Abstract
Sarcolipin (SLN) is a small molecular weight sarcoplasmic reticulum (SR) membrane protein expressed both in cardiac and skeletal muscle tissues. Recent studies using transgenic mouse models have demonstrated that SLN is an important regulator of cardiac SR Ca2+ ATPase 2a (SERCA2a). However, there is a paucity of information regarding the SLN protein expression in small versus larger mammals and its regulation during development and cardiac pathophysiology. Therefore, the major goal of this study was to generate a SLN specific antibody and perform detailed analyses of SLN protein expression during muscle development and in the diseased myocardium. The important findings of the present study are: (i) in small mammals, SLN expression is predominant in the atria but low in the ventricle and in skeletal muscle tissues, whereas in large mammals, SLN is quite abundant in skeletal muscle tissues than the atria (ii) SLN and SERCA2a are co-expressed in all striated muscle tissues studied except ventricle and co-ordinately regulated during muscle development and (iii) SLN protein levels are ∼3 fold upregulated in the atria of heart failure dogs and ∼30% decreased in the atria of hearts prone to myocardial ischemia. In addition we found that in the phospholamban null atria, SLN protein levels are upregulated.
Keywords: sarcolipin, SERCA2a, phospholamban, atria, skeletal muscle, heart failure, development
Introduction
Sarcolipin (SLN) is a small molecular weight protein (31 amino acids) originally identified to co-purify with the skeletal muscle sarcoplasmic reticulum Ca2+ ATPase (SERCA) [1, 2]. Subsequent studies at the mRNA level showed that SLN is expressed both in cardiac and skeletal muscle tissues of all mammals [2-5]. Within the cardiac muscle, SLN mRNA expression is more abundant in the atria compared to the ventricle [4, 5]. The expression pattern of SLN mRNA is different between small and larger mammals. In rodents, SLN mRNA is abundant in the atria and expressed at low levels in the ventricle and slow skeletal muscles [4, 5]. In contrast, in larger mammals including humans, SLN mRNA is more abundant in fast-twitch skeletal muscle tissues compared to the heart [2].
The importance of SLN as a regulator of SERCA pump was studied using adenoviral gene transfer [5] and transgenic mouse models [6-8]. Adenoviral mediated overexpression of N-terminally flagged-SLN (NF-SLN) into ventricular myocytes resulted in decreased myocyte contractility and Ca2+ handling [5]. Confocal imaging analyses of ventricular myocytes overexpressing NF-SLN showed that, SLN co-localizes with phospholamban (PLB) and SERCA2a in the SR membrane [5]. The functional significance of SLN in cardiac contractility either in the presence or absence of PLB was studied by overexpressing NF-SLN in the wildtype [6, 7] as well as in the PLB knockout mouse hearts [8]. Overexpression of NF-SLN in the mouse heart inhibits the SERCA pump activity by lowering its apparent Ca2+ affinity, resulting in decreased Ca2+ transient amplitude and rates of muscle relaxation [6-8]. Additionally these studies showed that the inhibitory function of SLN is independent of PLB and is relieved upon isoproterenol treatment [6, 8]. These studies suggest that SLN is an important regulator of SERCA pump activity and can mediate β-adrenergic response independent of PLB.
SLN mRNA expression is shown to be developmentally regulated [4, 5] and is altered by thyroid hormones [9, 10] and pathophysiological conditions [3, 4, 11-13]. Pressure-overloaded hypertrophy induced by transverse aortic constriction in mice or by monocrotaline administration in rats significantly decreased the expression of SLN, SERCA2a, and phospholamban mRNAs in the atrium [13]. In human, decreased expression of SLN mRNA has been reported in the atria of patients with atrial fibrillation [12]. Although SLN expression is low in the ventricle, it was shown to be up-regulated ∼50-fold higher in the hypertrophied ventricles of Nkx2-5 null mice [11]. However, most of these studies were carried out at the mRNA level and results could not be confirmed at the protein level due to the lack of SLN a specific antibody. In the present study, we generated an antibody highly specific to SLN and studied its expression during development and in cardiac pathology.
Materials and Methods
All experiments were performed in accordance with the National Institute of Health guidelines and approved by the Institutional Laboratory Animal Care and Use Committee at The Ohio State University. Sprague-Dawley rats, B6 mice, SLN transgenic mice [6] and PLB knockout mice [14] were used for this study. Developmental studies were done in timed pregnant Sprague-Dawley rats purchased from Harlan/Taconic farms. Pacing induced heart failure canine model [15] was described previously. The myocardial infarction model susceptible or resistant to ventricular fibrillation was generated as described previously [16, 17]. In brief, the anterior myocardial infarction was produced by ligation of the left anterior descending artery approximately one-third of the distance from its origin. A hydraulic occluder was also placed around the left circumflex coronary artery so that ischemia can be induced at a site distant to the previous injury. The susceptibility to ventricular fibrillation was tested by a two minute coronary occlusion during the last minute of a submaximal exercise test. The susceptible dogs had either ventricular fibrillation or ventricular tachycardia during this exercise plus ischemia test while the resistant dogs do not have arrhythmias. In this particular study, the animals were assigned to either a 10 week sedentary period or a 10 week exercise training (running on a treadmill) groups after the classification. The susceptible dogs have poorer ventricular function and an abnormal autonomic neural control as compared to the resistant animals.
Generation of SLN antibody
We utilized the 100% conserved C-terminal sequence of SLN to generate rabbit polyclonal antibody. A peptide corresponding to the last 6 residues of luminal domain (-VRSYQY) of SLN with an additional cysteine added to the C-terminus was conjugated to keyhole-limpet haemocyanin. Rabbits were immunized with these peptides and sera were collected. Sera from one of the four rabbits immunized with the above antigen recognize SLN across species. This SLN C-terminal peptide antibody (SLN-CTAb) was affinity purified and used for further studies. Specificity of the antibody was tested by Western blot analysis using bacterially expressed rat and human SLN. Generation and affinity purification of SLN antibody was carried out in Chemicon International, Inc.
Protein preparations
The following muscles were sampled in (i) rodents- atria, ventricle, soleus, quadriceps, diaphragm and tongue, (ii) rabbit- left and right atria, left and right ventricle, diaphragm, extensor digitorum longus (EDL) and soleus and (iii) dog- left and right atria, left and right ventricle, diaphragm, gastrocnemius muscle (a synergist of the soleus muscle in other species) and EDL. For developmental studies, atria, ventricle, tongue and quadriceps from rat embryos of different days of post coitum (dpc) and neonatal rats were used. Frozen tissues were homogenized in 8 volumes of homogenizing buffer containing (in mM) 50, Tris. Cl, pH 7.5, 150 NaCl, 1 Na2P2O7, 1 benzamidine, 5 Na3VO4, 10 NaF, and 0.5% Nonidet P-40 [6]. To express rat and human SLN in the bacteria, the coding sequence of rat or human SLN was cloned into the bacterial expression vector pET 23d (Novagen Inc). Cell growth, protein induction and protein extraction were followed precisely as described by the manufacturer's instructions. Protein concentrations were determined by the Bradford method using bovine serum albumin for the standard curve. The SR enriched microsomal fractions for rat atria and ventricles were prepared as described earlier [18].
Western blot analysis
Protein samples were analyzed by sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE) and loading was normalized for Western blot analysis. The homogenates were electrophoretically separated on (8% for SERCA2a, SERCA1a and CSQ, 14% for PLB) SDS- PAGE or 16% Tricine gel (for SLN) and transferred to nitrocellulose membrane. Membranes were immunoprobed with primary antibodies [Anti-rabbit SLN, 1:3000 (present study); anti-rabbit SERCA2a, 1:5000; anti-rabbit PLB, 1:3000; anti-rabbit calsequestrin (CSQ), 1:5000 [6]; anti-rabbit SERCA1a, 1:2000 (Custom made)] followed by HRP-conjugated secondary antibodies. Signals were detected by Super Signal WestDura substrate (Pierce) and quantitated by densitometry [6].
Statistics
Data shown are mean ± SEM. Statistical significance was estimated by a unpaired Student's t test. A value of p<0.05 was considered statistically significant.
Results
Generation and characterization of a SLN specific polyclonal antibody
In a recent study, the cytosolic region (N-terminal sequence) of mouse SLN was used to generate a SLN specific antibody [19]. This antibody reacts to mouse SLN but has a very low affinity for rabbit and pig SLN. This is most likely due to sequence diversity at the N-terminus of SLN between small and larger mammals and therefore the use of this antibody is restricted to rodents. In addition, this antibody was shown to cross-react to PLB [19]. In the present study, we took advantage of the highly conserved C-terminal amino acids (-VRSYQY) corresponding to the luminal domain of SLN [20] to generate a polyclonal antibody in rabbits. Using this SLN antibody (SLN-CTAb), we studied the SLN protein expression during muscle development and in cardiac pathophysiology.
The specificity of the SLN-CTAb was determined by Western blot analysis using bacterially expressed mouse and human SLN proteins. Results shown in Fig.1A indicate that SLN-CTAb recognized the bacterially expressed mouse and human SLN proteins with the predicted molecular weight of 3.6 kDa. Control experiments performed using rabbit preimmune serum and Western blot results were negative (Data not shown). Since SLN mRNA is abundant in the atria compared to the ventricle [4, 5], we also analyzed the SR enriched microsomal fractions prepared from rat atria and ventricles. As seen in Fig. 1A, SLN-CTAb recognizes a 3.6 kDa protein similar to that of bacterially expressed SLN protein and its level was several folds higher in the atria compared to that of ventricle. Further, our results show that unlike PLB, SLN did not form multimers. To further validate the antibody specificity, Western blot analysis was performed using total protein extract prepared from SLN transgenic mice atria and ventricles which express NF- SLN [6]. Results shown in Fig. 1B indicate that SLN antibody recognizes both endogenous SLN and transgenically expressed NF-SLN. SLN-CTAb does not cross-react with any other low molecular protein in particular PLB. SLN could not be detected with total ventricular protein extract because of its very low level suggesting that microsomal fractions should be used to detect SLN in the ventricle. These results together suggest that we successfully generated a rabbit-polyclonal antibody specific for SLN which recognizes SLN across species.
Figure 1.
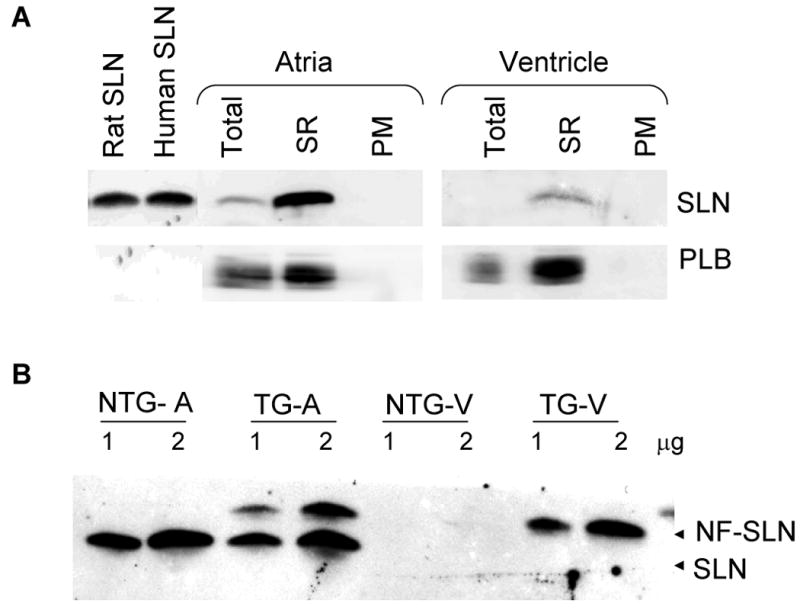
(A) Western blot analysis of sarcolipin (SLN) and phospholamban (PLB) in the total homogenate (Total), SR enriched microsomal fractions (SR) and post microsomal fractions (PM) from rat atria and ventricle. Bacterial extract containing rat and human SLN were used as control. 1 μg of each fraction were used. (B) Two different concentrations (1 and 2μg) of total homogenates from atria (A) and ventricle (V) of non-transgenic (NTG) and SLN- transgenic (TG) mice were resolved on a 16% tricine gel and immuno probed with SLN-CTAb. NF-SLN= N-terminally Flagged SLN
The expression pattern of SLN differs between small and larger mammals
To determine whether SLN protein expression follows its mRNA pattern [2-5], total protein prepared from various muscle tissues of mouse, rat, rabbit and dog were analyzed by Western blot analysis. Results shown in Fig. 2 point out that: 1) SLN protein is abundant in the atria regardless of species and 2) mouse and rat atria have higher levels of SLN protein when compared to atria from larger mammals. In rodents, SLN protein is also expressed at high levels in the tongue and at moderate levels in slow-twitch soleus muscle and diaphragm (Fig.2A & Table 1). In contrast, PLB expression was most abundant in the ventricle compared to the atria. SERCA2a protein expression, on the other hand, was significantly higher in the rodent atria (∼1.3 fold) compared to the ventricle (Fig.2A). In rodents, SERCA2a is also expressed at moderate levels in soleus and at low levels in diaphragm and tongue. Interestingly, the expression of SLN, SERCA2a and PLB proteins could not be detected in the fast-twitch quadriceps muscles (Fig. 2A). SLN protein was also not detected in smooth and non-muscle tissues (Data not shown).
Figure 2.
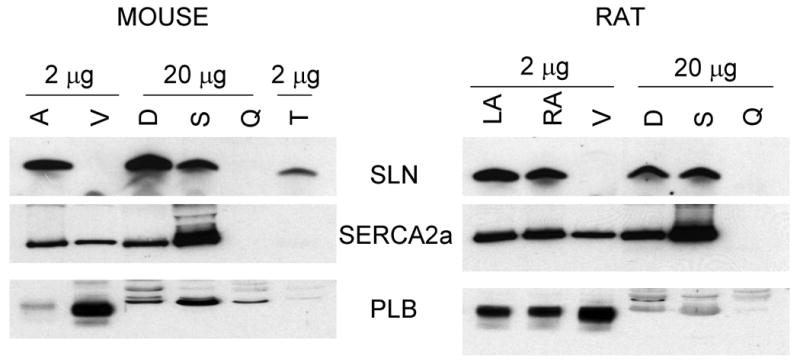
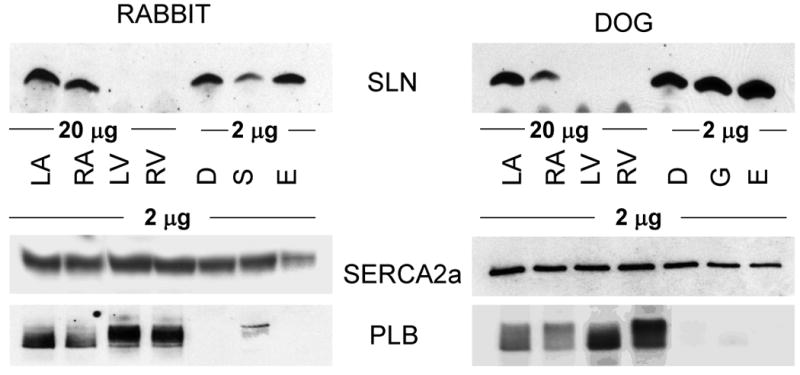
Expression analysis of SLN, PLB and SERCA2a proteins in striated muscle tissues of rodents (Panel A-mouse and rat) and larger mammals (Panel B- rabbit and dog). Total homogenates from atria (A), ventricle (V), diaphragm (D), soleus (S), quadriceps (Q), gastrocnemius (G) tongue (T) and EDL (E) from different species were resolved on polyacrylamide gel electrophoresis and probed with SLN-CTAb, PLB and SERCA2a specific antibodies as described in “methods” section. LA- left atria, RA-right atria, LV-left ventricle and RV-right ventricle. Data are representative of two independent experiments.
Table 1. Percent Expression of SLN and SERCA2a*.
| Species/Tissues | Percent expression of SLN | Percent expression of SERCA2a |
|---|---|---|
| RODENTS | ||
| Atria | 100** | 130 |
| Ventricle | 1 | 100** |
| Soleus | 10 | 60 |
| Quadriceps | ND | ND |
| Diaphragm | 22 | 15 |
| Tongue | 50 | 20 |
| RABBIT & DOG | ||
| Atria | 10 | 95 |
| Ventricle | 1 | 100** |
| Soleus | 70 | 35 |
| EDL | 100** | 35 |
| Diaphragm | 70 | 35 |
Values presented in the table are calculated from two independent experiments.
Percent expression taken as 100% and compared with other tissues.
ND- Not detectable
The expression pattern of SERCA, PLB and SLN were quite different between the skeletal muscle types in larger mammals (rabbit and dog). SLN protein levels were abundant both in slow- and fast-twitch skeletal muscles and diaphragm compared to that of atria (Fig. 2B & Table 1). Slow-twitch muscles (soleus and gastrocnemius) and diaphragm expressed high levels of SERCA2a and very low levels of PLB. Unlike rodents, the fast twitch muscle (EDL) expressed high levels of SERCA2a (Fig. 2B), whereas PLB was not detectable.
SLN expression is developmentally regulated in the rat atria and skeletal muscle tissues
We next determined whether SLN expression is developmentally regulated. Fig. 3 shows the temporal pattern of SLN expression in the rat atria, ventricle, quadriceps and tongue during embryonic and neonatal development. As shown in Fig.3A, the SLN protein was detected in the atria at 17.5 dpc and its level increases through out development, whereas in the ventricle its level was below detectable. On the other hand, the expression of PLB and SERCA2a proteins were detectable both in the atria and ventricle at 17.5 dpc and their expression remained high throughout development. At all developmental stages analyzed PLB levels were higher in the ventricle compared to the atrium.
Figure 3.
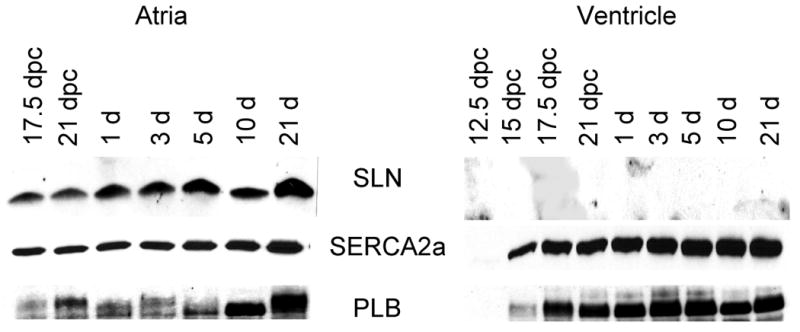
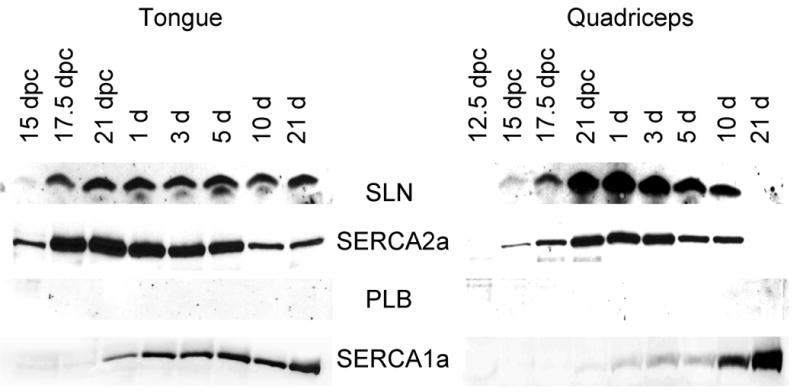
Western blot analysis of SLN, SERCA2a, PLB and SERCA1a proteins during muscle development. Two micrograms of total homogenates of atria, ventricular (Panel A), fast-twitch quadriceps muscle and tongue (Panel B) from rat embryos of different days of postcoitum (dpc) and from neonatal rats were separated on polyacrylamide gel electrophoresis and immunoprobed with specific antibodies. Data are representative of two independent experiments.
We next chose to examine SLN expression in rat tongue muscle. At 17.5 dpc, SLN and SERCA2a were expressed at high levels and remain high through out embryonic development (Fig. 3B). After birth, SERCA2a protein level declined gradually but continued to be expressed at a modest level in the adult. In contrast, SERCA1 expression which was detectable at 17.5 dpc increased after birth and became the predominant SERCA isoform in the adult tongue. Despite SERCA2a expression in the tongue, PLB expression was undetectable at all developmental stages studied.
The most striking finding was the pattern of SLN expression during fast-twitch skeletal muscle development in rats. At 15 dpc, SLN protein level was detectable in the quadriceps muscles and its level increased gradually during embryonic development along with SERCA2a. However, after birth SLN expression started to decline and completely disappeared by 21 days (Fig. 3B). The pattern of SERCA2a expression was also identical to that of SLN. These data indicate that in rodents, the expression of SLN and SERCA2a are co-ordinately regulated during fast-twitch skeletal muscle development, In contrast, SERCA1a that is expressed at low levels in quadriceps during embryonic development increased after birth and became the major SERCA isoform in the adult (Fig. 3B). Our studies failed to detect PLB protein expression in quadriceps at all developmental stages studied.
SLN expression is altered during cardiac pathophysiology
To determine whether SLN protein level is altered during cardiac pathophysiology, we analyzed SLN protein expression in the atrial tissues of (i) canine model of heart failure induced by tachypacing [15] and (ii) ischemic injured dogs either susceptible or resistant to ventricular fibrillation [17]. Results in Fig. 4A show that SLN protein levels were significantly increased (∼3 fold higher) in the atria of HF dogs. On the other hand, SLN protein was significantly downregulated in the atria of ischemic injured dogs either susceptible (73.25±3.7%) or resistant (71.75 ±8.1 %) to ventricular fibrillation (Fig. 4B). Interestingly, exercise training which improves the cardiac function in ischemic injury model [21-23] restored SLN expression to the levels of control atria (Fig.4B). However, the expression of SERCA2a, PLB and calsequestrin (CSQ) were not significantly altered in the HF or ischemic injured dog models (Fig. 4).
Figure 4.
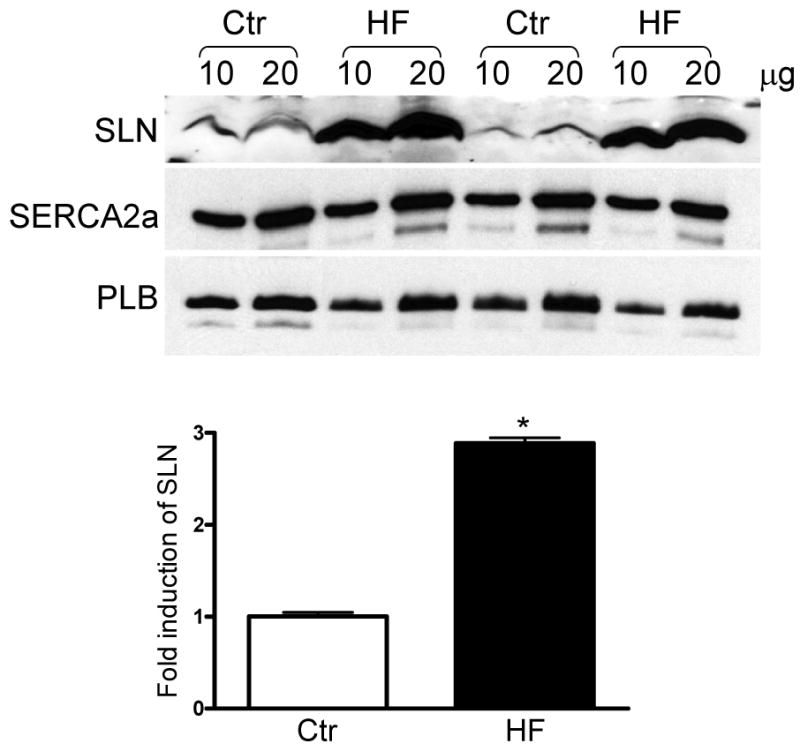
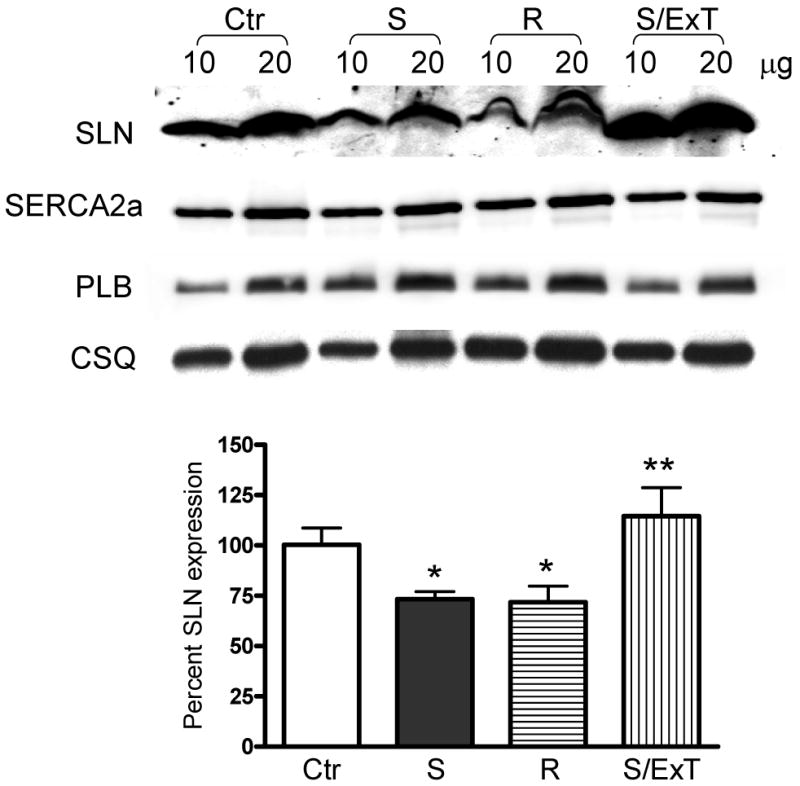
Quantitation of SLN in the atria of heart failure and ischemic injured dog models. (A) Representative Western blots showing the expression of SLN, SERCA2a and PLB in the atria of tachypacing induced heart failure dog model. SLN is upregulated in the heart failure (HF) atria. SERCA2a and PLB levels are unchanged between control (Ctr) and HF groups. Bar diagram showing the fold induction of SLN in HF atria. (B) Representative Western blots showing the protein levels of SLN, SERCA2a, PLB and CSQ in ischemic injured but either susceptible (S) or resistant (R) to ventricular fibrillation and exercise trained susceptible dogs (S/ExT). SLN expression is significantly downregulated in the atria of susceptible (S) and resistant groups but its level restored in the S/ExT groups to the levels of control atria (Ctr). SERCA2a, PLB and CSQ levels are unchanged between the groups. Bar diagram showing the percent expression of SLN in different groups. * indicates values significantly different from control groups and ** indicates values not significantly different from control groups.
SLN is upregulated in the plb-/- atria
To determine if loss of PLB alters the expression of SLN, we analyzed SLN protein levels in the plb-/- atria and ventricle. Results shown in Fig. 5 indicate that SLN levels were significantly increased in the plb-/- atria compared to that of wildtype controls (WT=100% vs. plb-/- = 152.4±10.8). The SLN levels were not detectable in WT as well as in plb-/- ventricles with total protein loading (data not shown). These results suggest that in the atria, loss of PLB could be compensated by SLN.
Figure 5.
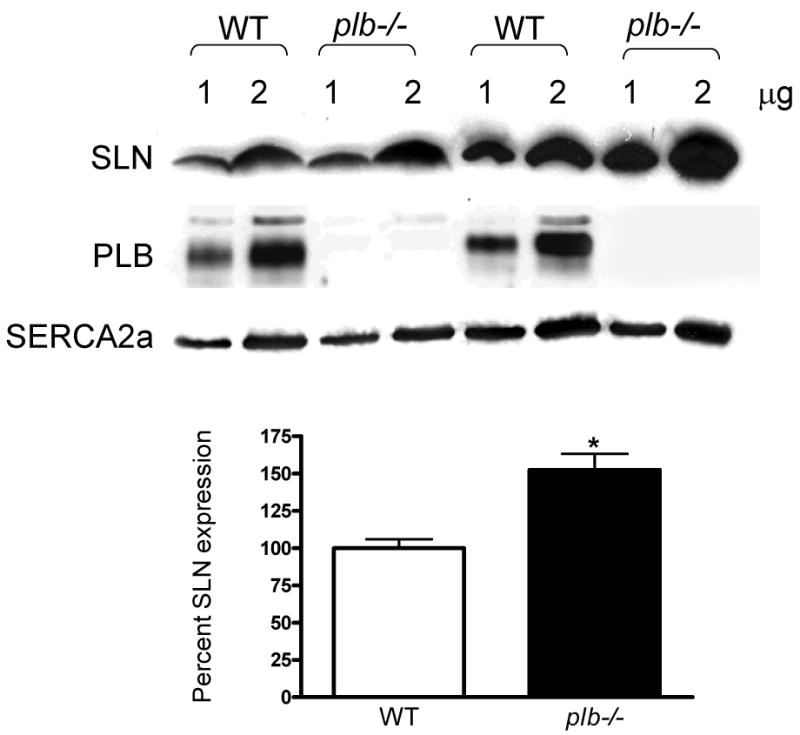
Increased SLN protein expression in plb-/- atria. Representative Western blots showing the expression of SLN and SERCA2a in the atria of plb-/- mice. SLN protein levels are 52% more in the plb-/- atria. SERCA2a levels are unchanged between WT and plb-/- atria. Bar diagram showing the percent expression of SLN in WT and plb-/- atria. * indicates values significantly different from WT atria.
Discussion
Our data clearly demonstrate that SLN exhibits both tissue and species-specific differences in its expression pattern. Within the heart, SLN protein expression is abundant in atria and confirmed the previous reports on the SLN mRNA expression pattern [4, 5]. Our data demonstrating differential expression of PLB in atria versus ventricle are also consistent with the previous report [24]. A critical question that remains to be answered is how SLN compares to PLB, whether they play complementary or distinct functional roles. The data showing the upregulation of SLN in the PLB null atria suggest that loss of PLB function may be compensated by SLN. The protein structure [25, 26] and expression analyses [present study] of SLN and PLB indicate that both are closely related proteins and the differential expression of these protein may play unique roles in the chamber specific regulation of Ca2+ transport function.
There are also species-specific differences when it comes to SLN expression in various skeletal muscle types. In rodents, the slow-twitch muscle expresses SLN, PLB and SERCA2a (slower isoform),whereas the fast twitch muscle express primarily SERCA1a (the isoform shown to transport Ca2+ with faster kinetics) and do not express either SLN or PLB that regulate SERCA pump activity. Based on our findings, we propose that differences in the expression of Ca2+ handling proteins (SERCA 1a, 2a and its regulators PLB and SLN) and contractile proteins in slow-twitch versus fast-twitch fibers [27- 29] could contribute to the unique contractile properties of these muscle types.
In larger mammals (rabbit and dog), SLN and SERCA2a are expressed at high levels both in soleus and EDL muscles. Unlike rodents, in larger mammals the fast-twitch muscles are composed of type I (slow) and type II (fast) fibers [29, 30] and therefore we predict that the expression of SERCA2a and SLN in the EDL muscle is due to the presence of slow-fiber types. Similarly, SLN and SERCA2a proteins are found at high levels in tongue, a tissue which has both slow- and fast -twitch skeletal muscle fibers [31]. Our results therefore suggest that the expression of SLN and SERCA2a are restricted to slow-twitch skeletal muscle fibers; though the ratio of SLN to SERCA2a may vary among different tissues. Based on these findings one might speculate that SLN is the major regulator of SERCA2a in skeletal muscle tissues. In skeletal muscle tissues, SERCA2a isoform is expressed primarily in slow-twitch fibers whereas in the fast twitch muscle, SERCA1a is the predominant isoform and plays a major role in maintaining SR Ca2+ stores needed for excitation-contraction coupling [reviewed in ref. 32]. Our findings suggest that SLN expression correlates well with SERCA2a but not with SERCA1a. Although in vitro studies have shown that SLN can inhibit SERCA1a pump activity when expressed together [33] the exact role of SLN in adult fast skeletal muscle needs to be explored.
An important finding is that SLN expression closely follows SERCA2a during cardiac and skeletal muscle development. We found that SERCA2a and SLN proteins are expressed at high levels in developing fast-twitch skeletal muscle but are absent in the adult muscle. During embryonic development, the fast-twitch muscle undergoes a sequence of transitions from slow- to fast-twitch fibers. This has been demonstrated by the continuous elimination of slow myosin from the fast-twitch fibers from birth to maturation [34-36]. Therefore, the expression of SLN and SERCA2a during fast-twitch muscle development recapitulates the slow-fiber phenotype. Similar pattern of co-expression of SERCA2a and SLN has been observed during tongue muscle development. These results suggest that SLN is a major regulator of SERCA2a in developing skeletal muscles.
The role of SLN in cardiac pathophysiology is less well understood. Recent studies showed that SLN mRNA is down-regulated in atrial myocardium of patients with chronic atrial fibrillation [12] and in the atria of pressure overloaded hypertrophy [13]. However, there are no protein data available to date to validate the importance of SLN in cardiac pathology. Here, for the first time we demonstrate that SLN protein levels are significantly upregulated in the atria of pacing induced canine model of chronic heart failure. This canine model mimics many aspects of chronic human heart failure [15] and exhibits decreased SR Ca2+ release characteristic of failing myocardium [37]. Based on the transgenic overexpression of SLN [6-8], we speculate that increased expression of SLN in the HF atria should inhibit SERCA pump activity and decrease Ca2+ transport and result in slowing of atrial function. The findings that SLN protein levels were significantly downregulated in the atria of ischemic hearts [16, 17] could be a compensatory mechanism to accelerate the SR Ca2+ uptake function during myocardial infarction. Exercise training program that improved cardiac function [21-23] also restored SLN protein levels further supports the view that SLN levels are critical for maintaining normal Ca2+ homeostasis.
In conclusion, we demonstrate that SLN has a distinct expression pattern compared to PLB. The findings that SLN and SERCA2a are co-expressed in the atria and skeletal muscle tissues along with functional studies reported from SLN transgenic mouse models [6-8] strongly suggest that SLN is an important regulator of SERCA2a isoform. The findings that SLN expression is modified in the atria during cardiac pathology further suggest that SLN levels could critically determine SERCA pump activity and Ca2+ homeostasis in the diseased myocardium.
Acknowledgments
We thank Dr. Litsa Kranias for providing us the phospholamban null mice. This work was supported by National Institute of Health Grant HL-64140 (to M. P.).
Abbreviations used
- SLN
Sarcolipin
- SR
Sarcoplasmic reticulum
- SERCA
SR Ca2+ ATPase
- PLB
Phospholamban
- NF-SLN
N-terminally Flagged-SLN
- SLN-CTAb
SLN C-terminal peptide antibody
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wawrzynow A, Theibert JL, Murphy C, Jona I, Martonosi A, Collins JH. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch Biochem Biophys. 1992;298:620–3. doi: 10.1016/0003-9861(92)90457-8. [DOI] [PubMed] [Google Scholar]
- 2.Odermatt A, Taschner PE, Scherer SW, Beatty B, Khanna VK, Cornblath DR, et al. Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics. 1997;45:541–53. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 3.Gayan-Ramirez G, Vanzeir L, Wuytack F, Decramer M. Corticosteroids decrease mRNA levels of SERCA pumps, whereas they increase sarcolipin mRNA in the rat diaphragm. J Physiol. 2000;524(Pt2):387–97. doi: 10.1111/j.1469-7793.2000.t01-2-00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minamisawa S, Wang Y, Chen J, Ishikawa Y, Chien KR, Matsuoka R. Atrial chamber-specific expression of sarcolipin is regulated during development and hypertrophic remodeling. J Biol Chem. 2003;278:9570–5. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- 5.Babu GJ, Zheng Z, Natarajan P, Wheeler D, Janssen PM, Periasamy M. Overexpression of sarcolipin decreases myocyte contractility and calcium transient. Cardiovasc Res. 2005;65:177–86. doi: 10.1016/j.cardiores.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Babu GJ, Bhupathy P, Petrashevskaya NN, Wang H, Raman S, Wheeler D, et al. Targeted overexpression of sarcolipin in the mouse heart decreases sarcoplasmic reticulum calcium transport and cardiac contractility. J Biol Chem. 2006;281:3972–9. doi: 10.1074/jbc.M508998200. [DOI] [PubMed] [Google Scholar]
- 7.Asahi M, Otsu K, Nakayama H, Hikoso S, Takeda T, Gramolini AO, et al. Cardiac-specific overexpression of sarcolipin inhibits sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs cardiac function in mice. Proc Natl Acad Sci U S A. 2004;101:9199–204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gramolini AO, Trivieri MG, Oudit GY, Kislinger T, Li W, Patel MM, et al. Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proc Natl Acad Sci U S A. 2006;103:2446–51. doi: 10.1073/pnas.0510883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minamisawa S, Uemura N, Sato Y, Yokoyama U, Yamaguchi T, Inoue K, et al. Post-transcriptional downregulation of sarcolipin mRNA by triiodothyronine in the atrial myocardium. FEBS Lett. 2006;580:2247–52. doi: 10.1016/j.febslet.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Trivieri MG, Oudit GY, Sah R, Kerfant BG, Sun H, Gramolini AO, et al. Cardiac-specific elevations in thyroid hormone enhance contractility and prevent pressure overload-induced cardiac dysfunction. Proc Natl Acad Sci U S A. 2006;103:6043–8. doi: 10.1073/pnas.0601072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–86. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 12.Uemura N, Ohkusa T, Hamano K, Nakagome M, Hori H, Shimizu M, et al. Down-regulation of sarcolipin mRNA expression in chronic atrial fibrillation. Eur J Clin Invest. 2004;34:723–30. doi: 10.1111/j.1365-2362.2004.01422.x. [DOI] [PubMed] [Google Scholar]
- 13.Shimura M, Minamisawa S, Yokoyama U, Umemura S, Ishikawa Y. Mechanical stress-dependent transcriptional regulation of sarcolipin gene in the rodent atrium. Biochem Biophys Res Commun. 2005;334:861–6. doi: 10.1016/j.bbrc.2005.06.186. [DOI] [PubMed] [Google Scholar]
- 14.Luo W, Grupp IL, Harrer J, Ponniah S, Grupp G, Duffy JJ, et al. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res. 1994;75:401–9. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 15.Nishijima Y, Feldman DS, Bonagura JD, Ozkanlar Y, Jenkins PJ, Lacombe VA, et al. Canine nonischemic left ventricular dysfunction: a model of chronic human cardiomyopathy. J Card Fail. 2005;11:638–44. doi: 10.1016/j.cardfail.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Billman GE, Schwartz PJ, Stone HL. Baroreceptor reflex control of heart rate: a predictor of sudden cardiac death. Circulation. 1982;66:874–80. doi: 10.1161/01.cir.66.4.874. [DOI] [PubMed] [Google Scholar]
- 17.Billman GE. A comprehensive review and analysis of 25 years of data from an in vivo canine model of sudden cardiac death: implications for future anti-arrhythmic drug development. Pharmacol Ther. 2006;111:808–35. doi: 10.1016/j.pharmthera.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Babu GJ, Wheeler D, Alzate O, Periasamy M. Solubilization of membrane proteins for two-dimensional gel electrophoresis: identification of sarcoplasmic reticulum membrane proteins. Anal Biochem. 2004;325:121–5. doi: 10.1016/j.ab.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 19.Vangheluwe P, Schuermans M, Zador E, Waelkens E, Raeymaekers L, Wuytack F. Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem J. 2005;389:151–9. doi: 10.1042/BJ20050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gramolini AO, Kislinger T, Asahi M, Li W, Emili A, MacLennan DH. Sarcolipin retention in the endoplasmic reticulum depends on its C-terminal RSYQY sequence and its interaction with sarco(endo)plasmic Ca(2+)-ATPases. Proc Natl Acad Sci U S A. 2004;101:16807–12. doi: 10.1073/pnas.0407815101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billman GE, Kukielka M. Effects of endurance exercise training on heart rate variability and susceptibility to sudden cardiac death: protection is not due to enhanced cardiac vagal regulation. J Appl Physiol. 2006;100:896–906. doi: 10.1152/japplphysiol.01328.2005. [DOI] [PubMed] [Google Scholar]
- 22.Billman GE, Kukielka M, Kelley R, Moustafa-Bayoumi M, Altschuld RA. Endurance exercise training attenuates cardiac beta2-adrenoceptor responsiveness and prevents ventricular fibrillation in animals susceptible to sudden death. Am J Physiol Heart Circ Physiol. 2006;290:H2590–9. doi: 10.1152/ajpheart.01220.2005. [DOI] [PubMed] [Google Scholar]
- 23.Billman GE, Kukielka M. Effect of endurance exercise training on heart rate onset and heart rate recovery responses to submaximal exercise in animals susceptible to ventricular fibrillation. J Appl Physiol. 2007;102:231–40. doi: 10.1152/japplphysiol.00793.2006. [DOI] [PubMed] [Google Scholar]
- 24.Luss I, Boknik P, Jones LR, Kirchhefer U, Knapp J, Linck B, et al. Expression of cardiac calcium regulatory proteins in atrium v ventricle in different species. J Mol Cell Cardiol. 1999;31:1299–314. doi: 10.1006/jmcc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 25.MacLennan DH, Asahi M, Tupling AR. The regulation of SERCA-type pumps by phospholamban and sarcolipin. Ann N Y Acad Sci. 2003;986:472–80. doi: 10.1111/j.1749-6632.2003.tb07231.x. [DOI] [PubMed] [Google Scholar]
- 26.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–77. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 27.Batkai S, Racz IB, Ivanics T, Toth A, Hamar J, Slaaf DW, et al. An in vivo model for studying the dynamics of intracellular free calcium changes in slow- and fast-twitch muscle fibres. Pflugers Arch. 1999;438:665–70. doi: 10.1007/s004249900100. [DOI] [PubMed] [Google Scholar]
- 28.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 29.Bicer S, Reiser PJ. Myosin light chain isoform expression among single mammalian skeletal muscle fibers: species variations. J Muscle Res Cell Motil. 2004;25:623–33. doi: 10.1007/s10974-004-5070-9. [DOI] [PubMed] [Google Scholar]
- 30.Lexell J, Jarvis JC, Currie J, Downham DY, Salmons S. Fibre type composition of rabbit tibialis anterior and extensor digitorum longus muscles. J Anat. 1994;185(Pt 1):95–101. [PMC free article] [PubMed] [Google Scholar]
- 31.McComas AJ. Oro-facial muscles: internal structure, function and ageing. Gerodontology. 1998;15:3–14. doi: 10.1111/j.1741-2358.1998.00003.x. [DOI] [PubMed] [Google Scholar]
- 32.Periasamy M, Kalyanasundaram A. SERCA pump isoforms: Their role in calcium transport and disease. Muscle Nerve. 2007;35:430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- 33.Odermatt A, Becker S, Khanna VK, Kurzydlowski K, Leisner E, Pette D, et al. Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem. 1998;273:12360–9. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 34.Lyons GE, Haselgrove J, Kelly AM, Rubinstein NA. Myosin transitions in developing fast and slow muscles of the rat hindlimb. Differentiation. 1983;25:168–75. doi: 10.1111/j.1432-0436.1984.tb01352.x. [DOI] [PubMed] [Google Scholar]
- 35.Periasamy M, Wieczorek DF, Nadal-Ginard B. Characterization of a developmentally regulated perinatal myosin heavy-chain gene expressed in skeletal muscle. J Biol Chem. 1984;259:13573–8. [PubMed] [Google Scholar]
- 36.Narusawa M, Fitzsimons RB, Izumo S, Nadal-Ginard B, Rubinstein NA, Kelly AM. Slow myosin in developing rat skeletal muscle. J Cell Biol. 1987;104:447–59. doi: 10.1083/jcb.104.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubalova Z, Terentyev D, Viatchenko-Karpinski S, Nishijima Y, Gyorke I, Terentyeva R, et al. Abnormal intrastore calcium signaling in chronic heart failure. Proc Natl Acad Sci U S A. 2005;102:14104–9. doi: 10.1073/pnas.0504298102. [DOI] [PMC free article] [PubMed] [Google Scholar]


