Abstract
Two monoclonal antibodies (MAbs) were raised against the Candida albicans cell-surface glycoprotein Als3 using the N-terminal domain of the protein as the immunogen. ELISA was used to demonstrate the specificity of the MAbs for the Als3 fragment, but not for the corresponding N-terminal domain fragments from other proteins in the Als family. The anti-Als3 MAbs immunolabeled the surface of germ tubes from a diverse collection of wild-type C. albicans isolates, but did not label yeast cells, an als3Δ/als3Δ deletion mutant strain, nor isolates of other Candida species associated with human disease. Als3 was visualized readily in fresh and formalin-fixed, paraffin-embedded kidney tissue from a murine model of candidiasis. The anti-Als3 MAbs were also useful for immunogold electron microscopy and Western blotting. Both MAbs blocked C. albicans adhesion to vascular endothelial cells and buccal epithelial cells. These versatile MAbs are a valuable addition to the reagents available to study C. albicans cell surface dynamics and interaction of the fungus with host cells.
Keywords: Als3, Als family, Candida albicans, cell-surface glycoprotein, monoclonal antibody
1. Introduction
Contact between Candida albicans and host cells is mediated primarily by cell-surface structures. As such, study of cell surface molecules is important to further our understanding of these interactions. The C. albicans genome sequence predicts many proteins with characteristics that suggest cell-surface localization (de Groot et al., 2003; Jones et al., 2004). Other investigations have revealed cell-surface localization for proteins that were not expected to occupy that location (reviewed in Chaffin, 2008). Despite increasing knowledge regarding the composition of the C. albicans cell surface, there is still a relative lack of reagents available to study cell surface proteins in a specific manner.
One group of C. albicans proteins with sequences that predict cell-surface localization is the Als family (reviewed in Hoyer et al., 2008). Als proteins share a common multi-domain structure and some of the proteins function in adhesion to host surfaces. Much of the work to characterize Als protein function has been done without the benefit of reagents that can monitor the presence of Als proteins on the cell surface. Here, we discuss one component of a larger effort to raise monoclonal antibodies (MAbs) against each of the Als proteins and describe two different MAbs that are specific for Als3. These MAbs are useful in various immunolabeling protocols, in Western blotting, and in blocking adhesion of C. albicans to host surfaces. Development of these reagents and similar ones against the other Als proteins will lend new insight into C. albicans cell surface structure and dynamics as well as Als protein function.
2. Materials and methods
2.1 Production of Als N-terminal protein fragments in heterologous systems
A fragment from the N-terminal domain of each Als protein was produced heterologously in Pichia pastoris GS115 (Invitrogen; Table 1). DNA-sequence-verified ALS gene fragments cloned in E. coli were altered by PCR mutagenesis to change CTG codons to TCT. These fragments were amplified with primers that added a BamHI site at the 5’ end of the fragment and a NotI site at the 3’ end. The reverse primer also included a hexa-His-encoding sequence. PCR products were cloned into BamHI-NotI-digested pPIC3 and transformed into P. pastoris as described previously (Hoyer and Hecht, 2001). The resulting P. pastoris strains secreted hexa-His-tagged Als N-terminal domain fragments using the native signal sequence. Culture of the recombinant P. pastoris strains followed the previous method (Hoyer and Hecht, 2001) except BMMY medium was modified to include only 0.6% yeast extract and 1.2% peptone. Strains were grown for 5 days with 0.5% methanol added daily. Recombinant Als protein was precipitated from the culture supernatant with ammonium sulfate, and following dialysis against binding buffer (20 mM NaCl, 500 mM NaPO4, pH 7.4), purified by His-Trap column chromatography according to the manufacturer’s instructions (GE Healthcare). When necessary, proteins were treated with Endoglycosidase H (Endo H; Roche) to remove N-linked carbohydrate. Final protein preparations were dialyzed in phosphate-buffered saline (PBS; 170 mM NaCl, 3.3 mM KCl, 10 mM Na2HPO4, pH 7.2) and protein concentration measured using the Bradford method (Bio-Rad). Subsequent assays of protein concentration used the Micro BCA Protein Assay kit (Thermo Scientific). Proteins were analyzed by mass spectrometry to verify their predicted molecular mass and visualized by SDS-PAGE and silver staining to confirm the presence of a single protein band.
Table 1.
Panel of Als N-terminal protein fragments used for raising and validating anti-Als MAbs
| Gene Sequencea | Protein Sequenceb | |||||
|---|---|---|---|---|---|---|
| Als Protein | GenBank Accession No. | Sequence Coordinates | No. Codon Corrections | Codon Positions | Amino Acids in Final Protein | Endo H Treated? |
| Als1 | XM712984 | 1 – 987 | 1 | 907 | 18 – 329 | No |
| Als2 | AF024582 | 1 – 984 | 1 | 841 | 18 – 328 | Yes |
| Als3 | U87956 | 1 – 987 | 0 | -- | 18 – 329 | No |
| Als4 | AF024586 | 1 – 987 | 2 | 739, 937 | 18 – 329 | No |
| Als5 | AF025429 | 327 – 1313 | 0 | -- | 18 – 329 | No |
| Als6 | AF075293 | 1 – 996 | 0 | -- | 19 – 332 | Yes |
| Als7 | AY296650 | 1 – 996 | 2 | 34, 634 | 19 – 332 | No |
| Als9-1 | AY269423 | 1 – 984 | 1 | 604 | 18 – 328 | Yes |
| Als9-2 | AY269422 | 1 – 984 | 0 | -- | 18 – 328 | Yes |
The GenBank accession number corresponds to the gene sequence used in the construct. The coordinates of the cloned fragment are shown. CTG codons were changed to TCT using site-directed mutagenesis so that a native C. albicans amino acid sequence would be produced in P. pastoris. The number of codons that were changed is specified, as well as their position within the cloned sequence.
Proteins produced in P. pastoris used the native C. albicans secretory signal sequence, which was cleaved upon export, resulting in the mature protein with the amino acid coordinates shown. N-linked glycosylation was present on four of the Als N-terminal proteins and was removed using Endoglycosidase H as indicated.
An N-terminal fragment of Als3 was also produced in S. cerevisiae. A PCR product encoding the N-terminal 433 amino acids of Als3 from C. albicans strain 1161 was produced with the forward primer ALS3Xho (5’ CCCCTCGAGATGCTACAACAATATACATTGTTACTC 3’) and the reverse primer ALS3Bgl2 (5’ CCCAGATCTTCAAGATGGAACTTGTACAATGACAGTGTC 3’). The PCR product was digested with XhoI and BglII and ligated into the XhoI-BglII-digested plasmid p138NB as described previously (Hoyer et al., 1998a). The resulting plasmid encoded the Als3 N-terminal fragment under control of the copper sulfate-inducible CUP1 promoter. Protein was produced and verified according to published protocols (Hoyer et al., 1998a).
2.2 Production of MAbs
MAbs against the P. pastoris-produced hexa-His-tagged fragments of Als3, Als5 and Als6 were raised at the Immunologic Resource Center (IRC), University of Illinois, Urbana. Five 2–3 month-old BALB/c mice were immunized intraperitoneally with 0.5 mg of Als protein emulsified in TiterMax for the first injection and 0.25 mg of protein in incomplete Freund’s adjuvant for subsequent injections. Injections were spaced at a three-week interval and the immune response against the immunogen monitored by enzyme-linked immunosorbent assay (ELISA; see below). Following spleen removal, splenic lymphocytes were extracted and fused with murine myeloma tumor cells (SP2/0) using polyethylene glycol (PEG). Clones that survived selection in hypoxanthine aminopterim thymidine (HAT) medium were screened for antibody production by ELISA. Positive clones were tested by ELISA for antibody specificity against a panel of all 9 Als protein N-terminal domains (Table 1). Antigen-specific clones were expanded into 24-well tissue culture plates for single-cell cloning by limiting dilution. The anti-Als3 MAb was named 3-A5 and the anti-Als5 and anti-Als6 MAbs were named 5-A5 and 6-A1, respectively.
A MAb was also raised against the S. cerevisiae-produced Als3 fragment using a method similar to that described above. For this immunogen, 4–6 week-old BALB/c mice were immunized subcutaneously with 50 mg of Als3 fragment emulsified in complete Freund’s adjuvant. Fourteen days post-inoculation, mice were immunized intravenously with 10 mg antigen in PBS. The spleen was removed from each mouse 3 days following the IV injection. Preparation of lymphocytes, myeloma fusion, and hybridoma selection and screening methods were similar to those described above. The resulting anti-Als3 MAb was named 113. MAbs were isotyped using the Monoclonal Antibody Isotyping Kit (Pierce); all were IgG1 with a kappa light chain.
Anti-Als5 and anti-Als6 MAbs were used as controls in various experiments reported here. Specificity of both MAbs was demonstrated by ELISA and by immunolabeling of fungal strains that overexpress the immunogen-encoding gene (data not shown).
2.3 ELISA
Different ELISA methods were used for initial testing and final evaluation of MAb specificity. The largest differences between the two methods were in composition of the buffers. The method for final evaluation is presented in greater detail here. Plastic wells (Immunlon 2HB Dividastrips, Fisher 14245-211) were coated with 50 µl of Als protein at a final concentration of 0.5 µg/ml diluted in a carbonate coating buffer (15 mM Na2CO3, 43 mM NaHCO3, pH 9.6) and incubated overnight at 4°C. The wells were washed and blotted 5 times with a 0.15 M PBS 0.05% Tween 20 (pH 7.4) solution between incubation steps except after incubation in blocking buffer; all subsequent steps were carried out at room temperature. Wells were incubated with 100 µl of a freshly prepared blocking buffer solution of PBS-Tween 20 as above mixed with 1.0% bovine albumin (Sigma A7030) for 60 min minimum. Fifty µl of purified anti-Als MAb diluted in blocking buffer to a concentration of 1 µg ml−1 or 50 µl of hybridoma culture supernatant was incubated in the wells for 2 h minimum followed by washing and incubation of 50 µl of a goat anti-mouse Ig H+L HRP antibody (Southern Biotechnology 1010-05) diluted 1:3000 in blocking buffer for 1.5 h at room temperature. Subsequently 100 µl of the enzyme substrate tetramethylbenzidine (TMB Substrate Solution, eBioscience 00-4201-56) was added and the reaction was stopped with 50 µl of 1M H3PO4 solution. Plates were read on a Dynatech MR5000 microplate spectrophotometer. Absorbance values were calculated as absorbance at 570 nm subtracted from absorbance at 450 nm. Experimental controls consisted of wells coated with Als protein without addition of primary or secondary antibodies and wells coated with protein with addition of secondary but not primary antibody.
2.4 Immunolabeling of cultured C. albicans cells
C. albicans strains were stored at –80°C and routinely cultured on YPD agar (per liter: 10 g yeast extract, 20 g peptone, 20 g glucose, 20 g Bacto agar) at 37°C. Cultures were stored at 4°C for no more than one week before a fresh plate was prepared. Starter cultures were grown to saturation by inoculating a single colony into 10 ml YPD liquid medium and incubating for 16 h at 37°C and 200 rpm. For yeast cell time course experiments, fresh YPD was inoculated at a density of 1 × 106 cells ml−1 and incubated at 30°C with 200 rpm shaking. Aliquots harvested at various time points were fixed in Dulbecco’s PBS without calcium or magnesium (DPBS), containing 3% paraformaldehyde. For germ tube assays, yeast cells from the saturated starter culture were washed in DPBS and suspended at a density of 5 × 106 cells ml−1 in pre-warmed RPMI 1640 culture medium (RPMI; Gibco catalog no. 11875). At various time points, cells were harvested by filtration over a 0.22 micron pore size filter and fixed in paraformaldehyde as described above. Germ tubes were also grown in YPD with 10% fetal calf serum and in the medium described by Lee et al. (1975; Lee medium). Starter cultures, cell harvesting and fixation methods were the same as described for RPMI 1640 medium above.
Fixed C. albicans cells were washed 3 times in DPBS prior to and between each step in the immunolabeling protocol. Cells were resuspended in 15 µl ml−1 normal goat serum for 15 min at room temperature to block non-specific antibody binding. Cells were incubated in 18 µg ml−1 purified antibody in DPBS for 60 min at 4°C on a rotating mixer and then in 3 µg ml−1 fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG F(ab’)2 fragment-specific antibody (Jackson ImmunoResearch 115-096-006) under the same conditions. Wet mounts of immunolabeled cells were examined using an Olympus BX50 microscope equipped with the Olympus Fluoview confocal system and Melles Griot Argon (488 nm) laser. An UP-PLAN 100X Oil objective with a Nomarski prism was used for all images that were captured with Olympus Fluoview software. Populations of immunolabeled cells were also analyzed by flow cytometry using a BD Biosciences LSR II equipped with a blue octagon 488-nm laser, a 505 longpass dichroic mirror, and a 530/30 bandpass filter for FITC detection. Fifty thousand events were collected at the low flow rate. Fluorescence analysis involved gating a region within a density plot of forward-scatter area (FSC-A) versus forward-scatter width (FSC-W) for a representative population of single cells. Data were analyzed using FCS Express Version 3 Research Edition software (De Novo).
2.5 C. albicans strain collections
Anti-Als immunolabeling was analyzed using various C. albicans strain collections. Strain CAI12 (Porta et al., 1999), which is a ura3/URA3 derivative of wild-type strain SC5314 (Gillum et al., 1984) was used for many experiments. C. albicans mutant strains were also immunolabeled with anti-Als MAbs. Strains included the als3Δ/als3Δ strain 1843 (Zhao et al., 2004) and the ALS3 reintegrant strain 2322 (Zhao et al., 2006), the als5Δ/als5Δ strain 2373, and the als6Δ/als6Δ strain 1420 (Zhao et al., 2007).
A set of diverse Candida species was assembled including Candida dubliniensis strains CD36 (from Derek Sullivan, Trinity College, Dublin), CM1 and 16F (from Richard Barton, University of Leeds, England) as well as isolates purchased from the American Type Culture Collection (Candida glabrata ATCC 2001, Candida krusei ATCC 14243, Candida parapsilosis ATCC 22109, Candida lusitaniae ATCC 42720, Candida tropicalis ATCC 201380, and Candida guilliermondii ATCC 6260). These isolates were grown for 16 h at 37°C in YPD (yeast forms) and in RPMI at 37°C for 1 h (germ tubes) to assess immunolabeling with anti-Als MAbs.
Another C. albicans collection included isolates of diverse origin and clade assignment (Table 2). Clade assignments were made using the consensus multilocus sequence typing (MLST) protocol for C. albicans (Bougnoux et al., 2003).
Table 2.
Collection of C. albicans strains of diverse origin and clade assignmenta
| Strain | Origin | Clade | Reference |
|---|---|---|---|
| SC5314 | Human blood (disseminated disease) | 1 | Gillum et al., 1984 |
| WO-1 | Human blood (disseminated disease) | 6 | Slutsky et al., 1987 |
| GC15 | Human vaginal (symptomatic) | 11 | Cheng et al., 2005 |
| GC23 | Human vaginal (asymptomatic) | 3 | Cheng et al., 2005 |
| 1–28 | Human oral (healthy) | 8 | Wrobel et al., 2008 |
| 2309 | Human oral (HIV-infected) | 1 | This study |
| SqF087 | Squirrel oral | 9 | Wrobel et al., 2008 |
| CrA038 | Crow oral | Sing. | Wrobel et al., 2008 |
| OKP77 | Human oral (healthy) | 4 | Blignaut et al., 2002 |
C. albicans isolates were from humans and animals, from diseased and asymptomatic individuals, and from various body sites. All clade assignments were made using MLST (Bougnoux et al., 2003). Diploid Sequence Types (DSTs) and genotype designations have been published for strains SC5314 and WO-1 (Odds et al., 2007), and for strains 1–28, SqF087 and CrA038 (Wrobel et al., 2008). Genotype designations (reported as AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, ZWF1b, and the strain DST) for the other strains are: GC15 (60, 13, 21, 4, 7, 11, 15; 1257); GC23 (13, 10, 85, 6, 7, 32, 15; 1258); 2309 (2, 3, 5, 12, 2, 82, 5; 1256); OKP77 (58, 14, 8, 4, 7, 3, 8). Clade assignments were made according to published methods (Wrobel et al., 2008).
2.6 Evaluation of monoclonal antibody-antigen binding kinetics
Antigen-antibody binding kinetics were measured on a Biacore 3000 Surface Plasmon Resonance System using an indirect capture method. A rabbit anti-mouse IgG antibody (BR-1008-38) was immobilized onto a Biacore CM5 chip (BR-1003-99) following manufacturer’s instructions. Purified anti-Als protein MAbs and Als N-terminal proteins were dialyzed into a running buffer solution (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.01% surfactant P20, BIAcore). Details of Biacore-based binding kinetics assays have been described elsewhere (Mistrik et al., 2004).
Anti-Als3 antibodies (3-A5 and 113) and the control anti-Als5 MAb (5-A5) were captured on parallel flow cells and the Als3 N-terminal domain simultaneously injected across both flow cells; 5 min association times and 30 min dissociation times were used. The antibody capture surface of each flow cell was regenerated between runs using 10 mM glycine pH 1.7. A minimum of six different concentrations of Als3 were included for analysis using BIAevaluation software version 4.1 (Biacore). Chi-square values were calculated for the average squared residual per data point to judge the closeness of fit of the data to the predicted model. Chi-square values were ≤ 2.0. The anti-Als5 MAb was used as a negative control in this assay because the amino acid sequences of the N-terminal domain immunogens for Als3 and Als5 are 79% identical.
2.7 Immunohistochemical staining of mouse tissues
Kidneys were collected from BALB/cByJ mice that were inoculated with 1 × 106 C. albicans cells of strain CAI12 (Porta et al., 1999) via the lateral tail vein. Kidneys were collected 27 h post-inoculation, fixed in 10% neutral buffered formalin (pH 7.2), placed in an automatic tissue processor for paraffin infiltration, cut into 2–3 µm thick sections, and mounted on glass slides. Mounted tissue sections were dried at 60°C for 30 min, deparaffinized, hydrated to water, placed in 3% H2O2 in methanol for 15 min to removed endogenous peroxidase activity, and rinsed two times in Supersensitive wash buffer (BioGenex HK583-5K). Epitope recovery consisting of treatment with 0.1% protease (Sigma P5147) diluted in Tris buffer pH 7.6 at 37°C for 20 min was followed by washing as above. Staining occurred in a BioGenex i6000 automated staining system with appropriate washes between the following incubation steps following manufacturer instructions: 10 min blocking solution (Powerblock, BioGenex HK085-5K), 30 min 35 µg/ml anti-Als3 MAb, 60 min anti-mouse EnVision+ System-HRP labeled polymer (DakoCytomation K4001), 5 min DAB substrate, and counterstained with Mayer’s hematoxylin (BioGenex HK100-9K) for 1 min. Staining procedures were conducted at room temperature unless otherwise indicated. Negative control tissues were treated identically except for omission of the anti-Als3 MAb incubation step.
2.8 Immunolabeling C. albicans cells dissected from a mouse kidney
Kidneys were removed from a BALB/cByJ mouse that had been inoculated with 5 × 105 cells of C. albicans strain SC5314 (Gillum et al., 1984). Kidneys were collected 24 h post-inoculation. Immediately following dissection from the mouse, kidney tissue was minced with a razor blade, deposited in a microfuge tube containing DPBS and homogenized by hand using a plastic homogenizer tip. A portion of the supernatant from this suspension was aspirated and transferred to another microfuge tube, pelleted, immunostained and visualized using the protocol for cultured cells described above.
2.9 Western blotting
SDS-PAGE gels (NuPAGE® Novex® 4–12% Bis-Tris; Invitrogen) with approximately 50 ng of Als N-terminal protein per lane were prepared for Western blotting by soaking in transfer buffer (25 mM Tris base, 192 mM glycine, 20% methanol). Proteins were transferred to Hybond-P PVDF membranes (Amersham) at 300 mA constant current for 3 h. Membranes were rinsed in water, dried, and stored at room temperature. Protein detection with anti-Als MAbs was performed using an ECL kit (Amersham RPN2132). Anti-Als MAbs were used at a concentration of approximately 5 µg ml−1. A control gel with 0.5 µg of each protein was silver stained using the Bio-Rad kit.
2.10 Electron microscopy analyses
Cells from liquid cultures were fixed in 3% paraformaldehyde (pH 7.0) for indirect immunogold labeling of the cell surface. The procedure is similar to that for immunofluorescent labeling except that the secondary antibody used was a 6 nm colloidal Gold-AffiniPure goat antimouse IgG + IgM (H+L) antibody (Jackson ImmunoResearch 115-195-068). Immunogold-labeled samples were then placed in Karnovsky’s fixative and submitted to the Center for Microscopic Imaging for embedding, sectioning and transmission electron microscopy. Methods can be found at http://treefrog.cvm.uiuc.edu/methods/methods_mainpage.html and are summarized briefly here. Following microwave fixation in Karnovsky’s solution, the tissue was washed in cacodylate buffer. Microwave fixation was also used with the secondary 2% osmium tetroxide fixative, followed by the addition of 3% potassium ferricyanide for 5 min. After washing with water, saturated uranyl acetate was added for enbloc staining for 1 h. The sample was dehydrated in a series of increasing ethanol concentration. Acetonitrile was used as the transition fluid between ethanol and epoxy. Infiltration was accomplished with a series of increasing concentration of acetonitrile:LX-112 epoxy (1:1, 1:4, pure LX-112). The resulting blocks were polymerized at 90°C overnight, trimmed and ultrathin sectioned with diamond knives. Sections were stained with uranyl acetate and lead citrate and examined with a Hitachi H600 transmission electron microscope.
2.11 C. albicans adhesion assays
MAbs for adhesion assays were purified by Protein G column chromatography, concentrated and dialyzed against DPBS using an Amicon filtration device. Protein concentration was measured using the Micro BCA Protein Assay Kit (Thermo Scientific). Methods for measuring antibody blocking of C. albicans adhesion to monolayers of human umbilical vascular endothelial cells (HUVECs) and to freshly collected human buccal epithelial cells (BECs) were described previously (Beucher et al., 2009). Anti-Als6 MAb was used as a negative control in the adhesion assays. Immunolabeling of wild-type C. albicans cells with the anti-Als6 MAb did not produce a detectable signal using the method described above.
3. Results
3.1 Validation of MAb specificity
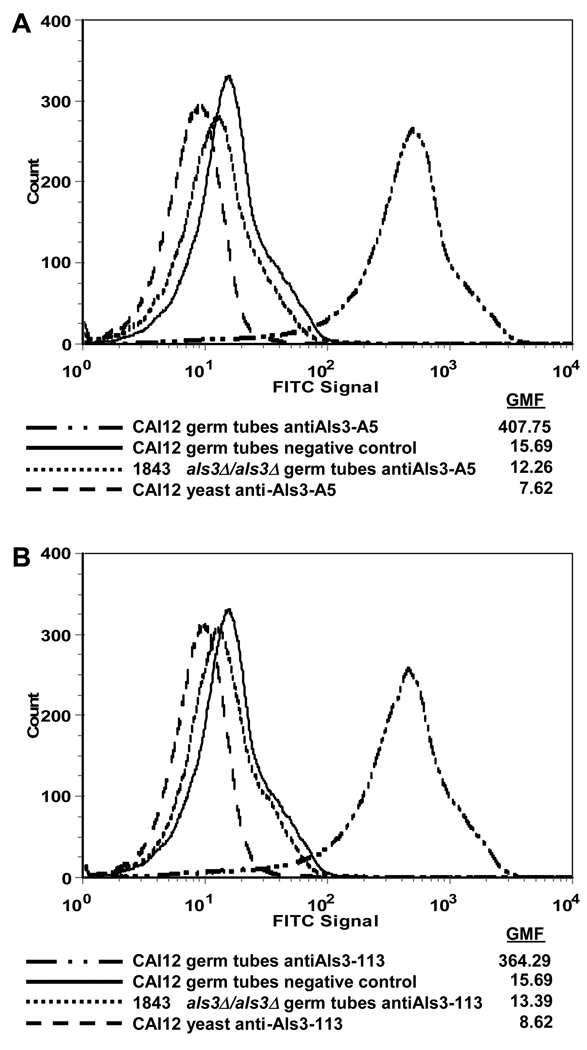
Initial testing of MAb specificity was accomplished by ELISA using the panel of nine Als N-terminal domain fragments (Table 1). Typical results showed absorbance readings of approximately 1.5 for reactions between the purified MAb and its immunogen, with typical readings of 0.05 for reactions between antibody and a non-immunogen Als protein. Typical results for testing hybridoma culture supernatant were absorbance readings of approximately 0.3 for reactions between the positive culture supernatant and the corresponding immunogen. ELISA-positive MAbs were further tested against wild-type, mutant and reintegrant C. albicans strains. Fig. 1 shows immunolabeling of C. albicans cells with anti-Als3 MAbs 3-A5 and 113. Fluorescence was detected on the surface of wild-type and reintegrant germ tubes, but not on als3Δ/als3Δ germ tubes or on yeast forms of any strain. These results were expected due to the large increase in ALS3 transcription that occurs in germ tubes and hyphae (Hoyer et al., 1998b). Immunolabeling of the same strains grown under other germ tube-inducing conditions (YPD with 10% serum, Lee medium) produced similar results (data not shown). MAb specificity was further demonstrated using flow cytometry (Fig. 2). Similar to the results in Fig. 1, flow cytometry showed a strong fluorescence on wild-type germ tubes with either anti-Als3 3-A5 or 113, but not on als3Δ/als3Δ germ tubes or on wild-type yeast forms.
Fig. 1.
C. albicans cells from strains CAI12, 1843 (als3Δ/als3Δ), and 2322 (als3Δ/als3Δ∷ALS3) labeled with either 3-A5 or 113 anti-Als3 MAb and a FITC-conjugated secondary antibody. Rows A, C, E, and G show Als3 detection on the surface of wild-type and reintegrant germ tubes grown in RPMI for 1 h. Als3 was not detected on mutant germ tubes grown in RPMI for 1 h (rows B and F) nor on yeast cells grown in YPD for 1 h (rows D and H). Left panels with a dark background have only laser (488 nm) illumination, center panels have both laser and white light illumination, and right panels have only white light illumination.
Fig. 2.
Flow cytometry of C. albicans germ tubes (1 h RPMI medium) and yeast (16 h YPD medium) labeled with anti-Als3 MAb and a FITC-conjugated secondary Ab with calculated geometric mean fluorescence for each peak (GMF). A) Cells labeled with anti-Als3 A5 MAb. B) Cells labeled with anti-Als3 113 MAb. For both A) and B), negative control germ tubes were incubated with only the secondary antibody.
Measurements of antibody binding kinetics using Biacore showed a dissociation constant (KD) of 0.4 nM for 3-A5 MAb (mean of two observations: 0.5 nm and 0.3 nm) and 15 nM for 113 (mean of three observations: 15 nm, 13 nm, 18 nm). Immunofluorescent labeling is specific for C. albicans and was not observed for either MAb when tested against other Candida species including three strains of C. dubliniensis, and one strain each of C. glabrata, C. krusei, C. parapsilosis, C. lusitaniae, C. tropicalis and C. guilliermondii. Each strain was grown for 16 h in YPD and also for 1 h in RPMI (data not shown). The pattern of immunostaining observed on strains derived from SC5314 was also observed on a diverse set of C. albicans isolates (Table 2). When inoculated into growth medium that promotes germ tube formation, germ tubes, but not mother yeasts, were immunolabeled. Growth of these strains under conditions that result in yeast forms failed to indicate any immunolabeling. Results were similar for both anti-Als3 3-A5 and 113.
3.2 Analysis of Als3 dynamics in vitro and in vivo
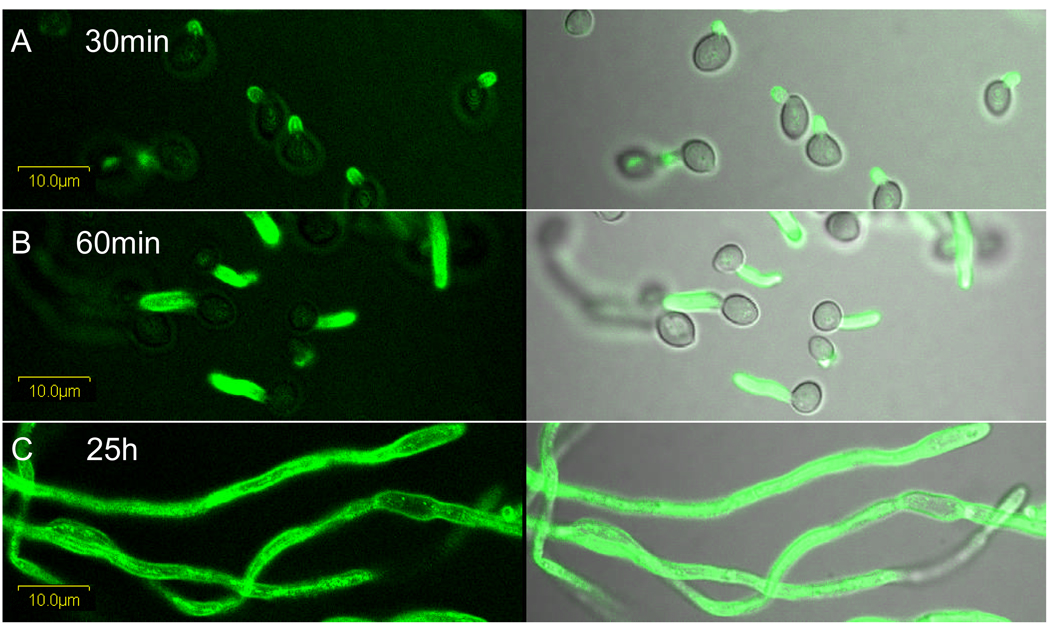
Immunolabeling of C. albicans hyphae was followed over a time course (Fig. 3). Als3 presence on the emerging germ tube surface was visible 30 min after inoculating yeast cells into RPMI medium, consistent with previous reports stating that transcription from the ALS3 promoter was detectable when germ tubes become visible microscopically (Zhao et al., 2004). This bright signal persists as the germ tube grows; intense immunolabeling of long hyphae suggests a consistent deposition of Als3 on the cell surface (Fig. 3). Loss of Als3 from the hypha surface, such as by shedding of the protein into the medium, was not apparent. Similar results were obtained with both anti-Als3 MAbs 3-A5 and 113 and for germ tubes grown in RPMI, YPD with 10% serum and in Lee medium.
Fig. 3.
Rows A–C: Germ tubes of strain CAI12 grown in RPMI medium sampled at 30 min, 60 min, and 25 h were labeled with anti-Als3 113 MAb and a FITC-conjugated secondary antibody. The left panels with a dark background have only laser (488 nm) illumination; the right panels have both laser and white light illumination.
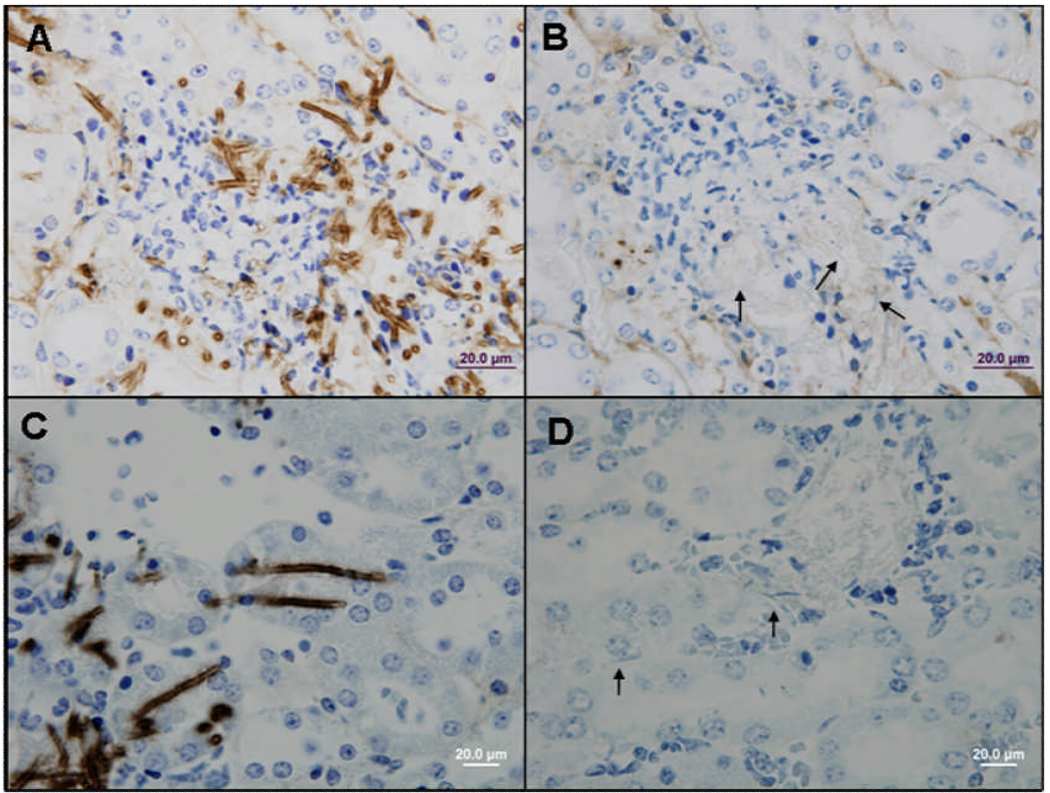
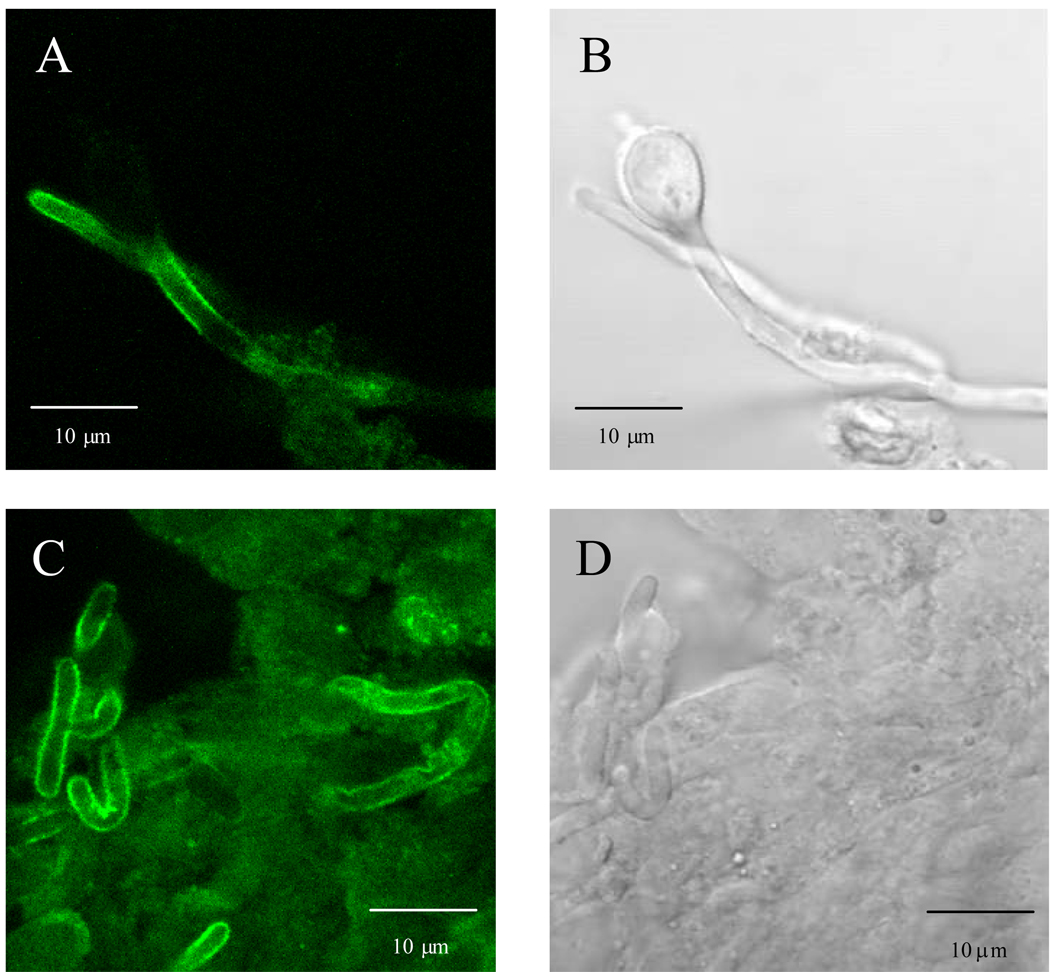
The consistent presence of Als3 on the germ tube and hypha surface suggests that Als3 should be detectable by immunohistochemical staining of tissue sections from disease models. This result was observed for kidney tissue taken from a mouse inoculated with C. albicans via the lateral tail vein. Tissue sections from 27 h post-inoculation showed an obvious Als3 presence on C. albicans cells in the kidney. Similar results were observed for both anti-Als3 MAbs (Fig. 4). The strong presence of Als3 on the C. albicans cell surface was also demonstrated on fungal cells dissected from an unfixed mouse kidney (Fig. 5).
Fig. 4.
Photomicrographs of kidney tissue from a mouse infected with C. albicans CAI12 showing hyphae labeled with anti-Als3 MAb and a peroxidase-conjugated secondary antibody, then counterstained with hematoxylin. A) Als3-A5 MAb; C) Als3-113 MAb; B and D) negative controls with only the secondary antibody. Arrows in B) and D) show unlabeled fungal hyphae.
Fig. 5.
Anti-Als3 immunolabeled hyphae from the kidney of a mouse inoculated with C. albicans strain CAI12. Hyphae were dissected from the kidney and treated with anti-Als3 A5 MAb and a FITC-conjugated secondary antibody. Panels A and C are illuminated with laser (488 nm) only; panels B and D are the same images, respectively, illuminated with white light only. Panel B shows a yeast cell that is not labeled with anti-Als3. Small amounts of disrupted renal tissue are visible in panel D with hyphae showing a diffuse distribution of Als3 protein on the hypha surface (panel C).
3.3 Utility of anti-Als3 MAbs for additional applications
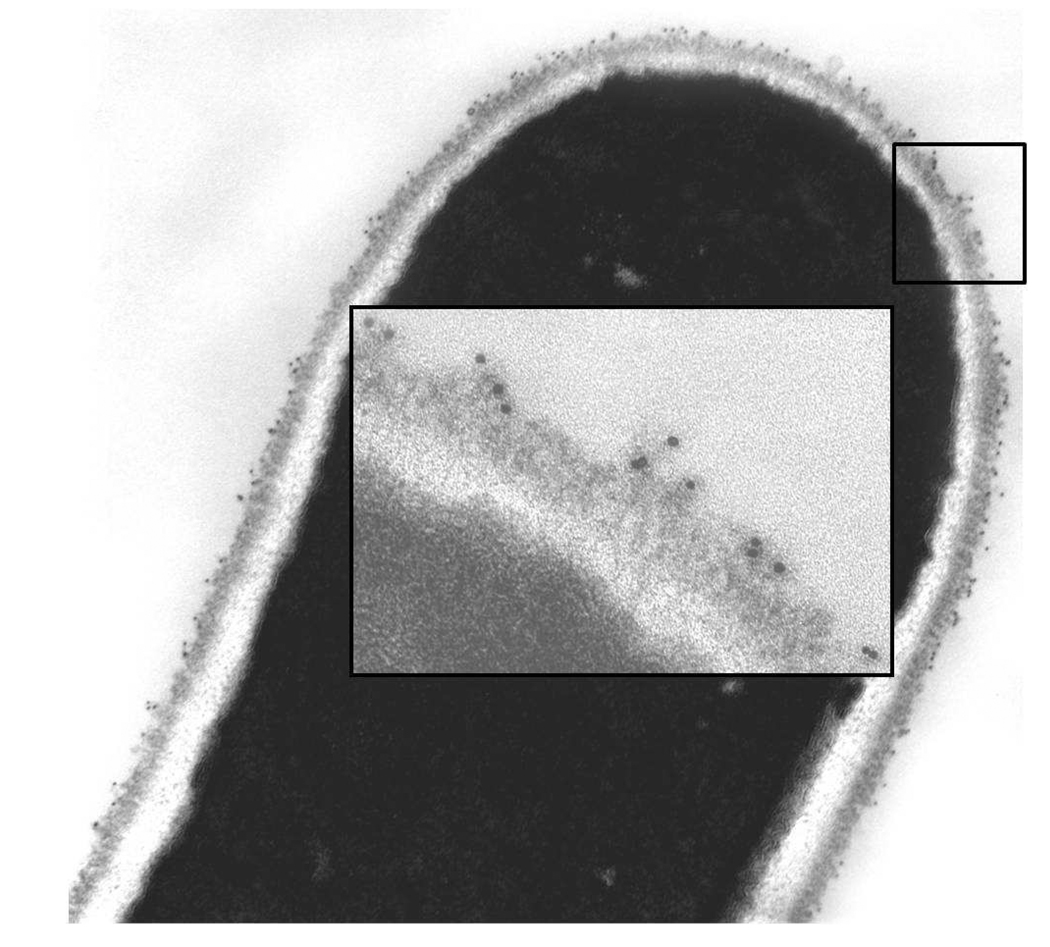
The anti-Als3 MAbs were also evaluated for their utility in other analyses such as immunogold electron microscopy (Fig. 6). Gold particles were distributed over the length of the germ tube and visible in the outermost flocculant layer of the cell surface. Similar results were obtained for both anti-Als3 MAbs 3-A5 and 113.
Fig. 6.
C. albicans CAI12 germ tubes grown in RPMI at 37°C for 45 min labeled with anti-Als3 113 MAb and a gold-conjugated secondary antibody. Gold particles were visible along the length of the germ tube. The inset shows a higher magnification image to visualize gold particles on the outermost layer of the C. albicans cell surface.
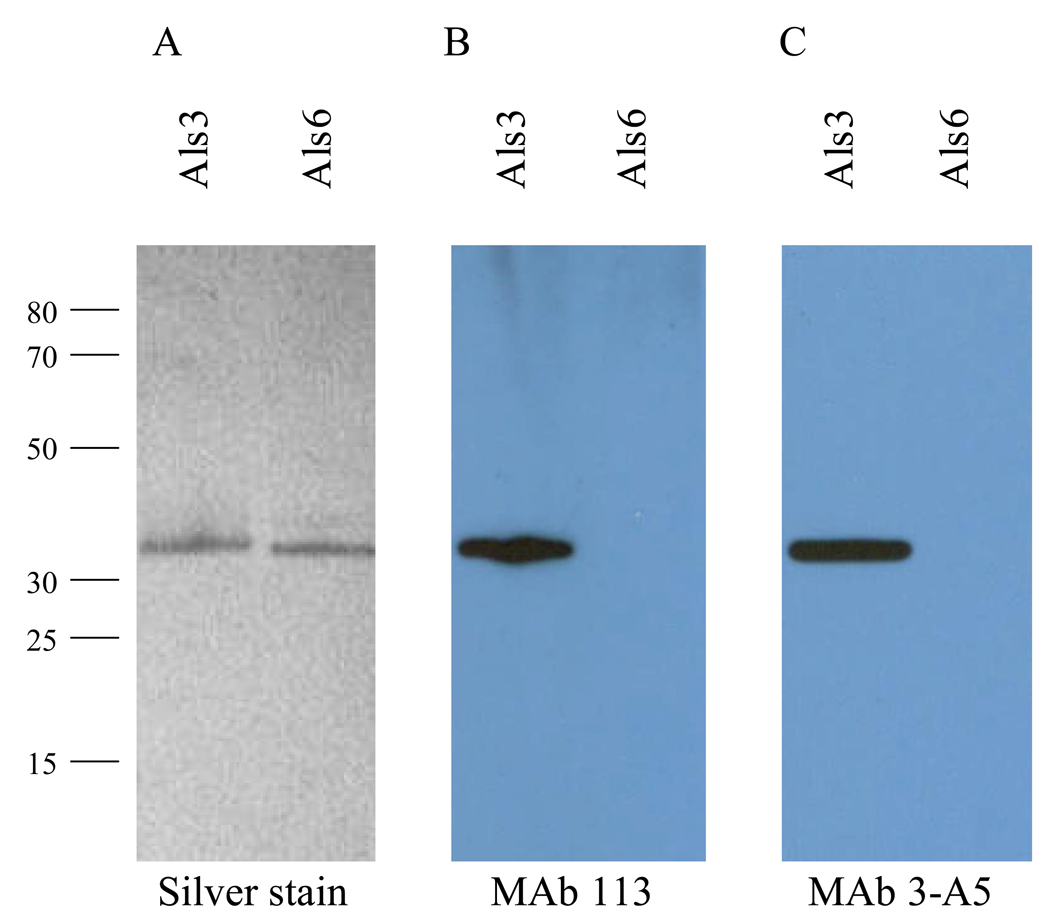
Both anti-Als3 MAbs were useful for detection of Als protein fragments on Western blots (Fig. 7). Lack of detection of the Als6 N-terminal domain indicated specificity of the MAbs for Als3.
Fig. 7.
Utility of anti-Als3 MAbs for Western blotting. Als3 and Als6 N-terminal domain proteins were run on SDS-PAGE gels. A portion of the gel was silver stained (A). Identical lanes were Western blotted with either anti-Als3 113 (B) or anti-Als3 3-A5 (C). Detection of Als3 demonstrates utility of the MAbs for Western blotting. Lack of detection of Als6 demonstrates anti-Als3 MAb specificity.
3.4 Anti-Als3 MAbs block adhesion of C. albicans to host cells
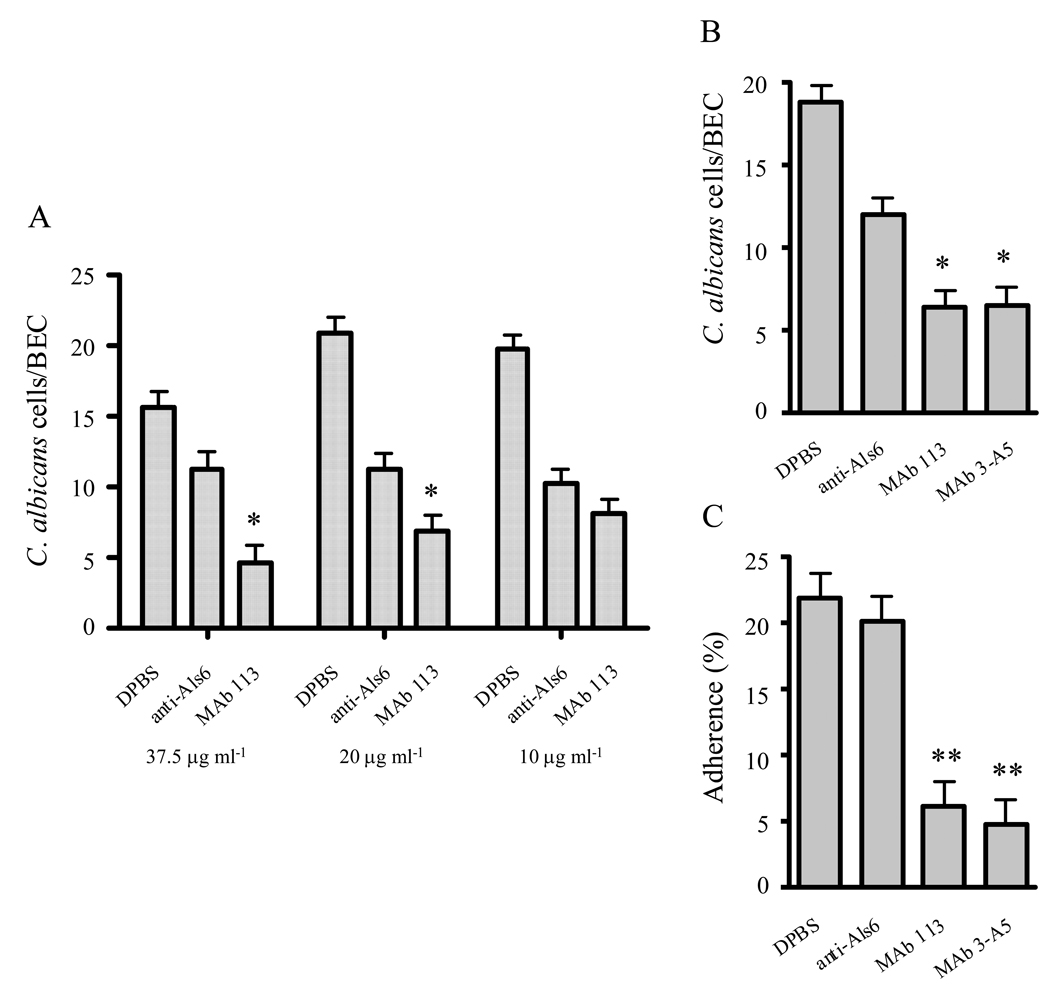
Both anti-Als3 MAbs were tested for their ability to block C. albicans adhesion to host cells (Fig. 8). A working concentration of MAb for adhesion assays was defined using anti-Als3 113 in a dilution experiment in the BEC adhesion assay (Fig. 8A). An anti-Als6 MAb was used as a negative control in these experiments. ALS6 is transcribed at a very low level in cultured C. albicans cells and cells recovered from clinical specimens (reviewed in Hoyer et al., 2008). At all concentrations tested (Fig. 8A and 8B), addition of the negative control MAb to the BEC adhesion assay resulted in significantly decreased adhesion of C. albicans to BECs (P = 0.0033 for DPBS vs. anti-Als6 at the 20 µg ml−1 concentration, for example). This non-specific effect of control protein was noted previously (Beucher et al., 2009) and does not occur in the assay that measures C. albicans adhesion to monolayers of vascular endothelial cells (Fig. 8C). Titration of MAb 113 concentration showed significant inhibition of adhesion at the 37.5 µg ml−1 (Fig. 8A; P = 0.0066) and 20 µg ml−1 concentrations (P = 0.0088), but not at a concentration of 10 µg ml−1 (P = 0.1825). The 20 µg ml−1 concentration was chosen for subsequent experiments (Fig. 8B and 8C). At 20 µg ml−1, both anti-Als3 MAbs 3-A5 and 113 were effective in blocking adhesion of C. albicans germ tubes to BECs (Fig. 8B) and to endothelial cell monolayers (Fig. 8C).
Fig. 8.
Anti-Als3 MAbs block C. albicans adhesion to HUVECs and BECs. (A) Anti-Als3 MAb 113 was titrated in the BEC adhesion assay. Adhesion was inhibited at concentrations of 20 µg ml−1 or higher. The 20 µg ml−1 concentration was selected for subsequent assays (*P < 0.01 for comparisons between anti-Als6 and either anti-Als3 MAb). (B) Both anti-Als3 MAbs were effective in blocking C. albicans adhesion to BECs at 20 µg ml−1. (C) Either anti-Als3 MAb at 20 µg ml−1 blocked C. albicans adhesion to HUVEC monolayers (**P < 0.0001 for comparisons between anti-Als6 and either anti-Als3 MAb).
4. Conclusions
We have demonstrated the utility of two anti-Als3 MAbs, 3-A5 and 113, in a variety of methods for studying the dynamics of C. albicans cell surface proteins and their role in host-pathogen interactions. Data presented here show that Als3 is a major presence on the C. albicans cell surface from the time when the germ tube is first visible to the stage when cells have grown into long hyphae. This persistent presence of Als3 on the germ tube/hypha surface has been suggested by other studies that documented that Als3 is the epitope recognized by immune-based reagents specific for C. albicans germ tubes and/or hyphae compared to yeast forms (Laforce-Nesbitt et al., 2008; Beucher et al., 2009). Data presented here show that the MAbs are specific for C. albicans and recognize C. albicans strains of diverse origin. Immunolabeling can be accomplished using cells grown in vitro or taken from animal disease models. Cells from disease models can be freshly isolated or formalin-fixed and paraffin-embedded. Detection of immunolabeling can be accomplished by fluorescent microscopy, flow cytometry or electron microscopy. Visualization by electron microscopy demonstrated the localization of Als3 N-terminal domain to the outer flocculant layer of the C. albicans cell surface. The anti-Als3 MAbs are also able to block C. albicans adhesion to buccal epithelial cells and monolayers of vascular endothelial cells. Results presented here suggest that the two anti-Als3 MAbs can be used interchangeably in the applications tested. The only demonstrated difference between the MAbs is the more favorable dissociation constant of 3-A5 (0.4 nM) compared to 113 (15 nM). Development and validation of these reagents represents an advance in the methods available to study C. albicans cell surface dynamics and interactions with host cells.
Acknowledgements
We thank Liping Wang and Rachel Breitenfeld of the University of Illinois Immunological Resource Center for producing anti-Als MAbs. Lou Ann Miller of the Center for Microscopic Imaging and Barbara Pilas and Ben Montez of the University of Illinois Flow Cytometry Facility also contributed to this work. This research was funded by grant DE14158 from the National Institute of Dental and Craniofacial Research, National Institutes of Health. The investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR16515-01 from the National Center for Research Resources, National Institutes of Health.
References
- Beucher B, Marot-LeBlond A, Billaud-Nail S, Oh SH, Hoyer LL, Robert R. Recognition of Candida albicans Als3 by the germ-tube-specific monoclonal antibody 3D9.3. FEMS Immunol. Med. Microbiol. 2009;55:314–323. doi: 10.1111/j.1574-695X.2008.00502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blignaut E, Pujol C, Lockhart S, Joly S, Soll DR. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J. Clin. Microbiol. 2002;40:826–836. doi: 10.1128/JCM.40.3.826-836.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougnoux ME, Tavanti A, Bouchier C, Gow NAR, Magnier A, Davidson AD, Maiden MCJ, d’Enfert C, Odds FC. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 2003;41:5265–5266. doi: 10.1128/JCM.41.11.5265-5266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin WL. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008;72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr, Trupin SR, Hoyer LL. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect. Immun. 2005;73:1656–1663. doi: 10.1128/IAI.73.3.1656-1663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot PWJ, Hellingwerf KJ, Klis FM. Genome-wide identification of fungal GPI proteins. Yeast. 2003;20:781–796. doi: 10.1002/yea.1007. [DOI] [PubMed] [Google Scholar]
- Gillum AM, Tasy EY, Kirsch DR. Isolation of the Candida albicans genes for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and pyrF mutations. Mol. Gen. Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Hecht JE. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast. 2001;18:49–60. doi: 10.1002/1097-0061(200101)18:1<49::AID-YEA646>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Hecht JE. Identification of Candida albicans ALS2 and ALS4 and localization of Als proteins to the fungal cell surface. J. Bacteriol. 1998a;180:5334–5343. doi: 10.1128/jb.180.20.5334-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3, and insights into the nature of the ALS gene family. Curr. Genet. 1998b;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh S-H, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforce-Nesbitt SS, Sullivan MA, Hoyer LL, Bliss JM. Inhibition of Candida albicans adhesion by recombinant human antibody single-chain variable fragment specific or Als3p. FEMS Immunol. Med. Microbiol. 2008;54:195–202. doi: 10.1111/j.1574-695X.2008.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC. An amino acid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Mistrik P, Moreau F, Allen JM. BiaCore analysis of leptin-leptin receptor interaction: evidence of 1:1 stoichiometry. Anal. Biochem. 2004;327:271–277. doi: 10.1016/j.ab.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li SY, Tavanti A, Maiden MCJ, Gow NAR, d’Enfert C. Molecular phylogenetics of Candida albicans. Eukaryot. Cell. 2007;6:1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta A, Ramon AM, Fonzi WA. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 1999;181:7516–7523. doi: 10.1128/jb.181.24.7516-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Whittington JK, Pujol C, Oh SH, Ruiz MO, Pfaller MA, Diekema DJ, Soll DR, Hoyer LL. Molecular phylogenetic analysis of a geographically and temporally matched set of Candida albicans isolates from humans and nonmigratory wildlife in Central Illinois. Eukaryot. Cell. 2008;7:1475–1486. doi: 10.1128/EC.00162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Oh S-H, Cheng G, Green CB, Nuessen JA, Yeater KM, Leng RP, Brown AJP, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Daniels KJ, Oh S-H, Green CB, Yeater KM, Soll DR, Hoyer LL. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology. 2006;152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Oh S-H, Hoyer LL. Deletion of ALS5 ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med. Mycol. 2007;45:429–434. doi: 10.1080/13693780701377162. [DOI] [PMC free article] [PubMed] [Google Scholar]