Abstract
Secretion of microneme proteins is essential to Plasmodium invasion but the molecular composition of these secretory organelles remains poorly defined. Here, we describe the first Plasmodium microneme proteome. Purification of micronemes by subcellular fractionation from cultured ookinetes was confirmed by enrichment of known micronemal proteins and electron microscopy. Quantitation of electron micrographs showed >14-fold microneme enrichment compared to the intact ookinete, such that micronemes comprised 85% of the identifiable organelles in the fraction. Gel LC-MS/MS of the most abundant protein constituents of the fraction identified three known micronemal proteins chitinase, CTRP, SOAP, together with protein disulphide isomerase (PDI) and HSP70. Highly sensitive MudPIT shotgun proteomics described a total of 345 proteins in the fraction. M1 aminopeptidase and PDI, the former a recognised target of drug development, were both shown to have a micronemal location by IFA. We further identified numerous proteins with established vesicle trafficking and signaling functions consistent with micronemes being part of a regulated secretory pathway. Previously uncharacterised proteins comprise the largest functional group of the microneme proteome and will include secreted proteins important to invasion.
Keywords: Invasion, Microneme, Plasmodium, Proteome, Subcellular fractionation
1 Introduction
Despite our best efforts malaria remains a disease of global significance with some 40% of the world's population at risk. Each year malaria kills up to 2.7 million people in Africa alone and global GDP losses are estimated to be of the order of $20 billion. Resistance of the parasite to drugs and of the mosquito to insecticides, requires that we search for new rational therapies. One approach has focused on the mechanisms by which the parasite invades host cells, an essential component of development in both man and the mosquito. The merozoite must recognise, enter and grow within the red blood cell; the sporozoite must lyse and traverse mosquito salivary gland cells, mammalian Kupffer cells and hepatocytes, prior to its final entry and development in the hepatocyte, and the ookinete has to lyse and traverse the mosquito midgut epithelium [1]. All invasive stages contain the apical complex of regulated secretory vesicles, which function in critical and sequential roles in the recognition and invasion of the host. Component vesicles include the micronemes (with roles in host cell adhesion and rupture); rhoptries (required for host cell modification and formation of the parasitophorous vacuole (PV)), and dense granules (required for late host cell modification) [2].
Despite the critical role that micronemes play in host invasion, very little is known of the proteins important to microneme biogenesis, signaling and exocytosis. Past efforts to determine the structure and function of these secretory vesicles have relied heavily on cell fractionation techniques notably in the readily accessible parasites Toxoplasma [3], Eimeria [4], Cryptosporidium [5], Sarcocystis [6, 7] and only one study in Plasmodium has described the merozoite rhoptry proteome [8]. Despite the elegance of these studies, there has always been the problem that the originating parasite contained more than one of the apical complex vesicle populations. Microscopic evidence suggests ookinetes possess neither rhoptries nor dense granules and therefore offers an excellent opportunity to fractionate and describe the proteome of just one class of secretory organelle – the microneme.
Many secreted microneme proteins are essential and thus are considered to be vaccine candidates e.g. thrombospondin-related anonymous protein (TRAP) [9], apical membrane antigen 1 (AMA-1) [10] and erythrocyte binding antigen 175 (EBA-175) [11]. Once secreted, microneme cargo proteins are important at distinct steps in ookinete invasion; circumsporozoite and TRAP related protein (CTRP) is the receptor component of the ‘glideosome’ complex which powers movement and invasion [12], secreted ookinete adhesive protein (SOAP) is important to host adhesion [13], the perforin-domain containing membrane attack ookinete protein (MAOP) is essential for host cell lysis [14] and cell traversal protein of Plasmodium ookinetes and sporozoites (CelTOS) is critical to host cell traversal [15].
Electron microscopy has shown that microneme biogenesis occurs at the Golgi apparatus [16, 17]. Pro-microneme vesicles containing cargo proteins such as AMA-1, are transported along subpellicular microtubules to the apical pole [16], where they reside. Microneme contents are discharged from the apical tip in response to a calcium mediated signal [18], possibly triggered by a specific host stimulus.
Bioinformatic searches for novel microneme proteins has been hindered by the lack of a clear micronemal targeting signal possibly because microneme proteins can be targeted by distinct escorter proteins [19]. To achieve this, the first biochemical characterisation of a Plasmodium microneme proteome, we capitalise on three advantages of the ookinete; it is large size relative to other Plasmodium invasive stages, the reported absence of rhoptries and dense granules and our recently developed ability to produce ookinetes in bulk. We identify a cell fraction in which known micronemal proteins are concentrated, undertake a morphological analysis which demonstrates that it is very highly enriched for micronemes and proteomic analyses of the fraction demonstrate enrichment of verified micronemal proteins. Putative novel micronemal proteins were verified by immunofluorescence assays (IFA), these included M1 aminopeptidase (A-M1) and the chaperone, protein disulphide isomerase (PDI). The derived microneme proteome contains proteins consistent with a eukaryotic regulated secretory pathway, together with the largest category of proteins, of unknown function, which is likely to include proteins important to host invasion.
2 Materials and methods
A schematic of the method followed is shown in Figure 1.
Figure 1.

The experimental protocol followed: Pb infected blood was harvested from ∼25 rats and ookinetes cultured. Ookinete purification by nycodenz gradient was followed by lysis by sonication. Remaining intact cells and nuclei were removed by centrifugation at 800×g. The post-nuclear fraction was separated on a sucrose gradient, 1 mL fractions were removed for analysis by Western blot. The 1.1 M sucrose fraction was characterised by TEM, abundant proteins were sequenced by LC-MS/MS and a global survey was conducted by MudPIT shotgun proteomics. The localisation of selected proteins were analysed by IFA.
2.1 Ookinete production, purification and microneme enrichment
Plasmodium berghei (Pb) strain ANKA 2.34 ookinetes were produced from rat blood as described [20] with significant adaptations. Adult female brown Norway rats were pre-treated with phenyl hydrazine and injected with blood stage parasites i.p. Approximately 2/3 of inoculated rats developed high blood parasitemia after 4 days. Infected blood was harvested from 20 to 25 rats and placed into culture such that the gametocytes developed into ookinetes after 22−24 h at 19°C [21]. Ookinetes were purified by ammonium chloride lysis of red blood cells and Nycodenz density gradient separation [12] to 95−99% purity, as assessed by Giemsa stained smears. Contamination by asexual and sexual blood stage parasites was less than 1 and 2%, respectively. The 5×108 purified ookinetes were resuspended in 2 mL HM buffer (250 mM sucrose, 1 mM EDTA, 5 mM triethanolamine, pH 7.5 with protease inhibitors) [4, 6] and sonicated at 2 μm on ice for 60 s in 10 s intervals, resulting in <98% cell lysis (minimising microneme membrane disruption). Intact cells and nuclei were removed by centrifugation at 800×g for 10 min. Parasite ‘post-nuclear’ fractions were applied to the top of 12 mL sucrose gradients (1.6−0.5 M). Samples were centrifuged at 70 000×g for 2 h at 4°C with no brake in a SW40Ti swing out rotor (Beckman). Fractions (1 mL) were collected and the sucrose concentrations were measured by refractometry. The protein content of each fraction was estimated using the bicinchoninic protein assay (Pierce).
2.2 Electron microscopy
The 1.1 M sucrose fraction was diluted 1:5 in HM buffer and the organelles pelleted at 130 000×g for 30 min. The pellet was fixed overnight in 2% glutaraldehyde, 250 mM sucrose in PBS (4°C) and prepared for transmission electron microscopy as described previously [21]. The organellar composition of the preparation was determined by morphometry. A regular point lattice was superimposed on random micrographs and the underlying organelles scored for 1029 random data points [22]. EM micrographs of intact ookinetes were similarly analysed for 2904 random points.
2.3 1-D PAGE analysis (GEL LC-MS/MS)
Thirty microlitres of the microneme-enriched fraction was separated on a 16 cm 12% acrylamide gel and stained with colloidal Coomassie [23]. Protein abundance was estimated by band density of scanned images and the five most abundant bands were excised and digested with trypsin and subjected to MS/MS analysis as described [23]. The resulting MS/MS spectra were submitted to MASCOT and searched against a locally installed protein database composed of Plasmodium falciparum (Pf) and Pb ORFs and annotated proteins (plasmoDB-5.2). The MASCOTscores and the number of peptides detected are shown in Supporting Information Table 1.
2.4 Multidimensional protein identification technology (MudPIT)
Four independent microneme-enriched preparations, originating from independent ookinete cultures, were separated into soluble and insoluble components which were analysed by MudPIT. Microneme-enriched samples (1.1 M sucrose) were solubilised in 2 M urea 10 mM Tris-HCl pH 8.5 and centrifuged at 16 000×g for 30 min at 4°C to separate soluble from insoluble fractions. Both fractions were digested with endoproteinase Lys-C and trypsin [24]. Peptide mixtures were analysed by MudPIT [25]. A quaternary Agilent 1100 series HPLC coupled directly to a Finnigan LTQ-IT mass spectrometer (Thermo, San Jose, CA, USA) equipped with a nano-LC ESI source [26] resolved peptide mixtures by strong cation exchange LC upstream of RP LC [27]. Fully-automated five step chromatography was carried out on each sample. Good quality spectra were filtered as described [28] and compared, using SEQUEST (version 27), to a database containing Pb and P. yoelli predicted proteins [29, 30] supplemented with a common contaminant NCBI database including rat proteins [31] and a reverse sequence database included to estimate the false positive rate. No enzyme specificity was considered for any search. All searches were parallelised and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs [28]. Results were assembled and filtered using DTASelect (version 2.0) to estimate the false positive rate (<5%). Ten soluble and eight insoluble fractions were analysed and combined to create a non-redundant catalogue (Supporting Information Table 2), exceeding the 10 MudPIT analysis necessary to achieve 95% coverage of complex mixtures [32].
2.5 In silico analysis of identified proteins
All proteins identified were manually assigned a functional classification using the MIPS functional catalogue database (FunCatDB) [33] as modified for Plasmodium [34] (Supporting Information Table 2). Proteins associated with the eukaryotic secretory pathway were identified by the presence of either a signal peptide or transmembrane domain(s), assessed by SignalP 3.0 [35] and ConPred [36], respectively. Recognising that the Pb genome is not fully sequenced and many gene predictions are incomplete, Pf orthologues (identified using InParanoid [37]) were also analysed. Secretory proteins retained in the ER were identified by the sequence motifs SDEL [38], KDEL [39] or HDEL [40], whilst those imported to the apicoplast were identified with PATS [41]. Cytoplasmic proteins imported to the nucleus or mitochondria were identified with NucPred [42] and PlasMit [43], respectively.
2.6 Immunoblotting
Equal volumes of each sucrose fraction were analysed for the micronemal proteins, CTRP and SOAP and the surface protein Pb28 by Western blots. Standard protocols were followed using the anti-SOAP mouse polyclonal antibody [13], 13.1 mAb to Pb28 [44] and a rabbit polyclonal antibody raised to recombinant C-terminus of CTRP (amino acids 1864−1904). Pb ookinetes were probed with an anti-peptide antibody (MAP1) to Pf M1 family zinc aminopeptidase (A-M1) [45] and another antibody to PfPDI [46]. Secondary antibodies used were either anti-rabbit or anti-mouse ECL™ IgG horse radish peroxidase (HRP), which were detected with the ECL plus Western blotting detection system (Amersham).
2.7 Immunofluorescence assays (IFA)
Cultured ookinetes were smeared on to glass slides in foetal bovine serum, fixed with 4% formaldehyde in PBS pH 7.4 for 10 min at room temperature, washed once with TBS, permeablised with 0.2% Triton X-100 for 5 min, then washed three times in TBS for 5 min each. Fluorescence was quenched with 0.1% sodium borohydride in TBS for 5 min. Slides were washed in TBS for 5 min, blocked with 3% BSA/5% Goat sera for 15 min and then incubated with either MAP1 antibody (1:200) or anti-PDI (1:500) together with 3A5 (mouse monoclonal to CTRP 1:100) for 2 h [12]. Primary antibodies were detected with anti-rabbit Alexa 488 and anti-mouse Alexa 546 (Molecular Probes each 1:1500) for 1 h in the dark. Slides were washed and mounted with Mowiol, 25 mg/mL DABCO and 25 μg/mL DAPI. Images were captured using a Zeiss confocal microscope with LSM510 software. Excitation lasers used were 405, 488 and 543 nm and the detection ranges were 420−480 nm, 505−550 nm and LP560 nm, respectively. Images were compiled in Adobe Photoshope 6.0.
3 Results
3.1 Enrichment of ookinete micronemes by subcellular fractionation
3.1.1 Co-sedimentation and enrichment of microneme proteins CTRP and SOAP
Subcellular fractionation of ookinetes resulted in co-sedimentation of two known micronemal markers, CTRP and SOAP in the 1.1 M sucrose fraction (Fig. 2), consistent with microneme sedimentation recorded in other Apicomplexa [5, 47]. Semi-quantitative analysis of scanned Western blots, suggests that 40% of the total CTRP and 24% of SOAP present on the entire gradient is found in the 1.1 M sucrose fraction (minimal estimations as Western blot exposures are non-linear). In contrast, protein assays show this fraction contains just 2−3.8% of the total protein (Fig. 2), indicating significant enrichment of the micronemal proteins CTRP and SOAP. The very abundant cell surface protein Pb28 [48], whilst detectable in the 1.1 M sucrose fraction, concentrates in the 0.75 M sucrose fraction (containing 23% of total Pb28 and 12% of the total protein) (Fig. 2).
Figure 2.
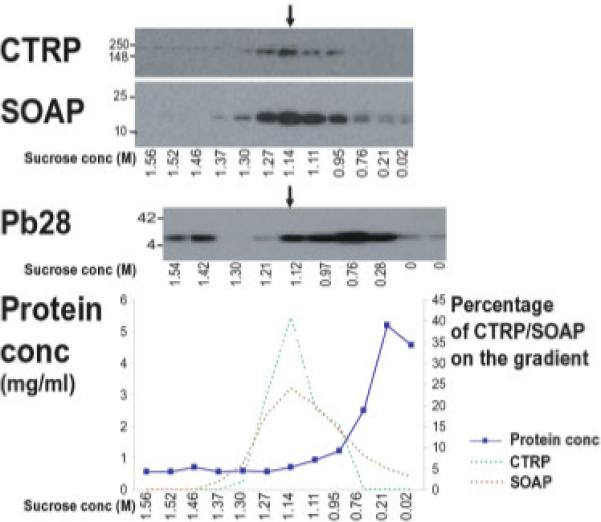
Micronemal proteins co-segregate and enrich at 1.1 M sucrose and separate from the surface protein Pb28: Western blot analysis of sucrose gradient fractions for the micronemal marker proteins CTRP and SOAP and the surface protein Pb28. CTRP and SOAP enrich at 1.1 M sucrose, whilst Pb28 enriches at 0.75 M sucrose but extends into the 1.1 M fraction. The graph shows the protein concentration profile of the post-nuclear fraction across the sucrose gradient and semi-quantitative analysis of the CTRP and SOAP Western blot bands (shown above), which are represented as a percentage of the total present across the gradient. The 1.1 M fraction contains over 40% of CTRP and 24% of SOAP but less than 4% of the total protein, demonstrating the enrichment of micronemal proteins.
3.1.2 Enrichment of microneme-like electron dense vesicles
Transmission electron microscopy of the 1.1 M sucrose fraction revealed significant microneme enrichment (Fig. 3). The bottle shaped electron dense vesicles (∼80 nm diameter) possess distinguishable bulb and neck regions [16]. Micro-nemes represent ∼85% (873:1029) of the total structures seen in this fraction. In contrast, examination of intact ookinetes found micronemes comprise only ∼6% (188:2904) of the ookinete cell mass-suggesting over 14-fold enrichment. Neither rhoptries, which can be distinguished from micronemes by their size (diameter ∼350 nm in Pf merozoites [49]) and containing intravesicular membranes [50, 51], nor dense granules were found in our preparations. Whilst a limited number of unidentified membranes are present in the preparation no other identifiable organelles were seen.
Figure 3.
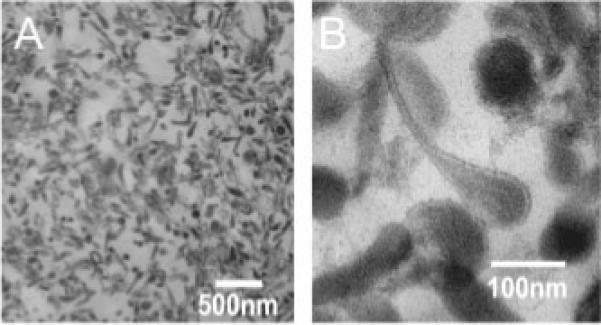
Electron dense vesicles enrich at 1.1 M sucrose: (A) TEM of the 1.1 M sucrose fraction demonstrates enrichment of microneme-like electron dense vesicles. (B) Higher magnification shows the diameter to be ∼80 nm consistent with microneme dimensions. Ookinete micronemes clearly have bulb and neck parts.
3.1.3 Abundant proteins are predominantly verified micronemal proteins
The five most abundant proteins in the 1.1 M sucrose fraction were visualised by SDS PAGE and Coomasie stain (Fig. 4) and were sequenced by LC MS/MS. The pre-dominant protein in each band is listed (Supporting Information Table 1). In order of protein band abundance, chitinase, SOAP together with the highly abundant surface protein Pb28, PDI, CTRP and heat-shock protein 70 (HSP70). Chitinase [52], SOAP and CTRP are previously verified micronemal proteins and we localise PDI to the micronemes in this study (Section 3.3, Fig. 6).
Figure 4.
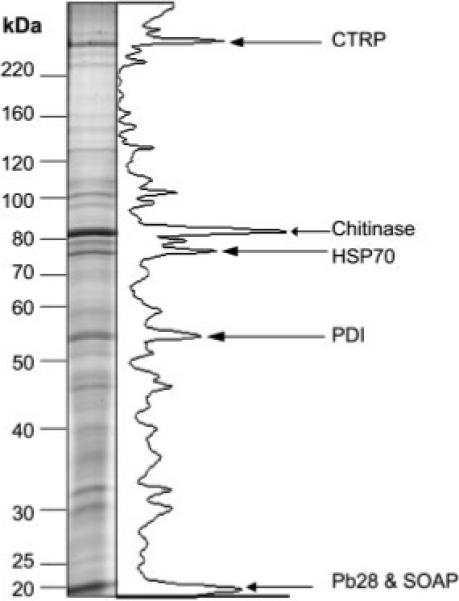
The major constituents of the microneme-enriched fraction are verified micronemal proteins: the microneme-enriched fraction was separated by SDS-PAGE and stained with colloidal coomasie. Protein band concentrations were estimated by densitometry and the greyscale is plotted. The abundant bands were analysed by LC-MS/MS and the results analysed by MASCOT and BLAST. CTRP, chitinase and SOAP are verified micronemal proteins confirming microneme enrichment. Identification of PDI suggests a micronemal localisation.
Figure 6.
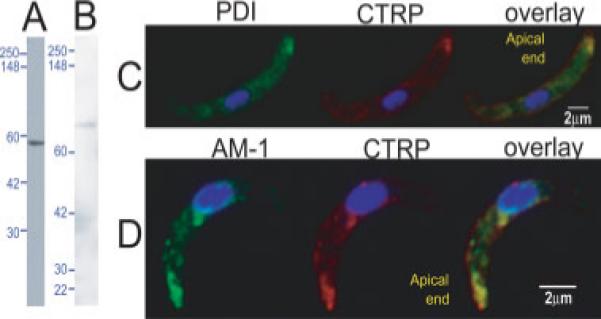
PDI and A-M1 locate at micronemes in ookinetes: ookinete lysate probed with (A) anti-PfPDI detects a protein of 51 kDa as expected for PDI and (B) anti-PfA-M1 (MAP1) detects two bands at 68 and 40 kDa, likely processed forms of PbA-M1, predicted to be 123 kDa. IFA of ookinetes probed with (C) PDI (green) co-stained with CTRP (red) and (D) A-M1 (green) co-labelled with CTRP (red). AM-1 and PDI clearly locate at the apical end of ookinetes and co-localise with CTRP. Additional perinuclear localisation of A-M1 and PDI is likely to be the ER and Golgi.
3.1.4 The microneme-enriched fraction is depleted of proteins from other organelles
MudPIT shotgun proteomics detected 345 proteins in the 1.1 M sucrose fraction (Supporting Information Table 2). Of the proteins detected 49% have signal peptides or transmembrane domains compared to 12% in the ookinete whole cell proteome. This suggests over four-fold enrichment of proteins from the secretory pathway.
Seven of the known ookinete micronemal proteins were detected in the microneme proteome i.e. CTRP, SOAP, chitinase, CelTOS [15], von Willebrand Factor A domain-related protein (WARP) [53] and PDI and A-M1 which we show to be micronemal in this study (Section 3.3 and Fig. 6). We did not find the other reported ookinete micronemal proteins subtilisin 2 (Sub 2) [54] or MAOP [14]. Interestingly, Sub 2 is present in the ookinete inside the mosquito midgut [54] but has not been demonstrated in ookinetes produced ‘in vitro’ (in our analysis or the Pb whole cell ookinete proteome [30]) and may be developmentally regulated to a different ookinete maturation point. Our lack of detection of MAOP may be due to technical limitations in achieving ∼95% coverage of protein detection in proteomic analyses [32].
Ribosomal proteins being highly abundant cellular proteins, are a common contaminant of sucrose gradient fractionations [4, 6], and unsurprisingly we detect 68 ribosomal proteins. We therefore recognise that not all the proteins present in the microneme-enriched fraction are bona-fide micronemal proteins. Thus we assessed the microneme-enriched proteome for co-enrichment of proteins located at the ER, apicoplast, mitochondria or nucleus. Of the 175 Pb predicted proteins containing ER retention signals SDEL [38], KDEL [39] or HDEL [40], we find only six in the microneme-enriched proteome. PATS predicts 22% of Pf proteins on chromosome 2 locate at the apicoplast [41] but predicts only 10% of the microneme-enriched fraction locates at the apicoplast (likely overestimated as includes the micronemal proteins CTRP and SOAP). PlasMit predicts 22% of Pf predicted proteins transit through mitochondria, but only 9% of the microneme-enriched proteome are similarly predicted [43]. NucPred predicts 5% of the ookinete whole cell proteome target to the nucleus, compared to 2% of the microneme-enriched proteome. Thus, the microneme-enriched fraction is depleted of ER, mitochondrial, apicoplast and nuclear proteins.
We observed 1% asexual contamination of the initial ookinete cell preparation and thus anticipated detection of highly abundant blood stage proteins. We detect the microneme protein AMA-1, however IFAs demonstrate it is absent from the ookinete (data not shown), suggesting the proteomic detection originates from merozoite contamination. Merozoite surface proteins were detected e.g. Small exported protein 1 (Sep1), despite IFAs showing Sep1 is absent from the ookinete (data not shown). We also identify rhoptry proteins, albeit at significantly lower spectral count numbers compared to the blood stage proteome (Supporting Information Table 3). IFA analyses of RhopH3 confirms it is absence in the ookinete (data not shown), strongly suggesting these proteins, being of common origin, are blood stage contaminants and not derived from ookinete micronemes.
3.2 Proteomic survey of the ookinete microneme-enriched fraction
We identified 345 proteins in the microneme-enriched proteome (Supporting Information Table 2 and Fig. 5). In addition to the identification of nine established micronemal proteins described above (Section 3.1.4), we detect a further 24 proteins with described roles in apical organelles or at the cell surface, in other malarial life cycle stages or other Apicomplexa. For example Plasmodium perforin-like protein 4 (PPLP4) from the same protein family as the micronemal protein MAOP, was readily detected. Micro-nemal proteins are known to include proteases, we detected A-M1, cathepsin c, falcilysin, falcipain-1 pepsinogen and plasmepsin. Micronemal proteins commonly form complexes and we found 23 chaperones, including PDI. However, the largest functional category of proteins detected were hypothetical proteins (n = 104), 59 of which have secretory features.
Figure 5.
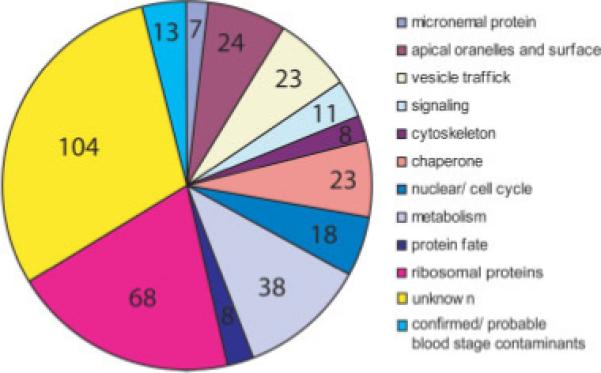
Functional categories of proteins identified in the microneme proteome by MudPIT: the microneme-enriched fraction was analysed by MudPIT shotgun proteomics. Homology to better characterised proteins allowed functional categorisation (Supporting Information Table 2) and the number of proteins in each category are depicted. Over a quarter of the proteins detected are of unknown function. Several proteins attributed a metabolic role include proteases which may play additional roles at micronemes.
A further 23 proteins with established vesicular trafficking roles were found, including sortilin, Rab11a, and the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs): Vesicle-associated membrane protein (VAMP) and Syn13, clathrin, dynamin, N-ethylmaleimide-sensitive factor NSF and EPS homology (EH) domain containing protein (with EPS15 and dynamin domains). Eleven putative signaling molecules were found, including calcium dependent protein kinase (CDPK) 4 and the hypothetical protein PB000924.02.0 (with EF hand domains). Proteins known to be present in the parasite actomyosin motor, namely actin, myosin A, aldolase, myosin A-tail interacting protein (MTIP) were also present in the fraction.
3.3 Intracellular localisation of proteins identified in the microneme-enriched fraction
PDI was the third most highly represented protein in the microneme-enriched fraction. Western blots of Pb ookinete whole cell lysate probed with anti-PfPDI antibody detected one band of the expected mobility, 51 kDa (Fig. 6A). Immunofluorescence assays show PDI locates apically in the ookinete and co-localises with the micronemal protein CTRP (Fig. 6C). PDI also shows a perinuclear and anteriad distribution consistent with ER and Golgi localisations, respectively.
PbA-M1 shares 67% sequence identity to its Pf orthologue, including complete sequence conservation at the active site (468−494aa of PbA-M1) and the aminopeptidase specific GAMEN motif (435−439aa of PbA-M1). Western blot analysis of Pb ookinetes with anti-PfA-M1 antibody, detected two bands at 68 and 40 kDa (Fig. 6B), suggesting proteolytic cleavage of the predicted 123 kDa protein. PfA-M1 similarly produces a 120 kDa precursor which is processed to 96 and 68 kDa [55]. IFA shows PbA-M1 co-localises with the micronemal protein CTRP at the apical pole (Fig. 6D), consistent with a micronemal localisation. PbA-M1 is additionally found in a perinuclear position, where it is possibly in transit through the ER.
4 Discussion
In compiling this microneme proteome we broaden our horizons of microneme biology. We identify putative microneme cargo with potentially critical roles in invasion including several proteases. Moreover we identify likely candidates in microneme biogenesis, signaling, exocytosis and fusion with the plasma membrane.
Microneme cargo includes proteins essential to host invasion and we identify 59 hypothetical proteins in the microneme proteome with secretory features, whose possible roles in invasion deserve to be investigated. Proteases are important to host invasion and we here demonstrate A-M1 to be a micronemal protein. PfA-M1 has been localised to the digestive vacuole in trophozoites and to spots in the schizont and a single spot in free merozoites [55], which our evidence now suggests may correspond to micronemes. PfA-M1 is reported to additionally locate at the nucleus in trophozoites [56] and we find it perinuclear in the ookinete. Inhibitor studies and the failure of attempts to ‘knock out’ PfA-M1 [56] suggest it is important to Plasmodium blood stage growth and thus is the target of drug development [57]. Similar to established micronemal proteases, A-M1 may regulate adhesion or micronemal protein processing critical to host invasion, essential for asexual parasite growth. The protease Rhomboid 1 (ROM1) has been described as a micronemal protein in Toxoplasma gondii [58] and alternatively in the novel mononeme vesicles in Pf merozoites [59]. Interestingly, we do not detect rhomboids in the ookinete microneme proteome and other proteomic analyses failed to detect ROM1 in the ookinete of either Pb [30] or P. gallinaceum [60]. However, proteins may remain undetected if less abundant, developmentally limited or not easily amenable to MudPIT analysis.
We detect a multitude of chaperones, the most abundant being PDI, which we demonstrate to be micronemal. PDI has previously been reported in Neospora caninum micronemes and is important to binding to the host cell [61]. In Pf schizonts, PDI also locates at peripheral punctate dots [46], which may correspond to micronemes. Moreover, PDI is found on the surface of T. gondii [62, 63] and PDI is a major constituent of the calcium induced secreted fraction [64], which is consistent with PDI secretion via the micronemes. PDI, an established ER chaperone, forms and maintains protein disulphide bridges. As many micronemal proteins have a high number of disulphide bridges, PDI may act as a chaperone and preserve their correct molecular structure. Consistent with this hypothesis, expression of disulphide rich Pfs25 in Pichia pastoris is enhanced by co-expression with PDI [65].
We detect proteins with established critical roles in regulated exocytosis. Proteins important to vesicle biogenesis, budding and fusion, including sortilin [66], clathrin and dynamin [67]. These proteins may play similar roles in microneme biogenesis. Additionally, EPS15 reportedly regulates clathrin mediated vesicle budding [68] and detection of the EH domain containing protein (with EPS15 and dynamin domains), implicates this protein in microneme biogenesis. In mammalian systems Rab11 binds myosin V and drives vesicle movement in regulated exocytosis. In T. gondii tachyzoites Rab11a locates at another secretory vesicle (the rhoptry) [3] and our detection of Rab11a in the microneme proteome suggests a conserved role in the secretion of apical organelles. In mammalian cells, NSF and SNAREs form complexes and are important to vesicle fusion [69, 70]. Located at micronemes, NSF and SNAREs may be important to fusion with the plasma membrane. Consistent with this role, PfNSF and mammalian NSF located at the regulated secretory synaptic vesicles, share similar membrane attachment properties [71]. SNAREs are reported to determine specificity to membrane fusion events and accordingly we detect only two SNAREs, VAMP and Syn13, from this large protein family (18 members in Pf). Plasmodium Syn13 is a highly divergent SNARE protein, with not one but two transmembrane regions [72], consistent with a role at specialised vesicles, the micronemes.
In the Apicomplexan T. gondii, microneme exocytosis is reportedly triggered by calcium signaling [18, 73]. We found calcium sensitive PbCDPK4, the orthologue of TgCDPK1 which has been implicated in microneme release [74]. CDPK4 has a putative N-myristoylation site [75] which may target it to microneme membranes, where it may sense the calcium flux, thus triggering exocytosis. Additionally, we identify a hypothetical protein, PB000924.02.0 with EF hand domains, which may point to a similar role [76].
Even though actin polymerisation is not required for microneme exocytoisis [77, 78], we detect glideosome proteins in the microneme proteome; CTRP, aldolase actin, myosin A and MTIP. Aldolase reportedly locates on the cytoplasmic face of micronemes in sporozoites and Toxoplasma tachyzoites [79, 80]. Also, myosin A [81], MTIP [82] and actin [83], all exhibit an apical distribution, consistent with a microneme associated localisation. Additionally, aldolase re-localises with the microneme TRAP-like receptor TgMIC2 during microneme exocytosis [80]. Thus it is possible that glideosome components are delivered to the plasma membrane/subpellicular cavity during the exocytosis of micronemes.
We note with interest that the micronemal protein AMA-1, which is critical to merozoite and sporozoite invasion, is absent from the ookinete (confirmed by IFA, data not shown). AMA-1 is essential to tight junction formation during host invasion [84]. However, it is not clear if the tight junction forms an anchor from which the parasite propels itself forward or if it is important to creation of the PV. Similar to other Apicomplexa the ookinete powers host cell entry, but a PVis not formed. This suggests AMA-1 and the tight junction are not necessary for host entry but may be important to PV formation.
This global approach to the identification of novel microneme proteins develops our understanding of microneme cell biology and identifies novel proteins with potential roles in invasion worthy of further investigation.
Supplementary Material
Table S1. Abundant bands identified by LC-MS/MS.
For each protein details are given including the number of matching peptides, sequence coverage, the Mascot score and the individual peptide sequences, including the charge state and any modification.
Table S2 Proteins detected in the microneme enriched fraction by MudPIT shotgun proteomics
Supplementary Table 3. Spectral counts of rhoptry proteins detected in the P. berghei ookinete microneme and mixed Asexual Blood Stage proteomes.
Acknowledgments
We extend our generous thanks to Isabelle Florent, Philippe Grellier and Chandra Ramakrishnan for use of antibodies and Ken Baker for technical help with preparing EM sections. Thanks to Mark Field and Tony Magee for helpful discussions and BBSRC grant BBS/B/03858 for support of this work. John R Yates III is supported by NIH P41 RR011823 and Judith Helena Prieto is supported by NIH-NIAID grant number R21 AI072615-01.
Abbreviations
- AMA-1
Apical membrane antigen 1
- A-M1
M1 aminopeptidase
- CDPK
Calcium dependent protein kinase
- CTRP
Circumsporozoite and TRAP related protein
- IFA
Immunofluorescence assays
- MAOP
Membrane attack ookinete protein
- MTIP
Myosin A-tail interacting protein
- MudPIT
Multidimensional protein identification technology
- NSF
N-ethylmaleimide-sensitive factor
- Pb
Plasmodium berghei
- PDI
Protein disulphide isomerase
- Pf
Plasmodium falciparum
- PV
Parasitophorous vacuole
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SOAP
Secreted ookinete adhesive protein
- TRAP
Thrombospondin-related anonymous protein
Footnotes
The authors have declared no conflict of interest.
5 References
- 1.Soldati D, Foth BJ, Cowman AF. Molecular and functional aspects of parasite invasion. Trends Parasitol. 2004;20:567–574. doi: 10.1016/j.pt.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Kats LM, Cooke BM, Coppel RL, Black CG. Protein trafficking to apical organelles of malaria parasites – building an invasion machine. Traffic. 2008;9:176–186. doi: 10.1111/j.1600-0854.2007.00681.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradley PJ, Ward C, Cheng SJ, Alexander DL, et al. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in toxoplasma gondii. J. Biol. Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 4.Kawazoe U, Tomley FM, Frazier JA. Fractionation and antigenic characterization of organelles of Eimeria tenella sporozoites. Parasitology. 1992;1:1–9. doi: 10.1017/s003118200006073x. [DOI] [PubMed] [Google Scholar]
- 5.Petry F, Harris JR. Ultrastructure, fractionation and biochemical analysis of Cryptosporidium parvum sporozoites. Int. J. Parasitol. 1999;29:1249–1260. doi: 10.1016/s0020-7519(99)00080-6. [DOI] [PubMed] [Google Scholar]
- 6.Dubremetz JF, Dissous C. Characteristic proteins of micronemes and dense granules from Sarcocystis tenella zoites (Protozoa, Coccidia). Mol. Biochem. Parasitol. 1980;1:279–289. doi: 10.1016/0166-6851(80)90061-4. [DOI] [PubMed] [Google Scholar]
- 7.Pohl U, Dubremetz JF, Entzeroth R. Characterization and immunolocalization of the protein contents of micronemes of Sarcocystis muris cystozoites (Protozoa, Apicomplexa). Parasitol. Res. 1989;75:199–205. doi: 10.1007/BF00931276. [DOI] [PubMed] [Google Scholar]
- 8.Sam-Yellowe TY, Florens L, Wang T, Raine JD, et al. Proteome analysis of rhoptry-enriched fractions isolated from Plasmodium merozoites. J. Proteome Res. 2004;3:995–1001. doi: 10.1021/pr049926m. [DOI] [PubMed] [Google Scholar]
- 9.Bejon P, Mwacharo J, Kai O, Mwangi T, et al. A phase 2b randomised trial of the candidate malaria vaccines FP9 ME-TRAP and MVA ME-TRAP among children in Kenya. PLoS Clin. Trials. 2006;1:e29. doi: 10.1371/journal.pctr.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heppner DG, Jr., Kester KE, Ockenhouse CF, Tornieporth N, et al. Towards an RTS,S-based, multi-stage, multi-antigen vaccine against falciparum malaria: Progress at the Walter Reed Army Institute of Research. Vaccine. 2005;23:2243–2250. doi: 10.1016/j.vaccine.2005.01.142. [DOI] [PubMed] [Google Scholar]
- 11.Pattnaik P, Shakri AR, Singh S, Goel S, et al. Immunogenicity of a recombinant malaria vaccine based on receptor binding domain of Plasmodium falciparum EBA-175. Vaccine. 2006;27:27. doi: 10.1016/j.vaccine.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 12.Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, et al. CTRP is essential for mosquito infection by malaria ookinetes. EMBO J. 1999;18:6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dessens JT, Siden-Kiamos I, Mendoza J, Mahairaki V, et al. SOAP, a novel malaria ookinete protein involved in mosquito midgut invasion and oocyst development. Mol. Microbiol. 2003;49:319–329. doi: 10.1046/j.1365-2958.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 14.Kadota K, Ishino T, Matsuyama T, Chinzei Y, Yuda M. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc. Natl. Acad. Sci. USA. 2004;101:16310–16315. doi: 10.1073/pnas.0406187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol. Microbiol. 2006;59:1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 16.Bannister LH, Hopkins JM, Dluzewski AR, Margos G, et al. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J. Cell Sci. 2003;116:3825–3834. doi: 10.1242/jcs.00665. [DOI] [PubMed] [Google Scholar]
- 17.Schrevel J, Asfaux-Foucher G, Hopkins JM, Robert V, et al. Vesicle trafficking during sporozoite development in Plasmodium berghei: Ultrastructural evidence for a novel trafficking mechanism. Parasitology. 2007;135:1–12. doi: 10.1017/S0031182007003629. [DOI] [PubMed] [Google Scholar]
- 18.Carruthers VB, Sibley LD. Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol. Microbiol. 1999;31:421–428. doi: 10.1046/j.1365-2958.1999.01174.x. [DOI] [PubMed] [Google Scholar]
- 19.Meissner M, Reiss M, Viebig N, Carruthers VB, et al. A family of transmembrane microneme proteins of Toxoplasma gondii contain EGF-like domains and function as escorters. J. Cell Sci. 2002;115:563–574. doi: 10.1242/jcs.115.3.563. [DOI] [PubMed] [Google Scholar]
- 20.Sinden RE, Butcher GA, Beetsma A. In: Malaria Methods and Protocols, Methods in Molecular Medicine. Doolan DL, editor. Humana Press Inc; Totowa, New Jersey: 2002. pp. 25–40. [Google Scholar]
- 21.Sinden RE, Hartley RH, Winger L. The development of Plasmodium ookinetes in vitro: An ultrastructural study including a description of meiotic division. Parasitology. 1985;91:227–244. doi: 10.1017/s0031182000057334. [DOI] [PubMed] [Google Scholar]
- 22.Weibel ER, Kistler GS, Scherle WF. Practical stereo-logical methods for morphometric cytology. J. Cell Biol. 1966;30:23–38. doi: 10.1083/jcb.30.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanderson SJ, Xia D, Prieto H, Yates J, et al. Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics. 2008;27:27. doi: 10.1002/pmic.200700804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: Single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 2002;219:245–251. [Google Scholar]
- 25.Link AJ, Eng J, Schieltz DM, Carmack E, et al. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 26.Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., III Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 27.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 28.Bern M, Goldberg D, McDonald WH, Yates JR., III Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20:i49–i54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 29.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 30.Hall N, Karras M, Raine JD, Carlton JM, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 31.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 33.Ruepp A, Zollner A, Maier D, Albermann K, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Florens L, Washburn MP, Raine JD, Anthony RM, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 35.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 36.Arai M, Mitsuke H, Ikeda M, Xia JX, et al. ConPred II: A consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 38.Kumar N, Koski G, Harada M, Aikawa M, Zheng H. Induction and localization of Plasmodium falciparum stress proteins related to the heat shock protein 70 family. Mol. Biochem. Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-z. [DOI] [PubMed] [Google Scholar]
- 39.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 40.Hager KM, Striepen B, Tilney LG, Roos DS. The nuclear envelope serves as an intermediary between the ER and Golgi complex in the intracellular parasite Toxoplasma gondii. J. Cell Sci. 1999;112:2631–2638. doi: 10.1242/jcs.112.16.2631. [DOI] [PubMed] [Google Scholar]
- 41.Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. Deciphering apicoplast targeting signals-feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280:19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 42.Brameier M, Krings A, MacCallum RM. NucPred-predicting nuclear localization of proteins. Bioinformatics. 2007;23:1159–1160. doi: 10.1093/bioinformatics/btm066. [DOI] [PubMed] [Google Scholar]
- 43.Bender A, van Dooren GG, Ralph SA, McFadden GI, Schneider G. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol. Biochem. Parasitol. 2003;132:59–66. doi: 10.1016/j.molbiopara.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Spano F, Matsuoka H, Ozawa R, Chinzei Y, Sinden RE. Epitope mapping on the ookinete surface antigen Pbs21 of Plasmodium berghei: Identification of the site of binding of transmission-blocking monoclonal antibody 13.1. Parassitologia. 1996;38:559–563. [PubMed] [Google Scholar]
- 45.Florent I, Derhy Z, Allary M, Monsigny M, et al. A Plasmodium falciparum aminopeptidase gene belonging to the M1 family of zinc-metallopeptidases is expressed in erythrocytic stages. Mol. Biochem. Parasitol. 1998;97:149–160. doi: 10.1016/s0166-6851(98)00143-1. [DOI] [PubMed] [Google Scholar]
- 46.Mouray E, Moutiez M, Girault S, Sergheraert C, et al. Biochemical properties and cellular localization of Plasmodium falciparum protein disulfide isomerase. Biochimie. 2007;89:337–346. doi: 10.1016/j.biochi.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Tomley F. Techniques for isolation and characterization of apical organelles from Eimeria tenella sporozoites. Methods. 1997;13:171–176. doi: 10.1006/meth.1997.0509. [DOI] [PubMed] [Google Scholar]
- 48.Carter R, Kaushal DC. Characterization of antigens on mosquito midgut stages of Plasmodium gallinaceum. III. Changes in zygote surface proteins during transformation to mature ookinete. Mol. Biochem. Parasitol. 1984;13:235–241. doi: 10.1016/0166-6851(84)90116-6. [DOI] [PubMed] [Google Scholar]
- 49.Bannister LH, Hopkins JM, Fowler RE, Krishna S, Mitchell GH. Ultrastructure of rhoptry development in Plasmodium falciparum erythrocytic schizonts. Parasitology. 2000;121:273–287. doi: 10.1017/s0031182099006320. [DOI] [PubMed] [Google Scholar]
- 50.Stewart MJ, Schulman S, Vanderberg JP. Rhoptry secretion of membranous whorls by Plasmodium falciparum merozoites. Am. J. Trop. Med. Hyg. 1986;35:37–44. doi: 10.4269/ajtmh.1986.35.37. [DOI] [PubMed] [Google Scholar]
- 51.Stewart MJ, Schulman S, Vanderberg JP. Rhoptry secretion of membranous whorls by Plasmodium berghei sporozoites. J. Protozool. 1985;32:280–283. doi: 10.1111/j.1550-7408.1985.tb03051.x. [DOI] [PubMed] [Google Scholar]
- 52.Li F, Templeton TJ, Popov V, Comer JE, et al. Plasmodium ookinete-secreted proteins secreted through a common micronemal pathway are targets of blocking malaria transmission. J. Biol. Chem. 2004;279:26635–26644. doi: 10.1074/jbc.M401385200. [DOI] [PubMed] [Google Scholar]
- 53.Yuda M, Yano K, Tsuboi T, Torii M, Chinzei Y. von Willebrand Factor A domain-related protein, a novel micro-neme protein of the malaria ookinete highly conserved throughout Plasmodium parasites. Mol. Biochem. Parasitol. 2001;116:65–72. doi: 10.1016/s0166-6851(01)00304-8. [DOI] [PubMed] [Google Scholar]
- 54.Han YS, Thompson J, Kafatos FC, Barillas-Mury C. Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: The time bomb theory of ookinete invasion of mosquitoes. EMBO J. 2000;19:6030–6040. doi: 10.1093/emboj/19.22.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allary M, Schrevel J, Florent I. Properties, stage-dependent expression and localization of Plasmodium falciparum M1 family zinc-aminopeptidase. Parasitology. 2002;125:1–10. doi: 10.1017/s0031182002001828. [DOI] [PubMed] [Google Scholar]
- 56.Dalal S, Klemba M. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum. J. Biol. Chem. 2007;282:35978–35987. doi: 10.1074/jbc.M703643200. [DOI] [PubMed] [Google Scholar]
- 57.Flipo M, Beghyn T, Leroux V, Florent I, et al. Novel selective inhibitors of the zinc plasmodial aminopeptidase PfA-M1 as potential antimalarial agents. J. Med. Chem. 2007;50:1322–1334. doi: 10.1021/jm061169b. [DOI] [PubMed] [Google Scholar]
- 58.Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc. Natl. Acad. Sci. USA. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh S, Plassmeyer M, Gaur D, Miller LH. Mononeme: A new secretory organelle in Plasmodium falciparum merozoites identified by localization of rhomboid-1 protease. Proc. Natl. Acad. Sci. USA. 2007;104:20043–20048. doi: 10.1073/pnas.0709999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patra KP, Johnson JR, Cantin GT, Yates JR, III, Vinetz JM. Proteomic analysis of zygote and ookinete stages of the avian malaria parasite Plasmodium gallinaceum delineates the homologous proteomes of the lethal human malaria parasite Plasmodium falciparum. Proteomics. 2008;8:2492–2499. doi: 10.1002/pmic.200700727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naguleswaran A, Alaeddine F, Guionaud C, Vonlaufen N, et al. Neospora caninum protein disulfide isomerase is involved in tachyzoite-host cell interaction. Int. J. Parasitol. 2005;35:1459–1472. doi: 10.1016/j.ijpara.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Meek B, Back JW, Klaren VN, Speijer D, Peek R. Protein disulfide isomerase of Toxoplasma gondii is targeted by mucosal IgA antibodies in humans. FEBS Lett. 2002;522:104–108. doi: 10.1016/s0014-5793(02)02911-3. [DOI] [PubMed] [Google Scholar]
- 63.Meek B, Back JW, Klaren VN, Speijer D, Peek R. Conserved regions of protein disulfide isomerase are targeted by natural IgA antibodies in humans. Int. Immunol. 2002;14:1291–1301. doi: 10.1093/intimm/dxf091. [DOI] [PubMed] [Google Scholar]
- 64.Zhou XW, Kafsack BF, Cole RN, Beckett P, et al. The opportunistic pathogen Toxoplasma gondii deploys a diverse legion of invasion and survival proteins. J. Biol. Chem. 2005;280:34233–34244. doi: 10.1074/jbc.M504160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai CW, Duggan PF, Shimp RL, Jr., Miller LH, Narum DL. Overproduction of Pichia pastoris or Plasmodium falciparum protein disulfide isomerase affects expression, folding and O-linked glycosylation of a malaria vaccine candidate expressed in P. pastoris. J. Biotechnol. 2006;121:458–470. doi: 10.1016/j.jbiotec.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 66.Hou JC, Pessin JE. Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr. Opin. Cell Biol. 2007;19:466–473. doi: 10.1016/j.ceb.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonanomi D, Benfenati F, Valtorta F. Protein sorting in the synaptic vesicle life cycle. Prog. Neurobiol. 2006;80:177–217. doi: 10.1016/j.pneurobio.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Tebar F, Sorkina T, Sorkin A, Ericsson M, Kirchhausen T. Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J. Biol. Chem. 1996;271:28727–28730. doi: 10.1074/jbc.271.46.28727. [DOI] [PubMed] [Google Scholar]
- 69.Igarashi M, Watanabe M. Roles of calmodulin and calmodulin-binding proteins in synaptic vesicle recycling during regulated exocytosis at submicromolar Ca21 concentrations. Neurosci. Res. 2007;58:226–233. doi: 10.1016/j.neures.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 70.Zhao C, Slevin JT, Whiteheart SW. Cellular functions of NSF: Not just SNAPs and SNAREs. FEBS Lett. 2007;581:2140–2149. doi: 10.1016/j.febslet.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayashi M, Taniguchi S, Ishizuka Y, Kim HS, et al. A homologue of N-ethylmaleimide-sensitive factor in the malaria parasite Plasmodium falciparum is exported and localized in vesicular structures in the cytoplasm of infected erythrocytes in the brefeldin A-sensitive pathway. J. Biol. Chem. 2001;276:15249–15255. doi: 10.1074/jbc.M011709200. [DOI] [PubMed] [Google Scholar]
- 72.Ayong L, Pagnotti G, Tobon AB, Chakrabarti D. Identification of Plasmodium falciparum family of SNAREs. Mol. Biochem. Parasitol. 2007;152:113–122. doi: 10.1016/j.molbiopara.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Chini EN, Nagamune K, Wetzel DM, Sibley LD. Evidence that the cADPR signaling pathway controls calcium-mediated microneme secretion in Toxoplasma gondii. Biochem. J. 2005;389:269–277. doi: 10.1042/BJ20041971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kieschnick H, Wakefield T, Narducci CA, Beckers C. Toxoplasma gondii attachment to host cells is regulated by a calmodulin-like domain protein kinase. J. Biol. Chem. 2001;276:12369–12377. doi: 10.1074/jbc.M011045200. [DOI] [PubMed] [Google Scholar]
- 75.Nagamune K, Sibley LD. Comparative genomic and phylogenetic analyses of calcium ATPases and calcium-regulated proteins in the apicomplexa. Mol. Biol. Evol. 2006;23:1613–1627. doi: 10.1093/molbev/msl026. [DOI] [PubMed] [Google Scholar]
- 76.Honore B, Vorum H. The CREC family, a novel family of multiple EF-hand, low-affinity Ca(21)-binding proteins localised to the secretory pathway of mammalian cells. FEBS Lett. 2000;466:11–18. doi: 10.1016/s0014-5793(99)01780-9. [DOI] [PubMed] [Google Scholar]
- 77.Carruthers VB, Sherman GD, Sibley LD. The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J. Biol. Chem. 2000;275:14346–14353. doi: 10.1074/jbc.275.19.14346. [DOI] [PubMed] [Google Scholar]
- 78.Bumstead J, Tomley F. Induction of secretion and surface capping of microneme proteins in Eimeria tenella. Mol. Biochem. Parasitol. 2000;110:311–321. doi: 10.1016/s0166-6851(00)00280-2. [DOI] [PubMed] [Google Scholar]
- 79.Buscaglia CA, Coppens I, Hol WG, Nussenzweig V. Sites of interaction between aldolase and thrombospondin-related anonymous protein in Plasmodium. Mol. Biol. Cell. 2003;14:4947–4957. doi: 10.1091/mbc.E03-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jewett TJ, Sibley LD. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- 81.Pinder JC, Fowler RE, Dluzewski AR, Bannister LH, et al. Actomyosin motor in the merozoite of the malaria parasite, Plasmodium falciparum: Implications for red cell invasion. J. Cell Sci. 1998;111:1831–1839. doi: 10.1242/jcs.111.13.1831. [DOI] [PubMed] [Google Scholar]
- 82.Green JL, Martin SR, Fielden J, Ksagoni A, et al. The MTIP-Myosin a complex in blood stage malaria parasites. J. Mol. Biol. 2006;355:933–941. doi: 10.1016/j.jmb.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 83.Siden-Kiamos I, Pinder JC, Louis C. Involvement of actin and myosins in Plasmodium berghei ookinete motility. Mol. Biochem. Parasitol. 2006;150:308–317. doi: 10.1016/j.molbiopara.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 84.Alexander DL, Mital J, Ward GE, Bradley P, Boot-hroyd JC. Identification of the moving junction complex of Toxoplasma gondii: A collaboration between distinct secre-tory organelles. PLoS Pathog. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Abundant bands identified by LC-MS/MS.
For each protein details are given including the number of matching peptides, sequence coverage, the Mascot score and the individual peptide sequences, including the charge state and any modification.
Table S2 Proteins detected in the microneme enriched fraction by MudPIT shotgun proteomics
Supplementary Table 3. Spectral counts of rhoptry proteins detected in the P. berghei ookinete microneme and mixed Asexual Blood Stage proteomes.


