Abstract
The E. coli signal peptide peptidase A (SppA) is a serine protease which cleaves signal peptides after they have been proteolytically removed from exported proteins by signal peptidase processing. We present here results of site-directed mutagenesis studies of all the conserved serines of SppA in the carboxyl-terminal domain showing that only Ser 409 is essential for enzymatic activity. Also, we show that the serine hydrolase inhibitor FP-biotin inhibits SppA and modifies the protein, but does not label the mutant S409A with an alanine substituted for the essential serine. These results are consistent with Ser 409 being directly involved in the proteolytic mechanism. Remarkably, additional site-directed mutagenesis studies showed that none of the lysines or histidine residues in the carboxyl-terminal protease domain is critical for activity, suggesting this domain lacks the general base residue required for proteolysis. In contrast, we found that E. coli SppA has a conserved lysine K209 in the N-terminal-domain that is essential for activity and important for activation of S409 for reactivity toward the FP-biotin inhibitor, and is conserved in those other bacterial SppA proteins that have an N-terminal domain. We also performed alkaline phosphatase fusion experiments that establish that SppA has only one transmembrane segment with the C-terminal domain protruding into the periplasmic space. These results support the idea that the E. coli SppA is a Ser-Lys dyad protease, with the Lys recruited to the amino-terminal domain that is itself not present in most known SppA sequences.
Almost all proteins that are exported to the cell surface of gram-negative and grampositive bacteria are synthesized in a higher molecular weight precursor form with an amino-terminal cleavable signal peptide. During protein export, the signal peptide is cleaved from the precursor by the membrane protease signal peptidase (1). The cleaved signal peptide can be further digested by signal peptide hydrolases (2, 3).
In 1984, the protease that degrades the lipoprotein signal peptide, named signal peptide peptidase A (SppA), was purified to homogeneity (4) and shown to correspond to the previously identified inner membrane protein, protease IV (5). SppA was subsequently cloned and sequenced, and the gene-encoded protein was shown to be a tetramer (6). Sequence analysis of E. coli SppA reveals three hydrophobic regions that are candidate membrane-spanning domains. SppA is inhibited by serine protease inhibitors such as chymostatin, leupeptin, antipain, and elastinal (4). However, unlike typical serine proteases, the SppA family of proteins does not have a conserved histidine residue, suggesting SppA is an atypical serine protease.
The E. coli SppA protease has been classified in the S49 family of proteases in the MEROPS protease database (7), specifically to the S49.001 subfamily. This group, like S49.004, contains, in addition to the carboxyl-terminal protease domain that is conserved in all the S49 family members, an amino-terminal domain. The other subfamily members--S49.002, S49.003, S49.005 and S49.006-- do not contain this amino-terminal domain. Members of these groups, respectively, include the sohB peptidase (8), protein C (9), protein 1510-N (10), and the archaeal signal peptide peptidase (11).
Recently, site-directed mutagenesis has been utilized (12) to identify the catalytically important residues of the archaeal SppA protease from T. kodakaraensis which contain only the C-terminal protease domain. They showed that Ser 162 and Lys 214 were critical for activity, suggesting that this protease may use a Serine-Lysine dyad for catalysis like signal peptidase 1 (13), LexA (14), UmuD (15), Tsp (16), Lon Protease (17) and VP4 protease (18). However, the E. coli SppA lacks a lysine at the homologous position as the Lys 214.
In this report, we have employed site-directed mutagenesis to identify possible active site residues for the E. coli SppA. We show within the carboxyl-terminal protease domain of the E. coli SppA that Ser 409, which is homologous to the critical Ser 162 of the T. kodakaraensis SppA (12), is essential for activity. The wild-type SppA, but not an S409 mutant where the serine is substituted for alanine can be covalently modified with the FP-biotin serine hydrolase inhibitor. These results support the notion that Ser 409 is the active site residue in the E. coli enzyme. None of the conserved amino acid residues within the protease domain that have ionizable side chains, including all the histidine and lysine residues, are critical for activity. Strikingly the invariant Lys 209 residue located in the amino-terminal domain is indispensable for activity and important for activation of S409. Alkaline phosphatase fusion approach suggests that the E. coli SppA, which was predicted to span the membrane three times by most topology programs, only spans the membrane once with its carboxyl-terminus localized to the periplasmic space. Taken together, the results provide evidence that the single-spanning E. coli SppA carries out catalysis using a Serine-Lysine dyad with the serine located in the conserved carboxyl-terminal protease domain and the lysine in the non-conserved amino-terminal domain.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes and T4 ligase were purchased from New England Biolabs. Pfu polymerase was obtained from Stratagene. Oligonucleotides were ordered from Integrated DNA Technologies, Inc. and the QIAprep Spin Miniprep Kit for the plasmid purification was purchased from Qiagen. TALON™ Metal Affinity Resins were ordered from Clontech. N-benzyloxycarbonyl-L-valine-p-nitrophenyl ester (Cbz-Val-ONP) was purchased from Research Organics.
Bacterial Strains and Plasmids
Escherichia coli strains BL21(DE3), DH5α, and MC1061 [DlacX74, araD139, D(ara, leu)7697, galU, galk, hsr, hsm, strA] were from our collection. Lin 205 [fhuA22, phoA8, ompF627(T2R), glpA24, fadL701(T2R), relA1, glpR8(glpc), glpR7 (glpn), glpD26, pit-10, spoT1] was obtained from E. coli Genetic Stock Center. The cloning vector pCR 2.1 was purchased from Invitrogen and the expression vector pET-28(a) was from our laboratory. The pING-1 vector with the araB promoter and the araC regulatory elements was from our collection (19).
Cloning and mutagenesis of E. coli SppA
The SppA gene was cloned by PCR method into the pET-28(a) expression vector. The E. coli chromosomal DNA from MC1061 [DlacX74, araD139, D(ara, leu)7697, galU, galk, hsr, hsm, strA] was used as the template. The PCR was run for 32 cycles (95°C for 1 min, 46°C for 1 min, and 72°C for 1 min and 30 s) with the sense primer; 5’- AAG TTG GGA GAA CAT ATG CGA ACC CTT TGG CG -3’, and the antisense primer; 5’- TCA GTA CAA AAG CTT ACG CAT GTT GGC GCA GGT C -3’ for SppA. The underlined sequences specify for the restriction sites for the endonuclease NdeI and HindIII, respectively. The PCR product was cloned into the cloning vector, pCR 2.1 (ampicillin resistant). SppA was then subcloned into the NdeI/HindIII sites of the pET-28(a) expression vector (kanamycin resistant). The pCR 2.1 plasmids bearing SppA gene were digested with NdeI and HindIII and the DNA fragment containing sppA was isolated by excision from an agarose gel. The pET-28(a) vector was prepared in the same way. The ligation product was transformed into E. coli host BL21(DE3) cells. The resulting DNA was sent to the Plant-Microbe genomics sequencing facility at the Ohio State University for confirmation.
Site-directed mutagenesis was carried out with the SppA-pET-28(a) vector as template using the QuickChange mutagenesis PCR method (Stratagene Inc) to incorporate different amino acid residues. Mutations were verified by DNA sequencing.
Purification of E. coli SppA
E. coli BL21(DE3) cells harboring 6-His tagged SppA encoded by the pET-28(a) vector were used for overexpression of SppA. Wild-type and mutants of SppA were expressed and purified using cobalt affinity chromatography. A single colony was used to inoculate 100 ml LB broth containing 25 µg/ml kanamycin. 20 ml of this overnight cell culture was diluted into 2 L of LB broth with the same concentration of kanamycin. The cultures were grown at 37°C until OD600 = 0.6 at which time they were induced with 1 mM IPTG (final concentration). After 5 h of further growth, the cells were harvested by centrifugation at 5,000 rpm for 10 min with JA-10 rotor. The cells were resuspended in lysis buffer (1 M NaCl, 25% sucrose, 10 mM sodium phosphate, pH 7.0) and lysed by ultrasonication. After removal of unbroken cells, the membrane fraction was spun in the centrifuge at 45,000 rpm for 3 h using a Ti-70 rotor. The membrane fraction was then resuspended in extraction buffer (0.5 M NaCl, 0.5% TX-100, 40% glycerol, 10 mM sodium phosphate, pH 7.0) and stirred at 4°C until the pellet was completely homogenized. After repeating the centrifugation step once more, the supernatant was isolated and applied to the affinity column. The Triton X-100 extract was loaded on the column with 5 ml bed volume of TALON cobalt resin, which was equilibrated with the extraction buffer. The column was washed with wash buffer (300 mM NaCl, 0.5% Triton X-100, 5 mM Imidazole, 50 mM sodium phosphate, pH 7.0). The fusion protein was eluted with elution buffer (300 mM NaCl, 0.5% Triton X-100, 5 mM imidazole, 50 mM sodium phosphate, pH 7.0) with increasing concentration of imidazole (50 mM, 100 mM and 150 mM). Following elution, proteins were assayed for purity using a 12% SDS-PAGE gel and selected fractions were dialyzed four times with 1 L of 50 mM sodium phosphate, pH 7.0 with 1% Triton X-100 buffer to remove the imidazole.
Assay of E. coli SppA activity
The activity of SppA, wild-type and mutants, was measured against the chromophoric substrate Cbz-Val-ONP as follows (5). A total of 10 µg of purified enzyme was preincubated with 0.2% Triton X-100 and 10 mM sodium phosphate (pH 7.2) for 10 min at room temperature, then 1 µl of 100 mM Cbz-Val-ONP was added. The reaction mixture (total volume of 1.0 ml) was incubated for 10 min at room temperature. The rate of p-nitrophenol release was monitored at 400 nm on a Perkin Elmer UV/VIS spectrophotometer.
FP-Biotin Reaction
A biotinylated fluorophosphonate (FP-Biotin) chemical reagent was used to inactivate SppA. The reaction of wild-type, S409A and K209A SppA mutants with FP-Biotin was performed as described in Liu et al., 1999 (20) with the following modifications. 20 µl of 0.1 mg/ml SppA was treated with 40 µM of FP-Biotin (final concentration) for 30 min at 37°C. The reaction mixture was then divided into two aliquots. One aliquot was used for the activity assay. The other aliquot was used for the immunoblotting study after the reaction was quenched by the addition of 2X SDS-PAGE loading buffer. The sample was analyzed on a 12% SDS-PAGE gel and the biotinmodified SppA was detected using the ECL Western blot detection kit (Amersham Biosciences). The SppA modified with biotin was identified with an avidin-horseradish peroxidase conjugate.
Construction of SppA-PhoA fusions
In order to produce the plasmid containing the full length SppA-PhoA fusion, the SppA gene was excised from SppA-pET-28(a) by digestion with NdeI and NotI. A pING plasmid bearing a gene encoding the mature phoA fusion protein was digested by the same restriction enzymes. The two were then ligated to yield the full length SppA-PhoA construct. Site directed mutagenesis was used to create a series of SppA-phoA fusions in which various lengths of the 3′ end of the sppA gene were deleted. For a negative control, we used a pING construct in which PhoA was attached immediately after the first transmembrane segment (H1) of leader peptidase in the cytoplasmic domain (21).
Alkaline phosphatase assay of the SppA-PhoA constructs and expression
The alkaline phosphatase activity of the fusion proteins was determined by a plate assay and by a spectroscopic enzyme assay. For the plating assay, cells transformed with the appropriate pING derived plasmids encoding the fusion protein were grown overnight in LB agar plate supplemented with 100 µg/ml ampicillin, 0.2% arabinose and 40 µg/ml 5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt. The enzyme assay was performed as described in (22). The cell culture were grown overnight in LB media supplemented with 100 µg/ml ampicillin, then back diluted to 2 ml of LB media and grown at 37°C until the absorbance was OD600 = 0.3. The cells were induced with 0.2% arabinose and grown for an additional 2 h at 37°C. The OD600 of the cells was measured and the cells were placed on ice to inhibit further growth. The 1 ml cell culture sample was pelleted and washed once with 1 ml of LB media and 1 ml of 1 M Tris-HCl, pH 8.0. The cells were permeabilized by adding one drop of 0.1% SDS and three drops of chloroform and incubating at 37°C for 5 min at which time the OD420 was measured. The reaction was initiated by adding 10 µl of p-nitrophenyl phosphate (40 µg/ml) and incubating at room temperature for 5 min. Absorption was measured using a Lambda 20 UV/VIS spectroscopy (Perkin Elmer) and the alkaline phosphatase activity was determined by using the following equations: AP total activity = ΔOD420/5min ×1000; normalized AP activity = AP total activity/OD600 of cells.
To measure the steady state cellular levels of the SppA-phoA fusion proteins, immunoblot analysis was performed using anti-phoA antiserum. Cell were normalized from 1 ml cultures and added directly to SDS-PAGE loading buffer. PhoA was detected by immunoblotting using the ECL Western blot detection kit (Amersham Biosciences).
RESULTS
Elucidating the catalytically important conserved residues of the E. coli SppA
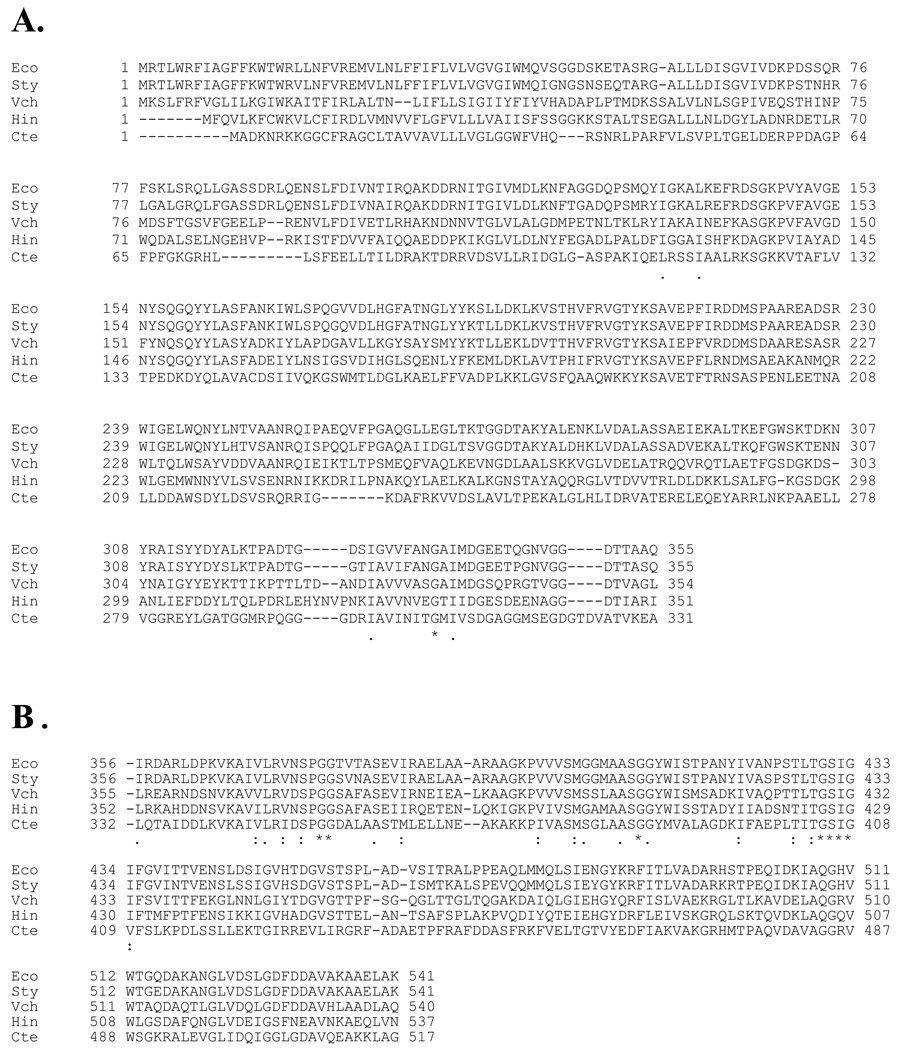
The serine protease SppA has a conserved carboxyl-terminal protease domain found in all S49 family members (Fig. 1B) and a non-conserved amino-terminal domain (Fig. 1A) found in the S49.001 and S49.004 groups. Recently, the archeon Thermococcus kodakaraensis SppA has been classified as a Serine-Lysine dyad protease (12). However, while the Ser residue found in the T. kodakaraensis lines up with the Ser409 residue in the E. coli SppA, the E. coli enzyme does not have a lysine at the homologous position as the archaeal protease. There are also no conserved histidine residues in the SppA family of proteases (Fig. 1; see sequence alignment), which could function as the general base in the Ser/His/Asp triad of conventional serine proteases. However, Arg 496 and Asp524 are absolutely conserved in the S49 family (Fig. 1B) and could be possible members of a variant catalytic triad or a novel Serine-Arginine dyad.
FIGURE 1. Multiple alignment of the amino acid sequences of SppA S49.001 members.
A. The amino-terminal domain within the S49.001 family. B. The carboxyl-terminal protease domain within the S49.001 family. Shown are the sequences for Eco, Escherichia coli; Sty, Salmonella typhimurium; Vch, Vibrio cholerae; Hin, Haemophilus influenzae; Cte, Chlorobium tepidum. Shown below the sequences are the consensus motifs for the entire S49.001 group. “*” denotes that the amino acids in the S49.001 are identical in all sequences in the alignment. “:” means that conserved substitutions in the S49.001 group have been observed, according to the nature of the amino acid (hydrophobic, hydrophilic, acidic or basic).“.” means that semi-conserved substitutions in the S49.001 group are found. The shaded residues in the carboxyl-terminal domain are invariant in the S49 family. The shaded residue in the amino-terminal domain is absolutely conserved when the amino-terminal domain is present.
To identify the serine residue that could function as the active site nucleophile in the E. coli SppA, we mutated the highly conserved Ser 374, Ser 402, Ser 409, and Ser 431 residues individually to Ala residues by site-directed mutagenesis. Mutant and wild-type sequence constructs with N-terminal 6-His tags were cloned into a pET-28a vector and high levels of expression observed. Expressed proteins were purified to homogeneity (Fig. 2) using cobalt affinity chromatography (see “Experimental procedures”). Protease activities were assayed using the chromophoric substrate N-benzyloxycarbonyl-L-valine-p-nitrophenyl ester (Cbz-Val-ONP) (Fig. 3). With the wild-type SppA protein, hydrolysis of the Cbz-Val-ONP substrate was rapid and linear with time (Fig. 3A). Activity was also observed with the Ser 374, Ser 402 and Ser 431 alanine mutants. In contrast, the Ser 409 mutant had background activity, indicating the Ser 409 is critical for enzymatic activity.
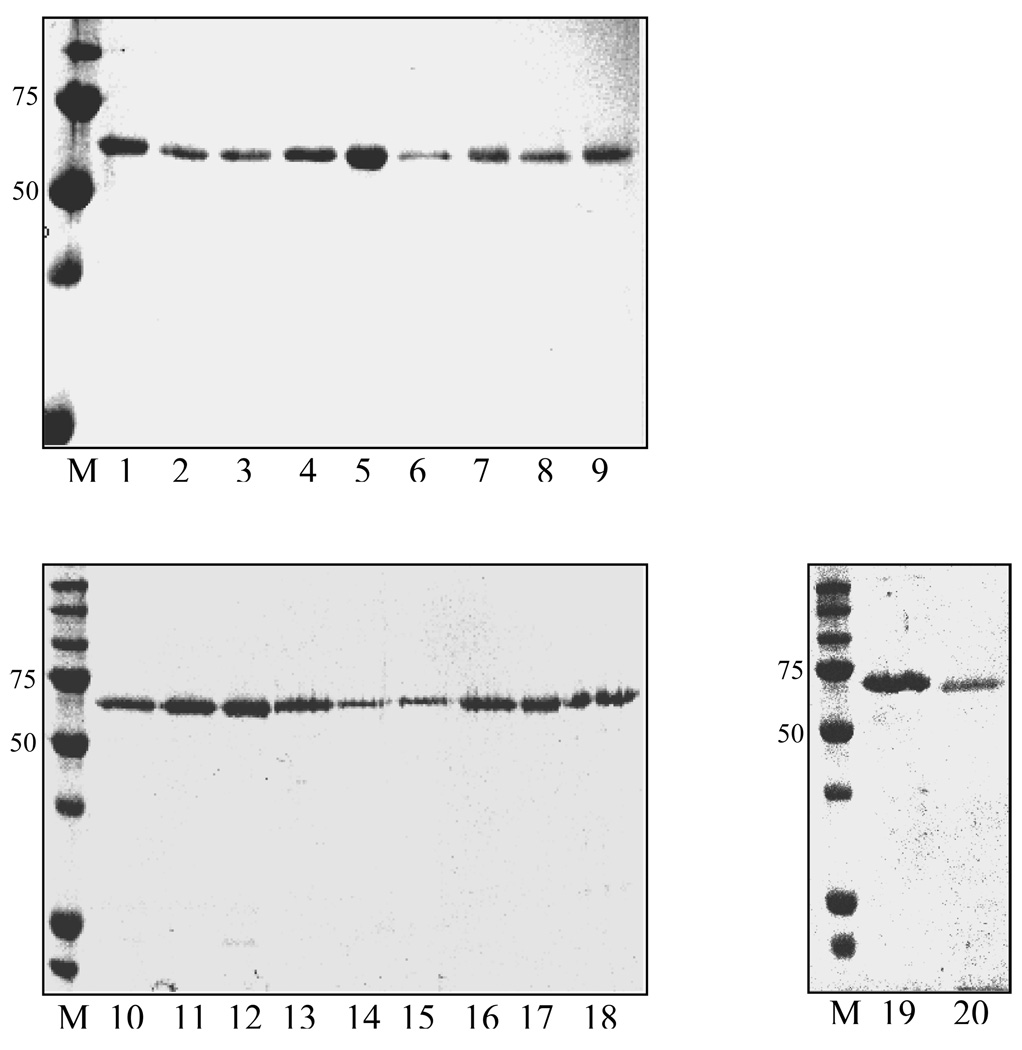
FIGURE 2. SDS-polyacrylamide gel electrophoresis analysis of the purified wild-type and mutant SppA.
The purified enzymes were run on 12% SDS-polyacrylamide gel and stained with Coomassie Blue R-250 M:Marker 1:WT 2:S374A 3:S402A 4:S409A 5:S431A 6:R496K 7:R496A 8:K366A 9:K397A 10:H451A 11:H497A 12:H510A 13: K364A 14: K535A 15:K486A 16:K505A 17:K518A 18:K541A 19:K209A 20 D524N
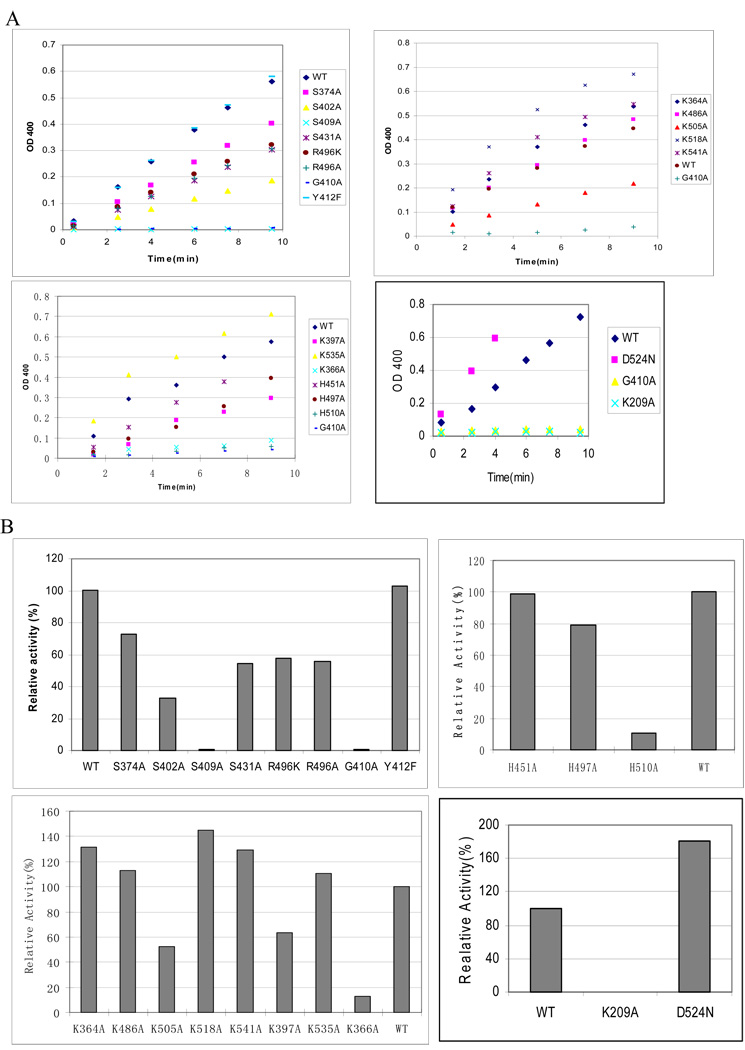
FIGURE 3. Activities of wild-type SppA and mutants.
A. The hydrolysis of the Cbz-Val-ONP substrate was measured with the wild-type and mutants SppA. The slopes correspond to the hydrolysis reaction velocity. B. Relative activity level of each mutant was calculated. The activity level of the wild-type SPPA was designated as 100%.
To assess the role in proteolysis of the invariant Arg 496 and Asp 524 amino acids of SppA, we altered these residues. Fig 3 shows that the R496 is not required for SppA activity; substitution with lysine or alanine does not lead to a marked reduction in activity. Neither is Asp 524 important for activity as the SppA D524N mutant had no reduction in activity (Fig. 3) In fact, the activity of this mutant was reproducibly higher than the wild-type SppA enzyme.
We also investigated the highly conserved Tyr 412 for its requirement for activity since it could act as a general base. Fig. 3 shows that Y412F SppA was fully active, ruling out a role of this residue as a general base. Interestingly, the Gly 410 adjacent to the catalytically important Ser 409 is critical for optimal activity. SppA with the glycine mutated to alanine severely perturbs the SppA activity. Substitution of the non-conserved histidine at positions 451 and 497 had only a small effect on activity whereas mutation of histidine at position 510 impairs activity roughly 20-fold, although it still exhibits measurable activity. The data together indicates that there is no histidine in the C-terminal domain of SppA that is essential for activity.
Since the T. kodakaraensis SppA has been proposed to use a Serine-Lysine mechanism, we assessed the catalytic role of lysine residues in the E. coli SppA by mutating the C-terminal domain Lys 364, Lys 366, Lys 397, Lys 486, Lys 505, Lys 518, Lys 535, and Lys 541 to Ala residues. As can be seen in Fig. 3B, SppA enzymes with these single mutations all maintained activity, indicating these residues are not essential for catalysis. Only the K366A mutation had large (5-fold) affect on catalysis. These results rule out that a lysine residue within the carboxyl-terminal domain is the general base residue.
We next tested whether the absolutely conserved Lys 209 in the amino-terminal domain of the E. coli SppA is crucial for activity. Strikingly, mutation of Lys 209 to alanine inactivates the SppA protease. The data shows that Lys 209 in the amino-terminal domain is essential for the proteolytic reaction and is consistent with the E. coli SppA employing a lysine in catalysis.
Inhibition of SppA by FP-Biotin
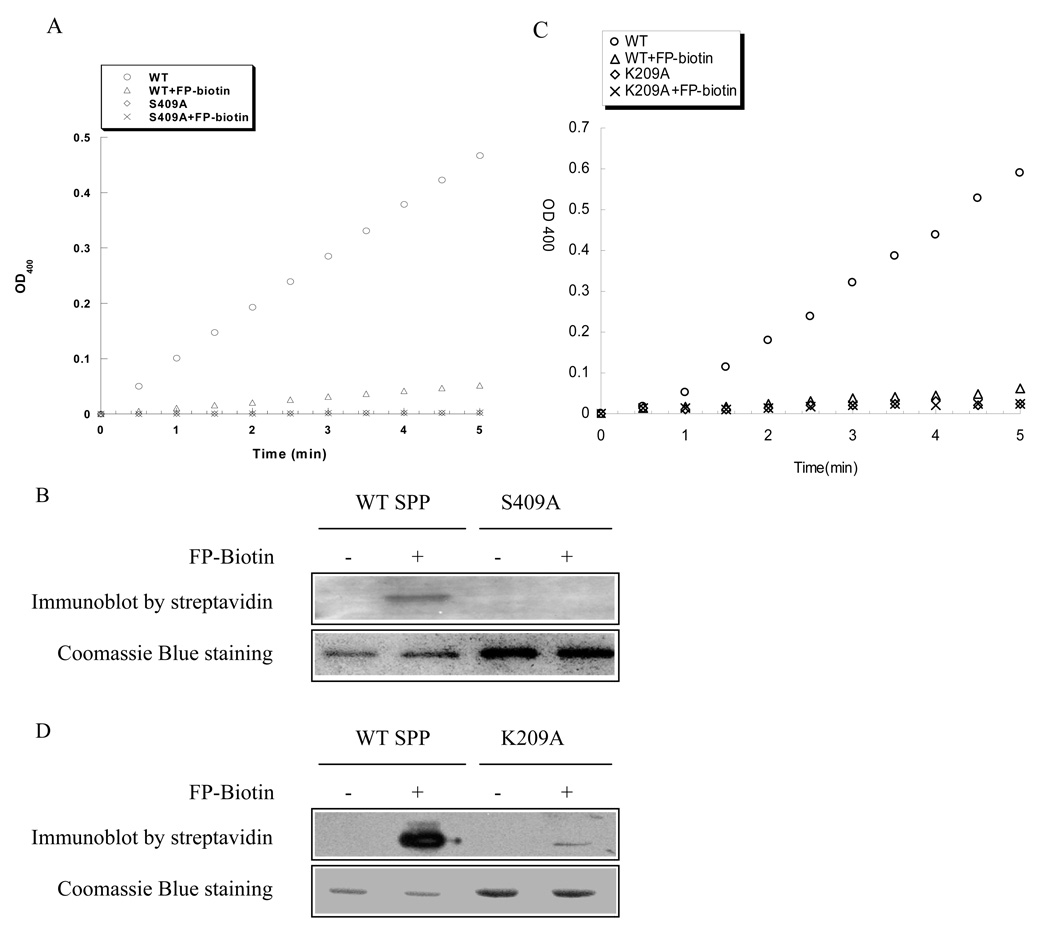
To provide further evidence that Ser 409 is the active site serine residue in the E. coli SppA enzyme, we tested whether the wild-type SppA, unlike the Ser409Ala mutant, is modified with the biotinylated fluorophosphonate inhibitor (FP-biotin), which has been shown to inhibit serine hydrolases (20). First, we tested whether the reagent FP-biotin inhibited the SppA enzyme. Fig. 4A shows that the addition of FP-biotin significantly decreased the activity of the wild-type enzyme compared with the mock treated enzyme. Next, we examined whether the FP-biotin treated enzyme is modified with the inhibitor, but remains unchanged when the candidate active site Ser 409 residue is mutated. As can be seen in Fig. 4B, a strepavidin reactive band is detected with the wild-type SppA, but not with the S409A SppA mutant where the active site serine has been substituted with an alanine. These results support the notion that serine 409 is indeed the active site residue. Likewise, we examined whether Lys 209 is critical for activation of the Ser such that it can react with the FP-biotin inhibitor. Fig. 4D shows that the K209A SppA mutant only poorly reacts with the inhibitor in comparison to the WT SppA.
FIGURE 4. Inhibition and modification of wild-type SppA with FP-biotin.
Purified wild-type, S409A and SppA proteins were incubated in the presence or absence of FP-biotin (final 40 µM) for 10 min, followed by activity assays (A and C) or analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with an avidin-horseradish peroxidase conjugate (B and D).
Topology of the E. coli SppA
Hydrophobicity analysis and topology programs do not provide a clear indication of the membrane orientation of the E. coli SppA. The topology program TMHMM predicts one transmembrane segment from residues 21–43 (23) while the TopPred2 program predicts 29–45, 398–414, and 421–441 are transmembrane segments (24). Both programs predict the amino-terminus of SppA is localized to the cytoplasm and the carboxyl-terminus protrudes into the periplasm.
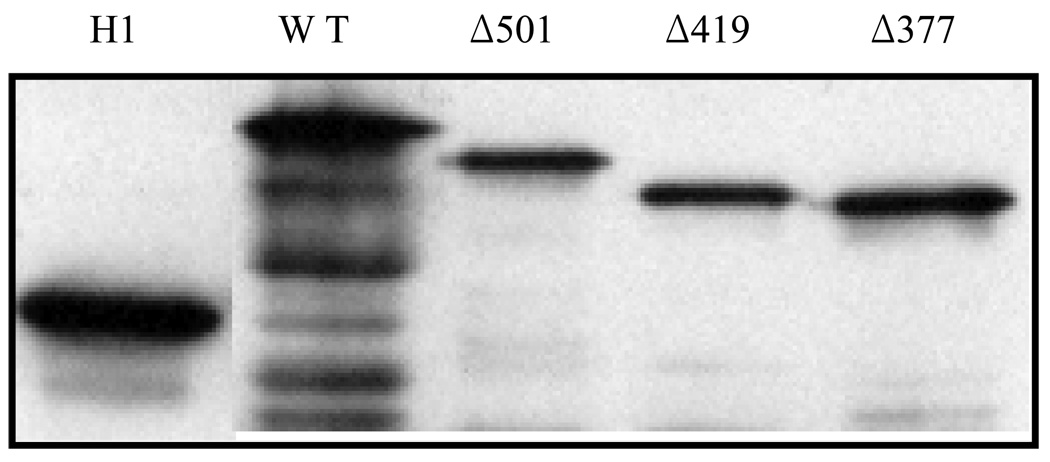
The membrane topology of SppA was experimentally probed using the well established PhoA fusion method (25). PhoA is enzymatically active only when it is exported to the periplasm; cytoplasmic phoA is inactive. Therefore, fusion of phoA to periplasmic domains of SppA would result in high phoA activity, whereas fusion to cytoplasmic loops of SppA would result in low phoA activity. In order to determine whether the C-terminal domain transmembrane segments predicted by TopPred2 were real, PhoA mature sequence was fused to SppA after residues 377, 419, 501, and 618. As a negative control, we used a construct where PhoA is fused after the first hydrophobic domain of leader peptidase. Previous studies with the H1Lep-phoA fusion (H1) showed that phoA is localized primarily to the cytoplasm although the first apolar domain (without the carboxyl-terminal positively charged residues present) can export a small percentage of phoA to the periplasmic space (21). All the PhoA fusion constructs were expressed in the phoA− strain to determine the expression level and the phoA activity of the phoA fusions. Fig. 5 confirms by immunoblotting that the various phoA constructs are expressed in the cell. We measured the activity of phoA in liquid using the chromophoric substrate p-nitrophenylphosphate. Table I summarizes the phoA activities of the SppA fusions. All the SppA-fusion proteins gave high enzymatic activities of alkaline phosphatase: the normalized AP activities are 21.5, 28.5, 16.9 and 23.3 units for the 618 (full-length), 501, 419, and 377 SppA-phoA constructs, respectively. The negative control (H1) gave normalized AP activity of 3.7. The results are consistent with all regions of the C-terminal domain of SppA being periplasmic. Because there are no other candidates for TM segments other than at the beginning of the protein, the data supports a model where SppA spans the membrane one time with its large carboxyl-terminal domain and most of its N-terminal domain protruding into the periplasmic space.
FIGURE 5. Expression of SppA-PhoA fusion proteins in the cell.
Lin 205 cells expressing the SppA-phoA constructs on the pING plasmid were grown and analyzed as described in the Experimental Procedures. The level of the fusion proteins were determined using anti-phoA antiserum.
Table 1.
Characteristics of SppA-phoA fusions.
| OD 600 | Total AP activity | Normalized AP activity | Predicted Location | |
|---|---|---|---|---|
| Full-length (WT) | 0.7877 | 17 | 21.5 | Periplasm |
| 501 | 0.6672 | 19 | 28.5 | Periplasm |
| 419 | 0.7647 | 12.9 | 16.9 | Periplasm |
| 377 | 0.8289 | 19.3 | 23.3 | Periplasm |
| H1(negative control) | 0.7303 | 2.7 | 3.7 | Cytoplasm |
DISCUSSION
In this report, we present two pieces of data that suggest that Ser 409 is directly involved in catalysis of the E. coli SppA. The first data is that the Ser 409 of the E. coli SppA, which is homologous to the proposed active site serine residue in the Thermococcus kodakaraensis SppA (12), is critical for enzymatic activity. Second, the FP-biotin serine hydrolase inhibitor modifies the wild-type SppA but does not label the protein when the Ser 409 is mutated, suggesting that Ser 409 is modified by the inhibitor (Fig. 4).
Surprisingly, we found that the Lys 209 is the best candidate for a general base residue for the E. coli SppA peptidase. This Lys 209 residue is localized to the non-conserved amino-terminal domain of the E. coli SppA peptidase. All other candidates for a general base residue were found not to be essential for catalytic activity (Fig. 3). Also we found that the mutagenesis of Lys 209 to an alanine markedly decreased the ability of the SppA to be modified with the FP-biotin inhibitor (Fig. 4). This is consistent with Lys 209 being important for activation of the Ser 409 nucleophile such that it can be reactive toward substrates and inhibitors.
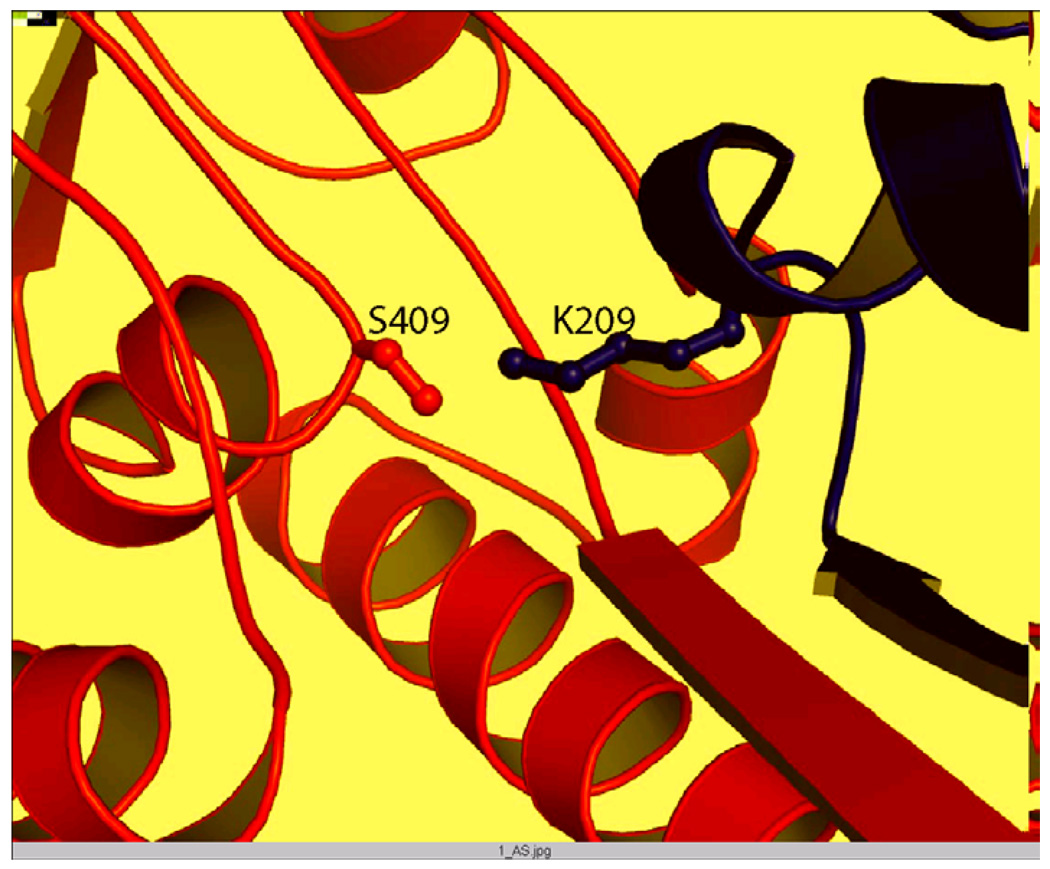
A serine and lysine residue is also implicated in the catalytic mechanism from the X-ray structure of the E. coli SppA that was solved in the course of this work (Fig.6; Kim, A. D., Oliver, D. and Paetzel, M., 2007, J. Mol. Biol., in press). The 2.4 Å structure revealed that Ser 409 and Lys 209 are indeed active site residues for the E. coli SppA peptidase. The structure reveals that the Ser 409 Oγ is within hydrogen bonding distance to the Lys 209 Nζ and that there are no other titratable functional groups within the vicinity of Ser 409 Oγ other than the Lys 209 Nζ. In fact, given that there is no sequence similarity and no similarity in protein fold, the active-site architecture of E. coli SppA is strikingly similar to that of E. coli signal peptidase. It appears that bacteria have converged on the same catalytic mechanism (Ser/Lys) to both cleave off the signal peptide (13) and hydrolyze the remaining signal peptide. In eukaryotic organisms these same processes are catalyzed by a Ser/His/Asp protease (within the signal peptidase complex (26)) and an aspartic protease mechanism (within signal peptide peptidase (27)).
FIGURE 6. Ser 409 and Lys 209 catalytic dyad at the active site of E. coli SppA.
Ribbon diagram of the E. coli SppA highlighting the active site Ser/Lys residues. This figure was adapted from Kim et al., 2007, in press.
This data, along with recent studies (12, 28), suggest that all SppA proteases are Ser-Lys dyad peptidases. This family of proteases all contains an active site Ser in the protease domain. They differ only in the location of the lysine residue. Two of the subfamilies (S49.001 and S49.004) have a highly conserved lysine in the amino-terminal domain (Fig. 1A) that we showed (Fig. 3) is crucial for activity of the E. coli SppA. The other subfamilies (S49.002, S49.003, S49.005, and S49.006) lack this domain and contain an absolutely conserved Lys residue in the protease domain located 42 residues downstream from the invariant Ser (see MEROPS protease database; (7)). Both the lysine and serine residues were shown to be critical for the activity of the T. kodakaraensis SppA (S49. 006) (12) and the structure of the Protein 1510-N from Pyrococcus horikoshii (S49.005) revealed a Ser and Lys residue at the active site region (28).
In addition to this work on the catalytic mechanism, we investigated the membrane topology of SppA. In contrast to what is predicted by several topology programs (23, 24) our results using alkaline phosphatase fusion analysis indicate that the E. coli enzyme spans the membrane only once and has its carboxyl-terminal domain, as well as the portion of the N-terminal domain containing the essential base Lys 209, in the periplasmic space. Although the active site Ser 409 of the E. coli SppA is located within a very hydrophobic region and the hydrophobic character of this region is conserved throughout evolution (Fig. 1B), this region is not membrane spanning.
ACKNOWLEDGMENT
This work was supported by the National Science Foundation Grant MCB-0316670 (to RED), the Canadian Institute of Health Research (to M.P.), the National Science and Engineering Research Council of Canada operating grant (to M.P.), the Michael Smith Foundation for Health Research Senior Scholar award (to M.P.), and the Canadian Foundation of Innovation grant (to M.P.).
Abbreviations
- ECL
enhanced chemiluminescence
- IPTG
isopropyl-β-D-thiogalactopyranoside
- LB
Luria broth
- M9
minimal medium
- PAGE
polyacrylamide gel electrophoresis
- SDS
sodium dodecyl sulfate
REFERENCES
- 1.Paetzel M, Karla A, Strynadka NC, Dalbey RE. Signal peptidases. Chem Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 2.Zwizinski C, Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980;255:7973–7977. [PubMed] [Google Scholar]
- 3.Hussain M, Ozawa Y, Ichihara S, Mizushima S. Signal peptide digestion in Escherichia coli. Effect of protease inhibitors on hydrolysis of the cleaved signal peptide of the major outer-membrane lipoprotein. Eur J Biochem. 1982;129:233–239. doi: 10.1111/j.1432-1033.1982.tb07044.x. [DOI] [PubMed] [Google Scholar]
- 4.Ichihara S, Beppu N, Mizushima S. Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity. J Biol Chem. 1984;259:9853–9857. [PubMed] [Google Scholar]
- 5.Pacaud M. Purification and characterization of two novel proteolytic enzymes in membranes of Escherichia coli. Protease IV and protease V. J Biol Chem. 1982;257:4333–4339. [PubMed] [Google Scholar]
- 6.Ichihara S, Suzuki T, Suzuki M, Mizushima S. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptide peptidase, of Escherichia coli. J Biol Chem. 1986;261:9405–9411. [PubMed] [Google Scholar]
- 7.Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 1999;27:325–331. doi: 10.1093/nar/27.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird L, Lipinska B, Raina S, Georgopoulos C. Identification of the Escherichia coli sohB gene, a multicopy suppressor of the HtrA (DegP) null phenotype. J Bacteriol. 1991;173:5763–5770. doi: 10.1128/jb.173.18.5763-5770.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Mushegian A. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J Bacteriol. 2004;186:4369–4375. doi: 10.1128/JB.186.13.4369-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama H, Matsui I. A novel thermostable membrane protease forming an operon with a stomatin homolog from the hyperthermophilic archaebacterium Pyrococcus horikoshii. J Biol Chem. 2005;280:6588–6594. doi: 10.1074/jbc.M411748200. [DOI] [PubMed] [Google Scholar]
- 11.Matsumi R, Atomi H, Imanaka T. Biochemical properties of a putative signal peptide peptidase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol. 2005;187:7072–7080. doi: 10.1128/JB.187.20.7072-7080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumi R, Atomi H, Imanaka T. Identification of the amino acid residues essential for proteolytic activity in an archaeal signal peptide peptidase. J Biol Chem. 2006;281:10533–10539. doi: 10.1074/jbc.M513754200. [DOI] [PubMed] [Google Scholar]
- 13.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor [see comments] [published erratum appears in Nature 1998 Dec 17;396(6712):707] Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 14.Slilaty SN, Little JW. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci U S A. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peat TS, Frank EG, McDonald JP, Levine AS, Woodgate R, Hendrickson WA. Structure of the UmuD' protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 16.Silber KR, Keiler KC, Sauer RT. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci U S A. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rotanova TV, Mel'nikov EE, Tsirul'nikov KB. Catalytic dyad Ser-Lys at the active site of Escherichia coli ATP-dependent Lon-proteinase. Bioorg Khim. 2003;29:97–99. doi: 10.1023/a:1022290705294. [DOI] [PubMed] [Google Scholar]
- 18.Feldman AR, Lee J, Delmas B, Paetzel M. Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J Mol Biol. 2006;358:1378–1389. doi: 10.1016/j.jmb.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 19.Johnston S, Lee JH, Ray DS. High-level expression of M13 gene II protein from an inducible polycistronic messenger RNA. Gene. 1985;34:137–145. doi: 10.1016/0378-1119(85)90121-0. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.San Millan JL, Boyd D, Dalbey R, Wickner W, Beckwith J. Use of phoA fusions to study the topology of the Escherichia coli inner membrane protein leader peptidase. J Bacteriol. 1989;171:5536–5541. doi: 10.1128/jb.171.10.5536-5541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 24.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 25.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 26.VanValkenburgh C, Chen X, Mullins C, Fang H, Green N. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J Biol Chem. 1999;274:11519–11525. doi: 10.1074/jbc.274.17.11519. [DOI] [PubMed] [Google Scholar]
- 27.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama H, Matsui E, Akiba T, Harata K, Matsui I. Molecular structure of a novel membrane protease specific for a stomatin homolog from the hyperthermophilic archaeon Pyrococcus horikoshii. J Mol Biol. 2006;358:1152–1164. doi: 10.1016/j.jmb.2006.02.052. [DOI] [PubMed] [Google Scholar]