Abstract
Germline and somatic PTEN mutations are found in Cowden syndrome (CS) and multiple sporadic malignancies, respectively. PTEN function appears to be modulated by subcellular compartmentalization, and mislocalization may affect function. We have shown that cellular ATP levels affect nuclear PTEN levels. Here, we examined the ATP-binding capabilities of PTEN and functional consequences, relevant to cancer-associated mutations. PTEN mutation analysis of CS patients and sporadic colorectal carcinomas and comparative aminoacid analysis were utilized to identify mutations in ATP-binding motifs. The ability of wild-type (WT) or mutant PTEN to bind ATP was assessed by ATP–agarose-binding assays. Subcellular fractionation, western blotting, confocal microscopy and growth assays were used to determine relative nuclear-cytoplasmic localization and function. Somatic colorectal carcinoma-derived PTEN missense mutations were associated with nuclear mislocalization. These mutations altered cellular proliferation, apoptosis and anchorage-dependent growth. Examination of PTEN's amino acid sequence revealed these mutations resided in previously undescribed ATP-binding motifs (c.60–73; c.122–136). In contrast to WT PTEN, both cancer-associated somatic and germline-derived PTEN missense mutations, which lie within the ATP-binding motifs, result in mutant PTEN that does not bind ATP efficiently. We also show that CS patients with germline ATP-binding motif-mutations had nuclear PTEN mislocalization. Of four unrelated patients with functional germline ATP-binding domain mutations, all three female patients had breast cancers. Germline and somatic mutations within PTEN's ATP-binding domain play important pathogenic roles in both heritable and sporadic carcinogenesis by PTEN nuclear mislocalization resulting in altered signaling and growth. Manipulation of ATP may represent novel therapies in tumors with such PTEN alterations.
INTRODUCTION
Germline mutations in PTEN (phosphatase and tensin homolog deleted on chromosome 10; MIM# 601728), encoding a lipid and protein phosphatase that mediates cell cycle arrest and apoptosis, were first described in Cowden syndrome [CS; MIM# 158350] (1). Germline PTEN mutations have been identified in 85% of CS probands, who carry an up to 50% risk of female breast cancer and 10% risk of epithelial thyroid cancer. Subsequently, 65% of patients with the developmental disorder Bannayan–Riley–Ruvalcaba syndrome and variable frequencies of individuals with several other related syndromes were also shown to carry germline PTEN mutations (2). Similarly, somatic mutations or deletions of PTEN have been identified in a broad range of sporadic tumor types (2).
Initial reports suggested that PTEN localized exclusively to the cytoplasm (3,4). However, it is now well established that a significant pool of PTEN protein is localized to, and functional within, the nucleus (4–8). Decreased nuclear (wild-type, WT) PTEN (WTPTEN) has been associated with more aggressive disease in patients with colorectal cancer (9,10), cutaneous melanoma (11,12), esophageal squamous cell carcinoma (13), pancreatic islet cell tumors (14) and cases of large B cell lymphoma (15). Nuclear PTEN appears to play a role in cell cycle regulation, MAPK phosphorylation and modulating cyclin D1 levels in various cell types (7,16–18). Furthermore, nuclear PTEN is involved in chromosome stability and DNA repair processes (19). Together, this indicates that the tumor suppressive function of PTEN is partly mediated through its nuclear localization and/or retention.
Despite ample evidence indicating that PTEN localizes in the nucleus (7,17,20), PTEN contains neither a traditional nuclear localization signal (NLS) nor a nuclear export signal (NES). Several methods have been implicated in modulating PTEN localization; however, these appear to be cell type dependent. These include passive diffusion (18), a putative cytoplasmic retention signal (6), transport mediated by either the Ran GTPase (7), or Major Vault Protein (MVP) (5,21), phosphorylation-dependent shuttling (7), monoubiquitination-dependent import (8) and S6K-mediated export (22). Recently, we have shown that intracellular localization of PTEN can be modulated by ATP levels (23). However, the exact mechanism remained unknown. Interestingly, other proteins whose subcellular localization is modulated by ATP, such as Hsc70 and nucleophosmin/B23, bind ATP through consensus ATP-binding domains (24–26).
We, therefore, hypothesize that since the intracellular localization of PTEN is modulated by ATP, PTEN can bind to ATP and that this interaction could influence its intracellular localization. In this study, we sought to examine the ATP-binding capabilities of PTEN, and to evaluate the importance of germline CS-related PTEN mutations and cancer-derived somatic PTEN missense mutations occurring within these ATP-binding sites in influencing nuclear PTEN localization and function.
RESULTS
Specific somatic misssense mutations affect PTEN localization, cellular growth and proliferation in vitro
We, and others, have shown that PTEN can be localized to both the cytoplasmic and nuclear compartment (6–8,17,23,27). Putative sites in PTEN have been postulated to be important in regulating nuclear localization, such as those shown in vitro to bind to MVP, one PTEN-interacting protein which mediates nuclear import (5,21), and those that mediate ubiquitination and genomic stability (8,19). However, naturally occurring PTEN mutations found within the latter two are extremely limited. We were surprised to note that PTEN was found to be mislocalized in the nucleus, by immunohistochemistry, in sporadic primary colorectal carcinomas harboring the K62R, Y65C and K125E missense mutations (9); yet, the three somatic mutations found in these primary tumors were not specifically located in any of the known regions within PTEN believed to be important for nuclear localization, as noted above.
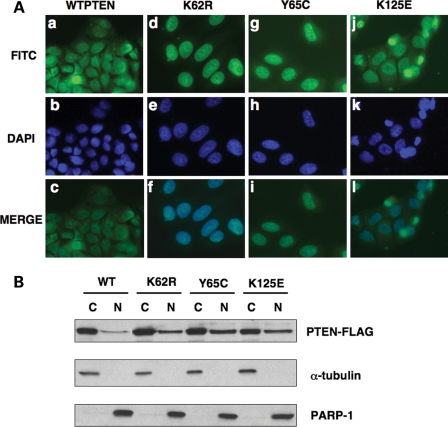
To explore the effects of these mutations further, we modeled these mutations in vitro. We generated FLAG-tagged protein as described in the Materials and Methods. Subcellular localization of these mutant proteins was performed utilizing both immunofluorescence and western analysis. We found that transiently transfected full-length over-expressed WTPTEN localized to both the cytoplasmic and nuclear compartments in MCF7 cells as previously described (Fig. 1A) (5,17). However, when we analyzed the sporadic carcinoma-derived somatic mutations in transfected MCF7 cells, we found that the K62R (pK62R-FLAG) and Y65C (pY65C-FLAG) mutant PTEN displayed a predominant nuclear accumulation, when compared with WTPTEN (Fig. 1A) as evidenced by DAPI counterstaining (Fig. 1A, panels c, f, i and l). The K125E mutant PTEN (pK125E-FLAG) also showed a preference towards nuclear accumulation. However, this localization was not as strong as that observed in cells expressing either the K62R or Y65C mutant PTEN (Fig. 1A). Nevertheless, the K125E mutant PTEN localized to the nuclear compartment to a greater extent than WTPTEN.
Figure 1.
Neoplasia-derived PTEN mutants results in increased nuclear PTEN localization. (A) MCF7 cells were transfected with either wild-type (WT) PTEN or individual PTEN missense mutants and subjected to direct immunofluorescence analysis using a α-FLAG antibody, as indicated. PTEN expression is detected as green fluorescence. Nuclei were also concurrently stained with DAPI, which was included in the mounting medium. Representative images showing the localization of WT and each mutant PTEN are shown. All experiments were carried out in duplicate. Approximately 100 cells were counted per individual transfection and representative images from three separate experiments are shown. All images were acquired at ×63 magnification. (B) MCF7 cells were transfected with either WT or mutant PTEN as indicated. Approximately 40–48 h post transfection, cells were harvested and subjected to subcellular fractionation as previously described (C, cytoplasmic protein; N, nuclear protein) and were analyzed by SDS–PAGE and western blotting.
Using biochemical techniques, we found that over-expressed WTPTEN (pWTPTEN-FLAG) can be detected in both cytosolic and nuclear fractions after western blot (Fig. 1B). Exogenously expressed K62R, Y65C and K125E PTEN mutants showed an increase in nuclear fraction protein levels, when compared with WTPTEN (Fig. 1B). In concordance with the immunofluoresence data, the K62R and Y65C PTEN mutants showed the highest amount of nuclear PTEN protein accumulation (Fig. 1B).
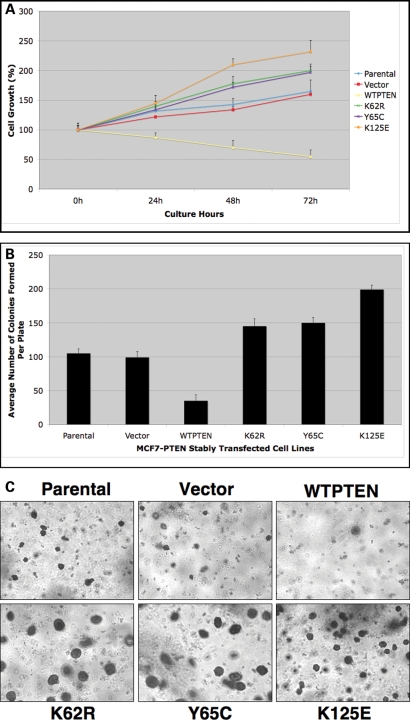
It has been suggested that proper cellular localization, or more accurately proper balance between nuclear and cytoplasmic localization, of PTEN regulates cellular proliferation (4,6,7,27). Thus, we examined the effect of overexpression of these PTEN mutants on proliferation. As expected, overexpression of WTPTEN, in MCF7 cells, markedly reduced the rate of cell proliferation, when compared with the parental cell line or to cells transfected with vector control (Fig. 2A and B). In contrast, MCF7 cells overexpressing the cancer-associated PTEN mutants, K62R, Y65C or K125E, had proliferation rates similar to, or higher than, that of the vector only control. MCF7 cells overexpressing the K125E mutant PTEN had the highest rate of proliferation (Fig. 2A and B). This suggests that these mutants do, indeed, have a functional effect, at least in vitro.
Figure 2.
Neoplasia-derived PTEN mutations result in a loss of growth suppressive activity. (A and B) The proliferation capacity of MCF7 cells stably expressing wild-type (WT) or mutant PTEN was assessed post-Tet release using the XTT (A) or soft agar (B) assay. Data shown corresponds to the average cell number (with SD). (C and D) Cell lines expressing WT or mutant PTEN were assessed for anchorage independent growth using a soft agarose colony assay. (C) A representative image of soft agarose colonies formed by MCF7 cells stably expressing either the WT or mutant PTEN (taken at ×4 magnification), data shown corresponds to the average number of colonies formed (with SD) after 2 weeks. (D) The average areas of individual colonies (in pixels) were calculated using the ImagePro Plus software and plotted as a box and whisker plot. The line inside each data box represents the mean colony area or size. The sizes of approximately 100–120 colonies were measured for each mutant (E–G) Neoplasia-derived PTEN mutations affect apoptosis and cell cycle. (E) After induction of PTEN expression, MCF7 cells were fixed, and DNA content was determined using flow cytometry with PI staining. Data from one of the three replicates of each experiment is shown. The three PTEN mutant overexpressing MCF7 cells show reduced G1/S ratio, as compared to the WTPTEN, (F and G) MCF7 cells stably overexpressing WT or mutant PTEN were grown in the absence of Tetracyclin (TetOff) to induce PTEN expression (for 96 h). Cells were fixed, and the number of cells undergoing apoptosis was determined using the TUNEL assay followed by flow cytometric analysis. Data from one of the three replicates of each experiment is shown.
We also analyzed the level of anchorage-independent growth, a hallmark of cellular transformation, utilizing colony formation in soft agar/agarose (6). MCF7 cells overexpressing WTPTEN had a significantly reduced ability to grow in soft agarose when compared with vector only and non-transfected parental MCF7 cells (Fig. 2C). In contrast, MCF7 cells expressing the K62R, Y65C or K125E PTEN mutant protein exhibited a capacity for anchorage-independent growth that was greater than that produced by the vector only expressing MCF7cells (Fig. 2C). Interestingly, cells expressing the K125E mutant PTEN protein produced the highest number of colonies (Fig. 2C). Cells expressing the Y65C mutant PTEN had the lowest level of anchorage-independent growth of the three mutant expressing lines, however colony formation was still higher than colony formation found in cells overexpressing WTPTEN. Interestingly, we also observed a difference in colony size in MCF7 cells overexpresing either K62R mutant PTEN or Y65C mutant PTEN when compared with the vector only and cells overexpressing K125E mutant PTEN (Fig. 2D).
It is well known that overexpression of WTPTEN induces G1 growth arrest and/or apoptosis in a cell type-dependent manner (4,17,27). Thus, we examined the effect of these three mutations on G1 arrest. While overexpression of WTPTEN resulted in a 40–50% increase in G1 arrest, when compared with parental or vector only transfected cells (Fig. 2E), MCF7 cells overexpressing any of the mutant PTEN proteins exhibited a decreased level of G1 arrest when compared with WTPTEN (Fig. 2E). Indeed, the level of G1 arrest exhibited in cells expressing the K125E mutant was even lower than those observed with the vector only control transfected cells (Fig. 2E). Using the TUNEL assay, we found that overexpression of WTPTEN led to a 5-fold increase in the number of apoptotic cells (Fig. 2F and G) compared with control cells. In contrast, cells overexpressing either K62R or Y65C mutant PTEN had reduced apoptotic capabilities (Fig. 2F and G), while the K125E mutant PTEN was unable to induce any apoptosis (Fig. 2F and G).
PTEN contains ATP-binding motifs in the N-terminal phosphatase domain
The above data indicate that the carcinoma-derived somatic PTEN mutations not only show increased nuclear retention, but also are highly pro-proliferative and induce a loss of growth-inhibitory activity, in contrast to those cells expressing WTPTEN. Further, this appears to be related to the nuclear localization of these mutant proteins. However, it is not readily apparent how these mutations affect nuclear localization. We have recently shown that PTEN localization can be modulated by intracellular ATP levels (23). Proteins that are regulated by ATP often contain consensus ATP-binding motifs (24,25,28,29). Upon examination, we found that PTEN's amino acid sequence contains two putative ATP-binding sequences (28,29), in the N-terminal phosphatase domain of the protein (Fig. 3A and B). A Type-A consensus ATP-binding sequence resides at amino acid residues 125–128 (Fig. 3A) and a Type-B consensus ATP-binding sequence resides at amino acid residues 60–73 (Fig. 3B). Both of these sites are located with PTEN's N-terminal phosphatase domain and interestingly the somatic-derived mutations that have altered PTEN nuclear localization occur within these regions.
Figure 3.
Consensus ATP-binding motifs in the N-terminal phosphatase domain of PTEN. Comparison of known consensus ATP-binding motifs to putative ATP-binding motif sequences in the N-terminal domain of PTEN. (A) A putative Type-A ATP-binding motif lies within exon 5, (B) A putative Type-B ATP-binding motif resides within exon 3. Germline (highlighted in red) and cancer-derived PTEN mutations analyzed in this study are highlighted in bold and underlined.
Endogenously expressed WTPTEN binds to ATP–agarose
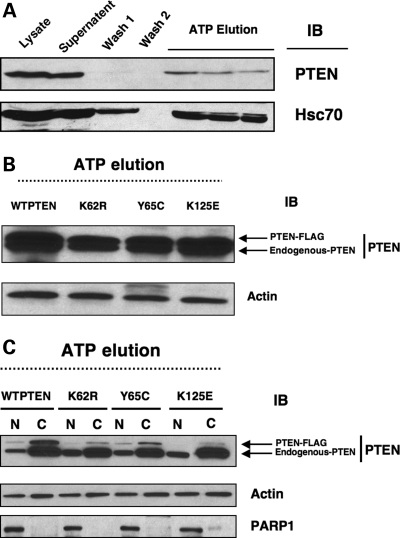
The binding of proteins to immobilized ATP has been taken as evidence for the association of various proteins with ATP (24,28,29) as has been well documented for the ATP-binding protein Hsc70 (28). We found that like Hsc70, WTPTEN binds to ATP (Fig. 4A). In ATP-binding assays, PTEN protein is not found in the ATP–agarose washes (Fig. 4A; W1 and W2), but was found eluted from the ATP–agarose with buffer containing ATP (Fig. 4A; ATP elution). This strongly suggests that, similar to a known ATP-binding protein (Hsc70), PTEN binds ATP.
Figure 4.
PTEN binds ATP that can be disrupted by missense mutations. (A) Total protein (1–2 mg) was extracted from MCF7 cells and subjected to ATP-binding assay as described in Materials and Methods. Dissociation of specifically bound protein to ATP was initiated by the addition of buffer A containing 1 mm ATP and 0.5 M KCl to the ATP–agarose. Samples were concentrated and analyzed by SDS–PAGE and western analysis for PTEN and a Hsc70. L, crude protein load; S, supernatant; W1, supernatant from wash 1 using buffer A only; W2, supernatant from wash 2 using buffer A + 0.5 M KCL; ATP elution; supernatant retained after wash with 1 mm ATP + 0.5 M KCl, (B) Cancer-associated PTEN mutants showed reduced ATP binding. Total protein from MCF7 cells stably expressing either the WTPTEN or mutant PTEN was used in an ATP-binding assay. Both endogenously expressed, and exogenously introduced (FLAG-tagged) WTPTEN showed strong ATP binding in comparison to the cancer-derived PTEN mutants, (C) MCF7 cells stably expressing WTPTEN or mutant PTEN were subjected to subcellular fractionation and used in ATP-binding assays. Exogenously expressed WTPTEN shows strong ATP binding in both the cytoplasmic and nuclear fractions; while the cancer-derived PTEN mutants showed significantly reduced ATP binding in both compartments. Note that K62R- and K125E-PTEN mutants show little to no ATP binding in the nuclear fractions (no bands in nuclear fraction-eluate) although they show predominant nuclear localization (see Fig. 1).
Carcinoma-associated PTEN mutations in the ATP-binding regions show reduced ATP binding
As alluded to above, the somatic PTEN mutations identified from primary sporadic colorectal tumors (K62R, Y65C and K125E) occur within PTEN's putative ATP-binding sites (Fig. 3A and B). Therefore, we examined whether these mutant PTEN proteins could bind to ATP, utilizing the various overexpressing cell lines utilized in the growth assays described earlier.
Each of the MCF7 cell lines expressed the FLAG-tagged PTEN (WT and mutants) protein at similar levels (data not shown). We found that in all the cell lines, endogenous PTEN bound efficiently to ATP, as expected (Fig. 4B). In contrast, we found that mutant PTEN protein did not bind to ATP as well as WTPTEN. There was less mutant PTEN detected in the ATP elute, when compared with the FLAG-tagged WTPTEN (Fig. 4B). These mutations, however, did not completely abolish ATP binding. In order to examine this in more detail, we performed ATP-binding assays on both cytosolic and nuclear fractions from cell lines expressing the mutant PTEN proteins. Endogenous nuclear and cytosolic WTPTEN efficiently bound to ATP (Fig. 4C). In addition, exogenously expressed nuclear and cytosolic FLAG-tagged WTPTEN bound to ATP (Fig. 4C). However, all exogenously expressed mutant PTEN showed reduced ATP binding, with the K62R and K125E mutant PTEN nuclear proteins showing little to no nuclear ATP-binding capability (Fig. 4C), despite the fact that these mutant proteins are predominately nuclear when compared with WTPTEN (Fig. 1A and B). This may suggest that ATP binding is required for PTEN nuclear export.
Germline PTEN mutations located in the ATP-binding domains identified in CS patients
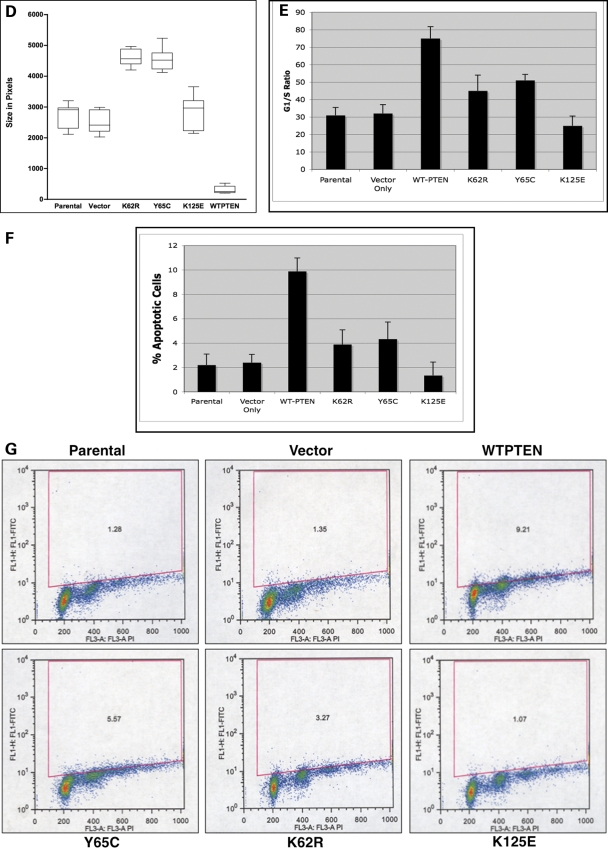
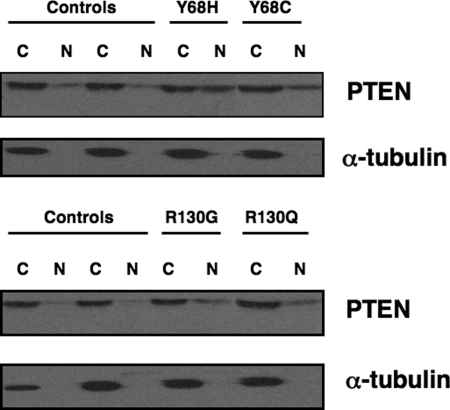
Our above data suggest that PTEN mutations located in the putative ATP-binding domains not only reduce the ability of PTEN to bind ATP, but also modulate PTEN protein in a detrimental manner. It stands to reason, then, that germline mutations in the ATP-binding sites may occur in patients with CS. Of the ∼400 classic CS patients, we found at least 4 CS patients with germline PTEN missense mutations that occur in the midst of these putative ATP-binding sites (Fig. 3A and B). It should be noted, however, that missense mutations in CS are less common than truncating ones, but of ∼20 different germline missense mutations in PTEN, ∼14 lie in these two putative ATP-binding sites. Two patients had germline mutations in the Type-A binding site: R130G and R130Q. Additionally, two patients had germline mutations in the B-Type ATP-binding site: Y68H and Y68C. Each of these mutations are novel missense mutations, which are highly predicted to be deleterious, although this has not been confirmed functionally. Using patient-derived lymphoblastoid lines from the four CS probands, we examined the subcellular localization of PTEN in these patient samples. Lymphoblastoid cell lines from patients with putative ATP-binding site mutations demonstrated an increase in PTEN nuclear protein when compared with control lymphoblastoid lines (Fig. 5). These CS patients consisted of three females and one male. Interesting, all three females had breast cancer.
Figure 5.
Lymphoblastoid lines from CS patients with germline ATP-binding PTEN mutations have altered PTEN localization. Lymphocytes from CS patients with germline PTEN mutations, occurring within the PTEN ATP-binding domains, were subjected to nuclear-cytoplasmic protein fractionation and then analyzed by SDS–PAGE and western blotting. Western blotting for PARP-1 (nuclear specific protein) or α-tubulin (cytoplasmic specific protein) is shown.
DISCUSSION
We recently reported that intracellular localization of PTEN is modulated by ATP levels (23). In our previous study, it was not readily apparent if ATP was modulating export or if there was an inhibitory phosphorylation event occurring. Our data here demonstrate that PTEN can bind ATP and somatic and germline-derived mutations in the ATP-binding sites result in PTEN being retained in the nuclear compartment, suggest that ATP is necessary for PTEN nuclear export.
In the ATP-binding domain of nucleophosmin/B23, the positively charged lysine residues, in particular lysine263, not only influences ATP-binding capabilities but also influences the subcellular localization of nucleophosmin/B23 (24,25,29). This suggests that positively charged residues are important in ATP binding. The PTEN consensus ATP-binding domain, identified here, contains several positively charged residues, including lysine residues (Fig. 3 and Supplementary Material, Fig. S1). Indeed, like nucleophosmin/B23, we found that naturally occurring germline and somatic mutations of these residues resulted in decreased ATP binding (Fig. 4 and Supplementary Material, Fig. S2) and altered localization of PTEN compared with WT protein (Fig. 5). Of importance, we found this in a variety of cell types (Supplementary Material, Fig. S3). Our data suggest that not only the positively charged amino acid residues (lysine 62, lysine 125 and arginine 130), but also the polar uncharged residues (tyrosine 65 and tyrosine 68) in PTEN's consensus ATP-binding domain are important for ATP binding.
Independent of our study, Gil et al. (7), and more recently, Denning et al. (6), have demonstrated that the amino acid residues in the N-terminal phosphatase domain of PTEN can potentially behave either like nuclear exclusion or cytoplasmic retention motifs. Additionally, mutation of the hydrophobic tyrosine residues within the N-terminal region of PTEN (residues 1–32) influence its nuclear localization in U87MG glioblastoma cells in a RanGTPase-dependent manner (7). However, a precise mechanism for this action was not fully elucidated. It is interesting to hypothesize, based on our data, that these regions contain specific charged residues that may be involved in ATP binding. Mutation of the lysine residue at amino acid 62 to arginine (K62R) leads to the introduction of an additional positive charge, which may require more energy for ATP to dissociate. This may explain the increased nuclear retention of the K62R-PTEN mutant in various cell lines tested (Fig. 1 and Supplementary Material, Fig. S3). Similarly, the change at residue 65 and 68 from tyrosine to cysteine may provide for the opportunity of an additional disulphide (-SH) bond, which could require more energy for cellular traffic. In contrast, mutation of PTEN at amino acid residues 130 (R130G and R130Q) leads to a loss of a positive charge versus a gain. Clearly, the physical chemistry involved in these mutations and its affect on localization mechanisms requires further study.
Previous work, by others and us, has suggested that nuclear PTEN is required for proper cellular proliferation and growth (4–8). More accurately, we believe that the balance of nuclear to cytoplasmic (WT) PTEN is important. Indeed, loss of nuclear WTPTEN has been correlated with unrestrained growth and tumor formation and progression. For example, when analyzing the subcellular localization of PTEN in melanoma, we found that 30/30 metastatic melanomas had loss of nuclear PTEN protein expression (11). This suggests that nuclear expression of WTPTEN is necessary for growth regulation and provides a protective effect in the cell, which has been supported in thyroid, breast and pancreatic islet models. Thus, it is interesting to note that in this study, mutations identified in colorectal cancer, alter PTEN: ATP binding and result in an increase in nuclear PTEN protein. This increase resulted in phenotypes, such as growth in soft agarose, which may be detrimental and not protective, as might have been expected. One hypothesis is that atypical increases in nuclear PTEN, which is also mutant, prevent normal cytosolic PTEN function. It is not unreasonable to consider the possibility that PTEN has unrecognized functions in the nucleus, as, until now, it was not known that PTEN could bind ATP. Our data indicate that proper regulation of PTEN: ATP binding is necessary for correct cellular growth. Whether this is simply due to alterations in nuclear transport or other signaling or protein:protein interactions remains to be determined. Undoubtedly, more remains to be elucidated regarding alternative PTEN functions, particularly those that require the binding of ATP. Nonetheless, PTEN regulation/function may not simply be a matter of localization, and while this may create a more confusing scenario it also provides other areas to target for novel therapeutics.
We have identified two putative ATP-binding sites in PTEN (Fig. 3). These sites are in areas that are highly conserved among a variety of species (Supplementary Material, Fig. S4), underlining the importance of these sites. Indeed, our data with sporadic- and germline-derived PTEN mutations strongly indicate that these mutations are pathologic. We found, that of four unrelated CS patients, that the three female patients all had breast cancer. While the sample size is small, overall functional, genetic and phenotypic data suggest that these mutations are pathogenic.
Taken together with our previous data, we demonstrate that PTEN binds ATP. The binding of PTEN to ATP not only controls nuclear localization, likely through the modulation of nuclear export, but also further regulates cellular functions such as abrogation of PTEN's tumor suppressor capabilities. This is supported not only by the somatic mutations identified in sporadic primary colorectal tumors, but also by the identification of four CS patients with germline ATP-binding domain missense mutations. In contrast, mutation of the bipartite NLS-like sequences which bind MVP do not result in alterations in phosphatase function (21). Because naturally occurring PTEN missense mutations outside the ATP-binding motifs do not result in abrogation of ATP binding nor nuclear mislocalization (Supplementary Material, Fig. S5 and S6), we believe that it may be both the presence of a mutation and the resulting nuclear mislocalization that are important. Whether it is any mutation in the ATP-binding motifs or whether it is the change in charge within the motifs that are important in nuclear mislocalization is not entirely known. Neutral mutations of our three somatic-associated ATP-binding motif mutations do result in some mislocalization to the nucleus (over WTPTEN), but slightly less so than the mutations which change the charge of those residues as well (Supplementary Material, Fig. S7 versus Fig. 1A). Thus, we may conclude that disruption of ATP binding is the major etiology for nuclear PTEN retention while charge plays a secondary role to determine the extent.
Initially, at a superficial level, our data may appear contradictory to previous observations that predominant nuclear expression of PTEN is seen in normal cells. However, it should be noted that normal cells have a predominance of nuclear PTEN and all of their PTEN is WT. WTPTEN in the nucleus signals down its protein phosphatase activity via cyclin D1 to downregulate MAPK signaling and elicit G1 cell cycle arrest and to stabilize the genome (16,19). Our current observations, therefore, suggest that mutant PTEN mislocalized in the nucleus prevents G1 arrest probably through upregulation of MAPK signaling and downregulation of cyclin D1. This hypothesis is corroborated by our cell cycle data presented here.
Missense mutations, as opposed to truncating mutations, are often conservatively interpreted as variants of unknown significance until proven otherwise. Thus, these observations should also aid in the genetic counseling of patients with germline PTEN missense mutations that lie anywhere in the ATP-binding domains. Together with the recent observation that germline variants in the genes encoding mitochondrial complex II subunits succinate dehyrogenase B and D (SDHB, SDHD) occur in PTEN mutation negative CS probands (30), our data here add to the importance of energy and energy production in both heritable and sporadic neoplasias where the PTEN pathways are relevant. These observations should provide a novel pathway which involves manipulation of ATP and mitochondrial function in the treatment of both these heritable and sporadic malignancies.
MATERIALS AND METHODS
Materials
Platinum Pfx polymerase, hygromycin, T4 DNA ligase, Prolong Gold anti-fade mounting medium and MAX efficiency DH5α competent cells were purchased from Invitrogen (Carlsbad, CA). The XTT cell proliferation kit was obtained from Roche (Indianapolis, IN). The mammalian expression vector pCMV-FLAG-5b, α-FLAG M2 FITC-conjugated antibody, α-FLAG antibody, ATP–agarose, protease inhibitors (cocktail), phosphatase inhibitors (cocktail 1 and 2) and paraformaldehyde (PFA) were purchased from Sigma Chemical Co. (Portland, OR). Restriction endonucleases were obtained from New England Biolabs (Ipswich, MA). The QuickChange in vitro site-directed mutagenesis system and XL-1 blue supercompetent cells were from Stratagene (La Jolla, CA, USA). DMEM and RPMI medium were obtained from GIBCO BRL (Hercules, CA, USA). The M-PER and NE-PER protein extraction kits and the BCA protein assay kit were from Pierce Biotechnology Inc. (Rockford, IL, USA). Sample concentration columns used were Millipore Biomax-30 kDa NMWL Centrifugal Filters (Millipore, Billerica, MA). Antibodies used were α-PTEN monoclonal antibody clone 6H2.1 (Cascade Biosciences, Portland, OR, USA), α-Hsc70, α-tubulin and PARP-1 (Cell Signaling Technologies, Danvers, MA, USA). All primers were obtained from Sigma Genosys (Portland, OR, USA). All other chemicals/reagents were obtained from common commercial sources.
Patients
We utilized peripheral blood samples accrued from four CS patients with germline mutations in the ATP-binding domain of PTEN. Classic CS was diagnosed when the operational diagnostic criteria of the International Cowden Consortium was met (1). We also utilized peripheral blood samples from four normal populational controls, which were anonymized prior to storage and analysis. Informed consent was obtained for all subjects (CS individuals and controls) in accordance with procedures and protocols approved by the respective Human Subjects Protection Committee of each participating institution. All subjects, whether CS or controls, participated on a voluntary (unpaid) basis.
Mutation analysis
Genomic DNA extracted from peripheral leukocytes, obtained from both CS patients and controls, were amplified by PCR and subjected to direct sequencing (ABI3730xl) of all PTEN exons and flanking introns, as previously described by us (31). All controls had no detectable PTEN sequence alterations.
Cell lines, culture conditions and reagents
Human immortalized lymphoblast cell lines obtained from CS patients and controls were cultured in high-glucose RPMI 1640 supplemented with 20% fetal bovine serum (FBS), 1% penicillin–streptomycin sulfate and maintained at 37°C and 5% CO2. MCF7 and MDA-MB-231 breast carcinoma cell lines and WM164 cutaneous melanoma cell lines were maintained in high-glucose DMEM supplemented with 10% FBS and 1% penicillin–streptomycin sulfate. The HT29 colorectal cancer cell line was maintained in high-glucose RPMI 1640 supplemented with 10% FBS and 1% penicillin–streptomycin sulfate. All cell lines were maintained at 37°C and 5% CO2. All cell lines were screened for PTEN mutations in the coding, flanking intronic and promoter regions using both DGGE and automated sequencing and contain the WTPTEN. MCF7 cells stably over-expressing either WTPTEN (TetWTPTEN) or mutant PTEN K62R (TetPTEN-K62R), Y65C (TetPTEN-Y65C) and K125E (TetPTEN-K125E) were generated as described below and grown in the presence of tetracycline (Tet), unless otherwise indicated. All plasmids generated contain a FLAG epitope at the C-terminus such that the expressed PTEN contains a C-terminal FLAG fusion.
ATP–agarose-binding assay
ATP–agarose was used in a binding procedure as previously described with minor modifications (28,29,32). Briefly, ATP–agarose (8 mg) was equilibrated with buffer A (40 mm HEPES, pH 7.5, containing 20 mm KCl, 5% glycerol and 5 mm MgCl2). Approximately 1–4 mg of protein was mixed with equilibrated ATP–agarose in a total volume of 1 ml buffer A. The protein–ATP–agarose mixture was then incubated for 24 h at 4°C with rotational mixing. The mixture was then centrifuged at 14 000 rpm for 10 min and the resultant supernatant (S) retained on ice for further analysis. The ATP–agarose was washed five times with 1 ml of buffer A each time. After each wash the mixture was centrifuged and the supernatant retained on ice (wash 1, total volume 5 ml). The agarose pellet was washed a further five times with high salt buffer (buffer A + 0.5 m KCl). The sample was centrifuged between washes and the supernatant was retained after each wash (wash 2, total volume 5 ml). Finally, dissociation of protein specifically bound to ATP was initiated by the addition of buffer A containing 1 mm ATP and 0.5 m KCl (ATP elution). The supernatants retained after each wash, or elution, were concentrated using the Millipore Biomax-30 kDa NMWL centrifugal filters (Millipore). Approximately 30 µl of concentrated sample was electrophoresed on 10% SDS–PAGE gels as previously described (23), and immunoblotted using the respective antibodies as indicated.
WTPTEN plasmid construction
Total RNA (1 µg) was reverse transcribed using the SuperScript One-Step RT–PCR for Long Templates system and amplified using high fidelity Pfx DNA polymerase essentially as described by the manufacturer (Invitrogen). The full-length PTEN open-reading frame was amplified by RT–PCR, prepared from total RNA, using the primers PTENAtgFwd (5′-CGCGAATTCGCCACCATGACAGCCATATC-3′) and PTENTgaRev (5′-TACCGGA TCCACTTTTGTAATTTG-3′) containing flanking EcoR1 and BamH1 restriction sites, respectively. The forward primer contained a Kozak sequence (GCCACC) to facilitate expression. The amplified PTEN cDNA sequence was then sub-cloned into the EcoR1 and BamH1 sites of the mammalian expression vector pFLAG-CMV-5b containing a 3′ FLAG tag which would be fused to the C-terminus of the expressed PTEN. Ligation was carried out using T4 DNA ligase at an insert: vector molar ratio of 3:1. Native and recombinant plasmids were transformed into supercompetent DH5α cells according to the manufacturer's instructions (Invitrogen). After plasmid midi/ maxi-preparation, plasmid DNA was digested with EcoR1 and BamH1 to validate the presence and correct size of the insert. The appropriate construction of the FLAG-tagged WTPTEN cDNA clone (pWTPTEN-FLAG) was verified by sequence analysis on both strands.
Preparation of PTEN mutant constructs
The WTPTEN cDNA clone in pFLAG-CMV-5b (pWTPTEN-FLAG) was used as the template to engineer each of the PTEN mutations (K62R, Y65C and K125E) previously identified by us in primary colorectal tumors (9). Each PTEN mutant (pK62R-FLAG, pY65C-FLAG and pK125E-FLAG) was engineered using the QuickChange in vitro site-directed mutagenesis system according to the manufacturer's instructions (Stratagene) using the following mutagenic primers: pK62R-FLAG-Fwd (5′GGTTT TTGGATTCAAAGCATAGAAACCATTACAAGATATACA ATC-3′) with pK62R-FLAG-Rev (5′-GATTGTATATCTTGTAATGGTTTCTATGCTTTGAA TCCAAAA ACC-3′); pY65C-FLAG-Fwd (5′-GGATTCAAAGCTAAAAACCATTGCAAGA TAT ACAATCTTTGTGC-3′) with pY65C-FLAG-Rev (5′-GCACAAAGATTGTATATCTTG CAATGGTTTTTATGCTTTGAATCC-3′) and pK125E-FLAG-Fwd (5′-ATGTTGCAGCAAT TCACTGTGAAGCTGGAAAGGGACGAACTGGTG-3′) with pK125E-FLAG-Rev (5′-CACCAGTT CGTCCCTTTCCAGCTTCACAGTGAATTGCTGCAACAT-3′). The corresponding mutagenic bases are indicated in the bold and are underlined type. The appropriate construction of the FLAG-tagged mutant PTEN plasmid(s) was verified by sequence analysis of both strands using the pFLAG-CMV-5b vector primers.
Transient transfection and cellular analysis
Cells were either grown on coverslips in 6-well plates (for subcellular localization assays) or cultured in 100 cm2 dishes (for western analysis) and were transfected with 3–6 µg of purified plasmid DNA (WTPTEN or PTEN mutant) using either LipofectAMINE 2000 or FuGENE6 and Opti-MEM according to the manufacturer's instructions. Briefly, the plasmid DNA in Opti-MEM was mixed with either LipofectAMINE or FuGENE6 and incubated at room temperature for 20 min to allow formation of lipid-DNA complexes. After incubation, the DNA-LipofectAMINE/ FuGENE6-OptiMEM lipid complex was gently added drop wise onto the cells on the coverslips, or in the culture dishes, and the cells were then overlayed with the appropriate culture medium.
Generation of stable Tet-Off cell lines
Stable cell lines with Tet-regulated PTEN expression were generated according to the manufacturer's protocol (Clontech Protocol No. PT3001-1). Briefly, the pTetOff vector containing the tTA regulatory element was transfected into the MCF7 parental line and placed under G418 selection. Resistant colonies were then isolated and expanded. Expanded clones were transiently transfected with pTRE2Hyg-luc in the presence or absence of Tet and luciferase levels were measured after 48 h. Clones showing an induction of luciferase activity in the absence of Tet were transfected with the WTPTEN cDNA construct cloned into the pTRE2Hyg vector (TetWTPTEN). Cells were grown under hygromycin selection in the presence of Tet and resistant colonies isolated and expanded. Tet-regulated expression of exogenous PTEN was confirmed by western analysis on total protein lysates collected from cells grown in the presence or absence of Tet. The TetWTPTEN construct was used as the template to engineer each of the individual mutations (TetPTEN-K62R-FLAG, TetPTEN-Y65C-FLAG and TetPTEN-K125E-FLAG) to be tested. Each mutant was engineered using the QuickChange in vitro site-directed mutagenesis system (Stratagene) according to the manufacturer's instructions using the primers described earlier. The appropriate construction of the FLAG-tagged mutant PTEN plasmids were verified by sequence analysis using vector primers as described above.
Direct immunofluoroscence and confocal microscopy
Approximately 40–48 h post transfection, cells grown on coverslips were fixed in a freshly prepared mixture of methanol:acetone (1:1 ratio) for 1 min at room temperature. Cells were washed thrice with Tris-buffered saline (TBS) and then incubated with the α-FLAG M2 FITC-conjugated antibody (1:500 dilution in TBS) at room temperature for 1 h. After further washing of the cells with TBS, the coverslips were mounted face down onto glass slides (Labtek Inc., Grand Rapids, MI) with a small drop of ProLong Gold antifade mounting medium containing DAPI. Cells were then examined at room temperature under a Leica DM RXE (TCS-SP/SP-AOSB) confocal microscope with HCX Plan 63× numerical aperture 1.4 oil immersion objective lens, using Leica confocal software version 2.5 (Leica, Wetzler, Germany). Confocal images were acquired as Z stacks or single XY sections. For merged images, projections of image stacks were prepared in Velocity version 3.7.0 (Improvision, Coventry, UK). All experiments were carried out in duplicate. Approximately 100 cells, from 10–15 fields, were counted and scored per transfection. Note should be made that visualization of relative intensities, by the human eye, of indirect immunofluorescence is different from that of standard immunohistochemistry. While relative intensity in indirect immunofluorescence to the human eye appears more linear (it is not linear in reality), relative intensity of staining from immunofluorescence is non-linear and the human eye is only able to detect an intensity increase or decrease only after quite a large (non-linear) increase or decrease. Because of the histological view of cells (which is two-dimensional) with the very much smaller nuclear area compared with the cytoplasmic area, relative nuclear expression of any protein will be very exaggerated (i.e. look very strong) when compared with an equal amount of that protein in the cytoplasm (with its much larger surface area). Hence, visualization of relative intensities between a histological approach and western blot will appear different to the human eye. Because total protein content is very much higher in the cytoplasm over that from the nucleus, the very reason why a two-dimensional imaged cell (compared with western blot) shows intense nuclear staining is also the reason why cytoplasmic proteins which mainly reside in one compartment (in PTEN's case, the cytosol) will always appear more strongly expressed over that in the nucleus. In each cell line studied, the majority of PTEN is localized to the cytoplasm with a small, but distinct, portion present in the nucleus.
Subcellular fractionation, protein isolation and western blot analysis
Total, nuclear and cytoplasmic proteins were isolated at the indicated time-points as previously described (21,23), using either the M-PER (total protein) or NE-PER (nuclear-cytoplasmic protein) extraction reagents according to the manufacturer's instructions (Pierce). Proteins obtained either from cell lines or cultured lymphocytes were fractionated on 10% SDS–PAGE gels using either the BioRad Mini-gel or the Criterion system and transferred to nitrocellulose. Equal protein loading was confirmed by staining the nitrocellulose blots with Ponceau S and by western analysis with α-tubulin (cytosolic protein) or α-PARP-1 (nuclear protein). Blots were probed with α-PTEN, α-FLAG, α-Hsc70 and α-actin, each at a 1:1000 dilution, followed by incubation with the appropriate secondary antibody and visualized using enhanced chemiluminescence detection.
Colony-forming assay
To evaluate the transforming potential of the PTEN mutants, anchorage-independent growth was measured by soft agarose colony assay as previously described, but with minor modifications (4). Briefly, the assay was performed in 60 mm dishes with a base of 2.5 ml of 2× DMEM medium containing 0.4% agarose and 5% FBS. Stably expressing, tetracyclin released (TetOff) WTPTEN and PTEN mutant expressing MCF7 cells were seeded, in duplicate, in 2 ml of 2× DMEM containing 5% FBS with 0.4% agarose at 2 × 104 cells/ml and layered onto the base. These embedded cells were then overlayed with 1 ml of 1× DMEM and 10% FBS. Culture dishes were incubated at 37°C and 5% CO2 for approximately 2 weeks (overlayed media was replenished as required), and the diameter of the tumor colonies formed were determined using a microscope equipped with an ocular scale in the eyepiece. The average area of individual colonies were calculated (in pixels) using the ImagePro Plus software as previously described (33). Approximately 100–120 colonies were counted and sized. All experiments were carried out in triplicate.
Cell proliferation assay
Stably expressing, Tet released (Tet-Off) WTPTEN and PTEN mutant expressing MCF7 cells were seeded, in triplicate, in 96-well cell culture plates at a density of 2 × 104 cells per well in 100 µl of culture medium and incubated at 37°C with 5% CO2. After 24–72 h days post-Tet release, cells were incubated in XTT solution for 4 h and reduction of the tetrazolium salt was quantified with an ELISA plate reader, as described by the manufacturer (Roche). The absorbance directly correlates with the cell number. All experiments were carried out in triplicate.
Flow cytometry, cell cycle analysis and TUNEL assay
For cell cycle analysis, at the indicated times, Tet released (Tet-Off) MCF7 cells stably expressing WT or mutant PTEN were harvested after trypsinization and pelleted by centrifugation. Cells were resuspended in 500 µl of cold PBS and fixed by addition (drop-wise) to 70% ice-cold ethanol. Fixed cells were washed with cold PBS, labeled with propidium iodide and analyzed by flow cytometry on a FACScan flow cytometer (Becton Dickinson, San Jose, CA). Cell cycle analysis was performed with the CELL-FIT program. All flow cytometric analyses were performed in triplicate. For the detection of apoptosis (by TUNEL), at the indicated times, cells were harvested by centrifugation after trypsinization, pelleted by centrifugation and washed with PBS. Cells were fixed in 1% PFA (prepared in PBS) for 15 min on ice. Fixed cells were then washed twice with PBS, resuspended in 500 µl cold PBS and fixed in 70% ice-cold ethanol. Samples were stored at −20°C until analysis. Flow cytometry was performed using a Beckman-Coulter elite flow cytometer using a 610 long pass filter for data collection. Data were then filtered, after which cell phases and apoptotic cells were quantified using the ModFit program (Verity Software, Bowdoin, ME). All experiments were carried out in triplicate.
WEB RESOURCES
The URL for data presented herein is Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was funded, in part, by the National Cancer Institute, Bethesda, MD (1R01CA118980-01A2 to C.E.) and the William Randolph Hearst Foundation (to C.E.). C.E. is an American Cancer Society Clinical Research Professor, a recipient of the Doris Duke Distinguished Clinical Scientist Award and holds the Sondra J. & Stephen R. Hardis Chair in Cancer Genomic Medicine at the Cleveland Clinic.
ACKNOWLEDGEMENTS
G.P.L would like to thank Bin Zhang, PhD and Jun Peng, PhD, for helpful discussions, Johannes von Lintig, PhD for critical review of the manuscript, and Unnikrishnan Chandrasekharan, PhD, for help with establishing the anchorage-independent growth assays and microscopy. The authors thank Frank Mularo for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M., et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 2.Zbuk K.M., Eng C. Cancer phenomics: RET and PTEN and illustrative models. Nat. Rev. Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- 3.Li D.M., Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 4.Liu J.L., Sheng X., Hortobagyi Z.K., Mao Z., Gallick G.E., Yung W.K. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol. Cell Biol. 2005;25:6211–6224. doi: 10.1128/MCB.25.14.6211-6224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung J.H., Ginn-Pease M.E., Eng C. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res. 2005;65:4108–4116. doi: 10.1158/0008-5472.CAN-05-0124. [DOI] [PubMed] [Google Scholar]
- 6.Denning G., Jean-Joseph B., Prince C., Durden D.L., Vogt P.K. A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene. 2007;26:3930–3940. doi: 10.1038/sj.onc.1210175. [DOI] [PubMed] [Google Scholar]
- 7.Gil A., Andres-Pons A., Fernandez E., Valiente M., Torres J., Cervera J., Pulido R. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell. 2006;17:4002–4013. doi: 10.1091/mbc.E06-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotman L.C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N.P., Carver B.S., Cordon-Cardo C., Erdjument-Bromage H., et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassif N.T., Lobo G.P., Wu X., Henderson C.J., Morrison C.D., Eng C., Segalov E. PTEN mutations are common in sporadic microsatellite stable colorectal cancer. Oncogene. 2004;23:617–628. doi: 10.1038/sj.onc.1207059. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X.P., Loukola A., Salovaara R., Nystrom-Lahti M., Peltomaki P., de la Chapelle A., Aaltonen L.A., Eng C. PTEN mutational spectra, expression levels and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002;161:439–447. doi: 10.1016/S0002-9440(10)64200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiteman D.C., Zhou X.P., Cummings M.C., Pavey S., Hayward N.K., Eng C. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int. J. Cancer. 2002;99:63–67. doi: 10.1002/ijc.10294. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X.P., Gimm O., Hampel H., Niemann T., Walker M.J., Eng C. Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am. J. Pathol. 2000;157:1123–1128. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana I., Smith J.S., Sato K., Hosek S.M., Kimmel D.W., Jenkins R.B. Investigation of germline PTEN, p53, p16(INK4A)/p14(ARF), and CDK4 alterations in familial glioma. Am. J. Med. Genet. 2000;92:136–141. doi: 10.1002/(sici)1096-8628(20000515)92:2<136::aid-ajmg11>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 14.Perren A., Komminoth P., Saremaslani P., Matter C., Feurer S., Lees J.A., Heitz P.U., Eng C. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am. J. Pathol. 2000;157:1097–1103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridberg M., Servin A., Anagnostaki L., Linderoth J., Berglund M., Soderberg O., Enblad G., Rosen A., Mutelin T., Jerkeman M., et al. Protein expression and cellular localization in two prognostic subgroups of diffuse large B-cell lymphoma: higher expression of ZAP70 and PKC-beta II in the non-germinal center group and poor survival in patients deficient in nuclear PTEN. Leuk. Lymphom. 2007;48:2221–2232. doi: 10.1080/10428190701636443. [DOI] [PubMed] [Google Scholar]
- 16.Chung J-H., Ostrowski M.C., Romigh T., Minaguchi T., Waite K.A., Eng C. The ERK1/2 pathway modulates nuclear PTEN-mediated cell cycle arrest by cyclin D1 transcriptional regulation. Hum. Mol. Genet. 2006;15:2553–2559. doi: 10.1093/hmg/ddl177. [DOI] [PubMed] [Google Scholar]
- 17.Ginn-Pease M.E., Eng C. Increased nuclear phosphatase and tensin homologue deleted on chromosome 10 is associated with G0-G1 in MCF-7 cells. Cancer Res. 2003;63:282–286. [PubMed] [Google Scholar]
- 18.Liu F., Wagner S., Campbell R., Nickerson J., Schiffer C., Ross A. PTEN enters the nucleus by diffusion. J. Cell Biochem. 2005;96:221–234. doi: 10.1002/jcb.20525. [DOI] [PubMed] [Google Scholar]
- 19.Shen W., Balajee A., Wang J., Wu H., Eng C., Pandolfi P.P., Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 20.Trotman L.C., Alimonti A., Scaglioni P.P., Koutcher J.A., Cordon-Cardo C., Pandolfi P.P. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;445:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minaguchi T., Waite K.A., Eng C. Nuclear localization of PTEN is regulated by Ca(2+) through a tyrosil phosphorylation-independent conformational modification in major vault protein. Cancer Res. 2006;66:11677–11682. doi: 10.1158/0008-5472.CAN-06-2240. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Mao Z., LaFortune T., Alonso M.M., Gallick G.E., Fueyo J., Yung W.K. Cell cycle-dependent nuclear export of phosphatase and tensin homologue tumor suppressor is regulated by the phosphoinositide-3-kinase signaling cascade. Cancer Res. 2007;67:11054–11063. doi: 10.1158/0008-5472.CAN-07-1263. [DOI] [PubMed] [Google Scholar]
- 23.Lobo G.P., Waite K., Planchon S., Romigh T., Houghton J.A., Eng C. ATP modulates PTEN subcellular localization in multiple cancer cell lines. Hum. Mol. Genet. 2008;17:2877–2885. doi: 10.1093/hmg/ddn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J.W., Lee S.B., Kim C.K., Lee K.H., Cho S.W., Ahn J.Y. Lysine 263 residue of NPM/B23 is essential for regulating ATP-binding and B23 stability. FEBS Lett. 2008;582:1073–1080. doi: 10.1016/j.febslet.2008.02.059. [DOI] [PubMed] [Google Scholar]
- 25.Jiang J., Prasad K., Lafer E.M., Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kose S., Furuta M., Koik M., Yoneda Y., Imamoto N. The 70-kD heat shock cognate protein (hsc70) facilitates the nuclear export of the import receptors. J. Cell Biochem. 2005;171:19–25. doi: 10.1083/jcb.200506074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung J.H., Eng C. Nuclear-cytoplasmic partitioning of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) differentially regulates the cell cycle and apoptosis. Cancer Res. 2005;65:8096–8100. doi: 10.1158/0008-5472.CAN-05-1888. [DOI] [PubMed] [Google Scholar]
- 28.Jakob U., Scheibel T., Bose S., Reinsein J., Buchner J. Assessment of the ATP binding properties of Hsp90. J. Biol. Chem. 1996;271:10035–10041. doi: 10.1074/jbc.271.17.10035. [DOI] [PubMed] [Google Scholar]
- 29.Chang J.H., Lin J.Y., Wu M.H., Yung B.Y. Evidence for the ability of nucleophosmin/B23 to bind ATP. Biochem. J. 1998;329:539–544. doi: 10.1042/bj3290539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni Y., Zbuk K.M., Sadler T., Patocs A., Lobo G., Edelman E., Platzer P., Orloff M.S., Waite K.A., Eng C. Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am. J. Hum. Genet. 2008;83:261–268. doi: 10.1016/j.ajhg.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mutter G.L., Lin M.C., Fitzgerald J.T., Kum J.B., Baak J.P., Lees J.A., Weng P., Eng C. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J. Natl. Cancer Inst. 2000;92:924–930. doi: 10.1093/jnci/92.11.924. [DOI] [PubMed] [Google Scholar]
- 32.Scheibel T., Neuhofen S., Weikl T., Mayr C., Reinstein J., Vogel P.D., Buchner J. ATP-binding properties of human Hsp90. J. Biol. Chem. 1997;272:18608–18613. doi: 10.1074/jbc.272.30.18608. [DOI] [PubMed] [Google Scholar]
- 33.Wolfman J., Planchon S., Liao J., Wolfman A. Structural and functional consequences of c-N-Ras constitutively associated with intact mitochondria. Biochim. Biophys. Acta. 2006;1763:1108–1124. doi: 10.1016/j.bbamcr.2006.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.