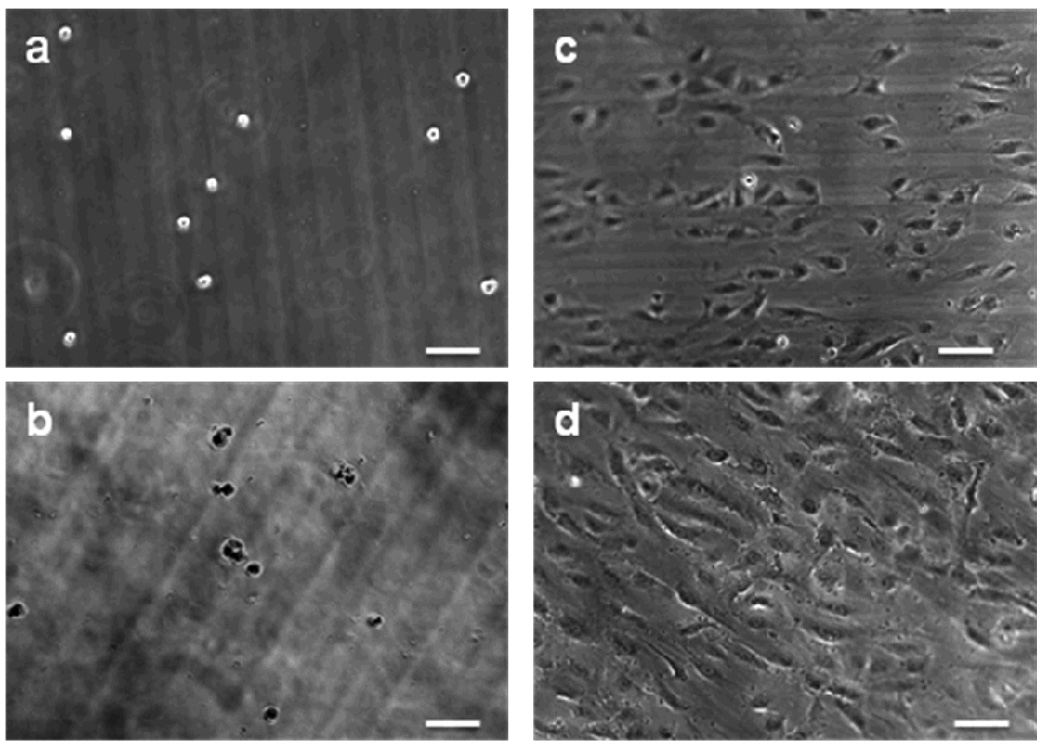
Human pulmonary artery ECs were used to test the cell responses to cRGD-PEGDA hydrogels. Cells were seeded on the material surfaces, which were made from 20% (w/v) macromers sterilized with a 0.2-µm syringe filter. Figure 6 shows phase-contrast micrographs of ECs seeded on the hydrogel surfaces at various time points. ECs seeded on the PEGDA hydrogels assumed a rounded morphology with little evidence of spreading 2 h after seeding (Figure 6a), and exhibited an apparent decrease in density 24 h post-seeding (Figures 6b), suggesting that ECs have only weak, non-specific interactions with these materials. ECs seeded on the cRGD-PEGDA hydrogels (cRGD density = 25.3 mM) showed higher initial cell attachment and cell spreading to some extent 2 h after seeding (Figure 6c), and cells were fully spreading 24 h after seeding (Figure 6d). Thus, the enhanced EC attachment and growth on cRGD-PEGDA hydrogels were attributed to the specific binding of cRGD ligands to the integrin receptors on EC surfaces.
Figure 6.
2D seeding and culturing of endothelial cells (ECs) on hydrogels. (a, b) Phase contrast of ECs 2 h and 24 h after seeding on PEGDA hydrogel, respectively; (c, d) Phase contrast of ECs 2 h and 24 h after seeding on cRGD-PEGDA hydrogel, respectively. The images show that ECs seeded on cRGD-PEGDA hydrogels exhibited higher initial cell attachment, greater cell spreading, and higher cell density than on PEGDA hydrogels. (Scale bar = 100 µm).
Effect of ligand binding affinity on cell growth
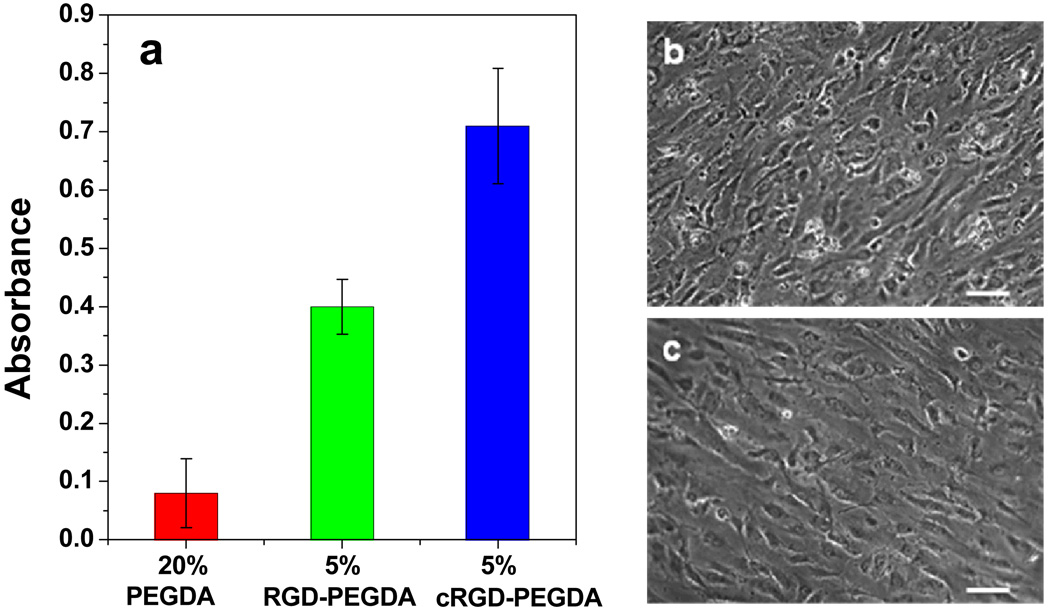
Quantitative modulation of adhesion strength can be achieved through variation of the affinity and surface density of peptide ligands. To amplify and compare the effects of ligand affinity on EC growth on hydrogels, we incorporated low ligand density into hydrogels to compare cell behaviors responding to cRGD and linear RGD ligands. Hydrogel disks from 5% (w/v) cRGD-PEGDA hydrogels (cRGD density = 6.3 mM) were fabricated by copolymerization with 15% (w/v) PEGDA. MTS cell proliferation assays were performed 96 h post-seeding of ECs on the hydrogel surfaces, as shown in Figure 7. Compared with the control PEGDA hydrogel, both 5% cRGD and RGD-modified PEGDA hydrogels exhibit significantly higher (p < 0.001) EC population than the control PEGDA hydrogel. This is due to the specific cell adhesion with peptide ligands presented on the hydrogel surfaces. Further analysis shows that 5% cRGD-PEGDA hydrogel shows significantly higher (p < 0.001) EC proliferation (cell population increased 44%) than 5% RGD-PEGDA hydrogel. This is attributed to the enhance cell adhesion resulted from the higher EC binding ability of cRGD peptides.
Figure 7.
Comparison of EC proliferation on linear RGD-PEGDA and cRGD-PETGDA hydrogels. (a) MTS assays performed at 96 hrs after EC seeding; (b) Phase contrast of ECs 96 h after EC seeding on 5% (w/v) cRGD-PEGDA hydrogel; (c) Phase contrast of ECs 96 h after EC seeding on 5% (w/v) RGD-PEGDA hydrogel. 5% cRGD-PEGDA hydrogel shows significantly higher (p < 0.001) EC proliferation (cell population increased 44%) than 5% RGD-PEGDA hydrogel. (Scale bar = 100 µm).
CONCLUSIONS
In summary, we have synthesized a new type of bioactive PEGDA macromers, cRGD-PEGDA 6, to mimic the cell-adhesive properties of ECM, aiming to create biomimetic scaffolds with controlled spatial organization and enhanced cell binding affinity for tissue engineering. To attach the cRGD peptide in the middle of PEGDA chain, a tailed cRGD peptide, c[RGDfE(SSSKK-NH2)] 1 was designed and synthesized with c(RGDfE) linked to a tail of SSSKK, which provides a hydrophilic spacer of SSS and a linker of KK for conjugation. cRGD-PEGDA possesses good ability of photopolymerization to fabricate hydrogel scaffolds under UV radiation. The resulting hydrogels were well-constructed with porous 3D structures and uniform distribution of cRGD ligands. Our results show that cRGD-PEGDA hydrogels facilitate EC adhesion and spreading on the hydrogel surfaces, and exhibit significantly higher EC population in comparison with linear RGD-modified hydrogels at low peptide incorporation. Since ligand presentation in biomimetc scaffolds plays an important role in controlling cell behaviors, cRGD-PEGDA has great advantages of controlling hydrogel properties and ligand spatial organization in the resulting scaffolds. Furthermore, cRGD-PEGDA is an attractive candidate for the future development of tissue engineering scaffolds with optimum cell adhesive strength and ligand density.
Acknowledgements
This work was supported by the National Institutes of Health (Grant R01EB002067). We gratefully thank Christopher Hoffman for his assistance with SEM and the facilities provided by Center for Cardiovascular Biomaterials.
LITERATURE CITED
- 1.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Niklason LE, Langer R. Prospects for organ and tissue replacement. J. Am. Med. Assoc. 2001;285:573–576. doi: 10.1001/jama.285.5.573. [DOI] [PubMed] [Google Scholar]
- 3.Vacanti CA. The history of tissue engineering. J. Cell Mol. Med. 2006;10:569–576. doi: 10.1111/j.1582-4934.2006.tb00421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaihara S, Vacanti JP. Tissue engineering: toward new solutions for transplantation and reconstructive surgery. Arch. Surg. 1999;134:1184–1188. doi: 10.1001/archsurg.134.11.1184. [DOI] [PubMed] [Google Scholar]
- 5.Ogle B, Cascalho M, Platt JL. Fusion of approaches to the treatment of organ failure. Am. J. Transplant. 2004;4:S74–S77. doi: 10.1111/j.1600-6135.2004.0347.x. [DOI] [PubMed] [Google Scholar]
- 6.Risbud M. Tissue engineering: implications in the treatment of organ and tissue defects. Biogerontology. 2001;2:117–125. doi: 10.1023/a:1011585117310. [DOI] [PubMed] [Google Scholar]
- 7.Hollister SJ. Porous scaffold design for tissue engineering. Nature Mater. 2005;4:518–524. doi: 10.1038/nmat1421. [DOI] [PubMed] [Google Scholar]
- 8.Hench LL, Polak JM. Third-generation biomedical materials. Science. 2002;295:1014–1017. doi: 10.1126/science.1067404. [DOI] [PubMed] [Google Scholar]
- 9.Griffith LG. Emerging design principles in biomaterials and scaffolds for tissue engineering. Ann. N. Y. Acad. Sci. 2002;961:83–95. doi: 10.1111/j.1749-6632.2002.tb03056.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: state of the art. Tissue Eng. Part B. 2008;14:61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, Leong KF, Du Z, Chua CK. The design of scaffolds for use in tissue engineering. Part I. Traditional factors. Tissue Eng. 2001;7:679–689. doi: 10.1089/107632701753337645. [DOI] [PubMed] [Google Scholar]
- 12.Hutmacher DW. Scaffolds in tissue engineering bone and cartilage. Biomaterials. 2000;21:2529–2543. doi: 10.1016/s0142-9612(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 13.Badylak SF. The extracellular matrix as a biologic scaffold material. Biomaterials. 2007;28:3587–3593. doi: 10.1016/j.biomaterials.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Kasemo B. Biological surface science. Curr. Opin. Solid State Mater. Sci. 1998;3:451–459. [Google Scholar]
- 15.Stoop R. Smart biomaterials for tissue engineering of cartilage. Injury. 2008;39S1:S77–S87. doi: 10.1016/j.injury.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 16.Cushing MC, Anseth KS. Hydrogel cell cultures. Science. 2007;316:1133–1134. doi: 10.1126/science.1140171. [DOI] [PubMed] [Google Scholar]
- 17.Lee KY, Mooney DJ. Hydrogels for tissue engineering. Chem. Rev. 2001;101:1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 18.Kretlow JD, Klouda L, Mikos AG. Injectable matrices and scaffolds for drug delivery in tissue engineering. Adv. Drug Deliv. Rev. 2007;59:263–273. doi: 10.1016/j.addr.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Chung C, Burdick JA. Engineering cartilage tissue. Adv. Drug Deliv. Rev. 2008;60:243–262. doi: 10.1016/j.addr.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP. Light-activated hydrogbel formation via the triggered folding and self-assembly of a designed peptide. J. Am. Chem. Soc. 2005;127:17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elisseeff J, Anseth K, Sims D, McIntosh W, Randolph M, Langer R. Transdermal photopolymerization for minimally invasive implantation. Proc. Natl. Acad. Sci. USA. 1999;96:3104–3107. doi: 10.1073/pnas.96.6.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buxton AN, Zhu J, Marchant RE, West JL, Yoo JU, Johnstone B. Design and characterization of poly(ethylene glycol) photopolymerizable semi-interpenetrating networks for chondrogenesis of human mesenchymal stem cells. Tissue Eng. 2007;13:2549–2560. doi: 10.1089/ten.2007.0075. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen KT, West JL. Photopolymerizable hydrogels for tissue engineering application. Biomaterials. 2002;23:4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 24.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature Biotech. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 25.Griffith LG, Naughton G. Tissue engineering-current challenges and expanding opportunities. Science. 2002;295:1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 26.Hirano Y, Mooney DJ. Peptide and protein presenting materials for tissue engineering. Adv. Mater. 2004;16:17–25. [Google Scholar]
- 27.Ito Y. Covalently immobilized biosignal molecule materials for tissue engineering. Soft Matter. 2008;4:46–56. doi: 10.1039/b708359a. [DOI] [PubMed] [Google Scholar]
- 28.Schaffner P, Dard MM. Structure and function of RGD peptides involved in bone biology. Cell Mol. Life Sci. 2003;60:119–132. doi: 10.1007/s000180300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 30.Kantlehner M, Finsinger D, Meyer J, Schaffner P, Jonczyk A, Diefenbach B, Nies B, Kessler H. Selective RGD-mediated adhesion of osteoblasts at surfaces of implants. Angew. Chem. Int. Ed. 1999;38:560–562. doi: 10.1002/(SICI)1521-3773(19990215)38:4<560::AID-ANIE560>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 31.Hern DL, Hubbell JA. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue engineering. J. Biomed. Mater .Res. 1998;39:266–276. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 32.Burdick JA, Anseth KS. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 33.Yang F, Williams CG, Wang D, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26:5991–5998. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Kinoshita T, Ishigaki Y, Nakano K, Yamaguchi K, Akita S, Nii S, Kawizumi F. Application of acrylate gel having poly(ethylene glycol) side chains to recovery of gold from hydrochloric acid solutions. Sep. Purif. Tech. 2006;49:253–257. [Google Scholar]
- 35.Tan G, Wang Y, Guang Y. Synthesis and properties of polyethylene glycol diacrylate/2-hydroxyethyl methacrylate hydrogels. Key Eng. Mater. 2008;368–372:1175–1177. [Google Scholar]
- 36.Tan G, Wang Y, Li J, Zhang S. Synthesis and characterization of Injectable photocrosslinking poly(ethylene glycol) diacrylate based hydrogels. Polym. Bull. 2008;61:91–98. [Google Scholar]
- 37.Zhu J, Beamish JA, Tang C, Kottke-Marchant K, Marchant RE. Extracellular matrix-like cell-adhesive hydrogels form RGD-containing poly(ethylene glycol) diacrylate. Macromolecules. 2006;39:1305–1307. [Google Scholar]
- 38.Chen W, Chang C, Gilson MK. Concepts in receptor optimization: targeting the RGD peptide. J. Am. Chem. Soc. 2006;128:4675–4684. doi: 10.1021/ja056600l. [DOI] [PubMed] [Google Scholar]
- 39.Leahy DJ, Aukhil I, Erickson HP. 2.0Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 40.Haubner R, Schmitt W, Holzemann G, Goodman SL, Jonczyk A, Kessler H. Cyclic RGD peptides containing β-turn mimetics. J. Am. Chem. Soc. 1996;118:7881–7891. [Google Scholar]
- 41.Haubner R, Gratias R, Diefenbach B, Goodman SL, Jonczyk A, Kessler H. Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin αvβ3 antagonists. J. Am. Chem. Soc. 1996;118:7461–7472. [Google Scholar]
- 42.Locardi E, Mullen DG, Mattern RH, Goodman M. Conformations and pharmacophores of cyclic RGD containing peptides which selectively bind integrin αvβ3. J. Pep. Sci. 1999;5:491–506. doi: 10.1002/(SICI)1099-1387(199911)5:11<491::AID-PSC218>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 43.Lambert JN, Mitchell JP, Roberts KD. The synthesis of cyclic peptides. J. Chem. Soc. Perkin Trans. 1. 2001;5:471–484. [Google Scholar]
- 44.Tang C, Kligman F, Larsen CC, Kottke-Marchant K, Marchant RE. Platelet and endothelial adhesion on flurosurfactant polymers designed for vascular graft modification. J. Biomed. Mater. Res. 2008 doi: 10.1002/jbm.a.31888. published online on Feb. 19, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, Marchant RE. Solid-phase synthesis of tailed cyclic RGD peptides using glutamic acid: unexpected glutarimide formation. J. Pep. Sci. 2008;14:690–696. doi: 10.1002/psc.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canal T, Peppas NA. Correlation between mesh size and equilibrium degree of swelling of polymeric networks. J. Biomed. Mater. Res. 1989;23:1183–1193. doi: 10.1002/jbm.820231007. [DOI] [PubMed] [Google Scholar]
- 47.Cruise GM, Scharp DS, Hubbell JA. Characterization of permeability and network structure of interfacially photopolymerized poly(ethylene glycol) diacrylate hydrogels. Biomaterials. 1998;19:1287–1294. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 48.Peppas NA, Bures P, Leobandung W, Ichikawa H. Hydrogels in pharmaceutical formations. Eur. J. Pharm. Biopharm. 2000;50:27–46. doi: 10.1016/s0939-6411(00)00090-4. [DOI] [PubMed] [Google Scholar]
- 49.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys. J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livnat M, Beyar R, Seliktar D. Endoluminal hydrogel films made of alginate and polyethylene glycol: physical characteristics and drug-eluting properties. J. Biomed. Mater. Res. 2005;75A:710–722. doi: 10.1002/jbm.a.30474. [DOI] [PubMed] [Google Scholar]
- 51.Kim JH, Kim JG, Kim D, Kim YH. Preparation and properties of PEG hydrogel from PEG macromonomer with sulfonate end group. J. Appl. Polym. Sci. 2005;96:56–61. [Google Scholar]
- 52.Ford MC, Bertram JP, Hynes SR, Michaud M, Li Q, Young M, Segal SS, Madri JA, Lavik EB. A macroporous hydrogel for the coculture of neural progenitor and endothelial cells to form functional vascular networks in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:2512–2517. doi: 10.1073/pnas.0506020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 54.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 55.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]