Abstract
Mutations affecting the BRCT domains of the breast cancer–associated tumor suppressor BRCA1 disrupt the recruitment of this protein to DNA double-strand breaks (DSBs). The molecular structures at DSBs recognized by BRCA1 are presently unknown. We report the interaction of the BRCA1 BRCT domain with RAP80, a ubiquitin-binding protein. RAP80 targets a complex containing the BRCA1-BARD1 (BRCA1-associated ring domain protein 1) E3 ligase and the deubiquitinating enzyme (DUB) BRCC36 to MDC1-γH2AX–dependent lysine6- and lysine63-linked ubiquitin polymers at DSBs. These events are required for cell cycle checkpoint and repair responses to ionizing radiation, implicating ubiquitin chain recognition and turnover in the BRCA1-mediated repair of DSBs.
DNA repair requires a series of molecular recognition steps that enable DNA damage response proteins to localize at and near DNA lesions. Failure of these responses results in genomic instability and predisposition to malignancy (1–3). Binding of the mediator of DNA damage checkpoint 1 (MDC1) protein to the phosphorylated tail of histone H2AX (γH2AX) (4–6) facilitates the formation of BRCA1 nuclear foci at DSBs (4, 7, 8). BRCT domain mutations also abrogate BRCA1 ionizing radiation (IR)–induced focus (IRIF) formation and localization to laser-induced DNA double-strand breaks (DSBs) (9). Thus, this protein-protein interaction domain participates in molecular recognition processes required for homing of BRCA1 to γH2AX- and MDC1-containing DNA repair sites. BRCA1 BRCT mutations disable BRCA1 interactions with two, known ligands, BACH1 (BRCA1-associated C-terminal helicase) (10) and CtIP (CtBP-interacting protein) (11). However, BACH1 and CtIP deficiency do not substantially alter BRCA1 localization to IRIF or laser-induced DNA DSBs (9), suggesting that a heretofore undetected BRCT-interacting protein recruits BRCA1 to DNA damage–induced foci.
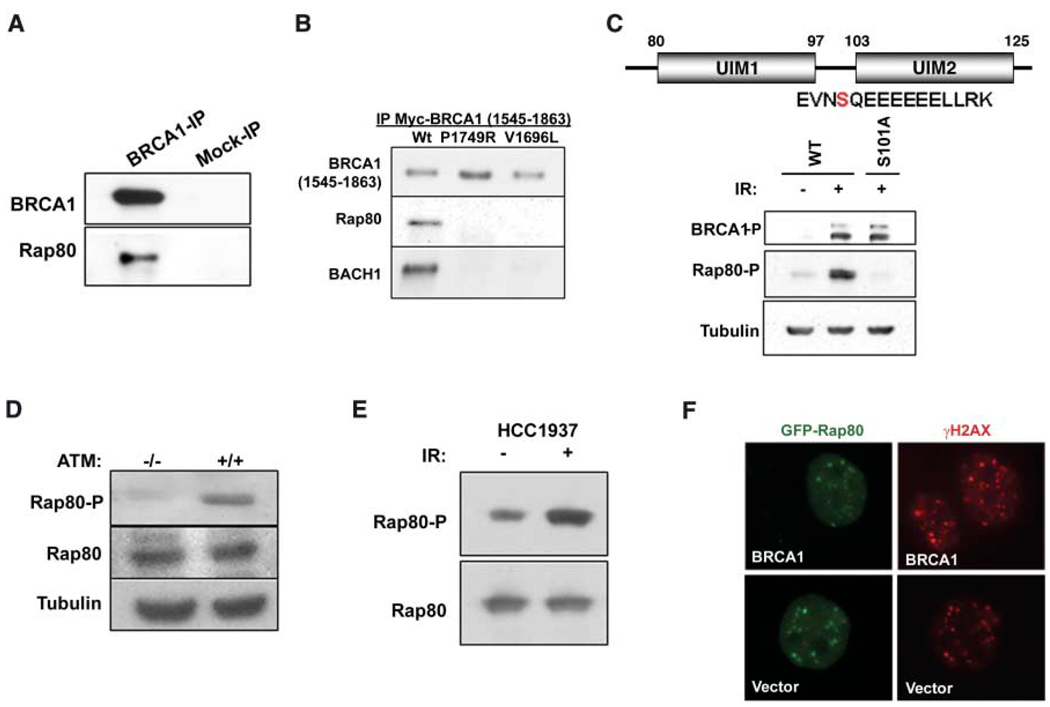
To address this hypothesis, we purified BRCA1-BARD1 complexes by double immunoaffinity chromatography of an epitope-tagged BARD1 protein (9). Multiple tryptic fragments derived from the tandem ubiquitin interaction motif (UIM) domain–containing protein, RAP80, were identified in this fraction by mass spectrometry (fig. S1A). Endogenous RAP80 coimmunoprecipitated (coIP) with endogenous BRCA1 (Fig. 1A). This interaction depended on the integrity of the BRCT motifs, because two BRCT-domain clinical missense mutations each disrupted RAP80 binding (Fig. 1B). Purification of tagged BARD1 after IR revealed the presence of a DNA damage–induced phosphorylation event between the two RAP80 UIMs at serine 101 (S101) (Fig. 1C). A rabbit polyclonal antibody specific for this phosphopeptide revealed IR-induced RAP80 phosphorylation (Fig. 1C). S101 represents a potential ATM (ataxia telangiectasia mutated) protein kinase phosphorylation site, given its contribution to a serine-glutamine (SQ) motif (12). Indeed, it was not phosphorylated in response to 10 gray (Gy) IR in ATM lymphoblasts (Fig. 1D). However, RAP80 IR-induced phosphorylation and DSB localization were independent of RAP80 interaction with BRCA1, as demonstrated in the BRCA1 BRCT–mutated HCC 1937 cell line (Fig. 1, E and F).
Fig. 1.
RAP80 interacts with the BRCA1 BRCT motif and responds to IR independently of BRCA1. (A) Co-immunoprecipitation (IP) between endogenous BRCA1 and RAP80. (B) Lysates were prepared from 293T cells transfected with WT or clinical mutant myc-BRCA1-BRCT domains. Myc antibody immunoprecipitated material was separated by SDS–polyacrylamide gel electrophoresis, and immunoblotting (IB) was performed. (C) HeLa S3 cells stably expressing epitope tagged-WT or S101A mutant RAP80 were IR-treated, and lysates were probed with an antibody specific to phosphorylated (P) RAP80-S-101. E, Glu; L, Leu; N, Asn; R, Arg; and V, Val. (D) ATM −/− and +/+ and (E) HCC1937 cells were gamma irradiated, and IB was performed on cell lysates as indicated. (F) HCC1937 cells reconstituted with vector or WT BRCA1 were transfected with GFP-RAP80 and gamma-irradiated, and, 1 hour later, IF was performed to assess colocalization with γH2AX.
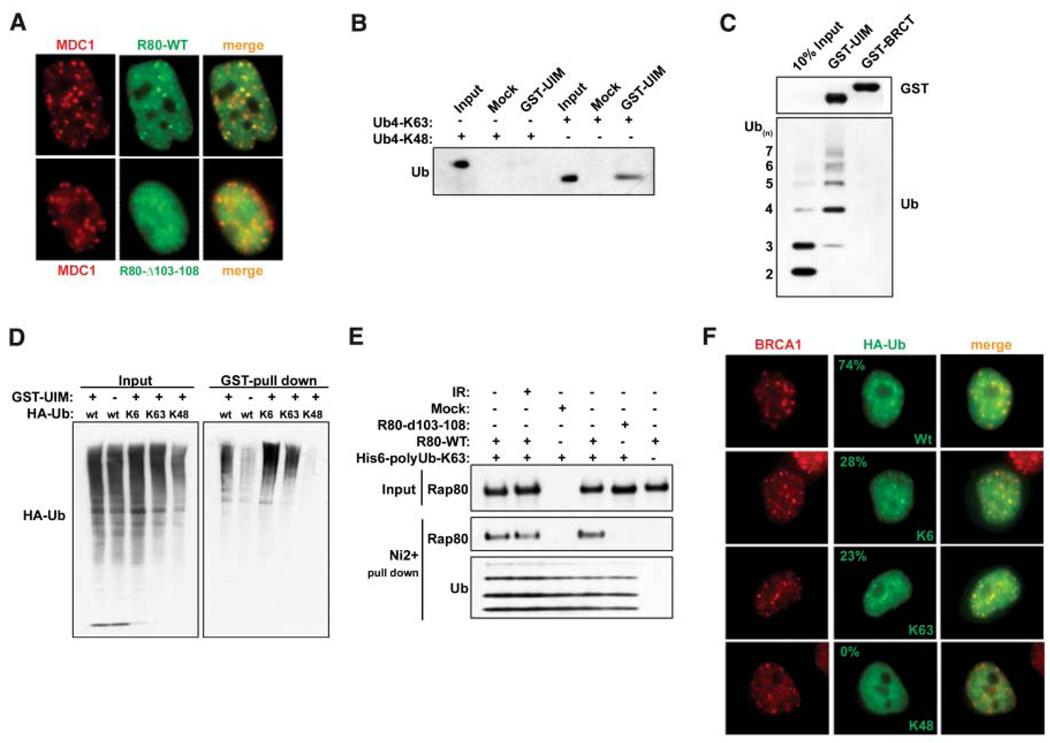
A series of green fluorescent protein (GFP)– RAP80 deletion mutants were monitored for IR-induced colocalization with MDC1 (fig. S2A). A six-amino-acid deletion mutant was generated that removed the stretch of conserved glutamates spanning residues 103 to 108 (Δ103–108), creating a hypomorphic UIM2 (103 to 125) domain. This mutant was expressed at equivalent levels to wild-type (WT) RAP80 in stable cell lines, and it coimmunoprecipitated with BRCA1 like WT RAP80 (fig. S2A). However, RAP80Δ103–108 demonstrated greatly reduced colocalization with MDC1 at IRIF formation 1 hour after 6 Gy (85% for WT and 2% for RAP80Δ103–108, n > 200) (Fig. 2A). Similar results were obtained for RAP80 UIM1 deletion mutants (fig. S2). These results suggest that RAP80 gains access to DSBs by cooperative UIM binding to ubiquitin or polyubiquitin. In this regard, a glutathione S-transferase (GST)– RAP80 fusion protein that included both UIM domains (residues 1 to 233) bound Lys63 (K63) isopeptide-conjugated tetra-ubiquitin with higher efficiency than K48-linked ubiquitin tetramers (Fig. 2B). A preference for polymers containing at least four ubiquitin molecules was observed for GST-RAP80 incubated with K63-linked ubiquitin polymers, each containing two to seven ubiquitin molecules (Fig. 2C).
Fig. 2.
RAP80 forms IRIF by binding to non-K48–linked ubiquitin. (A) HeLa cells stably expressing FLAG-HA– tagged WT or Δ103–108 RAP80 were treated with 6 Gy IR, and IF was performed 1 hour later. (B) K63- or K48-linked tetraubiquitin was incubated with the RAP80 GST-UIM domain. GST precipitations were analyzed by IB. (C) A mixture of K63-linked ubiquitin polymers containing two to seven molecules of ubiquitin (Ub2–7) was incubated with RAP80 GST-UIM or GST-BRCT. IB was performed as indicated after purification on glutathione-conjugated sepharose beads. (D) 293T cells transfected with the indicated HA- tagged ubiquitin expression vectors were treated with 10 µM MG132 for 2 hours before lysis and then incubated with RAP80 GST-UIM protein. The bound ubiquitin species were analyzed by IB. (E) FLAG-HA–tagged WT or Δ103–108 RAP80 were purified before and after IR from stably expressing HeLa-S3 cells by FLAG IP followed by FLAG-peptide elution. These proteins were then incubated with His-tagged, K63-Ub2–7, followed by purification on Ni2+- agarose beads. Ubiquitin-associated proteins were detected by IB. (F) HeLa cells were transfected with the same ubiquitin-expression vectors as in (D) and analyzed by IF 1 hour after IR. The percentage of transfected cells that display colocalization of epitope-tagged ubiquitin and BRCA1 is indicated (N > 200).
Ubiquitin can form polymers in vivo through any of its seven lysine residues (13). K6 isopeptides are the preferred linkage catalyzed by in vitro BRCA1 E3 activity (14–16). To test whether RAP80 can also interact with K6 linkages, we transfected human embryonic kidney 293T cells with expression vectors for ubiquitin species in which all lysine residues except one were mutated to arginine. This creates the in vivo synthesis of ubiquitin polymers containing defined isopeptide linkages. These chains, however, may also include endogenous ubiquitin and consequently may not be homogenous for a single, defined isopeptide linkage. In 293T cell lysates, GST-RAP80 bound to K6- and K63-linked polyubiquitin to a similar extent as to WT polyubiquitin, but did not recognize K48-linked polyubiquitin (Fig. 2D). These data indicate a preference for K63-, and possibly K6-, but not K48-linked ubiquitin polymers. Moreover, a positive correlation exists between ubiquitin binding and DSB localization, because WT RAP80 bound equally well to K63-linked ubiquitin before and after damage, whereas the UIM2 deletion mutant Δ103–108 demonstrated reduced binding to polyubiquitin (Fig. 2E). The RAP80 ubiquitin binding specificity observed in Fig. 2, B to E, appears to be relevant to the types of ubiquitin structures present at DNA damage sites. HeLa cells were transfected with vectors that produce different hemagglutinin (HA)-tagged ubiquitin chains, and colocalization of each ubiquitin species with BRCA1 at DNA damage sites was examined. BRCA1 colocalized with WT ubiquitin in about 75% of the cells examined and with either K6- or K63-linked ubiquitin in roughly 25% of the cells containing BRCA1 IRIF formation (Fig. 2F). K48-linked ubiquitin did not form foci at DNA damage sites (n = 500 transfected cells).
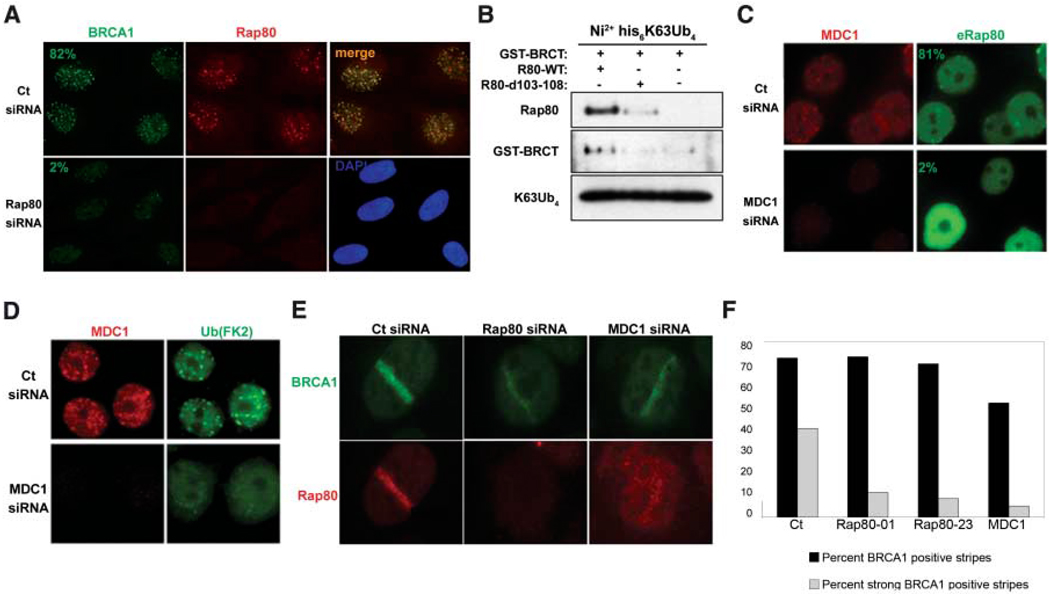
These results suggest that RAP80 is a candidate BRCT-interacting protein required to target BRCA1 to DNA damage sites. After small interfering RNA (siRNA) RAP80 knockdown in HeLa cells, strong BRCA1 foci were observed in just 8% of cells, compared to 82% in control (Ct) cells (n > 200). This indicates that RAP80 is largely required for BRCA1 IRIF formation (Fig. 3A). Moreover, WT RAP80 supported an interaction between the BRCT domain and ubiquitin, whereas the Δ103–108 mutant did not (Fig. 3B), providing in vitro evidence that RAP80 can target BRCA1 to ubiquitinated structures.
Fig. 3.
RAP80 targets BRCA1 to MDC1-dependent polyubiquitin structures at DSBs. (A) U2OS cells transfected with control (Ct) or RAP80-specific siRNA were fixed 6 hours after 10 Gy IR and analyzed by IF. (B) FLAG IP-purified WT or Δ103–108 RAP80 derived from HeLa S3 cells stably expressing these proteins was incubated with GST-BRCT and K63-linked tetraubiquitin. Ni2+-agarose precipitations were analyzed by IB as indicated. (C and D) HeLa cells transfected with Ct or MDC1-specific siRNA were fixed 6 hours after 6 Gy IR and analyzed by IF. (E) U2OS cells treated with the indicated specific siRNA were fixed at either 15 min or 4 hours after microirradiation by using a 337-nm laser and analyzed by IF. (F) Detection of the frequency and qualitative intensity of BRCA1 at γH2AX-positive stripes after treatment of U2OS cells with the indicated siRNA. Experiments were performed in duplicate at 15 min and 4 hours after laser-induced damage, and more than 125 stripes were counted per sample.
BRCA1 IRIF formation depends on the presence of γH2AX and MDC1 (4, 7). RAP80 also demonstrated a strong dependency on MDC1 expression for post-IR foci formation, with >80% of control cells showing eRAP80 foci and only 2% of MDC1-depleted cells exhibiting eRAP80 IRIF (Fig. 3C). Ubiquitin DSB localization was also reduced in cells depleted for MDC1 (n > 200) (Fig. 3D). As opposed to focus formation, BRCA1 homed to laser-induced DNA DSBs (stripes) at similar frequencies regardless of RAP80 or MDC1 expression (Fig. 3, E and F). However, knockdown of RAP80 or MDC1 (17) each reduced the intensity of BRCA1 immunostaining at stripes (Fig. 3, E and F), and RAP80 recruitment to laser stripes was also reduced in cells depleted of MDC1 (Fig. 3E). These results point to an MDC1-dependent pathway that is necessary to recruit RAP80-BRCA1 complexes to polyubiquitin structures at DSBs.
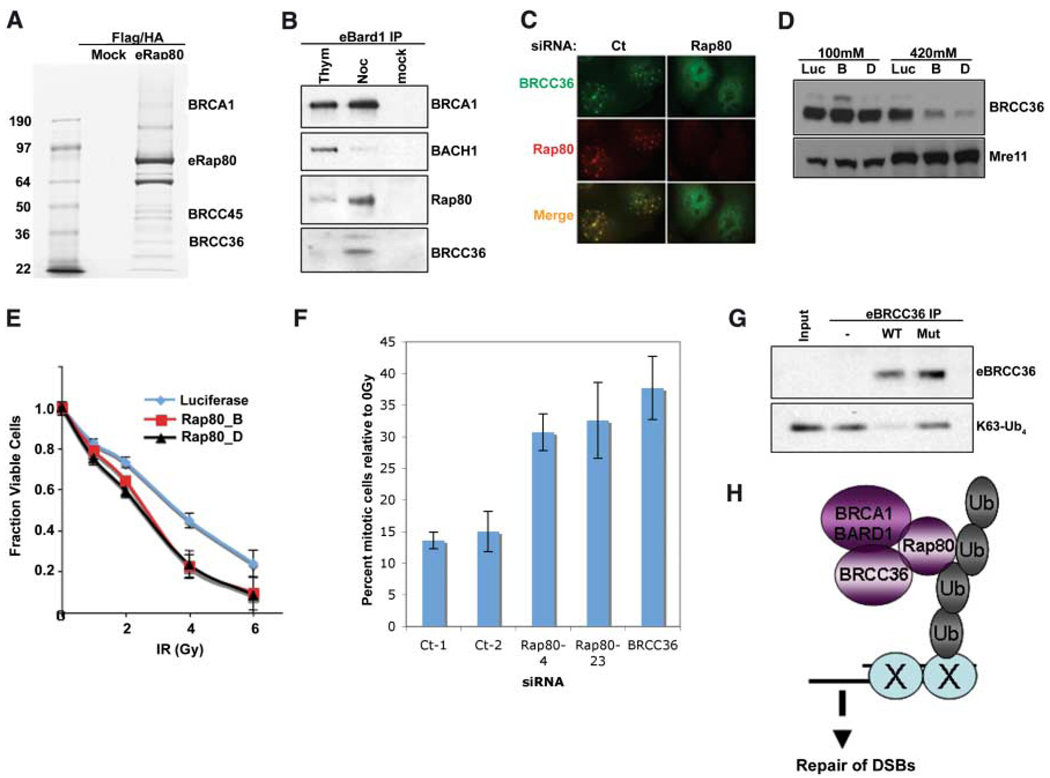
To gain insight into the functional interaction between BRCA1 and RAP80, we purified epitope-tagged RAP80 complex(es). Mass spectrometry revealed the presence of BRCA1 and the BRCA1-BARD1–interacting proteins BRCC45 and BRCC36 (18) (Fig. 4A). The abundance of a complex consisting of BRCA1-BARD1, RAP80, and BRCC36 increased in M phase–enriched cells during a nocodazole block compared with S phase–enriched cells that were collected after thymidine treatment (Fig. 4B). This suggests that RAP80 exists in a BRCA1-BARD1–associated complex containing BRCC36 that is different from the BRCA1-BARD1-BACH1 complex that occurs primarily in S phase. Consistent with this model, RAP80 was required for eBRCC36 subcellular localization. Specifically, RAP80 knockdown disrupted BRCC36 IRIF (Fig. 4C) and rendered BRCC36 more readily extractable from nuclear matrix and/or chromatin-rich fractions (Fig. 4D). Conversely, BACH1 IRIF appeared independent of RAP80, and BACH1 knockdown did not affect either RAP80 or BRCA1 stripe localization (fig. S4) (9). Moreover, RAP80 and BRCC36 demonstrated similar contributions to BRCA1-dependent DNA damage responses, implying that they contribute to similar DNA damage response functions. In this regard, stable knockdown of RAP80 using either of two different RAP80 short hairpin RNAs (shRNAs) produced dose-dependent IR supersensitivity (Fig. 4E), as previously reported for BRCC36 knockdown (19). In addition, depletion of RAP80 by two different siRNAs, like BRCC36 knockdown, resulted in a partial disruption of the G2 phase cell cycle checkpoint after 2 Gy IR compared with controls (Fig. 4F).
Fig. 4.
Interactions of RAP80, BRCC36, and BRCA1. (A) Coommassiestained gel of FLAG and HA double-purified RAP80 complexes from HeLa S3 cells stably expressing eRAP80. (B) HeLa S3 cells stably expressing eBARD1 were treated with 2 mM thymidine (Thym) or 0.5 µM nocodazole (Noc). FLAG-purified proteins were analyzed by IB. (C) U2OS cells expressing HA-tagged BRCC36 were transfected with shRNA specific to RAP80 or control and, 48 hours later, were monitored for IRIF at 1 hour after 6 Gy. (D) HeLa cells expressing stable siRNA vectors specific to luciferase (Luc) or RAP80 (B and D) were sequentially extracted with NETN buffer containing 100 mM NaCl and then NETN buffer containing 420 mM NaCl. Extracts were analyzed by IB. (E) HeLa cells expressing stable shRNA vectors specific to luciferase or RAP80 (B and D) were treated with the indicated doses of IR, and the number of viable cells were counted 96 hours later. The fraction of viable cells compared with a culture receiving 0 Gy is expressed graphically. Experiments were done in triplicate. Error bars indicate standard deviation. (F) The percentage of phosphohistone H3– positive cells (mitotic population) at 1 hour after 2 Gy IR compared with at 0 Gy (i.e., mock irradiation) was plotted for each siRNA-transfected U2OS population. Experiments were done in triplicate; error bars indicate standard deviation. (G) FLAG peptide eluates from FLAG IPs performed on extracts of HeLa S3 cells stably expressing FLAG-HA–tagged WT or a catalytically inactive BRCC36 mutant were incubated with K63-linked tetraubiquitin (K63-Ub4) for 30 min at 37°C. The reaction mixtures were then analyzed by IB. (H) Model for recruitment of a BRCA1-BARD1-BRCC36-RAP80 complex to sites of DNA damage by binding to non-K48–linked ubiquitin structures.
BRCA1 ubiquitin ligase activity is activated by DNA damage and is required for its G2 checkpoint function (16, 20). BRCC36 bears homology to the JAMM domain family of deubiquitinating enzymes (DUBs) (21), suggesting that, in addition to E3 activity, DUB activity may be involved in the checkpoint and repair functions of this specific BRCA1-BARD1 complex. Wild type and a double active site mutant, His122→Gln122 (H122Q) and H124Q BRCC36, were purified from HeLa S3 cells and examined for DUB activity on K63-linked tetraubiquitin (K63-Ub4). DUB activity was detected for WT BRCC36 but not for the mutant (Fig. 4G), establishing that BRCC36 has DUB activity toward K63-linked ubiquitin substrates, the same structures to which RAP80 binds.
These data support a model wherein MDC1-dependent, non–K48-linked ubiquitin chains at DNA damage sites are used as a targeting mechanism by specific BRCA1 complexes (Fig. 4H). Should this be a general phenomenon, RAP80 may represent the first in a new class of DNA repair proteins that uses tandem UIM domains as part of its recruitment to DSBs. In contrast to IRIF formation, incomplete BRCA1 localization at laser-induced DSBs still occurs in the absence of γH2AX (22), MDC1 (17), or RAP80 (Fig. 3, E and F). These findings may reflect the fact that BRCA1/BARD1 heterodimers are components of multiple distinct complexes (9) and that each may access DSBs by different mechanisms. Taken together, these findings strongly suggest an essential role for ubiquitin recognition by a specific BRCA1 complex in the response to DSB formation. In addition, the synthesis and turnover of certain polyubiquitinated structures by BRCA1 E3 and BRCC36 DUB activities, respectively, may contribute to BRCA1-dependent DSB repair.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/316/5828/1198/DC1
Materials and Methods
Figs. S1 to S4
References and Notes
References
- 1.Bassing CH, et al. Cell. 2003;114:359. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 2.Celeste A, et al. Cell. 2003;114:371. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Nat. Genet. 2005;37:750. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 4.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. Nature. 2003;421:961. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 5.Stucki M, et al. Cell. 2005;123:1213. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 6.Rogakou EP, Boon C, Redon C, Bonner WM. J. Cell Biol. 1999;146:905. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celeste A, et al. Science. 2002;296:922. doi: 10.1126/science.1069398. published online 4 April 2002 (10.1126/science.1069398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassing CH, et al. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8173. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg RA, et al. Genes Dev. 2006;20:34. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantor SB, et al. Cell. 2001;105:149. [Google Scholar]
- 11.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. J. Biol. Chem. 1998;273:25388. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 12.Shiloh Y. Nat. Rev. Cancer. 2003;3:155. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 13.Peng J, et al. Nat. Biotechnol. 2003;21:921. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 14.Wu-Baer F, Lagrazon K, Yuan W, Baer R. J. Biol. Chem. 2003;278:34743. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- 15.Morris JR, Solomon E. Hum. Mol. Genet. 2004;13:807. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 16.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. EMBO J. 2006;25:2178. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bekker-Jensen S, et al. J. Cell Biol. 2006;173:195. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Y, et al. Mol. Cell. 2003;12:1087. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK. Cancer Res. 2006;66:5039. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Fu S, Lai M, Baer R, Chen J. Genes Dev. 2006;20:1721. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambroggio XI, Rees DC, Deshaies RJ. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Celeste A, et al. Nat. Cell Biol. 2003;5:675. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]