Abstract
Mutations in the paired-domain transcription factor PAX9 are associated with non-syndromic tooth agenesis that preferentially affects posterior dentition. Of the 18 mutations identified to date, eight are phenotypically well-characterized missense mutations within the DNA-binding paired domain. We determined the structural and functional consequences of these paired domain missense mutations and correlated our findings with the associated dental phenotype variations. In vitro testing included subcellular localization, protein–protein interactions between MSX1 and mutant PAX9 proteins, binding of PAX9 mutants to a DNA consensus site and transcriptional activation from the Pax9 effector promoters Bmp4 and Msx1 with and without MSX1 as co-activator. All mutant PAX9 proteins were localized in the nucleus of transfected cells and physically interacted with MSX1 protein. Three of the mutants retained the ability to bind the consensus paired domain recognition sequence; the others were unable or only partly able to interact with this DNA fragment and also showed a similarly impaired capability for activation of transcription from the Msx1 and Bmp4 promoters. For seven of the eight mutants, the degree of loss of DNA-binding and promoter activation correlated quite well with the severity of the tooth agenesis pattern seen in vivo. One of the mutants however showed neither reduction in DNA-binding nor decrease in transactivation; instead, a loss of responsiveness to synergism with MSX1 in target promoter activation and a dominant negative effect when expressed together with wild-type PAX9 could be observed. Our structure-based studies, which modeled DNA binding and subdomain stability, were able to predict functional consequences quite reliably.
INTRODUCTION
Heritable tooth agenesis offers an insight into the genetic pathways that control the development of human dentition. Based on genetic, cellular and molecular studies, three genes play key roles in early tooth development (1–3). Two encode transcription factors expressed in mesenchymal cells, PAX9 and MSX1, both members of highly conserved, multigene families required throughout embryogenesis. Another is Bmp4 (Bone Morphogenetic Protein-4) which in the dental mesenchyme is the downstream target of PAX9 and MSX1. BMP4 encodes a pleiotropic growth and differentiation factor secreted by the mesenchyme to induce morphogenesis of the overlying epithelium (2). In the mouse, the human PAX9 ortholog (Pax9) is expressed early in tooth development and persists through the cap stage (3). In mice who have only one dentition, homozygous deletion of Pax9 leads to a downregulation of both MSX1 and Bmp4 expression and arrests tooth development at the bud stage while heterozygous littermates are unaffected (4). In marked contrast, all tooth agenesis-causing human PAX9 mutations are heterozygous, with most affecting only permanent teeth (2).
Functional haploinsufficiency resulting from frameshift, nonsense or deletion mutations has been hypothesized as the underlying cause of non-syndromic tooth agenesis for both PAX9 and MSX1 (5–7). Heterozygous null alleles, produced either by deletion of the entire PAX9 gene (8) or by mutation of the ATG start codon (9), lead to absence of both primary and permanent teeth, showing that PAX9 is dosage-sensitive in humans. However, for missense mutations, which substitute only a single amino acid residue rather than introduce a frameshift mutation, truncate or delete the protein, the alleles are likely hypomorphic. Given the role of PAX9 as a transcription factor, the mutations may affect multiple functions or processes, such as DNA-binding, nuclear translocation, transcriptional activation or synergistic protein–protein interactions with co-activators such as MSX1. We and others have previously characterized the DNA-binding properties of several of the known missense mutants by electrophoretic mobility shift assay (EMSA) using a variety of paired domain consensus sites. DNA-binding ranged from abolished for the PAX9 mutants L21P and I87F (10,11) or severely diminished for PAX9 S43K (12) to moderately affected (G6R) (12) with the CD19-2 (A-ins) consensus site, and for the non-binding proteins (L21P, I87F), the e5 consensus site as well. Another essentially non-binding missense mutant (R28P) was assayed with yet two other consensus sites (PRS4, PAX6CON) by Olsen and coworkers (13). Additional candidate functions have only been tested for limited subsets, hence the molecular mechanisms responsible for, or contributing to, the tooth agenesis phenotypes were not established until now. Furthermore, the structural basis for the loss of function has been hypothesized for a subset of the substitutions, but in the context of the crystal structure of the paired domain of PAX6, rather than homology models of PAX9 and the missense mutants.
Here, for eight of the nine missense mutations currently identified in the paired domain of PAX9 in humans with congenital tooth agenesis, we calculated structure-based homology models of the conserved wild-type and mutant domains, predicted the structural consequences of the single amino acid substitutions and determined their effect in parallel on DNA binding in vitro, subcellular localization, protein–protein interaction with Msx1, activation of target promoters in cell culture and synergistic activation with Msx1. Through our multifaceted survey of the panel of paired domain missense mutations, we found that the structure-based predictions were consistent with the underlying losses of function, which in turn correlated well with severity of tooth agenesis examined in the clinic.
RESULTS
Phenotype evaluation
To date, 18 distinct heterozygous mutations have been identified in PAX9. Nine of these are located within the DNA-binding paired domain of the transcription factor and result in single amino acid substitutions (7,11,13–16). One paired domain missense mutation (R47W) was omitted from our investigation because there were insufficient phenotype data. Although agenesis of posterior teeth dominates, different subsets of teeth are affected with variable frequency, hence phenotypes vary from mild (G6R, S43K) to moderate (L21P, G51S, K91E) and severe (R26W, R28P, I87F) (Table 1). The G6R mutation produces a mild phenotype; only one second premolar and two mandibular central incisors are affected in the individual with the sporadic G6R mutation, whereas molars, premolars and canines failed to develop in patients with the S43K mutation, although in relatively low numbers. L21P and K91E involve all types of teeth with an average of 8.4 –10 teeth missing. In K91E, the first molar was frequently unaffected and in one family member, only maxillary lateral incisors were missing. G51S shows a more anterior pattern resulting in oligodontia with preservation of first and second molar teeth. According to the average number of missing teeth in affected family members, the most severe phenotypes resulted from the R26W and R28P and I87F mutations with agenesis of molars and premolars as well as incisors and maxillary canines in the former two.
Table 1.
Summary of PAX9 paired domain mutations, functional and predicted structural consequences and tooth phenotypes
| Mutation | Subcellular localization | Protein stabilitya | DNA binding-EMSAb | Trans-activation | Trans-activation+MSX1 | MSX1 binding-colpc | Predicted structural alteration | Tooth phenotyped | Severity, average no. of missing teeth | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 219insG | Cytoplasm | Intact N-term SD only | – | Not detectable | Not detectable | N/D | Frameshift in linker; N-term SD only intact | Oligodontia M, P, Cl | Moderate 10 | Stockton et al. (2000); Mensah et al. (23) |
| Gly6Arg | Nucleus | +10.5% | ++++ | Moderate | Increased | + | Minimal, at surface | Hypodontia P, Cl | Mild 3 | Wang et al. (2008) |
| Leu21 Pro | Nucleus* | −0.9% | – | Negligible | Negligible | + | Steric clash with DNA | Oligodontia M, P, C, Cl, LI | Moderate 10 | Das et al. (7); Ogawa et al. (10) |
| Arg26TRP | Nucleus* | −17.8% | – | Negligible | Negligible | + | Instability and DNA contact loss | Oligodontia M, P, C, Cl, LI | Severe 11.3 | Lammil et al. (2003) |
| Arg28Pro | Nucleus* | −13.0% | −/+ | Negligible | Negligible | + | Instability, α2 kinked | Oligodontia M, P, C, Cl | Severe 13 | Jumlongras et al. (13) |
| Ser43Lys | Nucleus* | −7.4% | + | Negligible | Negligible | + | Instability, DNA clash | Oligodontia M, P, C | Mild 6.5 | Wang et al. (2008) |
| Gly51Ser | Nucleus | +2.5% | +++ | Enhanced | No increased | + | Context-dependent | Oligodontia P, C, Cl, LI | Moderate 9 | Mostowska et al. (15) |
| Ile87Phe | Nucleus* | −5.2% | −/+ | Negligible | Increased | + | C-term SD instability | Oligodontia M, P | Severe 11.5 | Kapadia et al. (11) |
| Lys91Glu | Nucleus | +0.2% | +++ | Negligible | Increased | + | Water/DNA binding | Oligodontia M, P, C, Cl, LI | moderate 8.4 | Das et al. (7) |
aProtein stability calculated by modeling routine as a total energy term for individual N- and C-terminal subdomains with Gly72 (C) and Gly73 (N) as terminal residues.
bEMSA, electrophoretic mobility shift assay; wild-type PAX9 binding would be +++++ by comparison.
ccolP, co-immunoprecipitation: wild-type PAX9 binding, +.
dCI central ineisor, LI laterla ineisor, C canine, P first or second premolar, M first or second molar—nomenclature and table after Jumlongras et al. (2004). Note the primary tooth development was unaffected by all of the missense mutation, however the 219insG frameshift mutation was associated with one or more missing primary molars. Nuclear localized mutation also detected in trace amounts in cytoplasmic fractions are designated with an asterisk.
Structure-based predictions of subdomain stability and DNA-binding activities
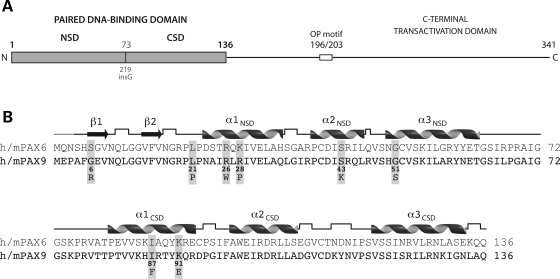
Human PAX9 is nearly 350 amino acid residues in length, comprised of an N-terminal bipartite or ‘paired’ DNA-binding domain, an octapeptide motif, no additional homeodomain and a C-terminal transcriptional regulatory domain, which is rich in alanine (13.7%), serine (13.2%), proline (12.7%) and glycine (8.3%) residues and likely intrinsically unstructured (Fig. 1A). The N-terminal paired domain of PAX9 may also lack appreciable structure in the free protein, as the paired domain of the paralogous PAX6 protein is relatively structureless in solution as judged by circular dichroism and one- and two-dimensional nuclear magnetic resonance spectroscopy (17,18). The PAX8 protein on the other hand has been found to have a defined residual helix-turn-helix structure of both paired subdomains in solution (19). However, interaction with DNA appears to be necessary to achieve the final homeodomain-like folded structures in both the N- and C-terminal subdomains, each composed of three α-helices, as determined from crystal structures of the paired domains of PAX6 (20) and PAX5 (21) bound to DNA (Fig. 1B).
Figure 1.
Paired DNA-binding domain of the PAX9 protein. (A) Overall structure of PAX9. The DNA-binding domain of PAX9 is divided into two independent subdomains, an N-terminal (NSD) and a C-terminal (CSD), joined by a short linker segment containing start site (Gly73) of the 219insG PAX9 frameshift mutation. The shift in reading frame produces a truncated polypeptide with a non-native C-terminal segment lacking the second DNA-binding subdomain (CSD), as well as the transcriptional repression (octapeptide motif) and activation domains. (B) Primary and secondary structures of the paired domains of human and mouse PAX6 and PAX9. The DNA-binding subdomains share a homeodomain-like fold, each comprised of three helices. A pair of strands folded into a β-hairpin structure precedes the α-helical homeodomain-like fold of the N-terminal subdomain. Six of the eight missense mutations in the paired domain are within the N-terminal subdomain, three of which are clustered near the N-terminus of the first helix. The secondary structures of the PAX6 crystal structure and PAX9 model (cartoon) were identical. Right angles represent hydrogen-bonded turns in the polypeptide backbone and the grey segment at the N-terminus unstructured residues not included in the crystal structure.
Sequence alignment with PAX9 orthologs from other organisms, as well as the paralogous members of the family (PAX1–8) revealed that the eight residues in the PAX9 paired domain mutated in human tooth agenesis are highly conserved. Between human and mouse PAX9 and PAX6, six of the eight are identical, one is conserved (Arg>Lys) and only one disparate (Gly>Ser), indicating that the structural and functional consequences of the missense mutations could be analyzed in the context of the crystal structure of the PAX6 paired domain. Even more, because the paired domains of the human and mouse PAX9 and PAX6 are 73% identical within the 133 amino acid residues of the PAX6 crystal structure and can be aligned without gaps (Fig. 1B), the experimentally determined structure of PAX6 provides a three-dimensional template for calculation of highly reliable structure-based homology models of wild-type and missense mutant PAX9 paired domains, along with estimates of relative stabilities (Fig. 2; Table 1).
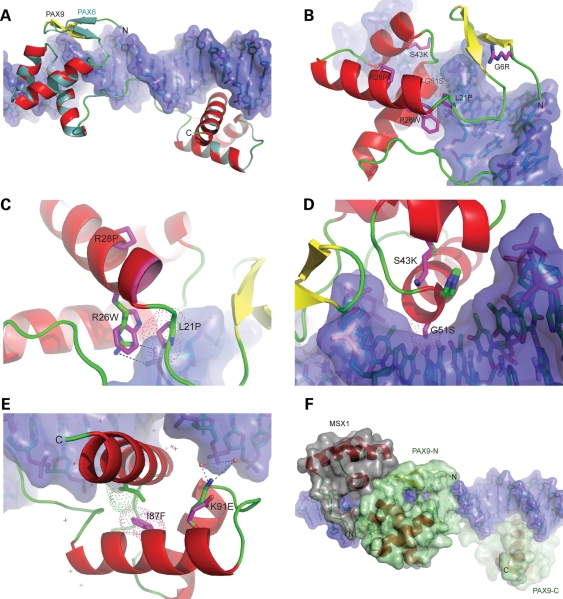
Figure 2.
Structure-based homology models of PAX9 missense mutant paired domains. (A) Superposition of wild-type PAX9 homology model on the PAX6–DNA complex crystal structure. The α-helical (red) and loop segments (green) of the backbone of the PAX9 model are nearly indistinguishable with those of the PAX6 structural template (turquoise). Note that although the defined secondary structures of model and template are identical (cf. Fig. 1B), the N-terminal β-hairpin of PAX9 (yellow) rendered dissimilarly by the graphics program. (B) Superposition of N-terminal subdomain PAX9 mutant models. The substituted sidechains of each model are depicted (magenta sticks). Similar to the serine sidechain of the PAX6 template, the arginine sidechain of the G6R mutant model extends away from the DNA surface, and without apparent conformational change in the subdomain. (C) Zoomed view of superimposed models of tryptophan and proline mutants in pre-loop and helix of α1 NSD. The molecular surface of the proline sidechain of the L21P mutant (magenta dots) overlaps that of the DNA backbone (violet surface). In the wild-type model, the guanido group of arginine 26 (blue, green stick) forms an ion pair (dashes) with a phosphate group of the DNA backbone that is abrogated by mutation to tryptophan, which is non-isosteric with arginine and alters packing of the subdomain. Introduction of a proline within α1 of the R28P mutant produced a displacement of the N-terminus of the helix (leading edge magenta). (D) Zoomed view of superposition of lysine and serine mutants in α2 and α3 NSD helices. In the S43K mutant model, the sidechain of histidine 50 (imidazole ring, side view) bulges towards the DNA surface to accommodate the lysine sidechain (magenta stick), extended over twice the length of the wild-type serine (green stick). The molecular surface of the serine sidechain of the G51S mutant (magenta dots) protrudes into the surface of the major groove primarily at a cytosine of a G:C base pair. (E) Superposition of C-terminal subdomain PAX9 mutant models. The molecular surface of the phenylalanine sidechain of the I87F mutant (magenta dots) overlaps that of the adjacent isoleucine 126 sidechain in the wild-type subdomain (green dots), resulting in repositioning of the bulky, branched isoleucine sidechain in the mutant (green stick). The wild-type interaction of the ε-amino group of lysine 91 (blue, green stick) with a phosphate group of the DNA backbone through a hydrogen-bonded network of bound water molecules (red spheres) is depicted by dashes. The substituted glutamate sidechain of the mutant model (magenta stick) and positions of additional bound waters (cross-marks) are also shown. (F) Hypothetical superposition model of a ternary complex of PAX9 and MSX1 bound to DNA. The crystal structure of the MSX1–DNA complex (PDB ID: 1IG7) was aligned with the PAX9–DNA model, maintaining the DNA backbones in register and maximizing interaction between the MSX1 and PAX9.
Based on comparison of the wild-type and mutant PAX9 models, two of the missense mutations (L21P, R26W), both located near the N-terminus of the α1 NSD helix (Figs 1B, 2A–C), were predicted to severely diminish DNA binding. The L21P substitution introduces a steric clash with the DNA backbone, the R26W disruption of a stabilizing interaction, as well as rearrangement of the subdomain core and destabilization of the protein (Fig. 3A; Table 1). Similarly, substitution at the adjacent residue (R28P), along with one in the N-terminal subdomain (S43K) and another in the C-terminal subdomain (I87F), was also calculated to destabilize the corresponding subdomains (Fig. 2C–E; Table 1). The S43K substitution, like the R26W, was anticipated to have dual roles in perturbing the stability of the N-terminal subdomain and interaction with DNA, yet in the latter case by a subtle shift of the contact surface rather than abolished interactions (Fig. 3A).
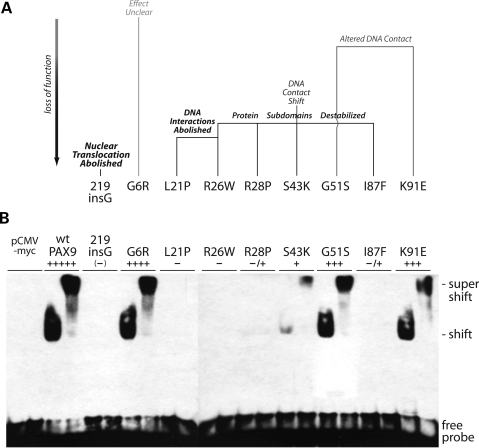
Figure 3.
Comparison of predicted functional consequences of missense mutations with paired domain consensus site DNA-binding activities. (A) Structure-based homology modeling, ranging from no apparent effect (grey) to severe loss of function (black, bold). The R26W and S43K mutations are predicted to affect both interaction with DNA and protein stability, the G51S mutation DNA interaction only. (B) Electrophoretic mobility shift assay (EMSA) of nuclear extracts from transiently transfected COS7 cells with a paired domain consensus oligonucleotide probe, CD19-2 (A-ins). Anti-myc antibody supershifts (right sample each pair) were performed with the tagged proteins to confirm specificity of labeled duplex oligonucleotide shifts. DNA-binding activities were ranked relative to wild-type PAX9 (+++++) and the 219insG null (–). Weak binding of the R26W and I87F mutant proteins (Table 1, −/+) was detected by prolonged autoradiography. pCMV-myc: nuclear extract from empty vector control transfection.
Two other missense mutations (G51S, K91E) were anticipated to have even more subtle effect on DNA binding by altering, not abolishing or displacing, DNA contacts. Near the N-terminus of the α3 NSD helix (Figs 1B, 2D), the G51S substitution introduces a small side chain into the major groove causing a slight steric clash, yet perhaps capable of hydrogen bonding with the paired bases at other binding sites and imparting stability, rather than instability. The K91E missense mutation, one of only two in the C-terminal subdomain, was predicted to alter the interactions with bound waters that mediate binding by the wild-type side chain, a hydrogen bond donor (Figs 2E and F). The mutant side chain, an acceptor, would likely interact less robustly through an altered arrangement of polar water molecules. Interestingly, one of the eight missense mutations (G6R), distal to the DNA surface in a structural element (β-hairpin) largely independent of the homeodomain-like helices of the N-terminal subdomain (Figs 1B, 2A and B), was not anticipated to affect DNA binding or subdomain stability. For this mutant, analysis with PAX6 as model structure could not predict the effect of G6R on DNA binding observed in other assays employed in this study. Among the missense mutations, the glycine is the only non-conserved residue suggestive of a specific structural role. Because the residue resides on the periphery of an extensive hydrophobic patch (P3, A4, F5, V8, V14, F15, V16, P20, P22, A24, I25) and a structural element required for protein–protein interactions (21,22), substitution with the bulky, charged sidechain of arginine might interfere with cooperative assembly of regulatory complexes in vivo requiring recruitment of protein partners in addition to MSX1.
In vitro DNA-binding activities with a paired domain consensus sequence
We and others have tested the DNA-binding ability of several of the mutants previously (see above); however, in order to compare the relative DNA-binding activity of the entire set of missense mutants, including the three previously uncharacterized ones (R26W, G51S, K91E), all of the proteins were prepared identically and assayed with one common consensus site, CD19-2 (A-ins). In the side-by-side comparison, a range of DNA-binding activity was observed that correlated well with the predicted functional consequences of the missense substitutions (Fig. 3A and B). Because the frameshift 219insG mutant is restricted to the cytoplasm as judged by immunofluorescence (22) and does not bind DNA consensus sites, this sample along with that from empty vector (pCMV-myc) served as negative control. The two mutations (L21P, R26W) predicted to disrupt DNA-binding did indeed (−), and the two solely predicted to destabilize the respective subdomains (R28P, I87F) led to severely diminished, but detectable binding upon longer exposures (−/+). Interestingly, the mutation predicted to destabilize the subdomain and shift contact with DNA (S43K) was more active than anticipated (+), perhaps due to a stabilizing interaction gained by lysine substitution and mediated by bound waters, as for the K91 side chain (Fig. 2D and E). More in line with prediction, the two mutations (G51S, K91E) with apparent altered DNA contacts only mildly affected binding (+++), and the freestanding mutation (G6R) with no clear defect had nearly indistinguishable effect (++++) relative to wild-type (+++++). None of the mutants was capable of inhibiting the DNA-binding of added wt PAX9.
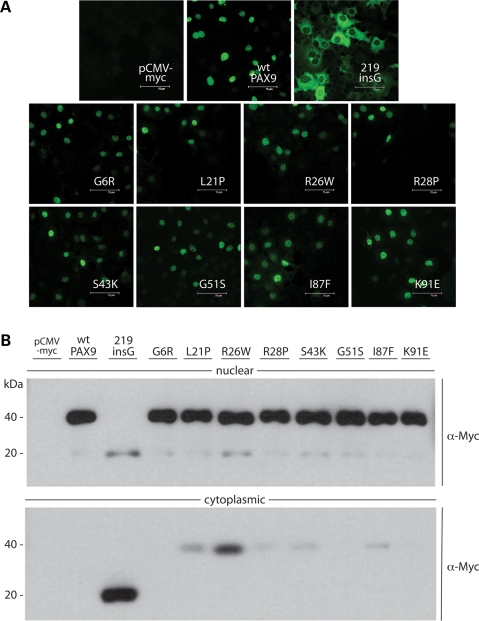
Nuclear localization
Although the in vitro DNA-binding activity of the 219insG mutant is abolished by the frameshift after the first paired subdomain, defective nuclear localization, judged by immunofluorescence, appears to be the root of the loss of function in cell culture and in vivo (23). To establish whether missense mutations might also affect nuclear localization of the transcription factor, resulting in, or contributing to, the tooth agenesis phenotypes, the subcellular localization of the entire set of missense mutants, including the four previously characterized ones (L21P, I86F, G6R and S43K) was determined by both immunofluorescence and western blot. In contrast to the 219insG mutant that produced intense cytoplasmic but negative nuclear fluorescence, each of the missense mutant proteins appeared to localize in the nuclei of transiently transfected cells like wild-type (Fig. 4A). Consistent with the fixed-cell immunofluorescence results, by western blot analysis the missense mutant proteins were detected predominantly in nuclear fractions (Fig. 4B). The G6R, G51S and K91E mutants, like wild-type, were undetectable in cytoplasmic fractions. However, several of the missense mutants did not partition completely, in particular the R26W protein calculated as most unstable (Table 1).
Figure 4.
Subcellular immunolocalization of missense mutant proteins. (A) Immunofluorescent detection of Myc-tagged PAX9 proteins in transfected COS7 cells. None of the missense mutations affect nuclear localization, in contrast to the 219insG frameshift that abrogates translocation. (B) Western blot analyses of nuclear and cytoplasmic extracts from transfected COS7 cells with α-Myc antibody. 219insG and small amounts of some missense mutant proteins predicted less stable than wild-type could be detected in the cytoplasmic fractions.
Our parallel analyses with both the methods reported here show that (1) the 219insG mutant allele is functionally a null, owing to abrogation first and foremost of nuclear localization and secondarily DNA binding, and that (2) in marked contrast, none of the eight missense mutations in the paired subdomains alter subcellular localization to a significant enough extent to play a role in the pathogenetic mechanisms leading to tooth agenesis.
Protein–protein interaction with MSX1
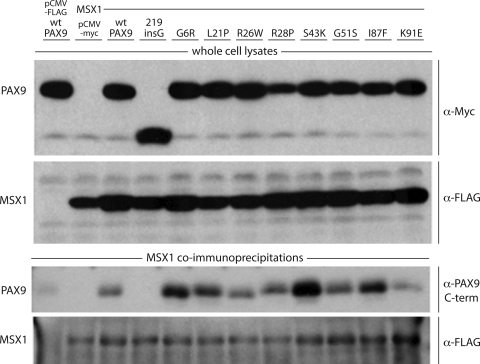
We have previously shown that the wild-type Pax9 protein and the missense mutant L21P can interact directly with Msx1 in vitro by co-immunoprecipitation of the full-length proteins (10,24). Since interaction of L21P with Msx1 was apparently unaffected, but DNA binding and transcriptional activation of L21P was severely impaired, we had ruled out disturbances of Pax9–Msx1 complex formation as a factor in pathogenesis. To determine whether any of the other known PAX9 missense mutations disrupt the Pax9–Msx1 complex, which would exacerbate the loss of DNA binding observed in vitro and further impair transcriptional activation of the effector (Bmp4), we assayed all eight in parallel for co-immunoprecipitation with Msx1.
Co-transfection of pCMV-Myc-Pax9 and pCMV-FLAG-Msx1, followed by immunoprecipitation with anti-FLAG antibody and detection with anti-Myc, showed that all of the missense mutant proteins were able to interact physically with wild-type Msx1, at least qualitatively (Fig. 5, third panel). Mutations resulting in more basic residues (R, K) appear to have a positive effect on binding while change to an acidic amino acid (E) seems to weaken interaction. However, since variability within samples was observed and interaction with Msx1 was at least qualitatively unaffected, we concluded that disruption of the Pax9–Msx1 complex by any of the missense mutations was not a significant factor in pathogenesis.
Figure 5.
Co-immunoprecipitation assay of protein–protein interaction with MSX1. (Upper panels) Western blot analyses of whole cell lysates from COS7 cells cotransfected with Myc-tagged PAX9 proteins and FLAG-tagged MSX1. (Lower panels) Western blot analyses of proteins pulled down with α-FLAG antibody from whole cell lysates. Immunoprecipitated MSX1 was detected with the α-FLAG antibody and co-immunoprecipitated wild-type and missense mutant PAX9 proteins with antibody against the C-terminal transactivation domain of PAX9.
Upregulation of msx1 and bmp4 target promoters
We sought to determine whether the loss of DNA-binding activity observed in vitro with a consensus site would correspond to a proportional reduction in activation of the two targets of Pax9, the Msx1 and Bmp4 genes, which are key effectors of tooth bud morphogenesis.
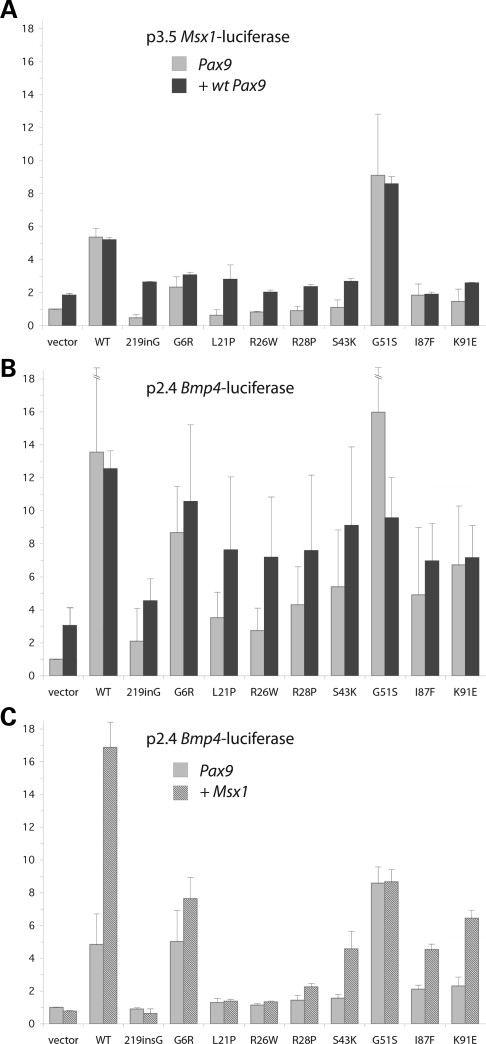
Towards that end, we have performed promoter reporter assays with two plasmid constructs, p3.5 Msx1-luciferase and p2.4 Bmp4-luciferase, composed of 3.5 and 2.4 kb of genomic DNA upstream of the first exon of the respective coding sequences and the luciferase gene. Either promoter construct is co-transfected with Pax9 expression vector in COS7 cells (10). With these gene activation assays, we compared the activities of the panel of Pax9 missense mutants with wild-type and the null mutant, 219insG (Fig. 6A and B). In all but one case, activation of the reporter gene was diminished or downregulated (light grey bars) to an extent proportional to the loss of DNA-binding activity observed in vitro. Only G51S, the mutation introducing a small hydrogen-bonding side chain into the major groove putatively capable of imparting greater stability, led to an increase in the activation of Msx1 and Bmp4 promoters (Fig. 6). None of the mutants showed dominant-negative activity when co-expressed with wild-type Pax9 (dark grey bars), with the exception again of G51S which shows diminished activation upon co-expression with wild-type protein.
Figure 6.
Luciferase reporter assays of target promoter upregulation and MSX1 synergy. (A, B) Upregulation of Msx1 and Bmp4 target promoters. Msx1 promoter–reporter (p3.5 Msx1-luciferase) or Bmp4 promoter–reporter (p2.4 Bmp4-luciferase) constructs were co-transfected with mutant Pax9 expression vectors alone (1.5 µg), or in combination with wild-type Pax9 (0.8 µg each). (C) Synergistic upregulation of Bmp4 promoter with MSX1. The Bmp4 promoter–reporter construct was co-transfected with Pax9 expression vector alone (1 µg Pax9, 0.5 µg vector), or in concert with the Msx1 expression vector (1 µg Pax9, 0.5 µg Msx1). Error maxima for wild-type and G51S upregulation in (B) were 20.46 and 25.13 relative units, respectively.
Synergistic upregulation of Bmp4 promoter with Msx1
To further mimic the in vivo function of Pax9 and hence better assess and correlate the effects of the mutations, we compared the promoter-activating ability of the panel of missense mutants with wild-type and the null mutant after co-expression of Msx1, which at certain concentration behaves as a positive cofactor to synergistically upregulate the Bmp4 reporter constructs in this system (10). As seen previously with the L21P mutant, no synergistic activation resulted from co-expression of Msx1 (dark hatched bars) with the R26W and R28P mutants, which alone (light grey bars) minimally activated expression of the Bmp4 reporter owing to loss of DNA-binding activity, despite interacting with Msx1 in vitro (Fig. 6C). Similar to our previous observation with wild-type Pax9, we did see synergistic effects with the G6R, S43K, I87F and K91E mutants. G51S Pax9 is the only mutant with fairly intact DNA-binding and transactivation ability that does not demonstrate a synergetic effect with MSX1.
DISCUSSION
A summary of our findings is presented in Figure 7 and Table 1.
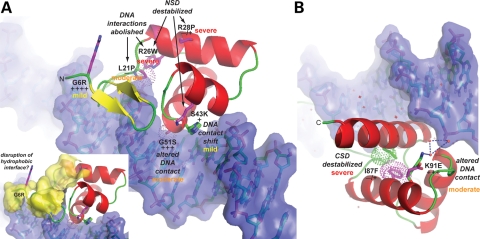
Figure 7.
Summary of structural and functional consequences of missense mutations and severity of effects on early odontogenesis. (A, B) Zoomed views of the N-terminal (NSD) and C-terminal (CSD) subdomains of PAX9. The structural and functional consequences of each missense mutation, shown schematically in Figure 3A, are indicated along with the observed reduction in in vitro DNA-binding activity relative to wild-type (+++++) as ranked in Figure 3B. In addition, the severity of the effect of each mutation (mild, moderate, severe) on early odontogenesis is indicated by color-coding (yellow, orange, red). The small inset depicts an extensive hydrophobic surface (yellow) that may serve as an interface for recruitment of binding partners to the PAX9:DNA complex. The G6R mutation, which introduces a charged and bulky, albeit relatively flexible sidechain, on the periphery of the hydrophobic patch, could potentially diminish interactions with other proteins within a transcriptional regulatory complex leading to detectable odontogenic affects.
Our analysis of the mutant proteins shows that for most of the mutations, the severity of the tooth agenesis phenotype correlates well with the level of residual DNA-binding and promoter-activating ability. Mutations resulting in total or near total abrogation of DNA-binding, such as L21P, R26W, R28P and I87F are associated with more severe phenotypes. Less severe phenotypes result from the S43K and K91G proteins, which retain DNA binding to some extent. The two sporadic tooth agenesis cases, G6R and G51S, produce proteins that retain excellent DNA-binding properties and G6R produces the mildest phenotype. The G51S mutation is exceptional in that it achieves equal or higher transcriptional activation of Msx1 and Bmp4 promoters than the wild-type protein but nevertheless manifests as tooth agenesis. Our studies indicate that a loss of responsiveness of G51S to MSX1 as co-activator for Bmp4 transcription may be responsible for the dental phenotype, which with its more anterior pattern resembles Msx1-induced tooth agenesis, however more studies are required to support this conclusion. In addition, the G51S Pax9 and the wild-type protein may inhibit each other as suggested by the mixing experiment (see above); thus contributing to the tooth agenesis in this heterozygous patient. Since MSX1 expression is enhanced by the G51S mutation in vitro, the original assumption (15) that G51S may act via a reduction of MSX1 expression is no longer tenable. On the other hand, over-expression of MSX1 can also lead to downregulation of Bmp4 promoter activity in vitro as we have previously shown (10).
Overall our findings suggest that haploinsufficiency is the likely pathogenic mechanism for more severe phenotypes, while protein products causing less severe phenotypes may arise from hypomorphic alleles that reduce transcriptional activation via impaired DNA-binding, or less frequently through hindrance of the synergistic action of the PAX9–MSX1 complex and an as yet unexplained mutual inhibition of wild-type and mutant protein. The considerable phenotype variability within families on the other hand, is most likely caused by polymorphisms in modifier genes such as other regulatory genes that are involved in tooth development.
The DNA sequences, which are recognized by paired domain and homeodomain proteins, are quite unspecific and require cooperative binding of one or several additional proteins to achieve highly selective biological effects. Other researchers have described the interaction of paired domain and homeodomain proteins in synergistic transactivation of target promoters (25), however MSX1 has been mostly found to repress transactivation owing to interaction of its homeodomain with diverse proteins, often without requiring any interaction with the TAAT core motif for DNA-binding (26–29). In contrast to the numerous studies that have attributed a repressor function to Msx1, one early study (30) hypothesized that MSX1 induces the expression of mesenchymal BMP4 as either ‘a transcriptional activator or as an accessory factor for a transcriptional activator’. We have previously shown that MSX1 cannot activate transcription from the BMP4 promoter by itself but that interaction of Msx1 with Pax9 leads to a strong synergistic enhancement of Bmp4 promoter activity in a dosage-dependent manner (10): Msx1 in relatively low concentration potentiates Pax9 transactivation of both Bmp4 and MSX1 promoters while higher doses of Msx1 may reduce Pax9-induced transactivation of both promoters. Thus, Msx1 may serve as a modulator of the Pax9 transcription factor activity, fine-tuning Bmp4 expression, and regulating its own expression in a feedback loop.
Although PAX9 interacts with non-homeodomain proteins (transcriptional repressor PLU-1/RBP2-H1/JARID1B) via amino acids located C-terminal of the paired domain (31), the complex formation with other homeo- or paired domain proteins is most likely a result of interactions between these domains themselves, in the same areas that are also responsible for DNA-binding. For the homeodomain protein Chx10 and the Pax6 protein which contains both a paired and a homeodomain motive, it was shown that mutations of single positively charged amino acids (arginine) in helix3 of the homeodomain disrupt the protein interaction, while mutations of negatively charged amino acids (glutamic and aspartic acid) in helix 1 and 2 of the C-terminal subdomain of the paired domain can strongly reduce but not abolish the interaction since they cannot sufficiently compromise the salt bridge formation at the interaction surface (32). A similar mechanism may explain our observation that all Pax9 paired domain mutants can still interact with Msx1. However, the protein–protein interaction between different homeo- and paired box domains does not always involve the same amino acid residues. A recent report shows that the interaction of the homeodomain of antennapedia (ANTP) with the paired box of eyless (EY) involves negatively charged amino acids (glutamic acid) of the first helix of the homeodomain (33). These will presumably interact with positively charged amino acids (arginine and lysine) of the paired domain. Several of the mutations that we investigated in our study are arginine and lysine mutations of the paired domain which could have an influence on the strength of the interaction with the Msx1 homeodomain. A more quantitative approach than co-immunoprecipitation together with structural elaboration of the Pax9–Msx1 protein–protein complex would be helpful to clarify this point.
The same authors (33) also report that the homeo/paired domain-mediated protein–protein interaction between ANTP and Ey result in phenotypic effects by binding and sequestering the partner protein without involving any regulation at the DNA or RNA level. Similarly, the Pax9–Msx1 complex isolated via co-immunoprecipitation may serve a function other than DNA-binding and transactivation. Since the structure of PAX proteins in solution has been described as poorly (18), or in a more recent study as moderately defined (18,19), the binding of Pax9 to Msx1 protein without involvement of DNA may be structurally and functionally different from a complex formed in the presence of DNA. Alternatively, a preformed complex may recognize a different DNA target sequence.
Concluding remarks
In vitro findings on functional properties of homeo- and paired domain proteins do not always reflect biological significance, because the network of in vivo factors determining DNA-binding specificity, protein interactions and their consequences is scarcely known. However, it has been shown that heterozygous mutations in the PAX6 gene cause developmental defects with a similar genotype–phenotype correlation (20): mutations that completely abolish PAX6N-subdomain function result in aniridia while missense mutations that retain partial DNA-binding activity show less severe phenotypic effects like Peters' anomaly and other congenital eye disorders (34).
The fairly good correlation between the in vitro assays used here and the in vivo tooth agenesis phenotype lends credence to the presented results and suggests that diminished or abolished DNA-binding of the PAX9 protein is the basis for the developmental dental defects in seven out of eight of the missense mutations while one of the mutations appears to affect the synergistic transactivation with MSX1, and its regulation in addition to a mutual inhibitory effect of wild-type and mutant protein.
MATERIALS AND METHODS
Structure-based homology modeling
Structure-based homology models of wild-type and missense mutant PAX9 paired domains were calculated by automated SwissModel routines input through the alignment interface followed by the project (optimize) mode in SwissModel Workspace (http://swissmodel.expasy.org/) (Biozentrum, Basel), with the DNA-bound human PAX6 paired domain (PDB ID: 6PAX, Chain A) serving as a three-dimensional template. A total energy term derived from force-field minimization (Gromos96, ETH, Zürich) of the models provided a measure of the relative stabilities of the PAX9 paired domains. Secondary structure assignments for the PAX6 crystal structure and wild-type PAX9 models were determined from the PDB files with the automated program DSSPcont (http://www.rostlab.org/services /DSSPcont/) (B. Rost, Columbia University). Models were viewed in stereo and figure graphics rendered with the program PyMOL (http://pymol.sourceforge.net/) (DeLano Scientific).
Plasmid constructs
Mouse Pax9 and Msx1 cDNAs were subcloned in-frame with N-terminal epitope tags downstream of the cytomegalovirus (CMV) immediate early promoter in pCMV-Tag3 and pCMV-Tag2 (Stratagene), yielding pCMV–Myc–Pax9 and pCMV–FLAG–Msx1, respectively (10,22). Site-directed mutagenesis was performed with the QuikChange mutagenesis kit (Stratagene) and the entire coding sequence of each mutant construct was verified by sequencing.
Cell culture and transient co-transfection
COS7 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and maintained at 37°C in the presence of 5% CO2. Transient co-transfections were performed with FuGENE 6 Transfection Reagent (Roche).
Immunolocalization
COS7 cells were transfected with wild-type or mutant pCMV–Myc–Pax9 and immunolocalization performed with monoclonal c-Myc primary and fluorescein isothiocyanate-conjugated secondary antibodies (Santa Cruz Biotechnology) as described previously (23). Cells were examined and digital images captured 24 h after transfection with a Leica DM IRBE confocal microscope. For immunoblot analyses, nuclear and cytoplasmic fractions were obtained from cells transfected for 24 h using the Nuclear Extract Kit (Active Motif) according to manufacturer's instructions followed by western blotting as described previously (23).
Electrophoretic mobility shift assays
EMSAs were performed in triplicate with the LightShift Chemiluminescent EMSA Kit (Pierce) using a 23 bp high-affinity paired domain binding site probe, CD19-2 (A-ins) (35). Briefly, the biotin-labeled double-stranded DNA probe was incubated at room temperature for 30 min with 1 µg of nuclear extract from COS7 cells transfected with the appropriate expression plasmid (wild-type or mutant pCMV–Myc–Pax9), followed by 0.1 µg of anti-Myc antibody (Santa Cruz Biotechnology) for another 5 min. The entire reaction mixture was loaded onto a non-denaturing 6% polyacrylamide gel for electrophoretic analysis.
Co-immunoprecipitation
Pax9 proteins were co-expressed with Msx1 by co-transfection of COS7 cells with wild-type or mutant pCMV–Myc–Pax9 and pCMV–FLAG–Msx1. After 24 h, cells were resuspended in lysis buffer (50 mm Tris–HCl, pH 8.0, 400 mm NaCl, 1% Triton X-100) with protease inhibitor cocktail tablets (Roche) and incubated for 30 min on ice. Cell lysates were added to 40 µl of anti-FLAG M2 affinity gel (Sigma) and rotated overnight at 4°C. The affinity gels were washed four times with lysis buffer and once with Tris-buffered saline then boiled in Laemmli buffer for sodium dodecyl sulphate–polyacrylamide gel electrophoresis. For detection on western blots, anti-Myc (9E10) monoclonal antibody (Santa Cruz Biotechnology) and mouse anti-FLAG M2 monoclonal antibody (Sigma) were diluted 1:400 and 1:1000, respectively.
Luciferase reporter assays
Pax9 and Msx1 expression vectors were co-transfected with the Msx1 or Bmp4 promoter–reporter constructs, p3.5 Msx1-Luc (0.5 µg) and p2.4 Bmp4-Luc (0.15 µg), in six-well plates as previously described (10). pCMV-SPORT β-galactosidase reporter plasmid (Invitrogen) served as an internal control to monitor transfection efficiency (0.25 µg/well). Cell extracts were prepared with Passive Lysis Buffer (Promega) 24 h after transfection and luciferase and β-galactosidase activities measured with commercially available kits (Promega, Invitrogen). The amounts of pCMV–Myc–Pax9 and pCMV–FLAG–Msx1 added per well varied between experiments (Fig. 6).
FUNDING
This work was supported by National Institutes of Health (DE16472 to R.N.D., DE16346 to H.K.).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J. Cell Sci. 2003;116:1647–1648. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- 2.Kapadia H., Mues G., D'souza R. Genes affecting tooth morphogenesis. Orthod. Craniofac. Res. 2007;10:237–244. doi: 10.1111/j.1601-6343.2007.00407.x. [DOI] [PubMed] [Google Scholar]
- 3.Peters H., Neubuser A., Balling R. Pax genes and organogenesis: Pax9 meets tooth development. Eur. J. Oral Sci. 1998;106(Suppl. 1):38–43. doi: 10.1111/j.1600-0722.1998.tb02151.x. [DOI] [PubMed] [Google Scholar]
- 4.Peters H., Neubuser A., Kratochwil K., Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Gene Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mostowska A., Biedziak B., Trzeciak W.H. A novel mutation in PAX9 causes familial form of molar oligodontia. Eur. J. Hum. Genet. 2006;14:173–179. doi: 10.1038/sj.ejhg.5201536. [DOI] [PubMed] [Google Scholar]
- 6.Nieminen P., Arte S., Tanner D., Paulin L., Alaluusua S., Thesleff I., Pirinen S. Identification of a nonsense mutation in the PAX9 gene in molar oligodontia. Eur. J. Hum. Genet. 2001;9:743–746. doi: 10.1038/sj.ejhg.5200715. [DOI] [PubMed] [Google Scholar]
- 7.Das P., Hai M., Elcock C., Leal S.M., Brown D.T., Brook A.H., Patel P.I. Novel missense mutations and a 288-bp exonic insertion in PAX9 in families with autosomal dominant hypodontia. Am. J. Med. Genet. 2003;118:35–42. doi: 10.1002/ajmg.a.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das P., Stockton D.W., Bauer C., Shaffer L.G., D'souza R.N., Wright T., Patel P.I. Haploinsufficiency of PAX9 is associated with autosomal dominant hypodontia. Hum. Genet. 2002;110:371–376. doi: 10.1007/s00439-002-0699-1. [DOI] [PubMed] [Google Scholar]
- 9.Klein M.L., Nieminen P., Lammi L., Niebuhr E., Kreiborg S. Novel mutation of the initiation codon of PAX9 causes oligodontia. J. Dent. Res. 2005;84:43–47. doi: 10.1177/154405910508400107. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa T., Kapadia H., Feng J.Q., Raghow R., Peters H., D'souza R.N. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J. Biol. Chem. 2006;281:18363–18369. doi: 10.1074/jbc.M601543200. [DOI] [PubMed] [Google Scholar]
- 11.Kapadia H., Frazier-Bowers S., Ogawa T., D'souza R.N. Molecular characterization of a novel PAX9 missense mutation causing posterior tooth agenesis. Eur. J. Hum. Genet. 2006;14:403–409. doi: 10.1038/sj.ejhg.5201574. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Wu H., Wu J., Zhao H., Zhang X., Mues G., D'souza R.N., Feng H., Kapadia H. Identification and functional analysis of two novel PAX9 mutations. Cells Tissues Organs. 2009;189:80–87. doi: 10.1159/000151448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jumlongras D., Lin J.Y., Chapra A., Seidman C.E., Seidman J.G., Maas R.L., Olsen B.R. A novel missense mutation in the paired domain of PAX9 causes non-syndromic oligodontia. Hum. Genet. 2004;114:242–249. doi: 10.1007/s00439-003-1066-6. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Hu Q., Chen Y., Luo S., Bao L., Xu Y. A novel missense mutation in the paired domain of human PAX9 causes oligodontia. Am. J. Med. Genet. 2007;143:2592–2597. doi: 10.1002/ajmg.a.31993. [DOI] [PubMed] [Google Scholar]
- 15.Mostowska A., Kobielak A., Biedziak B., Trzeciak W.H. Novel mutation in the paired box sequence of PAX9 gene in a sporadic form of oligodontia. Eur. J. Oral Sci. 2003;111:272–276. doi: 10.1034/j.1600-0722.2003.00036.x. [DOI] [PubMed] [Google Scholar]
- 16.Lammi L., Halonen K., Pirinen S., Thesleff I., Arte S., Nieminen P. A missense mutation in PAX9 in a family with distinct phenotype of oligodontia. Eur. J. Hum. Genet. 2003;11:866–871. doi: 10.1038/sj.ejhg.5201060. [DOI] [PubMed] [Google Scholar]
- 17.Epstein J., Cai J., Glaser T., Jepeal L., Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 18.Chi N., Epstein J.A. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 19.Codutti L., van Ingen H., Vascotto C., Fogolari F., Corazza A., Tell G., Quadrifoglio F., Viglino P., Boelens R., Esposito G. The solution structure of DNA-free Pax-8 paired box domain accounts for redox regulation of transcriptional activity in the pax protein family. J. Biol. Chem. 2008;283:33321–33328. doi: 10.1074/jbc.M805717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H.E., Rould M.A., Xu W., Epstein J.A., Maas R.L., Pabo C.O. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Gene Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garvie C.W., Hagman J., Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 22.Wheat W., Fitzsimmons D., Lennox H., Krautkramer S.R., Gentile L.N., McIntosh L.P., Hagman J. The Highly Conserved β-Hairpin of the Paired DNA-Binding Domain Is Required for Assembly of Pax-Ets Ternary Complexes. Mol. Cell. Biol. 1999;19:2231–2241. doi: 10.1128/mcb.19.3.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mensah J.K., Ogawa T., Kapadia H., Cavender A.C., D'souza R.N. Functional analysis of a mutation in PAX9 associated with familial tooth agenesis in humans. J. Biol. Chem. 2004;279:5924–5933. doi: 10.1074/jbc.M305648200. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa T., Kapadia H., Wang B., D'souza R.N. Studies on Pax9-Msx1 protein interactions. Arch. Oral Biol. 2005;50:141–145. doi: 10.1016/j.archoralbio.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Mikkola I., Bruun J.A., Holm T., Johansen T. Superactivation of Pax6-mediated transactivation from paired domain-binding sites by DNA-independent recruitment of different homeodomain proteins. J. Biol. Chem. 2001;276:4109–41018. doi: 10.1074/jbc.M008882200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Catron K.M., Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc. Natl Acad. Sci. USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shetty S., Takahashi T., Matsui H., Ayengar R., Raghow R. Transcriptional autorepression of Msx1 gene is mediated by interactions of Msx1 protein with a multi-protein transcriptional complex containing TATA-binding protein, Sp1 and cAMP-response-element-binding protein-binding protein (CBP/p300) Biochem. J. 1999;339(Pt 3):751–758. [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H., Habas R., Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 29.Lee H., Quinn J.C., Prasanth K.V., Swiss V.A., Economides K.D., Camacho M.M., Spector D.L., Abate-Shen C. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Gene Dev. 2006;20:784–794. doi: 10.1101/gad.1392006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., Bei M., Woo I., Satokata I., Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 31.Tan K., Shaw A.L., Madsen B., Jensen K., Taylor-Papadimitriou J., Freemont P.S. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 2003;278:20507–20513. doi: 10.1074/jbc.M301994200. [DOI] [PubMed] [Google Scholar]
- 32.Bruun J.A., Thomassen E.I., Kristiansen K., Tylden G., Holm T., Mikkola I., Bjorkoy G., Johansen T. The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res. 2005;33:2661–2675. doi: 10.1093/nar/gki562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaza S., Prince F., Adachi Y., Punzo C., Cribbs D.L., Gehring W.J. Cross-regulatory protein–protein interactions between Hox and Pax transcription factors. Proc. Natl Acad. Sci. USA. 2008;105:13439–13444. doi: 10.1073/pnas.0806106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H.K., Chao L.Y., Saunders G.F. Functional analysis of paired box missense mutations in the PAX6 gene. Hum. Mol. Genet. 1997;6:381–386. doi: 10.1093/hmg/6.3.381. [DOI] [PubMed] [Google Scholar]
- 35.Czerny T., Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol. Cell. Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]