Abstract
Mechanistic and structural studies have been carried out to investigate the molecular basis for the irreversible inhibition of human MAO-B by mofegiline. Competitive inhibition with substrate shows an apparent Ki of 28 nM. Irreversible inhibition of MAO-B occurs with a 1:1 molar stoichiometry with no observable catalytic turnover. The absorption spectral properties of mofegiline inhibited MAO-B show features (λmax ≃ 450 nm) unlike those of traditional flavin N(5) or C(4a) adducts. Visible and near UV circular dichroism spectra of the mofegiline-MAO-B adduct shows a negative peak at 340 nm with an intensity similar to that of N(5) flavocyanine adducts. The x-ray crystal structure of the mofegiline-MAO-B adduct shows a covalent bond between the flavin cofactor N(5) with the distal allylamine carbon atom as well as the absence of the fluorine atom. A mechanism to explain these structural and spectral data is proposed.
Keywords: Monoamine Oxidase, Mofegiline, MAO-B, MAO-B Inhibitor, Covalent Flavin Adduct
Introduction
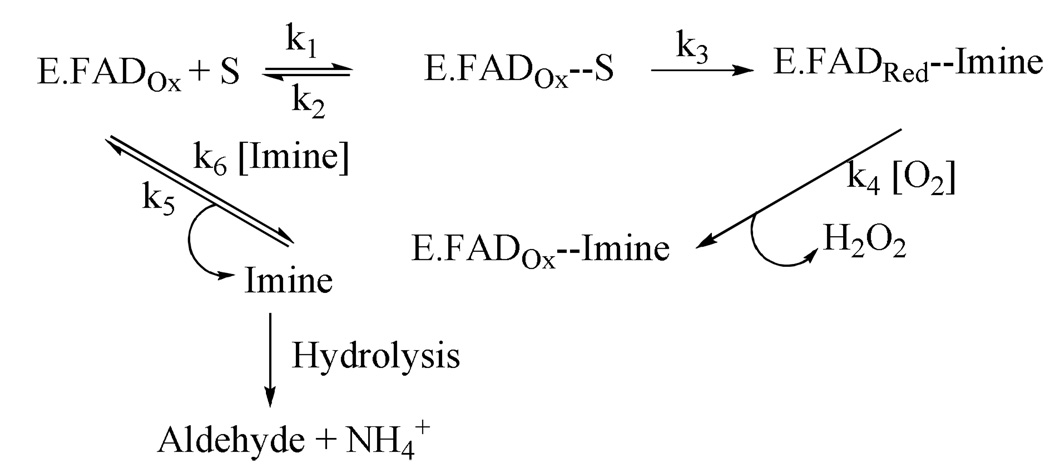
Monoamine oxidases (MAO) are flavin-dependent, outer mitochondrial membrane-bound enzymes that catalyze the oxidative deamination of amine neurotransmitters as well as dietary arylalkylamines (Scheme 1).1 Reduction of O2 to form H2O2 is concomitant with oxidation of the reduced cofactor. Mammals (including humans) contain the MAO-A and MAO-B isozymes that are separate gene products and exhibit differing substrate and inhibitor specificities.2,3 The isozymes share ~70% sequence identity and are differentially expressed in various tissues and in different amounts as a function of age.4 There is an increasing level of evidence supporting the suggestion that increased levels of MAO-B in neuronal tissue on aging in humans5 is related to the onset of neuro-degenerative diseases such as Parkinson’s or Alzheimer’s diseases. Current treatment invoking MAO-B specific inhibitors in combination with Levodopa have been shown to improve motor fluctuations in patients with Parkinson’s disease.6,7 Therefore, continued investigations of MAO-B specific inhibitors as neuroprotectants is important.
Scheme 1.
The reaction pathway for the oxidation of amines catalyzed by MAO.21
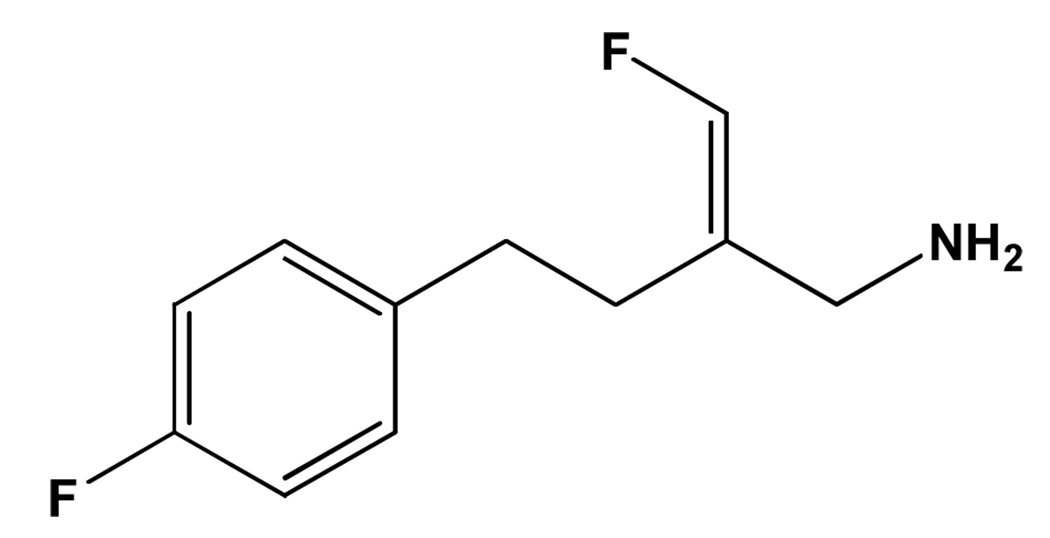
Mofegiline (Figure 1) is a mechanism-based inhibitor developed by the Merrell-Dow Research Institute in the 1980s for the treatment of Parkinson’s disease.8,9 This compound demonstrated nanomolar inhibitory properties for the inhibition of rat brain mitochondrial MAO-B and was shown to be highly specific for rat MAO-B with a 190-fold higher affinity for the MAO-B isoform. Additional pharmacological interest in mofegiline stems from its anti-inflammatory properties via the inactivation of the copper-dependent amine oxidase, semicarbazide-sensitive amine oxidase (SSAO/VAP-1).10 While information on the potency of mofegiline has been known for some time, the molecular mechanism of inhibition as well as the molecular basis for its high level of specificity remains unknown.
Figure 1.
The structure of mofegiline.
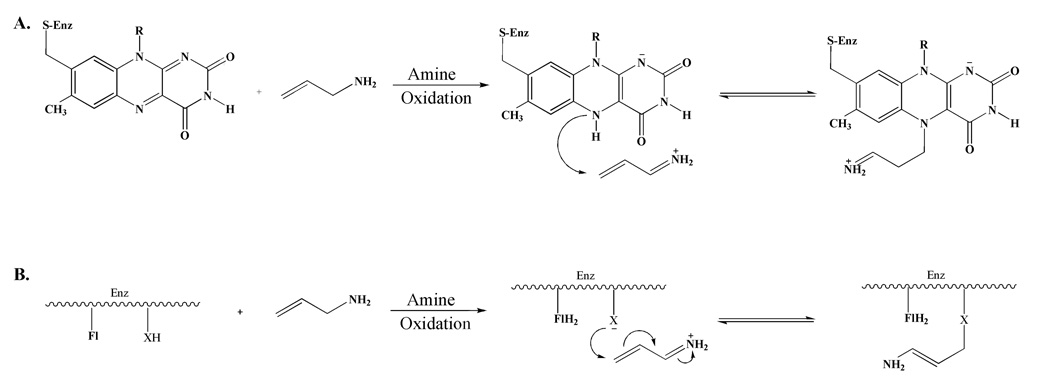
It was proposed that this compound acts as a mechanism-based inhibitor of MAO-B in line with the earlier observations by Rando and Eigner11 who demonstrated that chloroallylamine was a time-dependent, irreversible inhibitor of MAO. Their studies suggested that inhibition of rat liver MAO by allylamine occurs via oxidation of the amine moiety followed by covalent adduct formation with the N(5) position of the flavin. This proposal was supported by the observation that the absorption spectrum of the enzyme-bound flavin is “bleached” after inhibition and remains in this form after removal of excess reagent either by gel filtration or by dialysis (Scheme 2A). This suggestion was later challenged by Silverman and coworkers12 when they showed that tritium labeled allylamine remains attached to the protein after denaturation and demonstrated the flavin is reoxidized on denaturation. These observations led to the proposal that inhibition is achieved by catalytic oxidation of allylamine to its imine followed by Michael addition of an active site nucleophilic amino acid residue to the unsaturated imine (Scheme 2B). This proposed mechanism has prevailed in subsequent discussions describing the mode of MAO inhibition of allylamine inhibitor analogues, like mofegiline, during the last 20 years.8,13,14 To our knowledge, no studies on the molecular level have been published on the inhibition of human MAO-A or B with allylamine analogues. All previous studies have utilized either bovine or rat mitochondrial MAO.11,12
Scheme 2.
A. The proposed mechanism for allylamine inhibition of MAO proposed by Rando and Eigner.11 B. The proposed mechanism of allylamine inhibition of MAO proposed by Silverman.12
With access to purified recombinant human MAO-A15 and MAO-B16 as well as conditions for the crystallization of inhibitor-bound MAO-B,17 we initiated a collaborative study to further define the molecular details for the inhibition of human MAO-B with mofegiline. A combination of solution and crystallographic studies demonstrate that mofegiline inhibits human MAO-B via covalent flavin modification in a rapid and highly specific reaction.
Results
Inhibition of Human MAO-B with Mofegiline
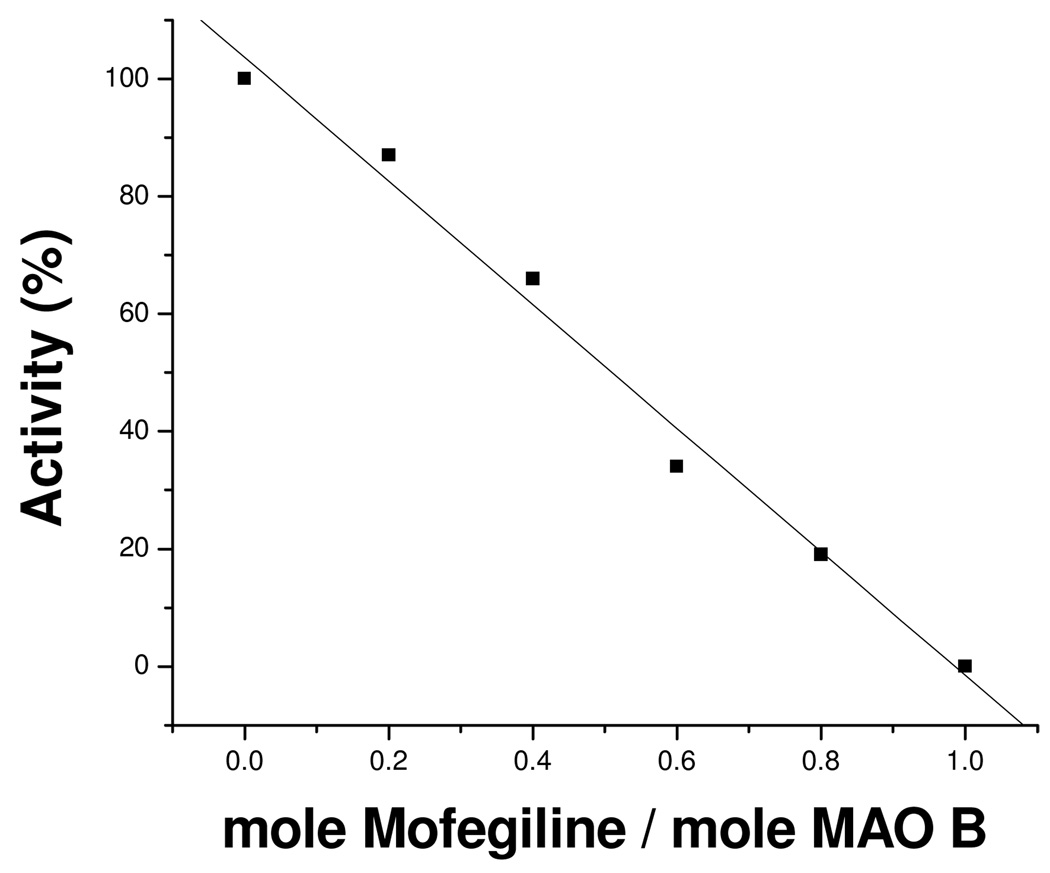
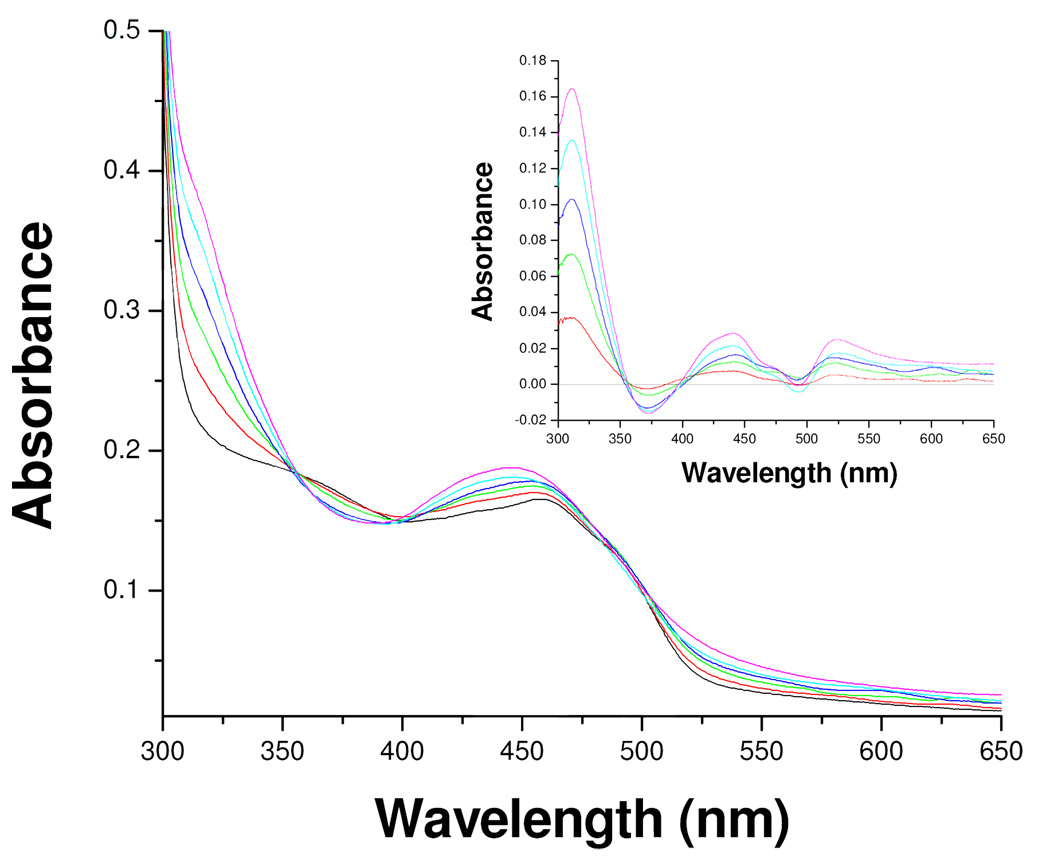
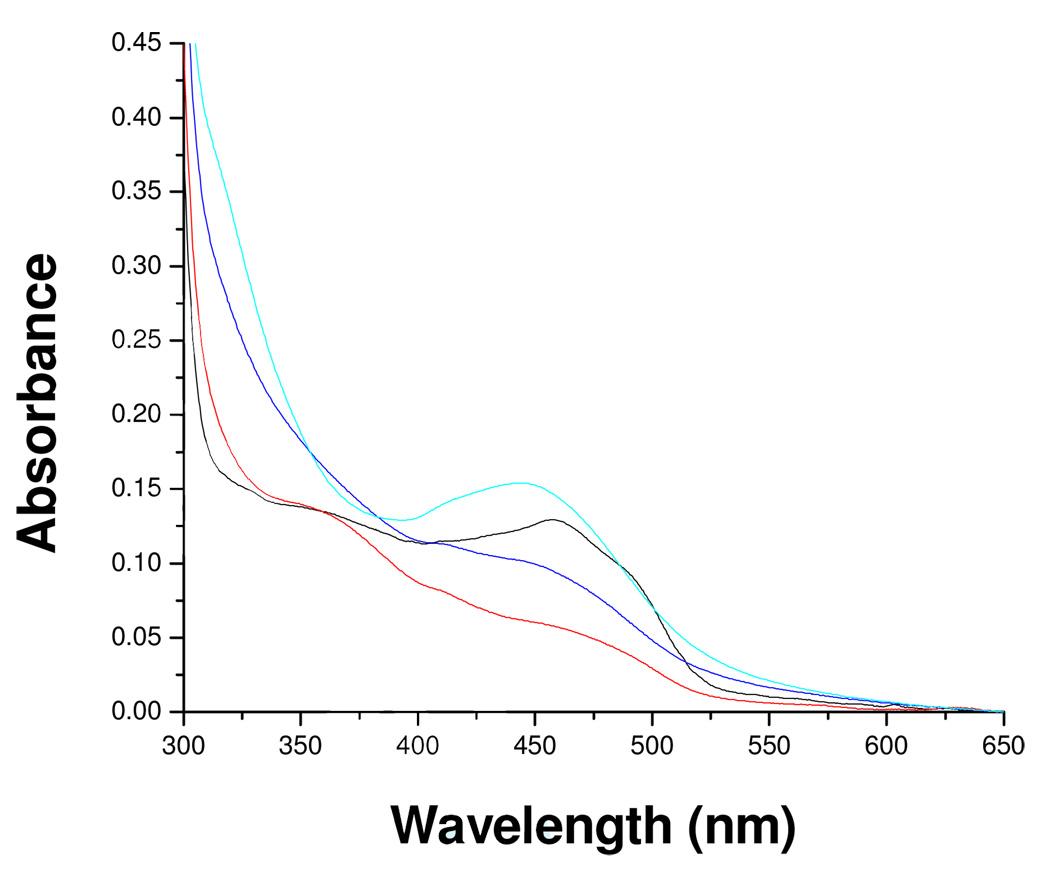
Incubation of purified human MAO-B with mofegiline leads to rapid and irreversible stoichiometric inhibition of catalytic activity. Reversible binding of mofegiline to the catalytic site of MAO-B prior to irreversible inactivation is shown from competitive inhibition experiments which show an apparent Ki of 28 nM (see supporting information); a value close to the IC50 value (3.6 nM) determined for membrane bound rat MAO-B.8 The data in Figure 2 show a linear loss of activity on titration of MAO-B with sub-stoichiometric amounts of mofegiline with complete inhibition observed at a stoichiometry of 1.0 mole mofegiline/mole E-FAD. Inhibition occurs rapidly and is complete by the time an enzyme sample can be withdrawn and assayed. Titration of enzyme samples with inactive protein in solution result in lower stoichiometries for complete inhibition demonstrating the requirement for functional enzyme involvement in the reaction. Ion-selective electrode experiments show the release of one mole of fluoride ion per mole of MAO-B inhibited which confirms that fluoride is eliminated during the inhibition reaction. When the visible absorption spectrum of the enzyme (due to the covalent FAD) is monitored during the titration, spectral perturbations are observed (Figure 3); however, no “bleaching” of the oxidized flavin absorbance at 455 nm is observed as expected if either flavin N(5) or C(4a) adducts were formed. For comparison, the spectral data in Figure 4 show spectral changes observed for phenethylhydrazine-inhibited MAO-B (N(5) adduct)18 and for tranylcypromine-inhibited MAO-B (C(4a) adduct).17 The flavin absorbance shifts in λmax from 455 nm to 445 nm on mofegiline inhibition of MAO-B along with a small increase in the molar extinction coefficient from 12,000 M−1-cm−1 to 12,900 M−1-cm−1 (Figure 3). Difference spectral data (inset, Figure 3) show maximal spectral changes on inhibition occur in the near UV region with a minimum at 370 nm, a maximum at 320 nm, and isosbestic points at 350 nm and 395 nm.
Figure 2.
The effect of substoichiometric levels of mofegiline on the activity of human MAO-B. The titration was carried out in 50 mM potassium phosphate, pH = 7.5 and 0.8% (w/v) β-D-octylglucoside.
Figure 3.
Effect of mofegiline inhibition of MAO-B on the absorption spectra in the visible and near UV region due to the flavin: ( ) 0.0, (
) 0.0, ( ) 0.2, (
) 0.2, ( ) 0.4, (
) 0.4, ( ) 0.6, (
) 0.6, ( ) 0.8, (
) 0.8, ( ) 1.0 (mol mofegiline/mol MAO-B). Inset. The difference absorption spectra observed on subtraction of the spectrum of uninhibited MAO-B from those observed during the titration: (
) 1.0 (mol mofegiline/mol MAO-B). Inset. The difference absorption spectra observed on subtraction of the spectrum of uninhibited MAO-B from those observed during the titration: ( ) 0.0, (
) 0.0, ( ) 0.2, (
) 0.2, ( ) 0.4, (
) 0.4, ( ) 0.6, (
) 0.6, ( ) 0.8, (
) 0.8, ( ) 1.0 (mol mofegiline/mol MAO-B).
) 1.0 (mol mofegiline/mol MAO-B).
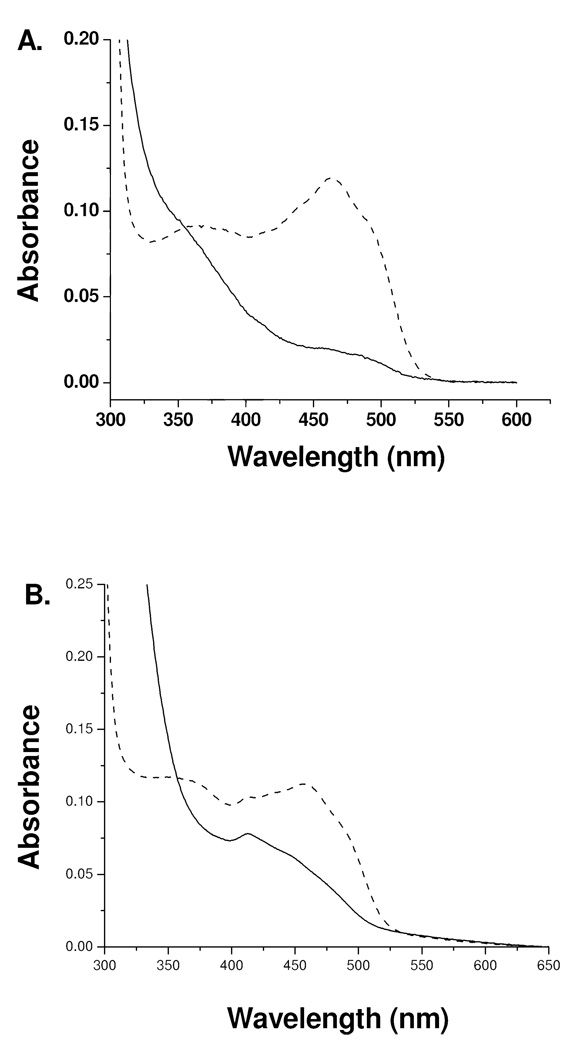
Figure 4.
The visible and near UV absorption spectra of flavin N(5) and of flavin C(4a) adducts of MAO-B. A. Oxidized MAO-B ( ), After phenylethylhydrazine inhibition (N(5) flavin adduct) (
), After phenylethylhydrazine inhibition (N(5) flavin adduct) ( ). B. Oxidized MAO-B (
). B. Oxidized MAO-B ( ), After tranylcypromine inhibition (C(4a) adduct) (
), After tranylcypromine inhibition (C(4a) adduct) ( ).
).
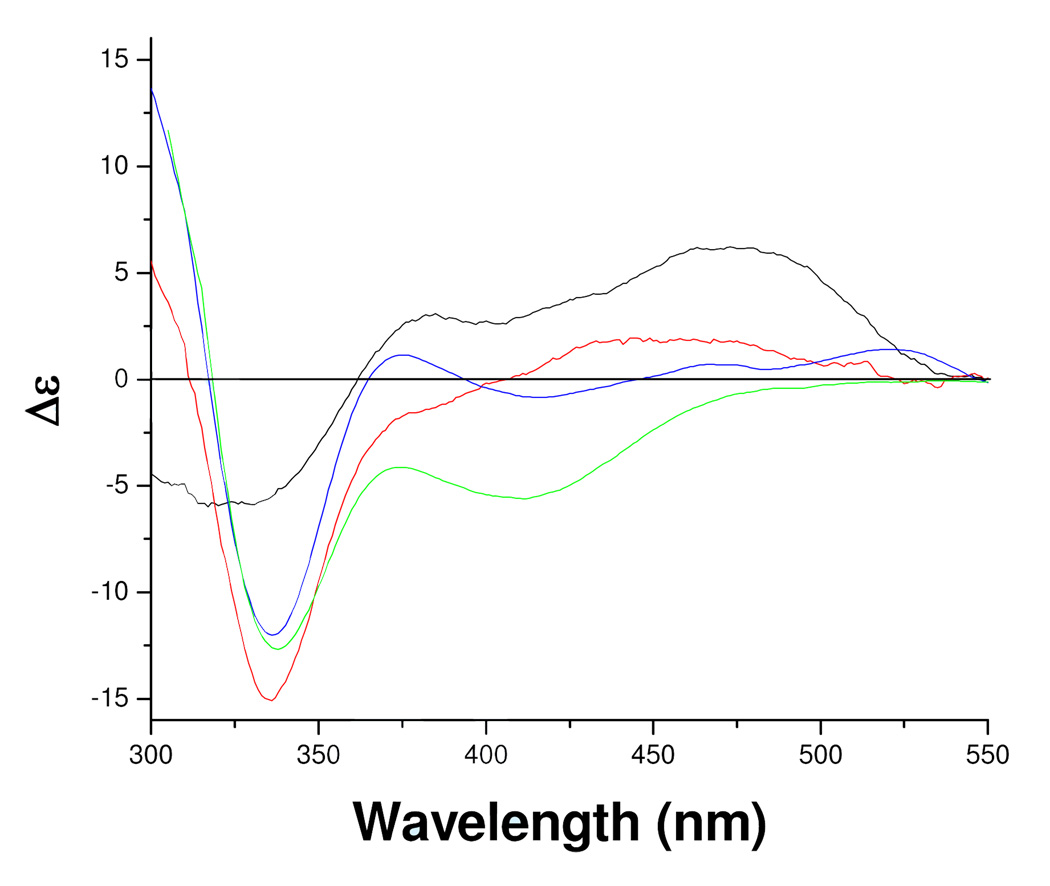
These spectral data suggest the inhibitor binds in the vicinity of the covalent FAD but do not suggest a covalent modification of the coenzyme. If that scenario were true, the FAD cofactor should be reactive to chemical reduction by sodium dithionite. The addition of excess dithionite to a sample of mofegiline-inhibited MAO-B shows no reduction of the flavin by the criteria that no observable bleaching in the 455 nm absorption of the flavin ring occurs. When dithionite is added to a sample of mofegiline-inhibited MAO-B complex in the presence of non-functional enzyme, only a partial bleaching of the 455 nm absorption occurs (data not shown). These experiments show that the flavin coenzyme in mofegiline-inhibited MAO-B is resistant to chemical reduction and that non-functional MAO-B is unreactive with mofegiline which further supports the conclusion that this compound is a mechanism-based inhibitor. The visible CD spectrum of the mofegiline-MAO-B complex (Figure 5) differs from that of free enzyme with an intense negative dichroic band at 340 nm which is similar in both position and intensity with those CD spectra of MAO-B inhibited by compounds forming covalent flavin N(5) adducts (Figure 5).
Figure 5.
Circular dichroism spectra of oxidized human MAO-B, mofegiline inhibited MAO-B, and of known MAO-B flavin N(5) adducts. Oxidized MAO-B ( ), After inhibition by mofegiline (
), After inhibition by mofegiline ( ), the MAO-B N(5) flavin adduct formed after inhibition by pargyline (
), the MAO-B N(5) flavin adduct formed after inhibition by pargyline ( ), and the MAO-B N(5) flavin adduct formed after inhibition by phenylethylhydrazine (
), and the MAO-B N(5) flavin adduct formed after inhibition by phenylethylhydrazine ( ). All spectral data were acquired in 50 mM potassium phosphate, pH 7.5 containing 0.8% (w/v) β-D-octylglucoside.
). All spectral data were acquired in 50 mM potassium phosphate, pH 7.5 containing 0.8% (w/v) β-D-octylglucoside.
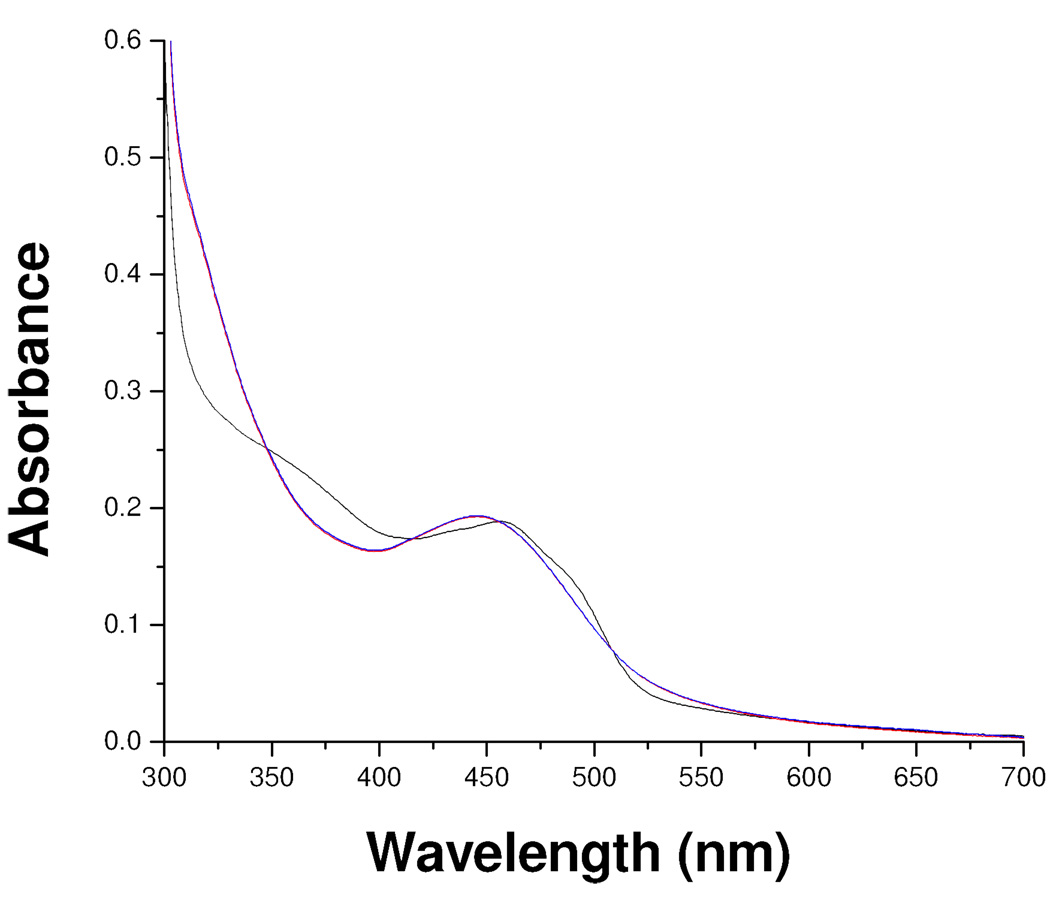
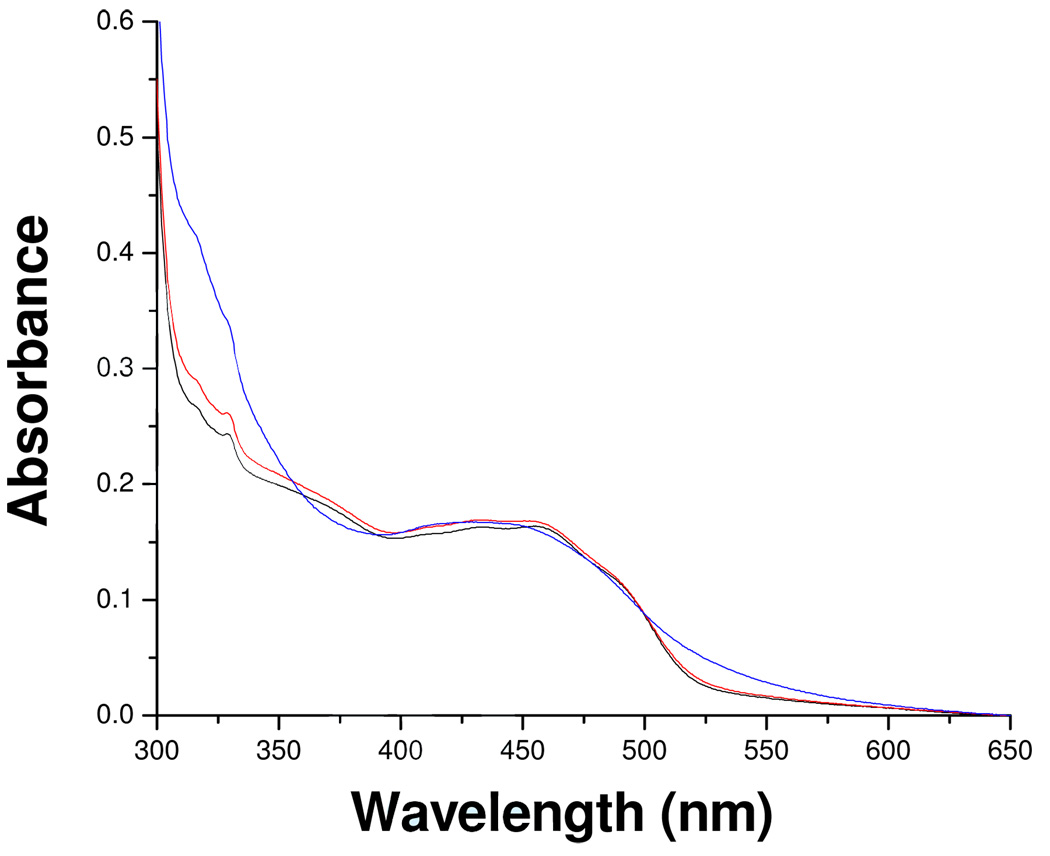
To determine whether catalytic turnover is involved in the inhibition reaction, the influence of oxygen in the inhibition process was examined. Inhibition reactions carried out in the presence of a coupled enzyme system that detects whether any H2O2 is produced (Amplex Red/Horseradish Peroxidase)19 show no detectable levels of H2O2. The addition of mofegiline to oxidized MAO-B under anaerobic conditions results in complete inhibition of the enzyme with spectral properties (Figure 6) identical to that of found during aerobic inhibition of the enzyme. Assay of anaerobically-withdrawn samples show complete inhibition. No spectral changes are detected on exposure of anaerobic mofegiline-inhibited MAO-B to air. These experiments demonstrate that no catalytic turnover occurs during the inhibition. If mofegiline is a mechanism-based inhibitor, inhibition occurs during a single turnover.
Figure 6.
The visible and near UV absorption spectra observed after anaerobic inhibition of MAO-B by mofegiline. Oxidized MAO-B ( ), mofegiline-MAO-B adduct under argon (
), mofegiline-MAO-B adduct under argon ( ), mofegiline-MAO-B adduct formed aerobically (
), mofegiline-MAO-B adduct formed aerobically ( ). The buffer used is the same as described in the legend of Figure 2.
). The buffer used is the same as described in the legend of Figure 2.
To probe whether reduced MAO-B is subject to inhibition by mofegiline, a sample of enzyme was reduced anaerobically with a stoichiometric amount of benzylamine. Mofegiline was then anaerobically added and the visible absorbance spectral properties of the enzyme followed. The data in Figure 7 show that the absorbance of the reduced enzyme increases on addition of mofegiline to level only ~50 % of that observed with mofegiline-inhibited MAO-B formed under aerobic conditions. The anaerobic removal and assay of an aliquot of enzyme from this experiment exhibits no activity. Upon exposure to oxygen, the spectrum exhibits properties identical to aerobically inhibited MAO-B. Attempts to determine the rate of MAO-B flavin reduction by mofegiline by stopped flow experiments show the rate is too fast to measure and occurs within the 1–2 ms dead time of the instrument.
Figure 7.
Absorption spectral changes in MAO-B after anaerobic reduction with a stoichiometric amount of benzylamine followed by the anaerobic addition of 1.5 molar equivalents of mofegiline. Exposure of the sample to oxygen results in additional spectral changes in the flavin absorption region to a spectrum that is identical to that observed with aerobically inhibited MAO-B with mofegiline. Absorption spectra of: Oxidized MAO-B ( ), Reduced MAO-B (
), Reduced MAO-B ( ), After the anaerobic addition of mofegiline (
), After the anaerobic addition of mofegiline ( ), Sample after exposure to air (
), Sample after exposure to air ( ). The buffer used is identical with that described in the legend to Figure 2.
). The buffer used is identical with that described in the legend to Figure 2.
Structure of Mofegiline-Inhibited MAO-B
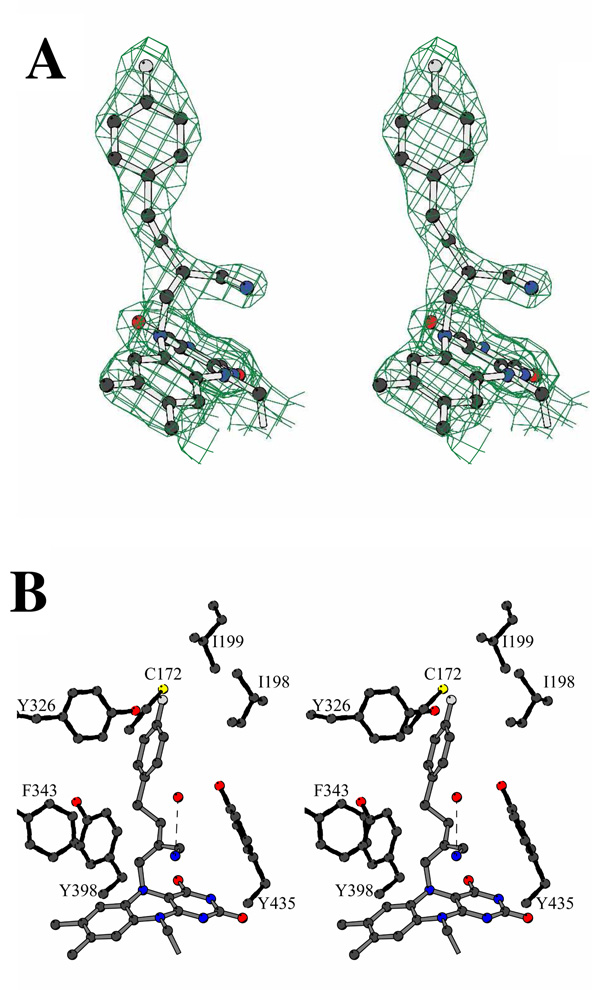
To provide conclusive evidence for suggested covalent modification of MAO-B on inhibition by mofegiline, the inhibited enzyme was crystallized and its structure determined by x-ray crystallography. Crystals of mofegiline-inhibited MAO-B diffract to 2.3 Å resolution. The structure of the complex about the flavin active site is shown in Figure 8. These structural data provide direct evidence that the inhibitor is bound to the enzyme as a flavin N(5) adduct (Figure 8). The inhibitor adopts an extended conformation spanning from the flavin ring to the entrance of the substrate cavity. The aromatic ring is located in the rear of the cavity and it is oriented perpendicular to the flavin ring as typically observed for most other MAO-B-inhibitor complexes.17 Ile199 adopts a conformation in which its side chain is positioned in the “open” conformation resulting in the fusion of the entrance and substrate cavities. This “open” conformation has been previously observed with other bulky inhibitors such as N-(2-aminoethyl)-4-chlorobenzamide and 1,4-diphenyl-2-butene, which also force the opening of the Ile199 gate.17 The bound mofegiline participates in numerous hydrophobic and van der Waals interactions with active site side chains (Cys172, Ile198, Ile199, Phe343, Tyr326, Tyr398, and Tyr 435). The electron density clearly defines the positions of several ordered water molecules, one of which forms a H-bond interaction with the mofegiline nitrogen atom. No extra electron density is found close to the covalent bond of the flavin N(5),which proves that the fluorine atom has been eliminated and supports the argument that the carbon (from mofegiline) is sp2 hybridized. The mofegiline nitrogen atom, which we interpret as an imine group, lies in front of the flavin ring between side chains of Tyr398 and Tyr435 that form the catalytically important aromatic cage20 (Figure 8b). These structural data support the idea that the aromatic cage can function as a recognition site for the substrate amine group.21
Figure 8.
Structure of human MAO-B in complex with the Mofegeline. A. Stereoview of the final 2Fo-Fc electron density map (contoured at 1.3 σ level). Carbons are in black, oxygen in red, nitrogen in blue, sulfurs in yellow, and fluorine in grey. B. The binding mode of mofegiline to the human MAO-B active site. Atom colors are as in Figure 8A. The inhibitor covalent bonds are depicted in grey. The H-bond between the inhibitor nitrogen atom and a water molecule is outlined by a dashed line. With respect to Figure 8A, the model has been rotated by about 60° around the vertical axis in the plane of the paper. The drawings were produced by using Bobscript34 and Molscript35.
Mofegiline Interactions with Purified Human MAO-A
A similar series of experiments were performed using recombinant human MAO-A to compare inhibition properties that might help explain why mofegiline exhibits a marked specificity for MAO-B. Mofegiline functions as a competitive inhibitor with MAO-A with a measured Ki value (1.1 µM) ~40-fold weaker than that observed with MAO-B. Dilution of the MAO-A-inhibitor mixture shows return of catalytic activity demonstrating that no irreversible inhibition occurs. Like MAO-B, inhibition reactions carried out in the presence of the Amplex Red/Horseradish Peroxidase coupled assay show no detectable levels of H2O2 produced; therefore, no catalytic oxidation of the amine moiety is observed for MAO-A. Similarly, the anaerobic addition of mofegiline to MAO-A shows no evidence for enzyme flavin reduction even at high concentrations of inhibitor (Figure 9). These experiments suggest the failure of MAO-A to oxidize the amine moiety of mofegiline may account for its resistance to inhibition.
Figure 9.
Spectral changes in the UV/Vis spectrum of human MAO-A on anaerobic addition of a 10-fold molar excess of mofegiline. Only small spectral changes in the flavin chromophore occur after incubation times of greater than 30 minutes. Absorption spectra of: Oxidized MAO-A ( ), After the anaerobic addition of mofegiline (
), After the anaerobic addition of mofegiline ( ), mofegiline and MAO-A incubated aerobically at 25 °C for 30 minutes (
), mofegiline and MAO-A incubated aerobically at 25 °C for 30 minutes ( ).
).
Anaerobic substrate reduction of MAO-A followed by the addition of mofegiline does not result in any irreversible inhibition but does show modest spectral changes from 300–350 nm in the UV/Visible spectrum (Figure 10). The CD spectrum of the mofegiline-MAO-A complex exhibits a mild perturbation of the flavin spectrum; however, the spectrum does not resemble those of MAO-A flavin-N(5) adducts (data not shown). These experiments demonstrate mofegiline is a weaker, reversible MAO-A inhibitor that is much more potent for MAO-B. The structural basis for this difference in behavior of mofegiline for the two isozymes for will be addressed in the discussion section.
Figure 10.
Absorption spectral changes on the anaerobic reduction of MAO-A with a stoichiometric amount of p-trifluoromethylbenzylamine followed by the anaerobic addition of 10-fold excess mofegiline (which does not result in any observed spectral changes). Exposure of the sample to oxygen shows re-oxidation of the flavin with minor changes in 300–350 nm region of the spectrum. Absorption spectra of: Oxidized MAO-B ( ), Reduced MAO-B (
), Reduced MAO-B ( ), After the anaerobic addition of mofegiline (
), After the anaerobic addition of mofegiline ( ), After exposure of the sample to O2 (
), After exposure of the sample to O2 ( ). The buffer used in this experiment is identical with that described in the legend to Figure 2.
). The buffer used in this experiment is identical with that described in the legend to Figure 2.
Discussion
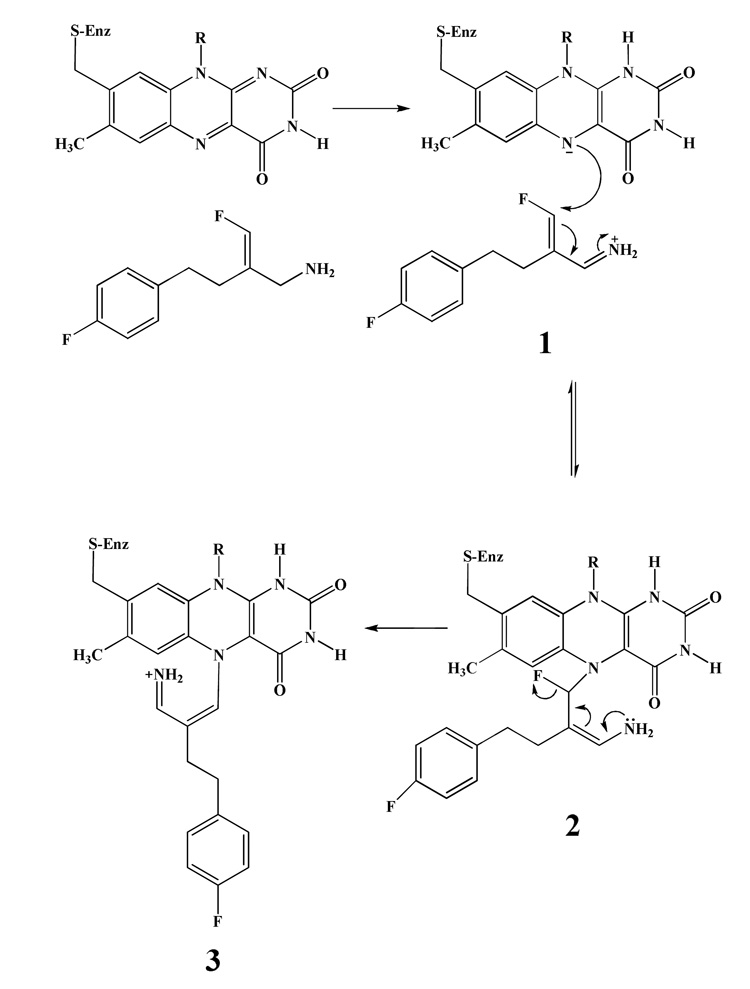
Model system studies have shown that reduced flavins are known to react with alkyl halides to form flavin N(5) adducts.22 Recent studies of the flavin dependent isopentenyl pyrophosphate isomerase23 have shown that fluorinated substrate analogues react with the reduced flavin coenzyme to form flavin N(5) alkyl adducts with elimination of the fluorine atom as F−. With the knowledge of the behavior of these flavin alkylating systems, we suggest the initial step of the inhibition of MAO-B by mofegiline involves flavin reduction concurrent with amine oxidation to the imine 1. The imine intermediate 1 then undergoes a Michael addition of the sp3 N(5) of the flavin hydroquinone. The instability of the enamine intermediate 2 results in elimination of F− to form the highly conjugated flavin N(5) adduct 3. (Scheme 3). This inhibition reaction is very rapid and occurs in a single catalytic turnover since no O2 uptake or H2O2 production is observed during inhibition.
Scheme 3.
Proposed mechanism for the irreversible inhibition of MAO-B by mofegiline with N(5) flavin alkylation to produce a highly conjugated flavocyanine adduct.
Preliminary interpretations of the UV/Vis profile of the mofegiline-MAO-B complex suggest that the flavin absorption bands are perturbed on mofegiline inhibition but that no covalent addition to the flavin moiety occurs. However, the x-ray crystal structure of the mofegiline-MAO-B complex with supporting CD data demonstrate that the flavin is indeed modified on mofegiline inhibition but in a manner with both differences and similarities with flavin adducts involving alkylation at N(5) or C(4a). The data collected from the reaction of reduced MAO-B with mofegiline (Figure 7) show that the initial adduct, while inactive, is not identical with the mofegiline adduct formed on aerobic inhibition. The incomplete spectral changes may be indicative of flavin re-oxidation by reduction of the vinyl group of mofegiline. The reduction of olefins by flavin containing enzymes has long been known.24,25 The observed increase in the absorption spectrum upon exposure to oxygen reflects full oxidation of the enzyme followed by mofegiline-adduct formation signaling the requirement for amine reduction to attain mofegiline-MAO-B adduct formation. These data provide additional support for the hypothesis that mofegiline inhibits MAO-B by initially reducing the flavin.
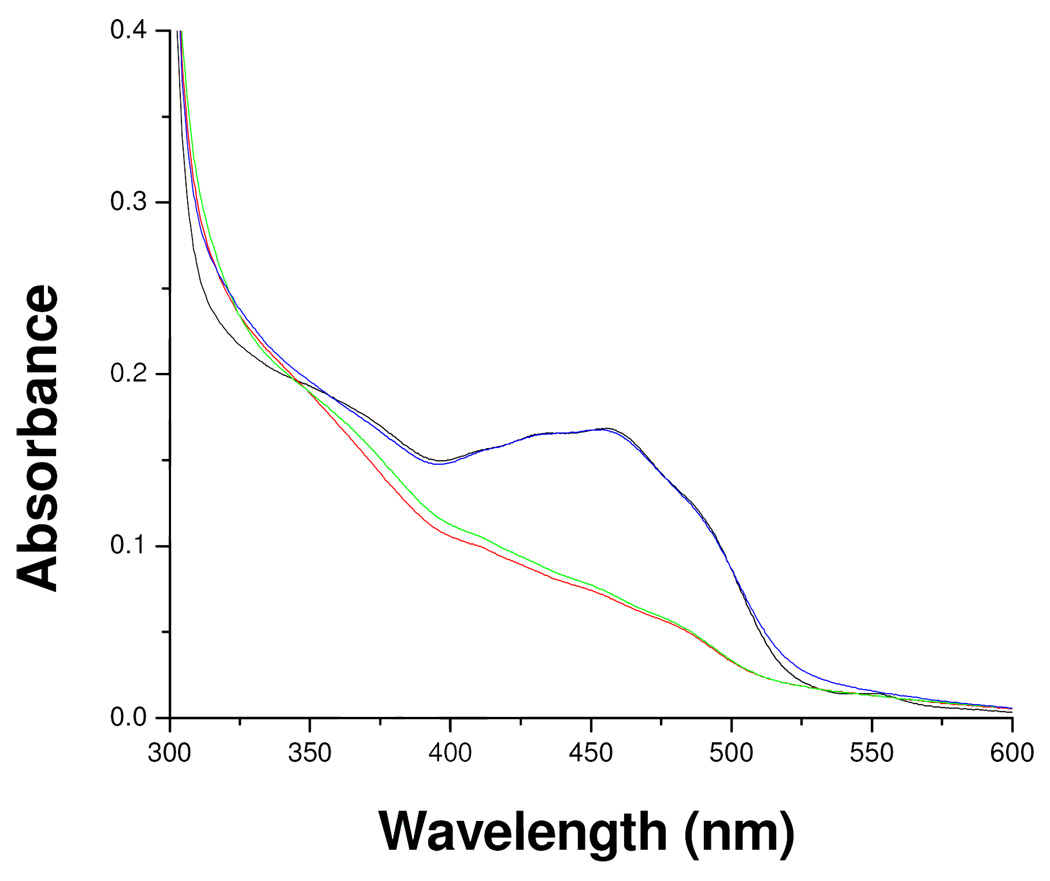
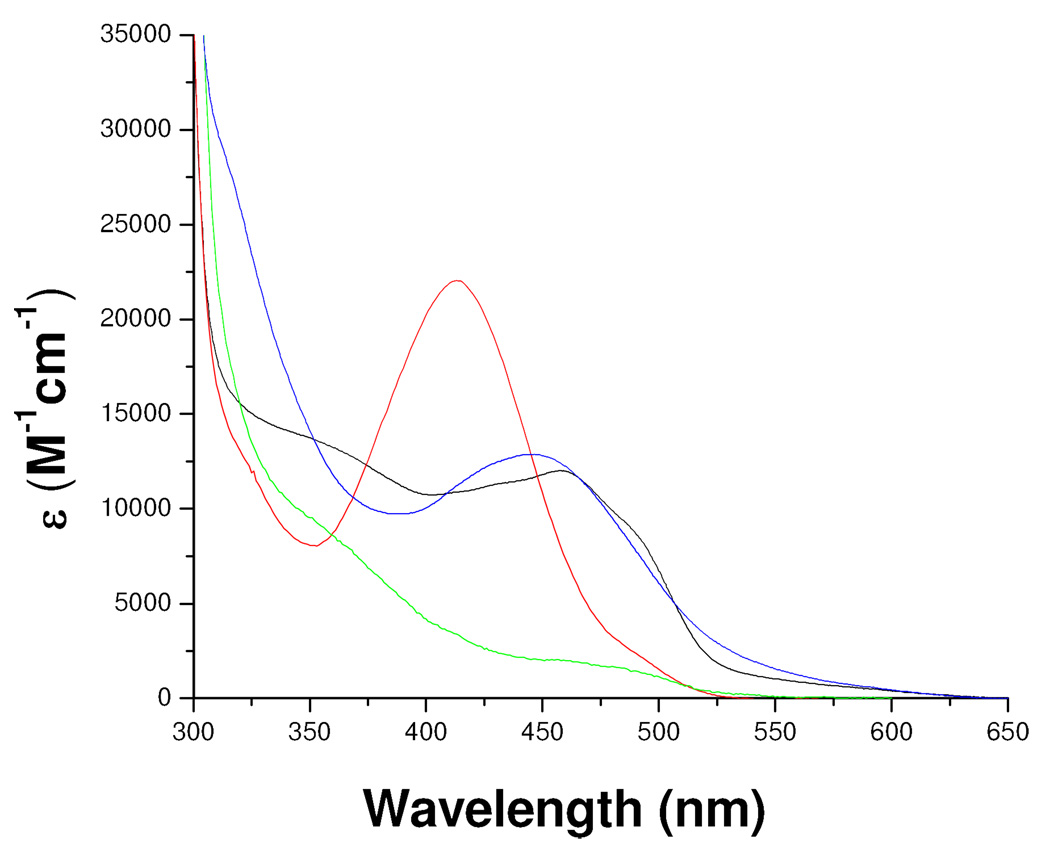
One interesting aspect that deserves further study is the description of factors that give the N(5) alkylated flavin its absorption spectral properties that differ marginally from those of the oxidized form of the flavin and differ extensively from those exhibited by other N(5) or C(4a) flavin adducts. The most reasonable explanation is that the extensive conjugation of the N(5) alkyl group (including the vinyl double bond and the imine double bond) with the flavin ring is sufficient to permit the spectral transitions found in the 450 nm region. Addition of acetylenic inhibitors to the MAO-B or MAO-A flavins result in N(5) flavocyanine adducts with maximal absorption at 415 nm and an εM of 21, 000 M−1cm−1 reflecting their extended conjugation.26 Geometric differences between these two types of MAO-B flavin N(5) adducts likely contribute to their observed spectral differences (Figure 11).
Figure 11.
Comparison of absorption spectral properties of MAO-B and the flavin adducts of MAO-B adducts with various inhibitors. Oxidized MAO-B ( ), Pargyline-inhibited MAO-B (
), Pargyline-inhibited MAO-B ( ), Phenylethylhydrazine- inhibited MAO-B (
), Phenylethylhydrazine- inhibited MAO-B ( ), and mofegiline-inhibited MAO-B (
), and mofegiline-inhibited MAO-B ( ).
).
Unlike MAO-B, mofegiline is a reversible competitive inhibitor of MAO-A and does not form a covalent adduct with the flavin. Addition of high concentrations of mofegiline to MAO-A results in only modest spectral changes in the 300–400 nm region of the flavin UV/Vis spectrum and no reduction of the flavin upon extended incubation (Figure 9). The CD spectral data do not support the formation of a flavin-N(5) adduct with MAO-A. An explanation for this differential behavior between MAO-A and MAO-B is found on comparison of their respective substrate specificities. Previous and current data show that 4-phenylbutylamine is a competitive inhibitor of human MAO-A (Ki = 31 µM)27 but is a substrate for human MAO-B (kcat = 109 ± 5 min −1 and Km = 19 ± 3 µM; kcat/Km = 5.8 min−1µM−1). Mofegiline is a close structural analogue of 4-phenylbutylamine, and thus is poorly suited to function as a substrate for MAO-A but does so for MAO-B. These considerations further support the notion that amine oxidation is a requirement for irreversible inhibition of MAO-B by mofegiline. The Michael addition of nucleophiles to a conjugated Michael acceptor occurs much more rapidly than addition to an unconjugated vinyl group, supporting the requirement for amine oxidation to an imine for inactivation to occur. In addition, steric factors are likely involved in the non-reactivity of the amine moiety of mofegiline for MAO-A. However, pre-reducing the flavin of MAO-A with a stoichiometric amount of substrate does not influence its reactivity toward mofegiline (Figure 10).
Conclusions
Mofegiline is a highly selective, stoichiometric inhibitor of MAO-B that forms an N(5) adduct with the covalent flavin coenzyme in a single catalytic turnover. X-ray structural data of the inhibited enzyme provide definitive evidence for the structure of the inhibited form. The conjugated iminium group is the site for nucleophilic attack by the N(5) of the reduced flavin which is followed by irreversible fluoride elimination. The resulting N(5) adduct is highly conjugated with spectral properties more like those of resting oxidized flavin or to N(5) flavocyanine adducts than to other N(5) or C(4a) alkyl flavins. The high level of mofegiline selectivity towards MAO-B is suggested to result from the nature of its alkyl side chain which is a feature that should be taken into consideration for the design of future MAO-A or MAO-B specific inhibitors.
Materials and Methods
Recombinant human liver MAO-B and MAO-A were expressed in Pichia pastoris and purified as described previously.15,16 Mofegiline was synthesized according to a published procedure.28,29 Standard MAO-A and MAO-B assays were performed spectrophotometrically using kynuramine and benzylamine, respectively at 25 °C in a 50 mM phosphate buffer (pH 7.5) and 0.5%(w/v) RTX-100.
Competitive Inhibition Assays
Ki values for mofegiline inhibition of MAO-A and MAO-B were determined by measuring the initial rates of substrate oxidation (six different concentrations) in the presence of varying concentrations of inhibitor (four different concentrations). All samples were incubated for 5 minutes at 25 °C prior to the addition of enzyme. Ki values were calculated by plotting the apparent Km values for each mofegiline concentration as a function of mofegiline concentration.
Fluoride Electrode Experiment
A sample of human MAO-B (10 µM, 10 nmol) was desalted by gel filtration over a G-25 Sephadex column in 50 mM potassium phosphate buffer (pH 7.5), 0.8 % n-octyl-β-D-glucopyranoside, and 20% glycerol. The enzyme sample was inhibited with one equivalent of mofegiline (2.3 µg, 10 nmol), immediately filtered over a PM-10 filter, and subsequently adjusted with total ion strength adjustment buffer (TISAB III with CDTA) to balance the ion concentration. The filtrate was analyzed by an Accumet combination fluoride specific electrode. The released fluoride concentration was verified by the addition of an internal fluoride standard. A control sample of human MAO-B in the absence of mofegiline was prepared by following the same experimental protocol.
Circular Dichroism Spectroscopy
An Aviv model 62DS circular dichroism spectrophotometer was utilized for the collection of the spectra reported. All spectra were collected from 550 nm to 300 nm with a 1 s dwell time in a 1 cm path length cell. Samples were analyzed in 3 mL quartz cells with concentrations ranging from 25 µM to 40 µM. A total of 5 repetitive scans were averaged, and the spectra were smoothed using an adjacent-point averaging function.
Crystallographic methods
Crystallographic studies were performed as described.17 Briefly, inhibited human MAO-B in 50 mM potassium phosphate pH 7.5, 8.5 mM Zwittergent 3–12 was crystallized by mixing equal volumes of protein sample and reservoir solution (12% PEG4000, 100 mM ADA buffer pH 6.5, 70 mM Li2SO4). X-ray diffraction data were collected at 100 K at the beam-line ID14-EH2 of European Synchrotron Radiation Facility in Grenoble. Data processing and scaling were carried out using MOSFLM30 and programs of the CCP4 package.31 The coordinates of the MAO-B-isatin complex17 were used as initial model after removal of all water and inhibitor atoms. Crystallographic refinements were performed with the programs REFMAC532 and Coot.33 Refinement statistics are listed in Table 1.
Table 1.
Data collection and refinement statistics
| MAO-B-mofegiline complex | |
|---|---|
| Space group | C222 |
| Unit cell (Å) | a = 132.0 |
| b = 223.7 | |
| c = 86.6 | |
| Resolution (Å) | 2.3 |
| Rsyma,b (%) | 11.2 (34.4) |
| Completenessb (%) | 97.7 (99.9) |
| Unique reflections | 55,954 |
| Redundancy | 4.0 (4.0) |
| I/σb,c | 12.0 (3.2) |
| Average B value for ligand atoms (Å2) | 32.2 |
| Rcrystd (%) | 18.7 |
| Rfreed (%) | 22.9 |
| Rms bond length (Å) | 0.008 |
| Rms bond angles (°) | 1.09 |
Rsym=Σ|Ii−<I>|/ΣIi, where Ii is the intensity of ith observation and <I> is the mean intensity of the reflection.
Values in parentheses are for reflections in the highest resolution shell.
The final model consists of residues 3–501 of MAO-B chain A, residues 3–496 of MAO-B chain B, two FAD-mofegiline adducts, two molecules of zwittergent 3–12, and 416 water molecules.
Rcryst=Σ|Fobs−Fcalc|/Σ|Fobs| where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. The set of reflections used for Rfree calculations and excluded from refinement was extracted from the structure factor file relative to the PDB entry 2vz2.
Supplementary Material
Supporting information is available online.
Acknowledgements
This work was supported by National Institutes of Health grant GM-29433 and National Institute of Neurological Disorders and Stroke award number F31NS063648 (to E.M.M.). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institutes of Health. We are grateful to Ms. Milagros Aldeco and to Dr. Jin Wang for their helpful discussions and valuable technical assistance through out this project.
Abbreviations
- MAO
monoamine oxidase
- SSAO
semicarbazide-sensitive amine oxidase
- TISAB
total ion strength adjustment buffer
- ADA
N-(2-acetamido)iminodiacetic acid
- PEG 4000
polyethylene glycol 4000
- CD
circular dichroism
Footnotes
Atomic coordinates have been deposited in the Protein Data Base with ID code: 2vz2.
References
- 1.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annu. Rev. Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weyler W, Hsu YP, Breakefield XO. Biochemistry and genetics of monoamine oxidase. Pharmacol. Ther. 1990;47:391–417. doi: 10.1016/0163-7258(90)90064-9. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez MJ, Saura J, Billett EE, Finch CC, Mahy N. Cellular localization of monoamine oxidase A and B in human tissues outside of the central nervous system. Cell and Tissue Res. 2001;304:215–220. doi: 10.1007/s004410100361. [DOI] [PubMed] [Google Scholar]
- 4.Bach AW, Lan NC, Johnson DL, Abell CW, Bembenek ME, et al. cDNA cloning of human liver monoamine oxidase A and B: molecular basis of differences in enzymatic properties. Proc. Natl. Acad. Sci. USA. 1988;85:4934–4938. doi: 10.1073/pnas.85.13.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler JS, Volkow ND, Wang GJ, Logan J, Pappas N, et al. Age-related increases in brain monoamine oxidase B in living healthy human subjects. Neurobiol. of Aging. 1997;18:431–435. doi: 10.1016/s0197-4580(97)00037-7. [DOI] [PubMed] [Google Scholar]
- 6.Olanow CW. Rationale for considering that propargylamines might be neuroprotective in Parkinson's disease. Neurology. 2006;66:S69–S79. doi: 10.1212/wnl.66.10_suppl_4.s69. [DOI] [PubMed] [Google Scholar]
- 7.Rascol O, Brooks DJ, Melamed E, Oertel W, Poewe W, et al. Rasagiline as an adjunct to levodopa in patients with Parkinson's disease and motor fluctuations (LARGO, Lasting effect in Adjunct therapy with Rasagiline Given Once daily, study): a randomised, double-blind, parallel-group trial. Lancet. 2005;365:947–954. doi: 10.1016/S0140-6736(05)71083-7. [DOI] [PubMed] [Google Scholar]
- 8.Palfreyman MG, McDonald IA, Zreika M, Cremer G, Haegele KD, et al. Mdl-72,974a - a selective MAO-B inhibitor with potential for treatment of Parkinson’s-Disease. J. Neural Transm. Suppl. 1993;40:101–111. [PubMed] [Google Scholar]
- 9.McDonald IA, Lacoste JM, Bey P, Palfreyman MG, Zreika M. Enzyme-activated irreversible inhibitors of monoamine-oxidase - phenylallylamine structure activity relationships. J. Med. Chem. 1985;28:186–193. doi: 10.1021/jm00380a007. [DOI] [PubMed] [Google Scholar]
- 10.Palfreyman MG, McDonald IA, Bey P, Danzin C, Zreika M, et al. Haloallylamine inhibitors of MAO and SSAO and their therapeutic potential. J. Neural Transm. Suppl. 1994;41:407–414. doi: 10.1007/978-3-7091-9324-2_54. [DOI] [PubMed] [Google Scholar]
- 11.Rando RR, Eigner A. Pseudo-irreversible inhibition of monoamine-oxidase by allylamine. Mol. Pharmacol. 1977;13:1005–1013. [PubMed] [Google Scholar]
- 12.Silverman RB, Hiebert CK, Vazquez ML. Inactivation of monoamine oxidase by allylamine does not result in flavin attachment. J. Biol. Chem. 1985;260:14648–14652. [PubMed] [Google Scholar]
- 13.Palfreyman MG, McDonald IA, Bey P, Schechter PJ, Sjoerdsma A. Design and early clinical evaluation of selective inhibitors of monoamine oxidase. Prog. Neuropsychopharmacol. Biol. Psychiatry. 1988;12:967–987. doi: 10.1016/0278-5846(88)90092-9. [DOI] [PubMed] [Google Scholar]
- 14.McDonald IA, Lacoste JM, Bey P, Wagner J, Zreika M, et al. Dual enzyme-activated irreversible inhibition of monoamine-oxidase. Bioorg. Chem. 1986;14:103–118. [Google Scholar]
- 15.Li M, Hubalek F, Newton-Vinson P, Edmondson DE. High-level expression of human liver monoamine oxidase A in Pichia pastoris: comparison with the enzyme expressed in Saccharomyces cerevisiae. Protein Expres. Purif. 2002;24:152–162. doi: 10.1006/prep.2001.1546. [DOI] [PubMed] [Google Scholar]
- 16.Newton-Vinson P, Hubalek F, Edmondson DE. High-level expression of human liver monoamine oxidase B in Pichia pastoris. Protein Expres. Purif. 2000;20:334–345. doi: 10.1006/prep.2000.1309. [DOI] [PubMed] [Google Scholar]
- 17.Binda C, Li M, Hubalek F, Restelli N, Edmondson DE, et al. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. USA. 2003;100:9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binda C, Wang J, Li M, Hubalek F, Mattevi A, et al. Structural and mechanistic studies of arylalkylhydrazine inhibition of human monoamine oxidases A and B. Biochemistry. 2008;47:5616–5625. doi: 10.1021/bi8002814. [DOI] [PubMed] [Google Scholar]
- 19.Zhou M, Panchuk-Voloshina N. A one-step fluorometric method for the continuous measurement of monoamine oxidase activity. Anal. Biochem. 1997;253:169–174. doi: 10.1006/abio.1997.2392. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Binda C, Mattevi A, Edmondson DE. Functional role of the "aromatic cage" in human monoamine oxidase B: Structures and catalytic properties of Tyr435 mutant proteins. Biochemistry. 2006;45:4775–4784. doi: 10.1021/bi051847g. [DOI] [PubMed] [Google Scholar]
- 21.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 22.Müller F, Hemmerich P, Ehrenber A, Palmer G, Massey V. Chemical and Electronic Structure of Neutral Flavin Radical as Revealed by Electron Spin Resonance Spectroscopy of Chemically and Isotopically Substituted Derivatives. Eur. J. Biochem. 1970;14 doi: 10.1111/j.1432-1033.1970.tb00277.x. 185-&. [DOI] [PubMed] [Google Scholar]
- 23.Rothman SC, Johnston JB, Lee S, Walker JR, Poulter CD. Type II isopentenyl diphosphate isomerase: Irreversible inactivation by covalent modification of flavin. J. Am. Chem. Soc. 2008;130:4906–4913. doi: 10.1021/ja7108954. [DOI] [PubMed] [Google Scholar]
- 24.Benson TE, Marquardt JL, Marquardt AC, Etzkorn FA, Walsh CT. Overexpression, Purification, and Mechanistic Study of Udp-N-Acetylenolpyruvylglucosamine Reductase. Biochemistry. 1993;32:2024–2030. doi: 10.1021/bi00059a019. [DOI] [PubMed] [Google Scholar]
- 25.Benson TE, Walsh CT, Massey V. Kinetic characterization of wild-type and S229A mutant MurB: Evidence for the role of ser 229 as a general acid. Biochemistry. 1997;36:796–805. doi: 10.1021/bi962220o. [DOI] [PubMed] [Google Scholar]
- 26.Hubalek F, Binda C, Li M, Herzig Y, Sterling J, et al. Inactivation of purified human recombinant monoamine oxidases A and B by rasagiline and its analogues. J. Med. Chem. 2004;47:1760–1766. doi: 10.1021/jm0310885. [DOI] [PubMed] [Google Scholar]
- 27.Nandigama RK, Edmondson DE. Structure-activity relations in the oxidation of phenethylamine analogues by recombinant human liver monoamine oxidase A. Biochemistry. 2000;39:15258–15265. doi: 10.1021/bi001957h. [DOI] [PubMed] [Google Scholar]
- 28.Evans JC, Goralski CT, Rand CL, Vosejpka PC. US: Application; 1996. Preparation of (E)-1-amino-2-(fluoromethylene)-4-(p-fluorophenyl)butane; (Merrell Dow Pharmaceuticals Inc., USA) 9 pp. [Google Scholar]
- 29.McDonald IA, Bey P, Zreika M, Palfreyman MG. MDL 72,974A Monoamine Oxidase Type B inhibitor. Drugs of the Future. 1991;16:428–431. [Google Scholar]
- 30.Leslie AGW. Integration of macromolecular diffraction data. Acta Crystallogr. Sect. D. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 31.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr. Sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. Sect. D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 35.Kraulis PJ. Molscript - a Program to Produce Both Detailed and Schematic Plots of Protein Structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information is available online.