Abstract
Phosphorylase kinase (PhK), the key enzyme that regulates glycogenolysis, has traditionally been thought to be expressed predominantly in muscle and liver. In this study, we show by two different database searches (Expressed Sequence Tag and UniGene) that PhK gene expression occurs in at least 28 – 36 different tissues, and that the genes encoding the α, β and γ subunits of PhK undergo extensive transcriptional processing. In particular, we have identified exon 6 of PHKG1 as a 3′ composite terminal exon due to the presence of a weak polyadenylation and cleavage site in intron 6. We have verified biochemically that transcriptional processing of PHKG1 does occur in vivo; mRNA corresponding to the alternate variant is expressed in skeletal muscle, brain, heart, and tongue. In silico translation of this mRNA yields a PhK γ subunit that contains the first 181 residues of the protein, followed by an additional 21 amino acids. The implication of this alternate processing is discussed within the context of γ catalysis and regulation.
Keywords: phosphorylase kinase, PHKG1, alternative splicing, polyadenylation, composite exon
INTRODUCTION
Phosphorylase kinase (PhK) is the regulatory enzyme responsible for catalyzing the rate limiting step in glycogen breakdown [1, 2]. PhK activates glycogen phosphorylase resulting in degradation of glycogen to glucose-1-phosphate. In liver, these reactions allow for the maintenance of blood glucose, and in muscle, they lead to energy production to sustain muscle contraction. PhK is one of the largest kinases known with a mass of 1.3 × 106 Da. It is comprised of four different subunits with a stoichiometry of (αβγδ)4; α,β, and δ are regulatory, while γ is catalytic. The α subunit is encoded by two different genes: PHKA1 located at Xq12–13, and PHKA2 also located on the X chromosome in region Xp22.2-p22.1 [3–5]. The β subunit is encoded by PHKB located on chromosome 16 in the region 16q12–13 [3]. PHKG1 and PHKG2 encode for two isoforms of γ PHKG1 is located on chromosome 7 in region p11.2 and PHKG2, is found in chromosomal region 16p11.2–12.1 [6, 7]. The smallest subunit of PhK, δ, is an intrinsic molecule of calmodulin (CaM) [8]. Three different CaM genes, all encoding the same protein, have been identified and are localized to chromosomal regions 14q24–q31, 2p21, and 19q13.2–q13.3 [9]. All three genes have been shown to encode for PhK δ yet the expression of these genes is not necessarily restricted to PhK.
Because liver and muscle are the primary storage sites for glycogen in mammals, PhK subunit expression has traditionally been thought to have a restricted tissue distribution profile in which PHKA1 and PHKG1 are considered muscle-specific genes and encode the αM and γM subunits, PHKA2 and PHKG2 are liver-specific and code for αL and γL, and PHKB is responsible for β expression in both tissues. There is mounting evidence however, that glycogenolytic regulating enzymes are expressed in other tissues, e.g. brain [10–14], lung [15], lymphocytes and erythrocytes [16–19], although it is not always known which of the two different α and γ isoforms are found in these tissues and whether or not they are alternatively processed. To date, only the α and β subunits of PhK have been shown to undergo alternative splicing [20]. An internal deletion of 59 residues in αM results in a cardiac and smooth muscle variant, α′. Additionally, many other variants of αM and αL exist as a result of elaborate differential splicing at a hypervariable region within the PHKA1 and PHKA2 genes, a region thought to be a hotspot for molecular evolution [21].
Structural and functional gene analysis has been dramatically accelerated through the wealth of genomic information available in public repositories and the advent of expressed sequence tag (EST) databases [22]. It is now possible to identify spatial relationships in gene expression patterns, as well as determine alternative splicing variants among genes in silico, thus eliminating years of tedious biochemical analysis. In this study, we have examined the expression profile for all five PhK genes that encode the α, β and γ subunits by computational sequence analysis of the human EST database and have determined the genomic location of each identified EST. We show that PhK is expressed in numerous tissues, and all five analyzed genes undergo some transcriptional processing. Moreover, we demonstrate that PHKG1 contains a composite 3′ terminal exon that results in a truncated transcript lacking the known C-terminal CaM binding domains. Given the fact that the catalytic activity of γ in the PhK complex is absolutely dependent on Ca2+ [23, 24], our findings suggest a potential alternative, Ca2+-independent γ activity in the cell.
MATERIALS and METHODS
Sequence analysis
cDNA reference sequences (RefSeq), corresponding to the following human PhK genes, were obtained from GenBank [25] and used for searching the human EST database [22]: PHKA1 [GI_4505778], PHKA2 [GI_4505780], PHKB-variant 1 [GI_4505782], PHKB-variant 2 [GI_73611905], PHKG1 [GI_94538351], and PHKG2 [GI_4505784]. The RefSeq database is a routinely updated set of sequence standards derived by merging multiple entry types for any given gene including chromosomal sequence, genomic molecules, mRNAs, RNAs, and proteins, and as such, provides non-redundant data representing the most current knowledge of each known gene sequence. The sequence analysis was performed by BLASTN [26] using the human dbEST database housed at the National Center for Biotechnology (NCBI) (www.ncbi.nlm.nih.gov). Search constraints were defined by the default parameters of the BLASTN program from NCBI. The results presented were obtained by using the dbEST file as of January 2006. The database contained 7,157,729 EST entries. The resulting EST data for each gene was categorized by tissue of origin as extracted from UniGene [27]. ESTs were scored for their alignment with other sequences and for their splicing patterns. ESTs showing sequence similarity to the PHK gene of interest using BLASTn were downloaded and further analyzed by aligning them to each other and to the full length genomic and cDNA reference sequences from which they were derived using the gapped alignment programs ClustalW [28] and Sim4 [29]. Genomic alignments generated by Sim4 were visualized using a program developed by E.C. Rouchka (http://kbrin.a-bldg.louisville.edu/~rouchka/estMapping2.html).
Amplification of truncated PHKG1
First strand cDNA for human brain, heart, tongue, small intestine, and skeletal muscle was obtained from Biochain Institute, Inc. and was used as the template for polymerase chain reaction (PCR). Full length and truncated PHKG1 were amplified using a forward primer corresponding to exon 1 of the PHKG1 open reading frame (5′-GCCTCCGGTCATCAAATAGCAA-3′), and a second reverse primer located in either exon 10 (5′-CCATAGATTCGGAAAGCGTAGG-3′) or intron 6 (5′-TGCAGATGGTTGATTGAGCACC-3′), respectively. Amplified DNA was resolved by agarose gel electrophoresis for molecular weight determination and proper sequences were verified by dideoxy sequencing using an ABI Hitachi 3130.
RESULTS
PhK is expressed in numerous tissues in humans
PhK gene distribution in human tissues was determined by in silico analysis of the human dbEST. Our results indicate that in addition to those tissues known to be glycogenolytic, numerous others express full complements of PhK genes (Table 1). Using the dbEST dated January 2006, 47 ESTs corresponding to PHKA1 were found expressed in 14 different tissues; the majority of these ESTs were in brain and pancreas. Fifty ESTs corresponding to PHKA2 were found in 19 total tissues of which 16% were from lymphocytes and 10% from brain. Seventy-eight ESTs matching PHKB were identified in 18 different tissues, most notably testis, eye, placenta and lung. Thirty ESTs were identified for PHKG1 from only 5 tissues, heart, brain, small intestine, prostate and tongue, while 52 ESTs corresponding to PHKG2 were found expressed in at least 13 different tissues with brain having the most ESTs, thus suggesting that PHKG2 is more ubiquitously expressed that PHKG1. Overall, 28 different tissues or organs were identified that express one or more PhK gene. Contrary to expectation, no EST representing PHKA1, PHKB, or PHKG1 was identified from skeletal muscle even though high expression of PhK in this tissue is well established. These results are likely due to the high stringency with which we performed our search in combination with the low percentage of skeletal muscle ESTs represented in the publicly available database (0.6% of the total EST pool). Furthermore, our results correlate very well to the EST expression profiles in the UniGene database, a more organized data resource that gives a view of the transcriptome by relating sets of sequences that originate from the same transcription locus. These sequences are in the form of well characterized genes, mRNAs and ESTs and are organized into transcription clusters [27]. By this analysis, PHK gene expression displays a spatial distribution where transcription of PHKB (36 tissues) > PHKA2 (35 tissues) > PHKG2 (31 tissues) > PHKA1 (29 tissues) > PHKG1 (17 tissues) (Table 2). The fact that the absolute numbers for tissue distribution are overall lower in our analysis is likely due to the high stringency with which our search was performed. Only sequences with 95% identity or greater to the query cDNA were selected for our results.
Table 1.
PhK tissue distribution by EST database analysis
| Tissue | Total ESTs identified | ||||
|---|---|---|---|---|---|
| PhKα1 | PhKα2 | PhKβ | PhKγ1 | PhKγ2 | |
| Adrenal gland | - | 1 | - | - | - |
| Bone | 2 | 2 | 5 | - | - |
| Brain | 7 | 5 | 4 | 9 | 9 |
| Breast | 2 | 1 | 2 | - | - |
| Cartilage/joint | - | - | 2 | - | - |
| Cervix | 1 | 1 | - | - | - |
| Colon | 2 | 4 | 4 | - | - |
| Eye | - | 1 | 10 | - | - |
| Head/neck | 1 | - | - | - | - |
| Heart | - | - | - | 14 | 14 |
| Intestine | - | - | - | 2 | 2 |
| Kidney | 3 | 2 | 4 | - | - |
| Liver | 2 | 2 | 3 | - | - |
| Lung | 4 | 4 | 7 | - | 4 |
| Lymphocyte | - | 8 | 6 | - | 4 |
| Nasopharnyx | - | 1 | - | - | - |
| Ovary | - | 1 | 1 | - | 1 |
| Pancreas | 7 | 1 | 3 | - | 1 |
| Pituitary | - | - | 2 | - | - |
| Placenta | 1 | 3 | 8 | 1 | 5 |
| Prostate | 1 | 1 | - | - | 3 |
| Skin | - | - | - | - | 3 |
| Stomach | 2 | 2 | 2 | - | 1 |
| Testis | - | 3 | 12 | - | 1 |
| Thymus | - | - | 2 | - | - |
| Thyroid | 2 | - | - | - | - |
| Tongue/papillary | - | 1 | - | 4 | 4 |
| Uterus | - | - | 1 | - | - |
| Total | 47 | 50 | 78 | 30 | 52 |
- indicates EST data was not found for the indicated tissue and cDNA
Table 2.
PhK tissue distribution by UniGene database analysis
| Tissue | Total ESTs identified | |||||
|---|---|---|---|---|---|---|
| PhKA1 | PhKA2 | PhKB | PhKG1 | PhKG2 | Total | |
| Adipose tissue | - | - | 1 (79) | - | 1 (79) | 2 |
| Adrenal gland | - | 2 (61) | - | 1 (30) | 2 (61) | 5 |
| Ascites | 1 (25) | 1 (25) | 2 (50) | - | 1 (25) | 5 |
| Bladder | - | - | 1 (34) | - | 1 (34) | 2 |
| Blood | 1 (8) | 3 (25) | 3 (25) | - | 22 (185) | 29 |
| Bone | 2 (28) | 1 (14) | 4 (56) | - | 4 (56) | 11 |
| Bone Marrow | - | - | 7 (147) | - | - | 7 |
| Brain | 16 (17) | 29 (31) | 16 (17) | 26 (25) | 28 (30) | 115 |
| Cervix | 1 (21) | 2 (42) | - | - | 4 (84) | 7 |
| Cochlea | 2 (126) | - | 1 (63) | - | - | 3 |
| Colon | 5 (28) | 15 (85) | 13 (73) | - | 6 (34) | 39 |
| Connective tissue | 1 (6) | 3 (20) | 15 (101) | 1 (6) | 4 (27) | 24 |
| Cranial nerve | 1 (55) | 2 (110) | - | - | 2 (110) | 5 |
| Eye | 1 (5) | 6 (30) | 19 (95) | 1 (5) | 9 (45) | 36 |
| Heart | 1 (11) | 2 (22) | 2 (22) | 22 (252) | 2 (22) | 29 |
| Kidney | 5 (24) | 8 (38) | 26 (124) | 1 (4) | 4 (19) | 44 |
| Liver | 4 (19) | 9 (44) | 7 (34) | - | 1 (4) | 21 |
| Lung | 7 (21) | 19 (57) | 14 (42) | 3 (9) | 11(33) | 54 |
| Lymph | - | - | 2 (45) | - | - | 2 |
| Lymph node | - | 14 (156) | 12 (134) | - | - | 26 |
| Mammary gland | 3 (20) | 9 (61) | 12 (81) | 3 (20) | 5 (34) | 32 |
| Mouth | 2 (30) | 4 (60) | 4 (60) | 11 (167) | - | 21 |
| Muscle | 4 (37) | 8 (75) | 7 (66) | 9 (85) | 4 (37) | 32 |
| Nerve | 1 (64) | 2 (128) | 1 (64) | - | - | 4 |
| Ovary | 1 (10) | 2 (20) | 1 (10) | 1 (10) | 8 (80) | 13 |
| Pancreas | 12 (56) | 7 (33) | 12 (56) | - | 5 (23) | 36 |
| Parathyroid | 5 (248) | 1 (49) | 1 (49) | - | 1 (49) | 8 |
| Pharynx | 5 (123) | 1 (24) | - | - | - | 6 |
| Pituitary gland | - | 1 (61) | 1 (61) | - | - | 2 |
| Placenta | 1 (3) | 7 (25) | 18 (65) | 1 (3) | 8 (29) | 35 |
| Prostate | 5 (27) | 9 (49) | 9 (49) | 1 (5) | 10(54) | 34 |
| Salivary gland | - | - | - | 1 (50) | - | 1 |
| Skin | 1 (5) | 2 (10) | 11 (56) | - | 8 (41) | 22 |
| Small Intestine | - | 1 (22) | 4 (89) | 3 (67) | 1 (22) | 9 |
| Spleen | - | - | 1 (18) | - | 1 (18) | 2 |
| Stomach | 1 (10) | 2 (21) | 4 (42) | - | 2 (21) | 9 |
| Testis | 7 (20) | 38 (108) | 36 (103) | 2 (5) | 12 (34) | 95 |
| Thymus | - | 6 (74) | 5 (49) | - | 2 (24) | 13 |
| Thyroid | 5 (109) | 2 (43) | 3 (65) | 2 (43) | 3 (65) | 15 |
| Trachea | - | 7 (134) | - | - | - | 7 |
| Uterus | 1 (4) | 4 (17) | 18 (78) | - | 6 (26) | 29 |
| Vascular | - | 2 (39) | 1 (19) | - | - | 3 |
| Total | 116 | 250 | 361 | 77 | 180 | |
- indicates EST data was not found for the indicated tissue; #’s in () are ESTs per million and normalize for variability among tissues
Genomic location of human PhK ESTs
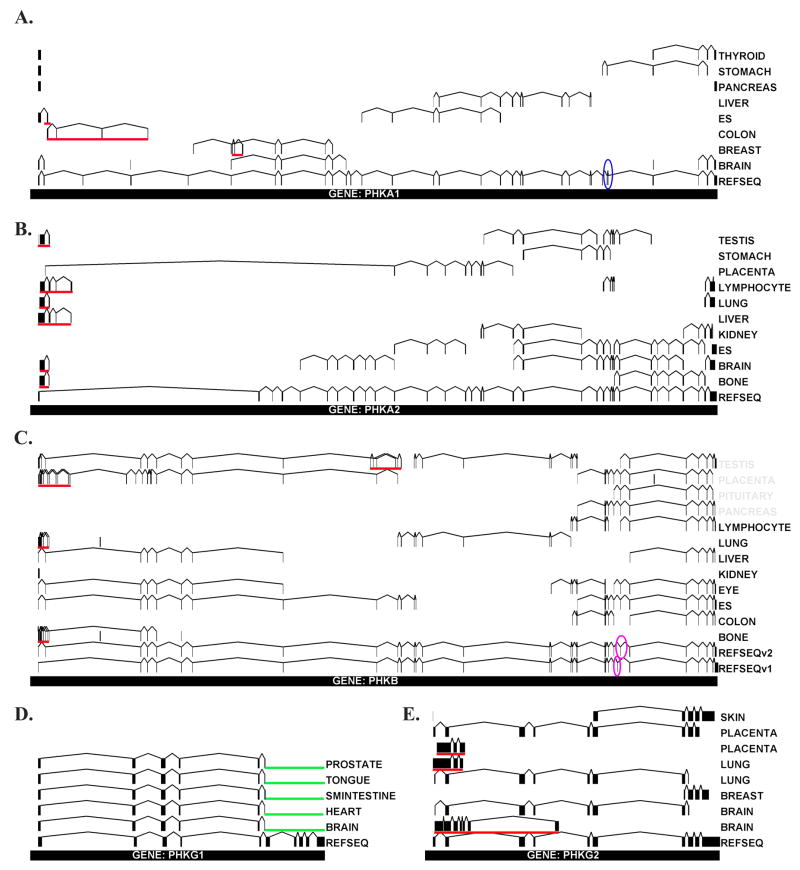
Unique EST sequences for each tissue were refined by sequence analysis using the ClustalW alignment tool [28]. ESTs from tissues that had at least two homologous sequences with a core region that was 90% identical or greater by ClustalW were subsequently aligned to the corresponding genomic sequence by Sim4 analysis [29]. These data are schematically represented in Figure 1. Based upon our results, PHKA1 is alternatively spliced in breast and colon (Fig. 1A). Three exons were identified in breast that are not found in same overlapping region of the reference sequence (RefSeq) [30], the sequence that provides the structure, i.e. intron/exon boundary, for each gene. Since the three exons were not identified in the known gene structure, they are likely novel and produced through splicing. These exons are encoded by nucleotides 38081–38220, 38585–35676, and 40096–40282 of the PHKA1 gene.
Figure 1. Genomic Alignment of EST Sequences.
The genomic location for each identified PHK EST sequence was determined as indicated in Materials and Methods. For each of the PHK genes shown (A–E), vertical lines and boxes represent exon regions while horizontal lines represent connecting intron regions. The tissue of origin for each identified EST is indicated on the right. Overlapping ESTs that were identified to encode for PHK exon regions were mapped to their genomic location by Sim4 analysis and are schematically represented here. The total number of nucleotides in each gene are as follows: PHKA1 – 133410 bp and Human Genome Locus (HGL) = NC_000023.9; PHKA2 – 91035 bp and HGL = NC_000023; PHKB – 237017 bp and HGL = NC_000016.8; PHKG1 – 12015 bp and HGL = NC_000007.12; PHKG2 – 8946 bp and HGL = NC_000016.8. Identified EST sequences are compared to the non-redundant RefSeq, the sequence that provides the intron/exon boundaries for each gene, and differences between ESTs and the RefSeq in a particular tissue represent alternative mRNA processing (underlined in red). Other highlighted regions include: exons 27–28 in PHKA1 previously reported to be alternatively spliced – blue circle; alternative splicing of PHKB in GenBank –purple circle; 3′ region of PHKG1 missing due to alternate polyadenylation - green. Two separate diagrams for PHKG2 expression in lung, placenta and brain are shown in order to emphasize the striking differential expression in the 5′ domain of this gene.
PHKA1 expressed in colon also appears to undergo alternative splicing at the 5′ region of the gene producing four novel exons encoded by genomic regions bps 1769–1969, 3620–3673, 12475–12645, and 21529–21606. The first of these exons, bps 1769–1969, also appears in embryonic stem cells. No alternate processing was observed in the genomic regions bps 111765–111880 and 112009–112047 which correspond to exons 27 and 28, the multiple phosphorylation domain of PHKA1 previously identified to be extensively spliced in skeletal muscle (Fig. 1A) [21]. PHKA2 undergoes differential splicing at the 5′ region of the gene in bone, brain, liver, lung, lymphocyte, and testis to produce four different exons not identified in the corresponding RefSeq (Fig. 1B). Additionally, a large region of the gene appears to be spliced out in placental tissue. According to the National Center for Biotechnology Information’s (NCBI) Entrez Gene database [27], PHKB encodes for two transcript variants as a result of differential splicing of exon 25 (Fig. 1C). In addition, our work indicates that PHKB undergoes extensive 5′ splicing in placenta, lung and bone as well as processing in genomic regions 116808–116830 and 117813–117862 in testis. The fact that of all the PHKB ESTs identified that overlap in the genomic region 202351–207888, the area identified by NCBI to be alternatively spliced, none contained the exon encoded by PHKB variant 1, suggests that the β isoform encoded by variant 2 is likely the predominant form. Previously, alternative splicing has been reported for the N-terminus of β in brain [20]; the lack of EST data from brain for PHKB did not allow us to confirm these results. Alternative processing of PHKG1 was found when compared to the RefSeq in that the last four terminal exons were not represented in any tissue analyzed in silico (Fig. 1D). PHKG2 is alternatively spliced in placenta, lung, and brain at the 5′ region (Fig. 1E).
The fact that in our analysis no EST was identified that contained any of the last 4 distal exons of PHKG1, led us to hypothesize that PHKG1 mRNA may undergo 3′ processing via an alternate stop codon and polyadenylation site located in intron 6, thus converting exon 6, an internal exon, into a 3′ terminal exon also called a composite terminal exon. Following searches of the genomic sequence for PHKG1, we did indeed identify an intronic polyadenylation site at nucleotide 106 of intron 6 (Fig. 2). The site was specified by the conserved hexanucleotide AAUAAA sequence followed by two possible CA cleavage sites 10 and 15 nucleotides downstream, and a U-rich region 25 nucleotides downstream. This site correlates well with consensus polyadenylation signal where the U-rich region is typically ~40 bps downstream of the cleavage site [31]. Analysis of this same region in other species indicates that the polyadenylation site is highly conserved across many species, although the downstream regulatory elements in mouse, rat, and zebrafish are less conserved. This is not necessarily surprising given that, in contrast to the AAUAAA sequence, downstream regulatory elements tend to be more variable in sequence composition, length and location [31, 32]. No putative polyadenylation sequence was identified in the chicken genome (data not shown). The identification of a second poly(A) site is not necessarily surprising given that ~ 50% of human genes have been found to have multiple poly(A) sites [33, 34]. We should note that it was surprising that no ESTs were identified that corresponded to the C-terminus of PHKG1, given that dbEST are typically 3′ ESTs, yet may reflect the fact that the number of ESTs mapping to the PHKG1 gene is comparatively small. As more ESTs become available, we expect to see this region represented in the database, and additional splice sites revealed.
Figure 2. Species Conservation of the PHKG1 Polyadenylation and Cleavage Site.
The DNA sequence of PHKG1 intron 6 is shown for six different species. The conserved hexanucleotide sequence is underlined; putative cleavage sites are indicated by arrows.
Identification of exon 6 as a composite terminal exon in PHKG1
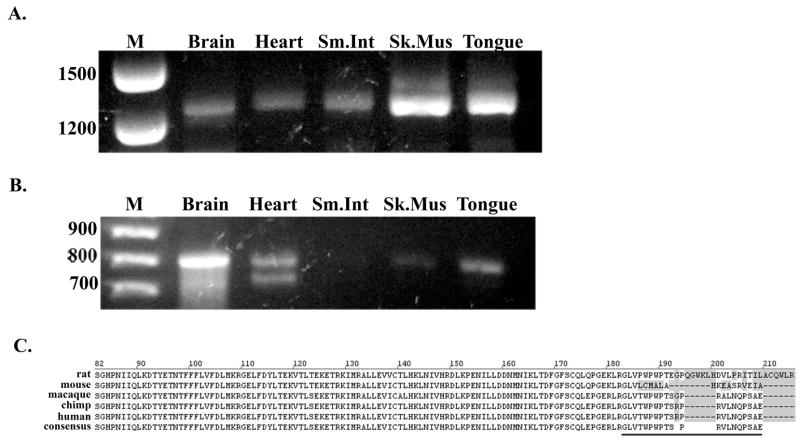
The two predicted PHKG1 transcripts were studied by RT-PCR in four of the five different human tissues that were identified to express the truncated PHKG1 variant in order to biochemically verify the newly identified composite terminal exon. When primer pairs corresponding to regions in either exon 1 and exon 10, or exon 1 and intron 6, were used for amplification of cDNA, all tissues were found to express the full length 1256 bp PHKG1 transcript, as well as a 795bp fragment corresponding the truncated product, respectively (Fig. 3A and B). The truncated PHKG1 did not amplify well from small intestine, yet was found to be present in skeletal muscle. These results confirm that alternative 3′ processing of PHKG1 does indeed occur in vivo. Translation of the alternate PHKG1 variant would result in an additional 21 amino acid residues (GLVTWPWPTSPRVLNQPSAE) being added past the end of the region encoded by exon 6 (Fig. 3C). These 21 residues are 95% conserved between humans and primates, varying at only a single glycine residue, yet diverge considerably across lower mammals. By PROSITE [35] database searching this region in the human sequence contains a putative protein kinase C phosphorylation site (TxR) at residue 191 that is not conserved across all species. The identification of this potential site for Ca2+ regulation in the truncated variant is interesting given that γ activity is normally regulated through Ca2+/CaM binding at its C-terminus, the region of the protein removed as a result of the alternative 3′ processing. Several other genes have been identified to use alternate composite terminal exons [36]; for many of these, the choice between isoforms discriminates between changes in tissue expression, developmental expression or secreted versus cytoplasmic location of the final protein. The functional significance of this newly identified PhK γ variant is currently being investigated.
Figure 3. PCR amplification of the full-length and spliced PHKG1 variants.
Human cDNA from brain, heart, small intestine, skeletal muscle, and tongue was used as the template for PCR as indicated in Materials and Methods. A. Amplification of full-length PHKG1 cDNA (1256 bp). B. Amplification of the 3′ truncated PHKG1 cDNA (795 bp). C. In silico translation of PHKG1 exon 6 with the additional 21 residues that result from alternate polyadenylation of intron 6 (underlined).
Discussion
In this study, we have used in silico methods to determine the spatial expression pattern for the five genes that encode the α, β, and γ subunits of PhK. We have determined that PhK transcripts are found in multiple tissues suggesting that the previous designation of PHKA1 and PHKG1 as muscle-specific, and PHKA2 and PHKG2 as liver specific, is not an accurate representation of their spatial distribution. While the absolute number of ESTs identified for each PhK gene varies among tissues, for all five PhK genes analyzed, more ESTs were identified in the brain than any other tissue, including liver and muscle. This result is likely an artifact given that the number of human brain ESTs in the database is 514,226, approximately 6% of the total, while skeletal muscle ESTs represent only 0.6% of the total (48,864 ESTs). Regardless, the fact that the EST number in a given library for the same gene is a rough indication of that gene’s expression in the tissue from which the library was derived, does indicate that relatively high levels of PhK are expressed in many different tissues. Such ubiquitous expression is not necessarily surprising since several studies suggest potential alternative roles for PhK outside of metabolism. PhK has been shown to mediate numerous interactions with proteins that play both a structural and/or a catalytic role in cytoskeletal architecture and dynamics [37, 38].
We have also shown that unique transcriptional processing occurs for all five PhK genes analyzed, and in particular, that PHKG1 undergoes terminal 3′ processing via a newly identified polyadenylation and cleavage site located in intron 6. Alternative polyadenylation is a common mechanism in human cells to increase the complexity of the transcriptome by producing mRNAs with either different 3′ untranslated regions and/or variable protein isoforms. Changing polyadenylation site usage can have enormous effects on overall amount, localization, translation efficiency and stability of the resultant gene product [36]. Generation of composite 3′ ends by competition between splicing and polyadenylation has been well studied [36] and is mediated by altering relative levels of polyadenylation, splicing and transcription termination factors usually resulting in an increase in the processing efficiency of weak polyadenylation sites. The splice site of PHKG1 intron 6 is relatively weak; the 5′ donor site lacks the conserved GUAAGU sequence and instead contains a GUCTGG, and the 3′ acceptor site contains 4 purines within the conical pyrimidine track, likely lowering the affinity of snRNP binding to these regions. It will be of interest to learn under what conditions, and in which tissues, PHKG1 undergoes alternative 3′ processing and the functional significance of this switch.
In addition to in silico identification, we have biochemically verified that the identified truncated variant of PHKG1 is expressed at the mRNA level and thus is likely to be translated. While transcription of a particular gene does not always correlate with translation, the fact that mRNA synthesis is the most regulated step in controlling protein concentrations, determining mRNA levels does typically provide a good approximation of the abundance of the corresponding protein [39]. Therefore, an important question that emerges is whether it is possible that this truncated variant of γ (γv), while considerably small, is catalytic. This alternative variant would contain the first 181 residues of γ followed by an additional 21 residues: GLVTWPWPTSPRVLNQPSAE. When compared to the solved crystal structure of the catalytic core of PhKγ (residues 1– 298) [40], γv would possess the N-terminal lobe responsible for metal and ATP binding [41], however, the C-terminal α-helical region of the kinase would be truncated in the activation loop (residues 178–185 of γ). The activation loop is the region identified in S/T kinases to contain a critical phosphorylatable residue leading to kinase activity [42]. In PhKγ, the conserved S/T is replaced by E182, and in the truncated γv this residue is changed to a G, thus preventing the polar associations with R (+2) in the peptide substrate, as shown to occur in the solved crystal structure [40]. Furthermore, while the key catalytic aspartate, D149, is present in the variant, several other residues identified to make contacts with the substrate peptide analog (V183, C184, G185 and S188) are changed in γv as a result of translation of intron 6, and thus key polar associations known to mediate substrate binding may be altered.
Alternatively, if γv is not catalytic itself, it may function to allosterically regulate catalysis of the PhK holoenzyme. When compared to the sequences of the α and β regulatory subunits, the additional 21 residues added to γv contain a 7 amino acid stretch (PTSPRVL) that is also partially conserved in the β subunit (residues 824–830). Given the fact that another region of β has previously been identified to inhibit the catalytic activity of γ (residues 420–436) [43], and that β makes numerous contacts with γ in the holoenzyme [44], it is reasonable to speculate thatthis portion of γv may also be inhibitory. Furthermore, the fact that γ has been shown to self-associate between resides 66–110 [37] and form dimers [40, 45], suggests a potential inhibitory mechanism through the formation of γ-γv heterodimers. Catalytic inhibition by an alternatively spliced protein variant is not unprecedented. The gene that encodes for endothelial nitric oxidesynthase (eNOS) is alternatively spliced in intron 13 to produce a 3′ truncated variant that serves as a dominant negative inhibitor for the activity of the full-length eNOS enzyme [46]. While the precise cellular function of γv is yet to be determined, such allosteric regulation of γ by γv would provide yet another mechanism to ensure that uncontrolled glycogenolysis did not occur in the absence of the regulatory α and β subunits.
Acknowledgments
We are grateful to Drs. G.M. Carlson and O. Nadeau for insightful discussion of the results, and Mr. Joseph Chavarria-Smith for graphical assistance. The project was supported in part by NIH Grant Number P20 RR-16481 from the IDeA Networks of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pickett-Gies CR, Walsh DA. In: The Enzymes. 3. Boyer PDaKEG., editor. Vol. 17. Orlando: Academic Press; 1986. pp. 395–459. [Google Scholar]
- 2.Brushia RJ, Walsh DA. Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front Biosci. 1999;4:D618–41. doi: 10.2741/brushia. [DOI] [PubMed] [Google Scholar]
- 3.Francke U, et al. Assignment of human genes for phosphorylase kinase subunits alpha (PHKA) to Xq12-q13 and beta (PHKB) to 16q12-q13. Am J Hum Genet. 1989;45(2):276–82. [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson JJ, et al. cDNA cloning of a liver isoform of the phosphorylase kinase alpha subunit and mapping of the gene to Xp22.2-p22.1, the region of human X-linked liver glycogenosis. Proc Natl Acad Sci U S A. 1992;89(6):2096–100. doi: 10.1073/pnas.89.6.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geng Y, et al. Mapping of a liver phosphorylase kinase alpha-subunit gene on the mouse X chromosome. Genomics. 1993;15(1):191–3. doi: 10.1006/geno.1993.1031. [DOI] [PubMed] [Google Scholar]
- 6.Jones TA, et al. Localisation of the gene encoding the catalytic gamma subunit of phosphorylase kinase to human chromosome bands 7p12-q21. Biochim Biophys Acta. 1990;1048(1):24–9. doi: 10.1016/0167-4781(90)90017-v. [DOI] [PubMed] [Google Scholar]
- 7.Burwinkel B, et al. Variability of biochemical and clinical phenotype in X-linked liver glycogenosis with mutations in the phosphorylase kinase PHKA2 gene. Hum Genet. 1998;102(4):423–9. doi: 10.1007/s004390050715. [DOI] [PubMed] [Google Scholar]
- 8.Cohen P, et al. Identification of the Ca2+-dependent modulator protein as the fourth subunit of rabbit skeletal muscle phosphorylase kinase. FEBS Lett. 1978;92(2):287–93. doi: 10.1016/0014-5793(78)80772-8. [DOI] [PubMed] [Google Scholar]
- 9.Berchtold MW, et al. Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics. 1993;16(2):461–5. doi: 10.1006/geno.1993.1211. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, et al. Phosphorylation of myelin basic protein by glycogen phosphorylase kinase. FEBS Lett. 1984;169(2):224–8. doi: 10.1016/0014-5793(84)80323-3. [DOI] [PubMed] [Google Scholar]
- 11.Ozawa E. Activation of phosphorylase kinase from brain by small amounts of calcium ion. J Neurochem. 1973;20(5):1487–8. doi: 10.1111/j.1471-4159.1973.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 12.Paudel HK. The regulatory Ser262 of microtubule-associated protein tau is phosphorylated by phosphorylase kinase. J Biol Chem. 1997;272(3):1777–85. [PubMed] [Google Scholar]
- 13.Paudel HK, Zwiers H, Wang JH. Phosphorylase kinase phosphorylates the calmodulin-binding regulatory regions of neuronal tissue-specific proteins B-50 (GAP-43) and neurogranin. J Biol Chem. 1993;268(9):6207–13. [PubMed] [Google Scholar]
- 14.Psarra AM, Sotiroudis TG. Subcellular distribution of phosphorylase kinase in rat brain. Association of the enzyme with mitochondria and membranes. Int J Biochem Cell Biol. 1996;28(1):29–42. doi: 10.1016/1357-2725(95)00117-4. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, et al. The testis isoform of the phosphorylase kinase catalytic subunit (PhK-gammaT) plays a critical role in regulation of glycogen mobilization in developing lung. J Biol Chem. 1996;271(20):11761–6. doi: 10.1074/jbc.271.20.11761. [DOI] [PubMed] [Google Scholar]
- 16.Bakker HD, et al. Hepatic phosphorylase b kinase deficiency with normal enzyme activity in leukocytes and erythrocytes. J Inherit Metab Dis. 1991;14(2):269–70. doi: 10.1007/BF01800604. [DOI] [PubMed] [Google Scholar]
- 17.Bashan N, et al. Phosphorylase kinase in leukocytes and erythrocytes of a patient with glycogen storage disease type IX. J Inherit Metab Dis. 1987;10(2):119–27. doi: 10.1007/BF01800035. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi M, et al. Enzymatic analysis in lymphocytes and erythrocytes from six patients with different phenotypes of phosphorylase kinase deficiency. J Inherit Metab Dis. 1988;11(3):315–8. doi: 10.1007/BF01800383. [DOI] [PubMed] [Google Scholar]
- 19.Morishita Y, et al. Hepatic phosphorylase kinase deficiency: a survey of phosphorylase kinase activity in erythrocytes. Kobe J Med Sci. 1978;24(4):211–22. [PubMed] [Google Scholar]
- 20.Harmann B, Zander NF, Kilimann MW. Isoform diversity of phosphorylase kinase alpha and beta subunits generated by alternative RNA splicing. J Biol Chem. 1991;266(24):15631–7. [PubMed] [Google Scholar]
- 21.Wullrich A, et al. The multiphosphorylation domain of the phosphorylase kinase alpha M and alpha L subunits is a hotspot of differential mRNA processing and of molecular evolution. J Biol Chem. 1993;268(31):23208–14. [PubMed] [Google Scholar]
- 22.Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST [mdash] database for [ldquo]expressed sequence tags[rdquo] Nat Genet. 1993;4(4):332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 23.Meyer WL, Fischer EH, Krebs EG. Activation of Skeletal Muscle Phosphorylase B Kinase by Ca. Biochemistry. 1964;3:1033–9. doi: 10.1021/bi00896a004. [DOI] [PubMed] [Google Scholar]
- 24.Brostrom CO, Hunkeler FL, Krebs EG. The relation of skeletal muscle phosphorylase kinase by Ca2+ J Biol Chem. 1971;246(7):1961–7. [PubMed] [Google Scholar]
- 25.Burks CCM, Cinkosky MJ, Cumella KE, Gilna P, Hayden JE, Keen GM, Kelley TA, Kelly M, Kristofferson D, Ryals J. GenBank. Nucleic Acids Res. 1991;19(Suppl):2221–2225. doi: 10.1093/nar/19.suppl.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SFGW, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler DLBT, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 29.Florea L, et al. A Computer Program for Aligning a cDNA Sequence with a Genomic DNA Sequence. Genome Research. 1998;8(9):967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007;35(suppl1):D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes and Development. 1997;11(21):2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 32.Gilmartin GM. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes and Development. 2005;19(21):2517–2521. doi: 10.1101/gad.1378105. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Lee J, Tian B. Biased alternative polyadenylation in human tissues. Genome Biology. 2005;6(12):R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Research. 2005;15(3):369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991;19(Suppl):2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Research. 1997;25(13):2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayers N. Biochemistry. University of Tennessee; Memphis: 1999. Interactions of the Alpha Subunit of Phosphorylase-b Kinase; p. 154. [Google Scholar]
- 38.Archila S, et al. The cytoskeletal organizing protein Cdc42-interacting protein 4 associates with phosphorylase kinase in skeletal muscle. Biochem Biophys Res Commun. 2006;345(4):1592–9. doi: 10.1016/j.bbrc.2006.05.073. [DOI] [PubMed] [Google Scholar]
- 39.Watson James DRMM, Caudy Amy A, Witkowski Jan A. Recombinant DNA. 3. New York: W.H. Freeman and Company; 2007. p. 474. [Google Scholar]
- 40.Lowe ED, et al. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. Embo J. 1997;16(22):6646–58. doi: 10.1093/emboj/16.22.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owen DJ, et al. Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product. Structure. 1995;3(5):467–82. doi: 10.1016/s0969-2126(01)00180-0. [DOI] [PubMed] [Google Scholar]
- 42.Taylor SSR-AE. Three protein kinase structures define a common motif. Structure. 1994;2(5):345–55. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez VE, Carlson GM. Isolation of an autoinhibitory region from the regulatory beta-subunit of phosphorylase kinase. J Biol Chem. 1993;268(24):17889–95. [PubMed] [Google Scholar]
- 44.Nadeau OW, Sacks DB, Carlson GM. Differential affinity cross-linking of phosphorylase kinase conformers by the geometric isomers of phenylenedimaleimide. J Biol Chem. 1997;272(42):26196–201. doi: 10.1074/jbc.272.42.26196. [DOI] [PubMed] [Google Scholar]
- 45.Skuster JR, Chan KF, Graves DJ. Isolation and properties of the catalytically active gamma subunit of phosphorylase b kinase. J Biol Chem. 1980;255(5):2203–10. [PubMed] [Google Scholar]
- 46.Lorenz M, et al. Alternative splicing in intron 13 of the human eNOS gene: a potential mechanism for regulating eNOS activity. The FASEB Journal. 2007:fj.06–7434com. doi: 10.1096/fj.06-7434com. [DOI] [PubMed] [Google Scholar]