Abstract
The completion of the genome sequence of Trypanosoma cruzi has been followed by several studies of protein expression, with the long-term aim to obtain a complete picture of the parasite proteome. We report a proteomic analysis of an organellar cell fraction from T. cruzi CL Brener epimastigotes. A total of 396 proteins were identified by LC-MS/MS. Of these, 138 were annotated as hypothetical in the genome databases and the rest could be assigned to several metabolic and biosynthetic pathways, transport, and structural functions. Comparative analysis with a whole cell proteome study resulted in the validation of the expression of 173 additional proteins. Of these, 38 proteins previously reported in other stages were not found in the only large-scale study of the total epimastigote stage proteome. A selected set of identified proteins was analyzed further to investigate gene copy number, sequence variation, transmembrane domains and targeting signals. The genes were cloned and the proteins expressed with a c-myc epitope tag in T. cruzi epimastigotes. Immunofluorescence microscopy revealed the localization of these proteins in different cellular compartments, such as endoplasmic reticulum, acidocalcisome, mitochondrion, and putative cytoplasmic transport or delivery vesicles. The results demonstrate that the use of enriched subcellular fractions allows the detection of T. cruzi proteins that are undetected by whole cell proteomic methods.
Keywords: Immunofluorescence, Organelles, Protein isoforms, Trypanosoma cruzi
1 INTRODUCTION
Trypanosomiases are diseases caused by parasites from the order Kinetoplastida, such as Trypanosoma cruzi (American trypanosomiasis or Chagas disease), Trypanosoma brucei (African sleeping sickness or African trypanosomiasis) and Leishmania major (leishmaniases). Chagas disease is endemic to Latin America, with 18 million people infected and over 40 million at risk [1]. The T. cruzi life cycle involves different stages. The epimastigote form in the insect vector differentiates into infective the metacyclic trypomastigote form, which will subsequently invade mammalian host cells, transform into intracellular amastigotes, and later on into bloodstream trypomastigotes [2].
There are only two drugs available for the treatment of Chagas disease, nifurtimox and benznidazole, but these are unsatisfactory due to their limited efficacy in the chronic stage of the disease and their toxic side effects [1]. Much effort is now focused on finding new inhibitors targeting pathways such as sterol biosynthesis [3], protein prenylation [4], peptidases [5], and pyrophosphate metabolism [6] that have unique features, are essential for the parasite, or are different from their counterparts in the host.
The completion of the genome sequence of T. cruzi [7], as well as those of two other related kinetoplastids (T. brucei and L. major) [8, 9], provided a wealth of new information that has greatly increased the pace of research into the biology of these parasites. The T. cruzi genome sequence showed that approximately 12,000 proteins are encoded per haploid genome. The actual number of gene products may be even higher due to missing regions in the genome assembly, alternative splicing and post-translational modifications. Putative function could be assigned to 50.8% of the predicted protein-coding genes on the basis of significant homology to previously characterized proteins or known functional domains [7]. This means that for more than 5,900 proteins, the function is unknown and further characterization is needed. Also, more than 50% of the genome consists of repeated sequences, such as retrotransposons, tandemly repeated genes, and genes for large families of surface molecules, which increases proteome complexity further.
Proteomics is the approach of choice to find novel gene products, expression data, and to validate genome annotations. Several T. cruzi proteomic studies have been reported [10–14]. In the case of organellar proteomics, the use of cell fractions reduces the complexity of the samples, and proteins present in smaller amounts and specific to the organelles are revealed [15]. In this way, fractionation techniques can make it possible to discover potentially important gene products that are expressed at low levels, or are masked by highly expressed genes.
T. cruzi has distinct organelles like the kinetoplast (a specialized region of the mitochondrion containing the mitochondrial DNA [16]; the acidocalcisomes (acidic organelles rich in phosphorus and calcium [17]; the reservosomes (lysosome-like organelles) [18]; and the glycosomes (peroxisome-like organelles containing several glycolytic enzymes [19], whose full protein content and functions are not completely known.
In this paper we report the proteomic analysis of an enriched organellar fraction from T. cruzi CL Brener epimastigotes. The objective of this work was to find novel expressed proteins and determine their localization by expressing them with a c-myc epitope tag in the parasite. The identification of a number of proteins that were not detected in previous whole cell proteomic analyses demonstrates the importance of analysing protein expression using enriched organellar fractions.
2 MATERIALS AND METHODS
2.1 Cell culture
Epimastigotes from T. cruzi CL Brener were grown at 28°C in RPMI 1640 (Sigma) supplemented with 5% FBS (Gibco), 0.5% Hepes (Amersham), 3 mM hemin (Sigma), 0.5% tryptone (Oxoid), 2 g/l sodium bicarbonate (Sigma), and streptomycin/ penicillin (Gibco), pH 7.2. Liver infusion tryptose (LIT) media [20] supplemented with 10% FBS (Gibco) and streptomycin /penicillin (Gibco), pH 7.3 was used for the transfection cultures
2.2 Cell Fractionation
We based our fractionation on the method described by Scott and Docampo ([21] that enriches mainly for acidocalcisomes and glycosomes along with other organelles. Briefly, late exponential epimastigote cultures were harvested at 3000 rpm, washed twice with PBS and once in hypotonic lysis buffer B (125 mM sucrose, 50 mM KCl, 20 mM Hepes, 4 mM MgCl2, 0.5 mM EDTA, 5 mM dithiotreitol (DTT), and a protease inhibitor cocktail (Sigma, P8340). The cells were homogenized in a mortar with 1.5 × wet weight silicon carbide, until 80–90% cell disruption could be observed under the microscope. The mixture was transferred to a clean tube and washed twice in buffer B to remove the silicon carbide. The supernatant was centrifuged twice at 2,000 rpm to separate nuclei and unbroken cells; and subsequently at 10,000 rpm. The resulting pellet was resuspended in buffer B and subjected to a 20–50% OptiPrep (Axis-Shield, Norway) discontinuous gradient ultracentrifugation for 1 hour at 20,000 rpm using a Sorvall AH629 swinging rotor [21].
2.3 Sample Preparation for One and Two Dimensional Gel Electrophoresis
The pellets obtained from several gradient ultracentrifugations were pooled together after being resuspended in a minimal volume of buffer S (7 M urea, 2 M thiourea, 1.2 % Chaps, 0.5% Triton X-100, 16 mM dithiotreitol (DTT), plus the protease inhibitor cocktail. This suspension was lightly sonicated in an ice bath and subjected to freeze/thaw cycles to disrupt membranes, centrifuged at 2500× g for 10 min, and the pellet washed twice in buffer S. The supernatants from these steps were combined and named sample A. The remnant pellet from this step was resuspended in 1% SDS, and became sample B.
Sample A was applied to a linear IPG (immobilized pH gradient) strip, pH 3–10, 24 cm in length (Amersham Biosciences) overnight. First dimension was run for 21:30 h at a stepwise increasing voltage, 300 V to 3500 V, 2 mA and 5 watts constant, in a Multiphor II apparatus (Amersham Biosciences). The IPG strip was incubated in an equilibration solution (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and 0.002% bromophenol blue), in the presence of DTT and iodoacetamide; applied on top of a 12% SDS-PAGE 20 × 24 cm format on a vertical Ettan-Dalt six unit (Amersham Biosciences), and stained with Colloidal Coomassie Blue. Scanned gel images were analysed for spot count with PDQuest 8.0 (BioRad) software. Sample B was mixed with Laemmli sample buffer and loaded onto a 4–20% linear gradient SDS-PAGE 20 × 24 cm and the gel was stained with Colloidal Coomassie Blue.
2.4 Nanoflow Liquid Chromatography – Tandem Mass Spectrometry (Nanoflow LC-MS/MS)
The mass spectrometry was performed at the Swegene Proteomics core facility at the Sahlgrenska Academy, Gothenburg University (Gothenburg - Sweden). Bands and spots from 1D and 2D gels, respectively, were excised with a scalpel, unstained 3 times in a mixture of 50% ammonium bicarbonate (25 mM) and 50% acetonitrile, and once more in 50% ammonium bicarbonate (25 mM) and 25% acetonitrile and 25% methanol. The samples were dried and digested for 15 min with 0.1–0.2 mg of trypsin in 20 ml of 50% ammonium bicarbonate (25 mM) and 50% acetonitrile. Ammonium bicarbonate (25 mM, pH 8) was added to cover the gels, and the incubation was continued for 12 h at 37 °C. The fragments were extracted with 10–50 ml of a mixture of 75% acetonitrile and 5% trifluoroacetic acid (in water). For the liquid chromatography an Agilent 1100 binary pump was used, together with a reversed phase column, 200 × 0.05 mm, packed in-house with 3 m particles Reprosil-Pur C18-AQ (Dr. Maisch, Ammerbuch, Germany). A split to approximately 100 nl/min reduced the flow through of the column. A 40 min gradient 10–50% CH3CN in 0.2% COOH was used for separation of the peptides. The nanoflow LC-MS/MS were done on a 7-Tesla (Linear Trap Quadrupole- Fourier Transform) LTQ-FT mass spectrometer (Thermo Electron) equipped with a nanospray source modified in-house. The spectrometer was operated in data-dependent mode, automatically switching to MS/MS mode. MS-spectra were acquired in the FTICR, while MS/MS-spectra were acquired in the LTQ-trap. For each scan of FTICR, the 3 most intense, doubly or triply charged, ions were sequentially fragmented in the linear trap by collision-induced dissociation.
2.5 MS/MS Analysis and Protein Identification
All the tandem mass spectra were compared using MASCOT (Matrix Science, London) locally against the T. cruzi whole genome predicted protein database. The search parameters were set as follows: MS accuracy, 15 ppm; MS/MS accuracy, 0.5 Da; one missed cleavage allowed; fixed propionamide modification of cysteine; and variable modification of oxidized methionine. Phosphorylations were also evaluated in a second search. A protein hit was set to a minimum of one peptide match, with an e-value of 10−3, only when this single peptide was: a) longer than 5 amino acids; b) had a peptide score above the threshold given by the Mascot with a 95% of confidence that this peptide is not a random event in the sample. When additional peptides with lower scores than the threshold were present in the match, they were also considered. Peptides with more than one peptide score in the sample were counted as one, being the highest scored considered.
2.6 Functional Classification and Bioinformatics Analysis
The classification was based on the gene ontology (GO) annotation for each gene on GenBank and/or InterPro or Pfam (http://www.sanger.ac.uk/Software/Pfam/) searches for each protein, and in most cases, through PubMed bibliography search. For the hypothetical proteins set, transmembrane helix regions (TMHs) and signal peptides were predicted using PolyPhobius [22], HMMTOP [23] and TMHMM [24], as well as the SignalP 3.0 Server [25]. Targeting signal predictions were performed using the following servers: Ptarget [26]; WOLF pSORT [27]; PeroxiP [28]; PENCE Proteome Analist (Lu, Szafron et al. 2004), targetP (Emanuelsson, Nielsen et al. 2000; Emanuelsson, Elofsson et al. 2003) and SLP-Local [29]. Gene copy numbers estimations were obtained using the database http://gemini1.cgb.ki.se/ek/cruzi/main.php [30].
2.7 Generation of Expression Vectors and Clones
pTEX-myc vector (courtesy of John Kelly, London School of Hygiene and Tropical Medicine, UK) was transformed into the C- and N-terminal tag destination vectors, using the Gateway Vector Conversion System (Invitrogen), according to the instructions of the manufacturer. Primers carrying the attB flanking regions were designed for each gene, with or without start/stop codons, for tagging the C- or N-terminal ends of the expressed recombinant proteins. The genes were amplified by PCR and purified from the gel using the Gel Extraction Kit (Qiagen). These products were ligated into the pDONR 221 vector (Invitrogen), by a recombination reaction using a Gateway® BP Clonase™ Enzyme Mix (Invitrogen), to generate the pENTR vector, carrying the specific inserts. These clones were subsequently recombined with the generated destination vectors, using the Gateway® LR Clonase™ enzyme mix (Invitrogen), to render the final pTEX-gene-myc (C-terminal tag) and pTEX-myc-gene (N-terminal tag) expression vectors. All reactions were performed as described in the manufacturers’ manuals. The constructs were confirmed by sequencing, using the DYEnamic ET dye terminator kit and a MegaBACE 1000 Sequencer (Amersham Biosciences).
2.8 T. cruzi Transfections
Approximately 4 × 107 parasites were collected for each transfection, washed twice in electroporation buffer (137 mM NaCl, 5 mM KCl, 5.5 mM Na2HPO4, 0.77 mM glucose, 21 mM Hepes, pH 7.2) and resuspended in 350 µl of the same buffer. The suspension was transferred into a 2 mm gap electroporation cuvette (BioRad) and incubated on ice with 50–60 µg of plasmid DNA, for 10 min. One pulse of 400 V, 500 µF was applied using a GenePulser electroporator (BioRad) followed by incubation on ice for 5 min. Subsequently, the cells were transferred to a flask containing 10 ml liver infusion tryptose (LIT) media with 10% newborn calf serum. Selection was applied after 48 h by addition of 250-µg/ml geneticin, and 48 h later 500 µg/ml drug were used. Control cells died after 3–4 weeks, and the transformed cultures expanded after 4–5 weeks.
2.9 Immunofluorescence Microscopy
Transfected exponential cultures were pelleted and washed twice in PBS and fixed in 4% paraformaldehyde, 0.1 M cacodylate buffer and 0.1 % glutaraldehyde in PBS, pH 7.2. Fixed cells were adhered to polylysine coated cover slides, permeabilized with Triton X-100 in PBS, neutralized in 50 mM NH4Cl and blocked with 3% BSA in PBS. Incubation with primary and secondary antibodies in 3% BSA-PBS was done for one hour, and visualization was done using a DeltaVision Restoration System microscope (Applied Precision Inc.) and the SoftWoRx image analysis software (Applied Precision Inc). The primary antibodies used were: mouse monoclonal supernatant anti-myc (E910 epitope) dilution 1:4 (obtained from the Developmental Studies Hybridoma Bank at the University of Iowa, Department of Biological Sciences); rabbit anti-BiP (gift from J. Bangs [31] dilution 1:600; rabbit anti-TbgGAPDH (gift from Fred Opperdoes) dilution 1:300; rabbit anti-TbVP1 (gift from Norbert Bakalara (Lemercier, Espiau et al. 2004)) dilution 1:800. The secondary antibodies were: goat Alexa anti-mouse or anti-rabbit 488 and 546 nm (Molecular Probes), dilutions 1:1000 and 1:800, respectively. Mitochondria were visualised using MitoTracker Red CMXRos and the plasma membrane was labelled using FM 1-43FX, both from Molecular Probes (Invitrogen).
3 RESULTS
3.1 Protein Electrophoresis and Mass Spectrometry
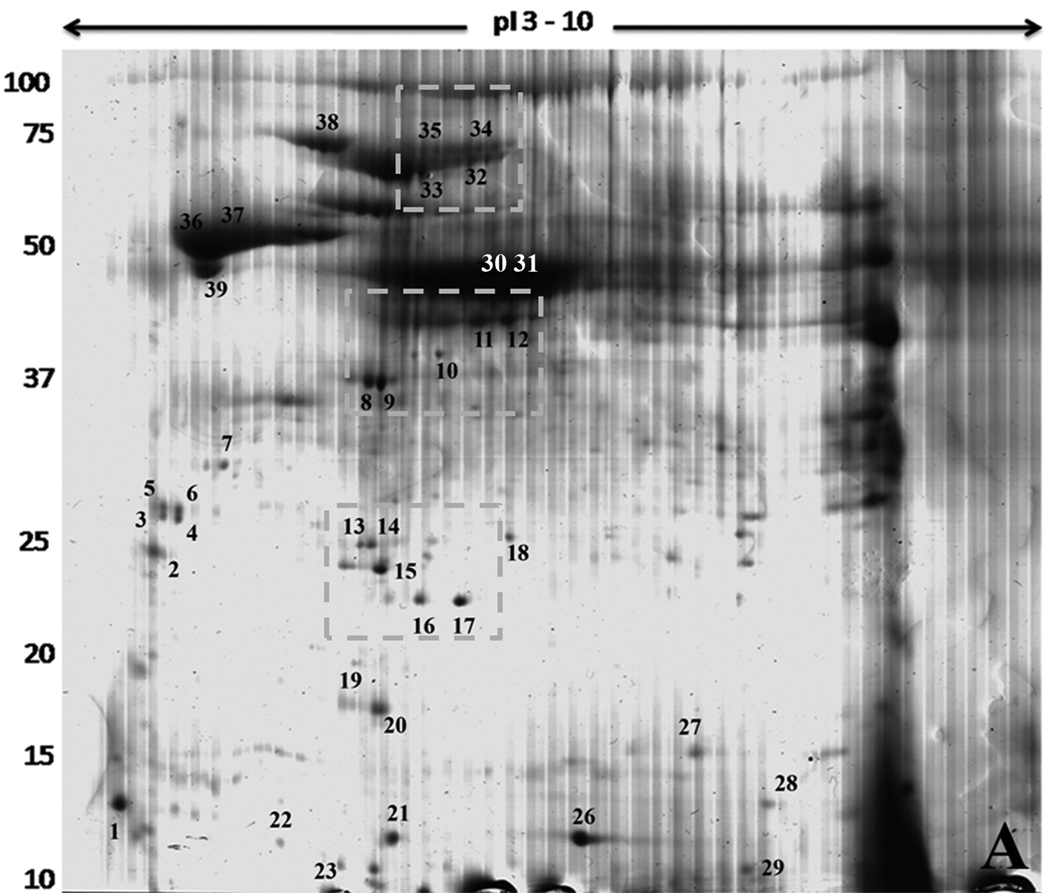
Samples A and B derived from the pellet of the density gradient ultracentrifugation, as explained in the experimental section, were resolved by two dimensional gel electrophoresis (2DGE) and one-dimensional gradient gel electrophoresis (1DGE), respectively (Figure 1 and Figure 2). Gels were stained with colloidal Coomassie blue (CBB) in order to detect proteins in amounts sufficient for mass spectrometry analysis. PDQuest analysis software detected over 180 spots on the 2D gel (Figure 1), distributed mainly in the acidic region of the gel. This pattern was always observed in the various sample preparations. A dark vertical region appearing towards the alkaline region was caused by a salt bridge generated during the first dimension run of this particular gel, and it was not used for spot excision. In order to improve the resolution of the dense high molecular weight zone, additional gels with smaller amounts of sample were run, resulting in the discrimination and excision of a few more spots. Thirty-nine visible spots were excised for a first analysis by LC-MS/MS, which generated a total of 125 hits in the T. cruzi translated genome; 92 were gene products annotated with described or putative identity to already known proteins and 33 were annotated as hypothetical proteins.
Figure 1.
Two Dimensional Gel Electrophoresis of T. cruzi proteins. The sample was electrophoresed on strips from pH 3 to 10 in the first dimension. Molecular weight markers are shown on the left and expressed in kDa. The gel was stained with Colloidal Coomassie Blue. Spots excised for mass spectrometry are numbered and located close to the respective spot. Dotted squares show the isoforms regions discussed in the text.
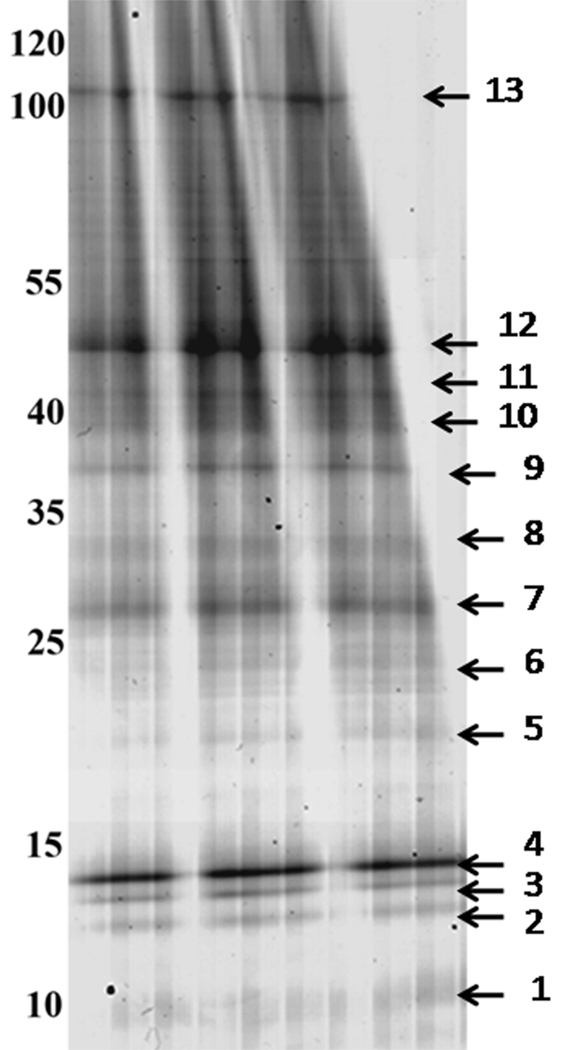
Figure 2.
One dimensional Gradient Gel Electrophoresis of Proteins from T. cruzi. The sample was extracted with SDS and run in a big format gradient gel (5–20%). Molecular weight markers are shown on the left side and expressed in kDa. Numbered arrows mark the 13 bands that were cut for mass spectrometry analysis. The gel was stained with Colloidal Coomassie Blue.
The 1D gel generated 13 distinguishable bands that could be excised for a first analysis (Figure 2) and run through LC-MS/MS, resulting in a total of 334 gene product identifications; 222 with identity to known proteins and 112 hypothetical ones. Proteins with isoelectric points over 10 were highly represented in this data set, such as histones (pI >11) and ribosomal proteins (pI 10 to 12), as well as membrane proteins.
A total of 63 proteins were present in both the 2D and 1D data set, resulting in a final number of total proteins in our sample of 396; 258 proteins with known identity and 138 hypothetical proteins.
Information for each hit that Mascot matched against the protein database, is available as Supplementary Data (Tables S1–S5), including: band or spot number; accession number; annotated protein name or description; molecular weight (Mr); protein score; number of peptides matched; e-values for each matched peptide; peptide sequences and number of additional proteins sharing those peptides (meaning proteins encoded by gene homologues or by additional copies of the gene). Only unique peptide matches are listed and only the highest scoring peptides are counted and reported in case of duplicated matches. A summary is shown in Table I. There was a list of additional peptides that could not be assigned to any gene product annotated in GenBank for T. cruzi.
Table 1.
Mascot Protein Identification: The table shows the main identified protein from each band (B) or spot (S) in the 1D and 2D gels, respectively. Mascot protein scores are indicated together with the number of unique peptides that matched to the sequence. An extended table with all the identifications and peptide sequences can be found in the supplementary material: Tables S1 to S5.
| Band/Spot no. | Accession no. | Protein Identity | Mr | Score | # Pept. |
|---|---|---|---|---|---|
| B1 | EAN87014.1 | kinetoplastid membrane protein KMP-11 | 10876 | 46 | 1 |
| B2 | EAN81533.1 | histone H4, putative | 11163 | 609 | 25 |
| B3 | EAN83533.1 | histone H2B, putative | 12353 | 532 | 25 |
| B4 | EAN84604.1 | histone H3, putative | 21714 | 412 | 21 |
| B5 | EAN90899.1 | ribosomal protein L21E (60S), putative | 18271 | 346 | 9 |
| B6 | EAN82805.1 | 40S ribosomal protein L14, putative | 20789 | 277 | 9 |
| B7 | EAN87845.1 | 40S ribosomal protein S4, putative | 30998 | 658 | 19 |
| B8 | EAN94839.1 | beta tubulin, putative | 50520 | 536 | 19 |
| B9 | EAN91944.1 | glyceraldehyde 3-phosphate dehydrogenase, putative | 39277 | 779 | 30 |
| B10 | EAN99374.1 | pyruvate phosphate dikinase, putative | 101851 | 714 | 23 |
| B11 | EAN99374.1 | pyruvate phosphate dikinase, putative | 101851 | 722 | 21 |
| B12 | EAN84978.1 | elongation factor 1-alpha (EF-1-alpha), putative | 49652 | 862 | 32 |
| B13 | EAN99374.1 | pyruvate phosphate dikinase, putative | 101851 | 2013 | 85 |
| S1 | EAN83392.1 | calmodulin, putative | 16814 | 261 | 8 |
| S2 | EAN98394.1 | p22 protein precursor, putative | 26158 | 363 | 12 |
| S3 | EAN83132.1 | hypothetical protein, conserved | 26313 | 44 | 2 |
| S4 | EAN84330.1 | mucin-associated surface protein (MASP), putative | 44673 | 21 | 1 |
| S5 | EAN83132.1 | hypothetical protein, conserved | 26313 | 144 | 2 |
| S6 | EAN84888.1 | tryparedoxin peroxidase, putative | 25774 | 239 | 6 |
| S8 | EAO00075.1 | cytochrome C oxidase subunit IV, putative | 39137 | 724 | 34 |
| S9 | EAN99021.1 | cytochrome C oxidase subunit IV, putative | 39097 | 1038 | 40 |
| S10 | EAN82883.1 | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor, | 39837 | 311 | 8 |
| S11 | EAO00025.1 | alcohol dehydrogenase, putative | 42442 | 596 | 18 |
| S12 | EAO00025.1 | alcohol dehydrogenase, putative | 42442 | 636 | 17 |
| S13 | EAN88322.1 | hslvu complex proteolytic subunit-like, putative | 23080 | 215 | 4 |
| S14 | EAN88322.1 | hslvu complex proteolytic subunit-like, putative | 23080 | 257 | 8 |
| S15 | EAN86560.1 | cytochrome c oxidase subunit V, putative | 22450 | 392 | 20 |
| S16 | EAN82242.1 | co-chaperone GrpE, putative | 24363 | 650 | 19 |
| S17 | EAN81815.1 | co-chaperone GrpE, putative | 24421 | 777 | 34 |
| S18 | EAN98352.1 | pyruvate phosphate dikinase, putative | 101833 | 438 | 15 |
| S19 | EAN97136.1 | ATP synthase, epsilon chain, putative | 20447 | 248 | 6 |
| S20 | EAN85671.1 | ribonucleoprotein p18, mitochondrial precursor, putative | 27768 | 169 | 6 |
| S21 | EAN85850.1 | 10 kDa heat shock protein, putative | 10694 | 236 | 35 |
| S22 | EAN82097.1 | hypothetical protein, conserved | 14352 | 247 | 7 |
| S23 | EAN93009.1 | hypothetical protein, conserved | 13620 | 136 | 6 |
| S24 | EAN93009.1 | hypothetical protein, conserved | 13620 | 114 | 4 |
| S25 | EAN81211.1 | cytochrome c oxidase subunit 10, putative | 13942 | 158 | 9 |
| S26 | EAN85850.1 | 10 kDa heat shock protein, putative | 10694 | 290 | 67 |
| S27 | EAN85850.1 | 10 kDa heat shock protein, putative | 10694 | 170 | 4 |
| S28 | EAN89116.1 | heat shock protein 20, putative | 16000 | 269 | 8 |
| S29 | EAN85850.1 | 10 kDa heat shock protein, putative | 10694 | 84 | 3 |
| S30 | EAN85498.1 | gamma-glutamyl carboxypeptidase, putative | 44258 | 714 | 34 |
| S31 | EAN85497.1 | gamma-glutamyl carboxypeptidase, putative | 44390 | 778 | 31 |
| S32 | EAN92318.1 | 69 kDa paraflagellar rod protein, putative | 70087 | 1273 | 37 |
| S33 | EAN92318.1 | 69 kDa paraflagellar rod protein, putative | 70087 | 1430 | 51 |
| S34 | EAN87979.1 | paraflagellar rod protein 3, putative | 69189 | 1246 | 48 |
| S35 | EAN87979.1 | paraflagellar rod protein 3, putative | 69189 | 1219 | 39 |
| S36 | EAN94839.1 | beta tubulin, putative | 50520 | 912 | 169 |
| S37 | EAN81053.1 | alpha tubulin, putative | 50549 | 1122 | 67 |
| S38 | EAN87966.1 | glucose-regulated protein 78, putative | 71414 | 2323 | 97 |
| S39 | EAN90720.1 | calreticulin, putative | 46396 | 906 | 29 |
3.2 Protein Isoforms and Allelic Genomic Versions
On the 2DGE, various common patterns of protein isoforms were observed (Figure 1 dashed boxes). We cut alternating spots from these areas to determine if they belonged to the same protein with differences in their isoelectric point (pI). This was the case for spots 8/9 (cytochrome C oxidase subunit IV, putative), 11/12 (alcohol dehydrogenase, putative), 13/14 (hslvu complex proteolytic subunit-like, putative), 16/17 (co-chaperone GrpE, putative), 23/24 (hypothetical protein), 30/31 (gamma-glutamyl carboxypeptidase, putative), 32/33 (69 kDa paraflagellar rod protein, putative) and 34/35 (paraflagellar rod protein 3, putative). In some cases, the source of isoelectric point (pI) variation could be assigned to amino acid sequence differences (Table 2 and Supplementary Table S5 and S6). In others, all identified peptides were shared, indicating that post-translational modifications as responsible for the migration pattern differences. By examining the 1DGE data set we found 43 cases with variations ranging from one amino acid change to several peptides present in one copy but not in the other or even the detection of more distant family members with lower homology (Supplementary Table S6). Of the 43 cases, 9 were hypothetical proteins linked to other hypothetical proteins and thus either homologous alleles or repeated copies.
Table 2.
Protein Isoforms: Spots presenting isoforms that differ in one or more sequence aminoacids are shown. (*) The peptides shared between the isoforms that carry the sequence differences are shown in bold. In italics are the peptides present in one isoform that do not match to the other copies. For an extended version of this table see Supplementary Table S6.
| Band/Spot No. | Accession no. | Protein Identity | # PEPT. | Sequence (*) |
|---|---|---|---|---|
| Spot 11–12 Spot 30–31 |
EAN85498.1 | glutamamyl carboxypeptidase, putative | 19 | R.EIAEEYR.R K.GGSHFWVR.V R.ALPNETVSK.M K.DYLEGLGVK.C K.LVAFDTTSR.N R.EIAEEYRR.N K.CTLLHNAER.N K.LREIAEEYR.R R.SMDHTQWLAK.L R.SMDHTQWLAK.L + Oxidation (M) K.WDSDPFTLTER.D R.SYVETQLLPAMK.A R.SYVETQLLPAMK.A + Oxidation (M) K.AEFEDAEIVITPR.N K.ANLWATLPGDGGVTK.G R.NETPSFEGSEEAPFTK.L K.GGIILSGHTDVVPVDGQK.W R.AEGCIIGEPTGMTVVIAHK.G + Oxidation (M) R.SYVETQLLPAMKAEFEDAEIVITPR.N + Oxidation (M) |
| EAN82710.1 | glutamamyl carboxypeptidase, putative | 18 | R.DETPSFEGSEEAPITK.L | |
| EAN82317.1 | glutamamyl carboxypeptidase, putative | 16 |
R.EIAEEYR.R K.DYLEGLGVK.C K.LVAFDTTSR.N R.EIAEEYRR.N K.CTLIHNAER.N K.LREIAEEYR.R R.SMDHTQWLAK.L R.SMDHTQWLAK.L + Oxidation (M) K.WDSDPFTLTER.D R.SYVETQLLPAMK.A R.SYVETQLLPAMK.A + Oxidation (M) K.AEFEDAEIVITPR.N K.ANLWATLPGDGGVTK.G K.GGIILSGHTDVVPVDGQK.W R.AEGCIIGEPTGMTVVIAHK.G + Oxidation (M) R.SYVETQLLPAMKAEFEDAEIVITPR.N + Oxidation (M) |
|
| EAN88605.1 | acetylornithine deacetylase-like, putative | 4 |
R.LIAFDTTSR.N M.PLDSVEWLR.R R.IDDFVAATAQK.M K.WDSDPFTLTER.D |
|
| EAN96060.1 | glutamamyl carboxypeptidase, putative | 4 |
K.LVSFDTTSR.N K.AYVNDTLLPSMK.K + Oxidation (M) R.YLPEAEAEKFEER.I K.DYLEGLGVK.C |
|
| Spot 8 | EAO00075.1 | cytochrome C oxidase subunit IV, putative | 29 | R.HITPEAIK.A |
| Spot 9 | EAN99021.1 | cytochrome C oxidase subunit IV, putative | 30 | R.HITSEAIK.A |
| Spot 11–12 | EAO00025.1 | alcohol dehydrogenase, putative | 14 |
K.EVQIPSGFEQLGMK.E + Oxidation (M) K.EVQIPSGFEQLGMKEK.D + Oxidation (M) |
| EAN97413.1 | alcohol dehydrogenase, putative | 14 |
K.EVQIPPGFEQLGMK.E + Oxidation (M) K.EVQIPPGFEQLGMKEK.D + Oxidation (M) |
|
| Spot 16 | EAN82242.1 | co-chaperone GrpE, putative | 16 |
K.VSAEEIESNK.N K.VSAEEIESNKNLSSIHTGVK.L |
| Spot 17 | EAN81815.1 | co-chaperone GrpE, putative | 16 |
K.VSTEEIESNK.N K.VSTEEIESNKNLSSIHTGVK.L |
3.3 Comparison to Whole Cell Proteomics and Novel Expression Data
A large proteomic study of whole cell preparations from the four main life cycle stages of T. cruzi [13] was reported at the same time of the publication of the trypanosomatid genomes. The data is available through TcruziDB (http://www.tcruziDB.org), where the number of peptides and spectra found for each protein in each parasite stage, as well as the percentage of sequence coverage given by Mascot searches, is present. When comparing our total protein list to that of TcruziDB, 34% (135/396) of the proteins in our data set were not detected there in any stage, and another 9.6% (38/396) were not found in the epimastigote stage (See Supplementary Table S7 and S8C). Only for 8 proteins not found in TcruziDB there is evidence available from other reports indicating expression in the epimastigote stage. Around 50% of the hypothetical proteins detected in our sample were not found in the T. cruzi total whole cell proteomic study (Supplementary Table S8C). Therefore, the use of enriched subcellular fractions for proteome analysis resulted in the validation of additional 173 proteins being expressed in epimastigotes, and not present in the T. cruzi protein expression database.
3.4 Identified Proteins and Functional Classification
The total number of identified expressed genes from the 1D and 2D data was 396, of which 258 were annotated (http://www.ncbi.nlm.nih.gov/Genbank/) as homologues of previously described genes and 138 were annotated as hypothetical. Not all the annotated gene products (http://www.ncbi.nlm.nih.gov/Genbank/) had an assigned function and most of the annotated with a function were labelled as putative. We therefore searched the literature and compared to other kinetoplastids or to other species to group the proteins into different functional categories (Supplementary Table S7). In agreement with the enrichment of organelles in this fraction, we found well established markers of T. cruzi acidocalcisomes (vacuolar-type proton translocating pyrophosphatase, EAN91609.1) [21], vacuolar ATPase, subunit C, EAN86373.1 [32], and vacuolar-type Ca2+ ATPase (EAN95492.1) [32]; glycosomes (solanesyl diphosphate synthase, EAN80692.1 [33], hexokinase, EAN94612.1 [34], pyruvate phosphate dikinase, EAN86096.1 [35], phosphoenolpyruvate carboxykinase, EAN88964.1 [36], and malate dehydrogenase, EAN90616.1 [37]); mitochondria (glutamate dehydrogenase, EAN87724.1 [38], aspartate aminotransferase, EAN86462.1 [39], alanine aminotransferase, EAO00085.1 [39], malate dehydrogenase, EAN87359.1 [37], and kinetoplast DNA-associated protein, EAN85210.1 [40]); reservosomes (glutathione-S-transferase, EAN88371.1 [41], cysteine peptidase, EAN83138.1 [42], and serine carboxypeptidase, EAN81557.1 [43]); endoplasmic reticulum (40S ribosomal protein S4, EAN87845.1 [44], and calreticulin, EAN82340.1 [45]); and flagellum and cytoskeletal components (paraflagellar rod proteins, EAN92318.1, EAN87979.1, EAN81200.1, EAN83974.1, EAN99876.1 [46], flagellar calcium-binding protein, EAN86963.1 [47], beta, EAN94839.1), and alpha (EAN81053.1) tubulins [48], actin, EAN85411.1 [49], and kinetoplastid membrane protein KMP-11, EAN87014.1 [50]). Among the only five proteins that were known to localize exclusively to the plasma membrane we detected two that belong to families with large numbers of members (mucin-associate surface protein, EAN84550.1 [14], and trans-sialidase EAN86366.1 [51]). Most of the presumably cytoplasmic proteins detected were, however, ribosomal proteins and proteins involved in protein translation that associate to the membrane of the endoplasmic reticulum, chaperones, microtubule proteins, or proteins involved in trafficking that usually associate with different types of vesicles. Some nuclear histones (histone H4, EAN81533.1 [52], H2B, EAN83533.1 [53], H2A, EAN85330.1 [54], and H3, EAN84604.1 [55]) were also detected, indicating some contamination with nuclear proteins.
3.5. Hypothetical proteins
The set of proteins identified by LC-MSMS was searched for transmembrane regions (TMHs) and putative signal peptides, together with target signal predictions, to infer some probable cellular location in this parasite and were used for updated protein blast (http://www.ncbi.nlm.nih.gov/blast) searches (Supplementary Tables S8A, S8B and S8C). The majority of the proteins (117/138) did not have a signal peptide and were predicted to be non-secreted proteins. Of this set, 14/117 contained one transmembrane region and 12/117 had 2 to 14 TMHs. The fraction of probable secreted proteins with a signal peptide prediction was 20/138, where 50% of them had no TMH and the other half contained more than 1 transmembrane region (Supplementary Table S8A). There was only one case where none of the servers used agreed, EAN85286.1. The SignalP 3.0 server classified a set of 14 proteins as carrying a signal that was not cleaved, thus indicating that the proteins possess an anchor signal (Supplementary Table S8A).
The hypothetical proteins were also searched for other import signals, which could direct them to specific organelles. The different location predictor servers did not coincide in most of the cases, but a probable single location could be predicted when two or more results coincided for 74/138 cases (Supplementary Table S8B). The majority of the predictions showed nuclear (20/138), mitochondrial (20/138), secretory pathway (17/138) and cytoplasmic (13/138) locations. Dual localization proteins were cytoplasmic-nuclear (25/138), nucleus-mitochondrion (1/138) and cytoplasm-glycosome (1/138). Almost 26% of the hypothetical proteins could not be assigned to any compartment, as there was no agreement for the six servers used (Supplementary Table S8B). There was a lack of a good glycosomal target prediction server, as only two proteins were tentatively assigned to this organelle.
Protein Blast searches showed that the majority of the hypothetical proteins were represented in all three trypanosomatids (T. cruzi, T. brucei and L. major) and only a few hits with lower similarity to proteins of other species were found (Supplementary Table S8C). In 16 cases, while the annotation for T. cruzi was that of hypothetical proteins, the genes had homologues in other trypanosomatids that were annotated as known gene products, although with low expected values (Supplementary Table S8C).
3.6 Subcellular localization of Five Hypothetical Proteins
Five proteins were selected for further characterization. The selection was based on target prediction, with the aim to locate hypothetical proteins to organellar compartments, and to find novel transporters localized to the acidocalcisome. Thus, three of the proteins selected have more than one transmembrane domain. The T.cruzi vacuolar proton pyrophosphatase, V-H+-PPase, was used both as a control for acidocalcisome location of the tagged proteins, and as control for stable transfections. The genes inserted into the pTEX expression vector converted by the Gateway System, carried an NH2-terminal or COOH-terminal fusion of the human c-myc epitope 9E10. Both C- and N-terminal tagged constructs were generated, sequenced, and their fusion frame confirmed, but the N-terminal clones did not render stable transfected cultures and only the transfectants tagged in the C-terminal were used.
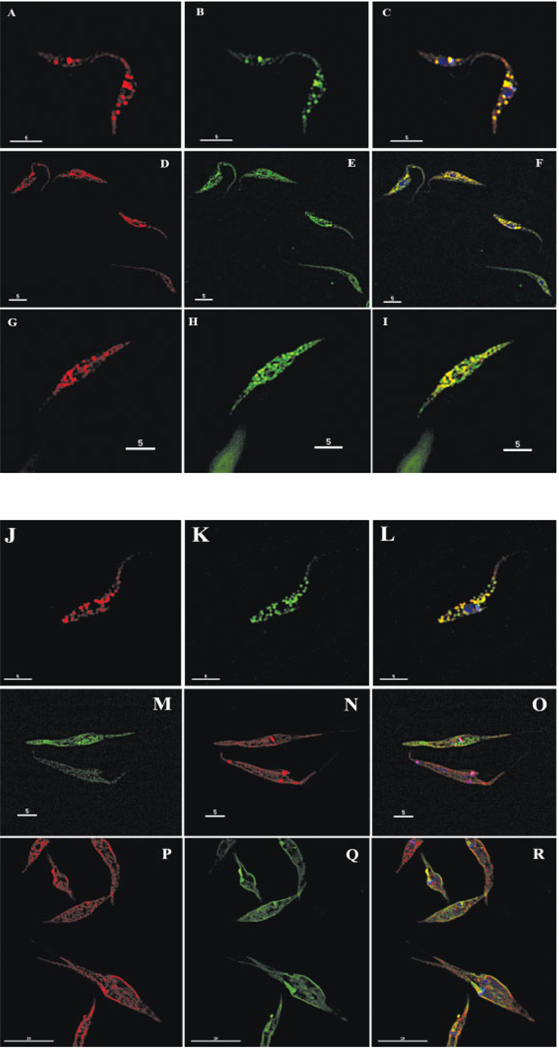
The over-expressed V-H+-PPase-myc fusion protein was located to the acidocalcisome as expected [56], as shown by immunofluorescence using both the mouse monoclonal anti-myc tag and the specific rabbit polyclonal anti-TbVP1 primary antibodies. These results support the idea that the tag does not interfere with the final destination of the protein (Figure 4A–C).
Figure 4.
Immunofluorescence microscopy: The images show the results of the colocalization assays performed with the transformed epimastigote cultures. An anti-myc antibody was used to detect the respective overexpressed fusion protein. Panel A–C: EAN91609.1 acidocalcisome V–H+-PPase: A. anti-myc; B. anti-TbVP1; C. merged + DAPI staining - Panel D–F: EAN91782.1 hypothetical ER protein: D. anti-myc; E. anti-BiP; F. merged + DAPI staining - Panel G–I: EAN86484.1 hypothetical partially ER protein: G. anti-myc; H. anti-BiP; I. merged - Panel J–L: EAN89594.1 metal ion transporter, acidocalcisome: J. anti-myc; K. anti-TbVP1; L. merged + DAPI staining - Panel M–O: EAN83261.1 hypothetical mitochondrial protein: M. anti-myc; N. Mitotracker; O. merged + DAPI staining - Panel P–R: EAN81429.1 hypothetical vesicular protein: P. anti-myc; Q. FM1-43FX; R. merged + DAPI staining.
3.6.1 Endoplasmic Reticulum (ER)
We identified several proteins predicted to belong to the ER and the secretory pathway in our proteomic data, such as protein disulfide isomerase, COP-associated proteins, and calreticulin, involved in the correct folding and transport of proteins, and essential for parasite survival [31, 57–60], as well as several hypothetical proteins.
We here add two novel proteins to the ER network of T. cruzi; EAN91782.1 (Figure 4D–F) and EAN86484.1 (Figure 4G–I). EAN91782.1 [Band 8; 4 peptides matched] is a 327 amino acid protein (35.9 kDa/pI 10) with a signal peptide and one predicted transmembrane region. Three prediction servers located it to the ER and the rest to the secretory pathway. This hypothetical protein was not detected previously in whole cell epimastigote stage proteomics. Its allelic variant is EAN89907.1, and it has orthologs in L. major and T. brucei. It contains a Pfam domain (PF00106) for short chain dehydrogenases, a protein family with a wide range of substrate specificities. A protein Blast search showed weaker similarity to other hypothetical proteins and dehydrogenases, being the highest hit emb|CAL52554.1| 17 beta-hydroxysteroid dehydrogenase type 3, HSD17B3 (ISS) [Ostreococcus tauri] (Supplementary Table S8C). Steroid biosynthesis has been shown to localize to the ER [61].
The second ER protein, EAN86484.1 [Band 9; 4 peptides matched], showed partial localization to the ER (Figure 4G–I). It has a length of 458 amino acids (50.5 kDa/pI 10), a signal peptide and 3 transmembrane regions (TMH). No specific location could be assigned with the servers used. The gene is also present in T. brucei but the L. major counterparts are partial sequences and of lower similarity. It was predicted in-silico to be a mitochondrial protein, but when we performed co-localization studies with Mitotracker, no association to the mitochondria was seen (data not shown).
3.6.2 Acidocalcisome
We found a novel protein localized in the acidocalcisome (Figure 4J–L), which was not detected previously by whole cell proteomics. EAN89594.1 [Bands 10 and 11; 2 peptides] annotated as a putative metal transporter, is a protein of 464 amino acids (49.9 kDa/pI 6.4) with no signal peptide but five transmembrane spanning regions. Pfam detected a cation efflux domain (PF01545). Its ortholog in L. major is annotated as a zinc-transporter like protein. The gene is single copy in the genome annotation and both alleles have the same length and features.
3.6.3 Mitochondrion
Another hypothetical protein, EAN83261.1 [Band 7; 16 peptides matched], is a 29.6 kDa protein (pI 9.3) and it was predicted not to have transmembrane regions or a signal peptide. In-silico predictions were not able to locate it to any particular compartment in the cell. From the cloning and transfection experiments we determined its location to the mitochondrion of epimastigotes of T. cruzi (Figure 4M–O). The protein shows a uniform distribution throughout the mitochondrion, but does not intensely label the kinetoplast area. This protein contains the Pfam domain PF01459: Porin-3 (IPR001925: Eukaryotic porin). During the cloning and sequencing process of this gene product in the pTEX-myc vector, we detected more than 5 distinct gene variants. An analysis of the shotgun read coverage of this gene in the genome assembly [30] estimated its copy number to 13.
3.6.4 Undefined or Putative Cytoplasmic Vesicles
Another selected protein, EAN81429.1 [Band 10 and 13; 2 peptides], is a hypothetical protein with 14 trans-membrane domains and no signal peptide predicted. This hypothetical protein contains a Pfam domain PF06813.3: nodulin-like, which represents a conserved region within plant nodulin-like proteins. Members of the family are mainly transporters. Our localization studies showed that it is not located to any previously known compartment in the cell (Figure 4P–R). It was located to the plasma membrane by three prediction servers. It shows a punctuated pattern, distributed along the cell body that is not consistent with the plasma membrane, acidocalcisome, glycosomes, mitochondrion, Golgi, ER or any other known compartment. The size of these dots is smaller than that of vesicles of the endocytic pathway.
4 DISCUSSION
This study describes a comprehensive proteomic analysis of an organellar cell fraction from T. cruzi. We have previously reported [21] the yield of different markers and the 60- and 5-fold purification of acidocalcisomes and glycosomes, respectively, in this fraction, as detected by enzymatic analysis. As pointed out in [62], the concept of “pure” organelles is untenable and the high sensitivity of the proteomic approach usually leads to identification of multiple contaminants in any given fraction.
The fractionation method used enriched the sample for characteristic organelles, such as acidocalcisomes, glycosomes, mitochondria, reservosomes, flagellum, and endoplasmic reticulum, reducing the level of contamination from highly abundant proteins in the cell, and from plasma membrane proteins, as shown by liquid chromatography-tandem mass spectrometry analyses (Table S7). The large number of identifications coming from the small number of bands and spots analyzed was due to proteins that co-migrated in the gel as a result of similar pI and/or molecular weight, incomplete focussing, and/or the presence of proteolytic degradation products contaminating other areas of the gels.
For several genes, different protein variants were detected. It is known that post-translational modifications and sequence variations in protein isoforms alter the mass to charge ratio of the protein, generating a characteristic migration pattern in 2DGE. The hits obtained from the Mascot searches detected amino acid differences between copies of the same protein. Even though post-translational modifications were not analyzed in detail, it is likely that such modifications were one of the reasons for the presence of different protein isoforms. The peptide differences varied from a single amino acid substitution to entire peptides present in one but not in the other copy. This can be explained as either large allelic differences between gene homologs because of the hybrid nature of the T. cruzi CL Brener strain [63], or to differences between closely related gene copies present as tandem or dispersed repeats. A large proportion of genes in T. cruzi have been shown to be repeated in tandem and for certain genes there are significant levels of nucleotide sequence differences between copies. There is no information in the T.cruzi genome database concerning the presence of additional copies of hypothetical gene products. Our mass spectrometry data showed that some hypothetical proteins shared the whole set of peptides identified for each hit, and that others had amino acid sequence variants.
The subcellular localization of many of the proteins identified has not previously been experimentally demonstrated. However, a number of the proteins identified could be assigned to the mitochondria, endoplasmic reticulum, acidocalcisomes, plasma membrane, and vesicular compartments. Only highly expressed cytoplasmic proteins appeared in our proteomic analysis, although most of them presumably attached to the endoplasmic reticulum, trafficking vesicles, or tubulin. Alpha and beta tubulin were present in most of the spots and bands analyzed, as they are the major components of the cytoskeleton of trypanosomes. We also searched for putative signals among the hypothetical proteins, which would identify their localization, and immunofluorescence microscopy was used to confirm these presumptions using epitope tagged proteins. The proteins investigated were located in the endoplasmic reticulum, acidocalcisomes, mitochondria and small vesicles. Of the two proteins associated with the endoplasmic reticulum, one completely co-localized with the ER chaperone marker BiP. The second protein might be part of the smooth ER, as it does not co-localize completely with BiP, but is a part of the ER network of tubules and vesicles [61]. A putative metal transporter protein located to the acidocalcisome. To date only a few proteins have been localized to the acidocalcisome [17]. This is a very important finding since the acidocalcisome in trypanosomatids is responsible for several functions, including its role as storage organelle, and in calcium homeostasis, maintenance of intracellular pH homeostasis, and osmoregulation [17]. Furthermore, the absence of known targeting signals to this organelle has made it impossible to detect more acidocalcisome proteins by traditional computational genome screening [17].
Another interesting finding was the localization of a protein with 14 transmembrane domains to vesicles distributed all along the cell. The presence of this large number of hydrophobic domains makes a free cytoplasmic distribution very unlikely. We can thus only speculate as to its exact location. One possibility is that the protein is located in transport vesicles of the secretory pathway. Another possible location is in small vesicles in the cell that translocate proteins to the plasma membrane in response to environmental signals or stress [64, 65]. Lipid droplets (adiposomes) have recently been described as important in managing the availability of proteins, and they have been proposed to serve as generic sites of protein sequestration [66]. The T. cruzi 2-aminoethylphosphonate:pyruvate aminotransferase, another protein found in our data set, had also been described to localize to an unidentified compartment in the cell [67], showing a similar distribution to this hypothetical protein.
Among the proteins detected, there were two proteins thought to be present in the plasma membrane, a trans-sialidase (EAN84330.1) and a mucin-associated surface protein (MASP, EAN86366.1). The detection of only two surface proteins suggests that either there is little contamination of our preparations with plasma membrane, or that these proteins were in traffic to the plasma membrane and sedimented with our organellar fraction.
EAN91861.1 corresponds to a putative pyrroline-5-carboxylate reductase. The activity of this enzyme has been detected in T. brucei bloodstream forms and is important for the differentiation of slender to stumpy forms [68]. Proline has also been described as important for differentiation of T. cruzi amastigotes to trypomastigotes [69], and from epimastigotes to metacyclic trypomastigotes [70]. The detection of this protein suggests that proline oxidation might serve in energy production in insect-stage trypanosomes [71].
The early demonstration of the aerobic fermentation of glucose in T. cruzi epimastigotes raised the question of whether the tricarboxylic acid cycle and the respiratory chain were operative [72]. Our proteomic analysis detected the expression of almost all the enzymes of the Krebs cycle, including the citrate synthase (EAO00028.1) and the iron-sulfur subunit of succinate dehydrogenase (EAN84275.1) that were not detected in a previous whole cell proteomic study [13]. Although we did not detect a fumarase, this enzyme was reported before in the proteome of epimastigotes [13]. We also validated the expression of a NADH dehydrogenase, a Rieske iron-sulfur protein, cytchrome c1, and cytochrome c oxidase subunits VI and VIII, as well as the alpha subunit of the ATP synthase in epimastigotes. These results are in agreement with the reported operation of the respiratory chain and oxidative phosphorylation in these stages [73]. The glycolytic pathway was also well represented in our proteomic study in agreement with the enrichment in glycosomes of the fraction studied. Interestingly, we confirmed the expression of phosphoglycerate kinase, which is present both in the cytosol and in glycosomes.
We validated the expression of several enzymes of the isoprenoid pathway that were expected to occur in epimastigotes (isopentenyl-diphosphate isomerase, squalene monoxygenase, lanosterol synthase) and confirmed the expression of other (lanosterol 14α-demethylase [74], solanesyl diphosphate synthase [33]). We also confirmed the expression of a fatty acid desaturase [75] and a fatty acid elongase [76].
Finally, we ratified the expression of a vacuolar transporter chaperone (VTC1) homologue to the recently described TbVTC1 present in acidocalcisomes of T. brucei [77], in agreement with the enrichment of acidocalcisomes in this fraction.
Some gene products could not be assigned to any metabolic process, pathway or structure of T. cruzi. This was the case for trichohyalin (EAN81577.1). Trichohyalin has been described in eukaryotes as an intermediate filament-associated protein of the hair [78], but its presence in other organisms has no precedence. However this protein has annotated gene orthologs in the T. brucei and L. major genomes.
A comparison with previous, mainly whole cell, large-scale protein expression studies carried out in T. cruzi, showed that cell fractionation resulted in the detection of a surprisingly large amount of previously undetected proteins. Indeed, close to half of the proteins had not been detected previously in epimastigotes. In this way, valuable information regarding protein expression in T. cruzi epimastigotes can be added to the databases, and since this is only a first analysis, more data can be expected from organellar samples in the future. Clearly, several studies of this nature are required before the T. cruzi proteome is fully characterized, and approaches where fractions or cellular components are individually analyzed are essential for this purpose.
Supplementary Material
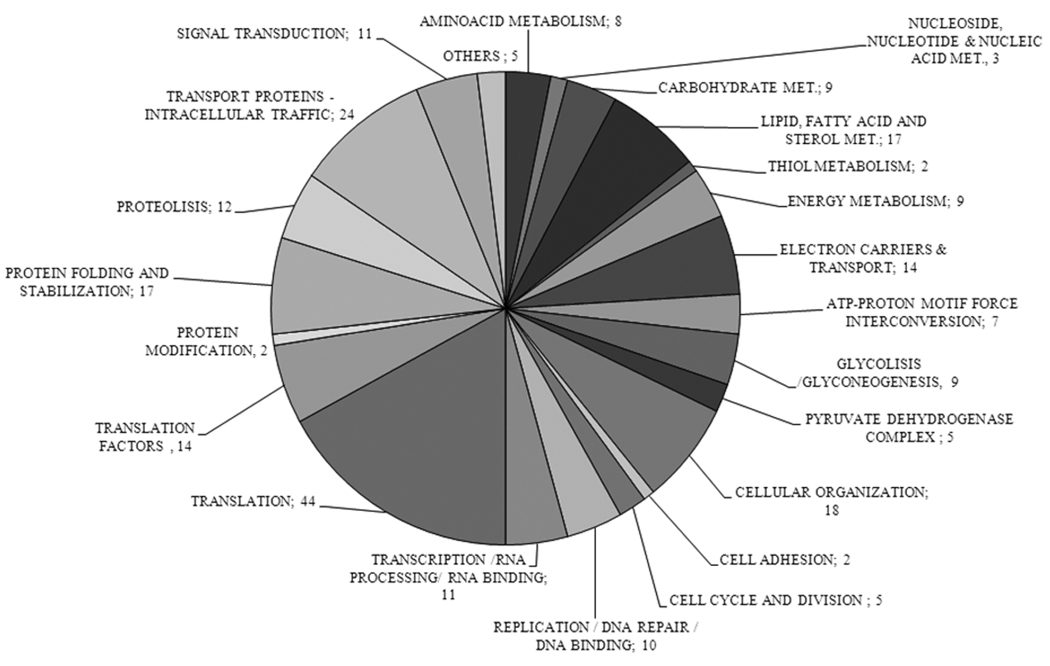
Figure 3.
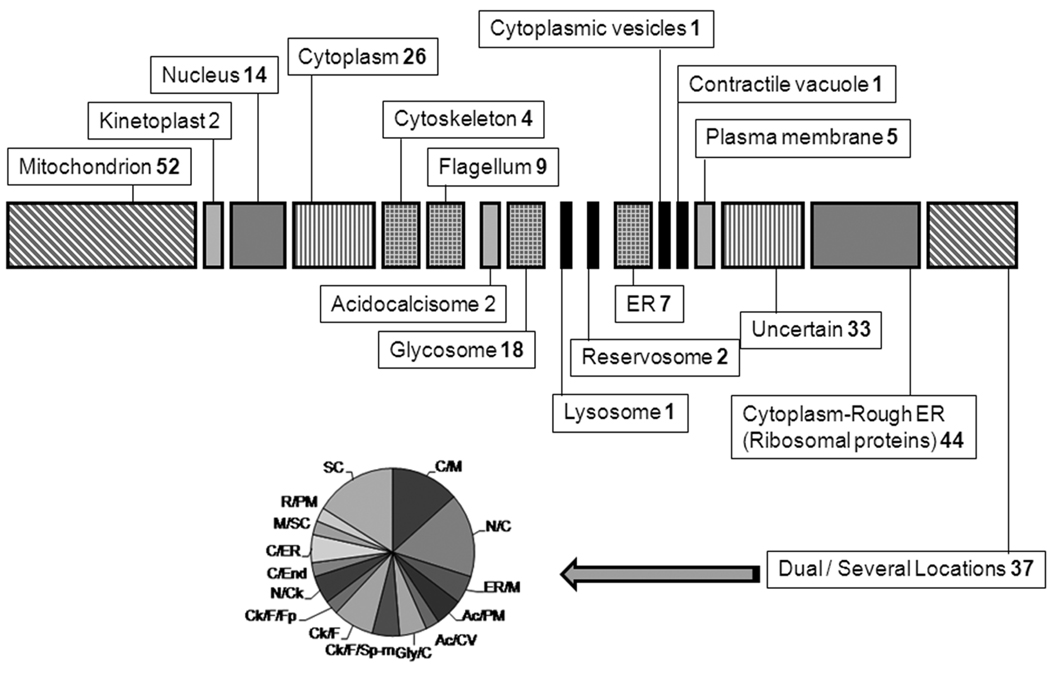
A. Functional Classification of Proteins. The chart shows the different metabolic pathways or groups to which the 258 identified proteins with known or predicted function can be assigned. Numbers represent the number of proteins for each group. Hypothetical proteins are not represented here. B. Sub-cellular Distribution of Identified Proteins. The drawing shows the distribution to different cell compartments of the same set of proteins from A. Numbers indicate the proteins to that particular location. For the dual localization proteins a sub-graph indicates the distribution. SC: several compartments; C/M: cytoplasm/mitochondrion; N/C: nucleus/cytoplasm; ER/M: endoplasmic reticulum/mitochondria; Ac/PM: acidocalcisoma/plasma membrane; Ac/CV: acidocalcisome/contractile vacuole; Glyc/C: glycosome/cytoplasm; Ck/F/Sp-m: cytoskeleton/flagellum/sub-pellicular microtubules; Ck/F: cytoskeleton/flagellum; Ck/F/Fp: cytoskeleton/flagellum/flagellar pocket; N/Ck: nucleus/cytoskeleton; C/End: cytoplasm/endosomes; C/ER: cytoplasm/rough ER; M/SC: mitochondrion/several other compartments and R/PM: reservosome/plasma membrane.
ACKNOWLEDGMENTS
We are thankful to Dr Daniel Hirschberg who helped in the pilot mass spectrometry analysis; to Professor Thomas Bergman for his constructive criticisms to our manuscript; to Drs. J. Bangs (University of Wisconsin), Fred Opperdoes (DeDuve Institute) and N. Bakalara (University of Montpellier), for antibodies; to Dr Kildare Miranda for valuable discussions and help with the microscopy; to Dr Celso Sant’ Anna for technical assistance and to Dr John Kelly and Dr Martin C Taylor (London School of Hygiene and Tropical Medicine) for pTex-myc plasmid.
Funding: This work was supported in part by a grant from the U.S. National Institutes of Health (AI68647) to R.D. M.F. was supported in part by a training grant from the Ellison Medical Foundation to the Center for Tropical and Emerging Global Diseases.
Abbreviations
- LC-MS/MS
Liquid Chromatography coupled to tandem Mass Spectrometry
- TMHs
transmembrane helices
- 1DGE
one dimensional gel electrophoresis
- 2DGE
two dimensional gel electrophoresis
- IPG
immobiline dry-strip gel.
REFERENCES
- 1.Urbina JA, Docampo R. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 2003;19(11):495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 2.de Souza W. Cell biology of Trypanosoma cruzi. Int Rev Cytol. 1984;86:197–283. doi: 10.1016/s0074-7696(08)60180-1. [DOI] [PubMed] [Google Scholar]
- 3.Docampo R, Schmunis GA. Sterol biosynthesis inhibitors: potential chemotherapeutics against Chagas disease. Parasitol Today. 1997;13(4):129–130. doi: 10.1016/s0169-4758(97)01021-1. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama K, et al. The effects of protein farnesyltransferase inhibitors on trypanosomatids: inhibition of protein farnesylation and cell growth. Mol Biochem Parasitol. 1998;94(1):87–97. doi: 10.1016/s0166-6851(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 5.Doyle PS, et al. A cysteine protease inhibitor cures Chagas' disease in an immunodeficient-mouse model of infection. Antimicrob Agents Chemother. 2007;51(11):3932–3939. doi: 10.1128/AAC.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Docampo R, Moreno SN. Bisphosphonates as chemotherapeutic agents against trypanosomatid and apicomplexan parasites. Curr Drug Targets Infect Disord. 2001;1(1):51–61. doi: 10.2174/1568005013343191. [DOI] [PubMed] [Google Scholar]
- 7.El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 8.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 9.Ivens AC, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paba J, et al. Proteomic analysis of Trypanosoma cruzi developmental stages using isotope-coded affinity tag reagents. J Proteome Res. 2004;3(3):517–524. doi: 10.1021/pr034075o. [DOI] [PubMed] [Google Scholar]
- 11.Paba J, et al. Proteomic analysis of the human pathogen Trypanosoma cruzi. Proteomics. 2004;4(4):1052–1059. doi: 10.1002/pmic.200300637. [DOI] [PubMed] [Google Scholar]
- 12.Parodi-Talice A, et al. Proteome analysis of the causative agent of Chagas disease: Trypanosoma cruzi. Int J Parasitol. 2004;34(8):881–886. doi: 10.1016/j.ijpara.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Atwood JA, 3rd, et al. The Trypanosoma cruzi proteome. Science. 2005;309(5733):473–476. doi: 10.1126/science.1110289. [DOI] [PubMed] [Google Scholar]
- 14.Atwood JA, 3rd, et al. Glycoproteomics of Trypanosoma cruzi trypomastigotes using subcellular fractionation, lectin affinity, and stable isotope labeling. J Proteome Res. 2006;5(12):3376–3384. doi: 10.1021/pr060364b. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SW, Fahy E, Ghosh SS. Global organellar proteomics. Trends Biotechnol. 2003;21(2):82–88. doi: 10.1016/S0167-7799(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro TA, Englund PT. The structure and replication of kinetoplast DNA. Annu Rev Microbiol. 1995;49:117–143. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- 17.Docampo R, et al. Acidocalcisomes - conserved from bacteria to man. Nat Rev Microbiol. 2005;3(3):251–261. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- 18.Vieira M, et al. Role for a P-type H+-ATPase in the acidification of the endocytic pathway of Trypanosoma cruzi. Biochem J. 2005;392(Pt 3):467–474. doi: 10.1042/BJ20051319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michels PA, et al. Metabolic functions of glycosomes in trypanosomatids. Biochim Biophys Acta. 2006;1763(12):1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Bone GJ, Steinert M. Induced change from culture form to blood-stream form in Trypanosoma mega. Nature. 1956;178(4529):362. doi: 10.1038/178362a0. [DOI] [PubMed] [Google Scholar]
- 21.Scott DA, Docampo R. Characterization of isolated acidocalcisomes of Trypanosoma cruzi. J Biol Chem. 2000;275(31):24215–24221. doi: 10.1074/jbc.M002454200. [DOI] [PubMed] [Google Scholar]
- 22.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338(5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17(9):849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 24.Krogh A, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 25.Bendtsen JD, et al. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Guda C, Subramaniam S. pTARGET [corrected] a new method for predicting protein subcellular localization in eukaryotes. Bioinformatics. 2005;21(21):3963–3939. doi: 10.1093/bioinformatics/bti650. [DOI] [PubMed] [Google Scholar]
- 27.Horton P, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuelsson O, et al. In silico prediction of the peroxisomal proteome in fungi, plants and animals. J Mol Biol. 2003;330(2):443–456. doi: 10.1016/s0022-2836(03)00553-9. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda S, et al. A novel representation of protein sequences for prediction of subcellular location using support vector machines. Protein Sci. 2005;14(11):2804–2813. doi: 10.1110/ps.051597405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arner E, et al. Database of Trypanosoma cruzi repeated genes: 20,000 additional gene variants. BMC Genomics. 2007;8:391. doi: 10.1186/1471-2164-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bangs JD, et al. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei Divergent ER retention signals in a lower eukaryote. J Cell Sci. 1993;105(Pt 4):1101–1113. doi: 10.1242/jcs.105.4.1101. [DOI] [PubMed] [Google Scholar]
- 32.Lu Z, et al. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20(4):547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 33.Ferella M, et al. A solanesyl-diphosphate synthase localizes in glycosomes of Trypanosoma cruzi. J Biol Chem. 2006;281(51):39339–39348. doi: 10.1074/jbc.M607451200. [DOI] [PubMed] [Google Scholar]
- 34.Taylor MB, et al. Subcellular localization of some glycolytic enzymes in parasitic flagellated protozoa. Int J Biochem. 1980;11(2):117–120. doi: 10.1016/0020-711x(80)90243-8. [DOI] [PubMed] [Google Scholar]
- 35.Acosta H, et al. Pyruvate phosphate dikinase and pyrophosphate metabolism in the glycosome of Trypanosoma cruzi epimastigotes. Comp Biochem Physiol B Biochem Mol Biol. 2004;138(4):347–356. doi: 10.1016/j.cbpc.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Linss J, et al. Cloning and characterization of the gene encoding ATP-dependent phospho-enol-pyruvate carboxykinase in Trypanosoma cruzi: comparison of primary and predicted secondary structure with host GTP-dependent enzyme. Gene. 1993;136(1–2):69–77. doi: 10.1016/0378-1119(93)90449-d. [DOI] [PubMed] [Google Scholar]
- 37.Cannata JJ, Cazzulo JJ. Glycosomal and mitochondrial malate dehydrogenases in epimastigotes of Trypanosoma cruzi. Mol Biochem Parasitol. 1984;11:37–49. doi: 10.1016/0166-6851(84)90053-7. [DOI] [PubMed] [Google Scholar]
- 38.Barderi P, et al. The NADP+-linked glutamate dehydrogenase from Trypanosoma cruzi: sequence, genomic organization and expression. Biochem J. 1998;330(Pt 2):951–958. doi: 10.1042/bj3300951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duschak VG, Cazzulo JJ. Subcellular localization of glutamate dehydrogenases and alanine aminotransferase in epimastigotes of Trypanosoma cruzi. FEMS Microbiol Lett. 1991;67(2):131–135. doi: 10.1016/0378-1097(91)90343-9. [DOI] [PubMed] [Google Scholar]
- 40.Zavala-Castro JE, et al. Stage specific kinetoplast DNA-binding proteins in Trypanosoma cruzi. Acta Trop. 2000;76(2):139–146. doi: 10.1016/s0001-706x(00)00079-6. [DOI] [PubMed] [Google Scholar]
- 41.Ouaissi MA, et al. Trypanosoma cruzi: a 52-kDa protein sharing sequence homology with glutathione S-transferase is localized in parasite organelles morphologically resembling reservosomes. Exp Parasitol. 1995;81(4):453–461. doi: 10.1006/expr.1995.1138. [DOI] [PubMed] [Google Scholar]
- 42.Souto-Padron T, et al. Cysteine proteinase in Trypanosoma cruzi: immunocytochemical localization and involvement in parasite-host cell interaction. J Cell Sci. 1990;96(Pt 3):485–490. doi: 10.1242/jcs.96.3.485. [DOI] [PubMed] [Google Scholar]
- 43.Parussini F, et al. Characterization of a lysosomal serine carboxypeptidase from Trypanosoma cruzi. Mol Biochem Parasitol. 2003;131(1):11–23. doi: 10.1016/s0166-6851(03)00175-0. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez R, et al. The deduced primary structure of a ribosomal protein S4 from Trypanosoma cruzi. Biochim Biophys Acta. 1998;1395(3):321–325. doi: 10.1016/s0167-4781(97)00224-8. [DOI] [PubMed] [Google Scholar]
- 45.Furuya T, et al. TcSCA complements yeast mutants defective in Ca2+ pumps and encodes a Ca2+-ATPase that localizes to the endoplasmic reticulum of Trypanosoma cruzi. J Biol Chem. 2001;276(35):32437–32445. doi: 10.1074/jbc.M104000200. [DOI] [PubMed] [Google Scholar]
- 46.Fouts DL, et al. Evidence for four distinct major protein components in the paraflagellar rod of Trypanosoma cruzi. J Biol Chem. 1998;273(34):21846–21855. doi: 10.1074/jbc.273.34.21846. [DOI] [PubMed] [Google Scholar]
- 47.Engman DM, et al. A novel flagellar Ca2+-binding protein in trypanosomes. J Biol Chem. 1989;264(31):18627–18631. [PubMed] [Google Scholar]
- 48.De Lima AR, et al. Tight binding between a pool of the heterodimeric alpha/beta tubulin and a protein kinase CK2 in Trypanosoma cruzi epimastigotes. Parasitology. 2006;132(Pt 4):511–523. doi: 10.1017/S0031182005009352. [DOI] [PubMed] [Google Scholar]
- 49.Cevallos AM, et al. Trypanosoma cruzi: allelic comparisons of the actin genes and analysis of their transcripts. Exp Parasitol. 2003;103(1–2):27–34. doi: 10.1016/s0014-4894(03)00066-3. [DOI] [PubMed] [Google Scholar]
- 50.Puerta C, et al. Molecular characterization of the histone H2A gene from the parasite Trypanosoma rangeli. Parasitol Res. 2000;86(11):916–922. doi: 10.1007/s004360000270. [DOI] [PubMed] [Google Scholar]
- 51.Frasch AC. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol Today. 2000;16(7):282–286. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- 52.da Cunha JP, et al. Post-translational modifications of Trypanosoma cruzi histone H4. Mol Biochem Parasitol. 2006;150(2):268–277. doi: 10.1016/j.molbiopara.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Toro GC, et al. Presence of histone H2B in Trypanosoma cruzi chromatin. Biol Res. 1993;26(1–2):41–46. [PubMed] [Google Scholar]
- 54.Maranon C, et al. Control mechanisms of the H2A genes expression in Trypanosoma cruzi. Mol Biochem Parasitol. 1998;92(2):313–324. doi: 10.1016/s0166-6851(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 55.Bontempi EJ, et al. Genes for histone H3 in Trypanosoma cruzi. Mol Biochem Parasitol. 1994;66(1):147–151. doi: 10.1016/0166-6851(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 56.Scott DA, et al. Presence of a plant-like proton-pumping pyrophosphatase in acidocalcisomes of Trypanosoma cruzi. J Biol Chem. 1998;273(34):22151–22158. doi: 10.1074/jbc.273.34.22151. [DOI] [PubMed] [Google Scholar]
- 57.Maier AG, et al. The coatomer of Trypanosoma brucei. Mol Biochem Parasitol. 2001;115(1):55–61. doi: 10.1016/s0166-6851(01)00268-7. [DOI] [PubMed] [Google Scholar]
- 58.Conte I, et al. The interplay between folding-facilitating mechanisms in Trypanosoma cruzi endoplasmic reticulum. Mol Biol Cell. 2003;14(9):3529–3540. doi: 10.1091/mbc.E03-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubotham J, et al. Characterization of two protein disulfide isomerases from the endocytic pathway of bloodstream forms of Trypanosoma brucei. J Biol Chem. 2005;280(11):10410–10418. doi: 10.1074/jbc.M409375200. [DOI] [PubMed] [Google Scholar]
- 60.McConville MJ, et al. Secretory pathway of trypanosomatid parasites. Microbiol Mol Biol Rev. 2002;66(1):122–154. doi: 10.1128/MMBR.66.1.122-154.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan DP, et al. Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem Soc Trans. 2006;34(Pt 3):356–358. doi: 10.1042/BST0340356. [DOI] [PubMed] [Google Scholar]
- 62.Hu ZZ, et al. Comparative Bioinformatics Analyses and Profiling of Lysosome-Related Organelle Proteomes. Int J Mass Spectrom. 2007;259(1–3):147–160. doi: 10.1016/j.ijms.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sturm NR, et al. Evidence for multiple hybrid groups in Trypanosoma cruzi. Int J Parasitol. 2003;33(3):269–279. doi: 10.1016/s0020-7519(02)00264-3. [DOI] [PubMed] [Google Scholar]
- 64.Rohloff P, Docampo R. A contractile vacuole complex is involved in osmoregulation in Trypanosoma cruzi. Exp Parasitol. 2008;118(1):17–24. doi: 10.1016/j.exppara.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nielsen S, et al. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci U S A. 1995;92(4):1013–1017. doi: 10.1073/pnas.92.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17(8):363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Sarkar M, Hamilton CJ, Fairlamb AH. Properties of phosphoenolpyruvate mutase, the first enzyme in the aminoethylphosphonate biosynthetic pathway in Trypanosoma cruzi. J Biol Chem. 2003;278(25):22703–22708. doi: 10.1074/jbc.M302419200. [DOI] [PubMed] [Google Scholar]
- 68.Hamm B, et al. Differentiation of Trypanosoma brucei bloodstream trypomastigotes from long slender to short stumpy-like forms in axenic culture. Mol Biochem Parasitol. 1990;40(1):13–22. doi: 10.1016/0166-6851(90)90075-w. [DOI] [PubMed] [Google Scholar]
- 69.Tonelli RR, et al. L-proline is essential for the intracellular differentiation of Trypanosoma cruzi. Cell Microbiol. 2004;6(8):733–741. doi: 10.1111/j.1462-5822.2004.00397.x. [DOI] [PubMed] [Google Scholar]
- 70.Krassner SM, et al. Further studies on substrates inducing metacyclogenesis in Trypanosoma cruzi. J Protozool. 1990;37(2):128–132. doi: 10.1111/j.1550-7408.1990.tb05882.x. [DOI] [PubMed] [Google Scholar]
- 71.Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149(1):1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 72.Cazzulo JJ. Intermediate metabolism in Trypanosoma cruzi. J Bioenerg Biomembr. 1994;26(2):157–165. doi: 10.1007/BF00763064. [DOI] [PubMed] [Google Scholar]
- 73.Stoppani AO, et al. Effect of inhibitors of electron transport and oxidative phosphorylation on Trypanosoma cruzi respiration and growth. Mol Biochem Parasitol. 1980;2(1):3–21. doi: 10.1016/0166-6851(80)90044-4. [DOI] [PubMed] [Google Scholar]
- 74.Buckner FS, et al. Cloning and analysis of Trypanosoma cruzi lanosterol 14alpha-demethylase. Mol Biochem Parasitol. 2003;132(2):75–81. doi: 10.1016/j.molbiopara.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Maldonado RA, et al. Trypanosoma cruzi oleate desaturase: molecular characterization and comparative analysis in other trypanosomatids. J Parasitol. 2006;92(5):1064–1074. doi: 10.1645/GE-845R.1. [DOI] [PubMed] [Google Scholar]
- 76.Livore VI, Tripodi KE, Uttaro AD. Elongation of polyunsaturated fatty acids in trypanosomatids. Febs J. 2007;274(1):264–274. doi: 10.1111/j.1742-4658.2006.05581.x. [DOI] [PubMed] [Google Scholar]
- 77.Fang J, et al. Ablation of a small transmembrane protein of Trypanosoma brucei (TbVTC1) involved in the synthesis of polyphosphate alters acidocalcisome biogenesis and function, and leads to a cytokinesis defect. Biochem J. 2007;407(2):161–170. doi: 10.1042/BJ20070612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steinert PM, Parry DA, Marekov LN. Trichohyalin mechanically strengthens the hair follicle: multiple cross-bridging roles in the inner root shealth. J Biol Chem. 2003;278(42):41409–41419. doi: 10.1074/jbc.M302037200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.