Abstract
Purpose
Patients with early-onset breast and/or ovarian cancer frequently wish to know if they inherited a mutation in one of the cancer susceptibility genes, BRCA1 or BRCA2. Accurate carrier prediction models are needed to target costly testing. Two widely used models, BRCAPRO and BOADICEA, were developed using data from non-Hispanic Whites (NHW), but their accuracies have not been evaluated in other racial/ethnic populations.
Methods
We evaluated the BRCAPRO and BOADICEA models in a population-based series of African-American, Hispanic and NHW breast cancer patients tested for BRCA1 and BRCA2 mutations. We assessed model calibration by evaluating observed versus predicted mutations and attribute diagrams, and model discrimination using areas under receiver operating characteristic curves (AUCs).
Results
Both models were well-calibrated within each racial/ethnic group, with some exceptions. BOADICEA over-predicted mutations in African Americans and older NHWs, and BRCAPRO under-predicted in Hispanics. In all racial/ethnic groups, the models over-predicted in cases whose personal and family histories indicated greater than 80% probability of carriage. The two models showed similar discrimination in each racial/ethnic group, discriminating least well in Hispanics. For example, BRCAPRO’s AUCs were 83% (95% confidence interval 63–93%) for NHWs, compared to 74% (59–85%) for African Americans and 58% (45–70%) for Hispanics.
Conclusions
The models’ poor performance for Hispanics may be due to model misspecification in this racial/ethnic group. However, it also may reflect racial/ethnic differences in the distributions of personal and family histories among breast cancer cases in the Northern California population.
Keywords: African-American, BOADICEA, BRCA mutation, BRCAPRO, breast cancer, carrier prediction models, Hispanic, two-stage sampling
INTRODUCTION
Since identification of the BRCA1 and BRCA2 cancer susceptibility genes, predictive models have been developed to identify individuals likely to carry inherited deleterious BRCA mutations. These models assign a patient a probability of mutation carriage using the cancer histories of the patient and her first-, second-degree, and, in some cases, more distant relatives, and estimates of BRCA mutation prevalence and penetrance. The BRCAPRO model assumes that all genetic susceptibility to breast cancer is due to BRCA mutations (1–3). The BOADICEA model considers the simultaneous effects of BRCA1, BRCA2 and the residual familial clustering of breast cancer not explained by these genes, which is assumed to be polygenic (4, 5). Multiple studies have validated BRCAPRO’s performance, and most have found it to discriminate as well as or more accurately than other models (2, 3, 6–18), although some studies suggest that BOADICEA performs slightly better in certain high-risk populations (1, 6, 19). Most evaluations of BRCAPRO and BOADICEA have included primarily non-Hispanic White (NHW) breast cancer patients, with a few exceptions (13, 17, 18, 20–22); consequently, little is known about the performance of these models across different racial/ethnic groups.
The BRCAPRO model was developed and has been validated with data from patients presenting for clinical genetics evaluation because of strong family cancer history; the BOADICEA model was developed using population-based data. As we and others have observed (23), a substantial portion of population-sampled young breast cancer patients with BRCA mutations do not have a family history of breast or ovarian cancer. Many women with early-onset breast cancer but no family history wish to know their mutation status, because results may guide subsequent management of breast and ovarian cancer risk, and family planning; such patients increasingly present for evaluation at cancer genetics clinics. An accurate and precise risk prediction algorithm based on personal and family history could identify individuals who lack a known deleterious mutation, but who, if tested, would have a 6–13% chance of receiving ambiguous and anxiety-producing results (24). We report an analysis of the performance of the BRCAPRO and BOADICEA mutation carriage prediction models in African-American, Hispanic and NHW female breast cancer patients from a population-based study in the San Francisco Bay Area.
METHODS
Study Population
To increase the precision of our inferences while maintaining their population-based properties, we identified breast cancer patients diagnosed at age <65 years through the population-based Greater San Francisco Bay Area Cancer Registry, which ascertains all incident cancers as part of the Surveillance, Epidemiology and End Results (SEER) Program and the California Cancer Registry, and invited them to participate in the Northern California Breast Cancer Family Registry (NC-BCFR) (25–27). We recruited patients using a two-stage sampling design, with over-sampling of patients having characteristics that suggest an inherited basis for their cancers (25–27). In stage one of sampling, we administered a brief telephone interview to all patients and assessed self-identified race/ethnicity and family history of breast and ovarian cancer. Based on age at diagnosis and personal and family history, patients were classified into either Category A (patients whose cancers are likely to be hereditary) or Category B (all other patients with cancers less likely to be hereditary). Category A patients were those who met at least one of the following criteria: (1) breast cancer diagnosis before age 35 years; (2) bilateral breast cancer, with first diagnosis before age 50 years; (3) prior ovarian or childhood cancer; or (4) at least one first-degree relative with breast or ovarian cancer. Categories A and B were designed with the aim of reducing the variance of overall prevalence estimates in the entire population-based series of patients. In stage two, we invited all patients in Category A and a random sample of patients in Category B to enroll in the family registry. Participants completed questionnaires on family history of cancer and breast cancer risk factors and provided a biospecimen sample. This two-stage sampling design provides unbiased estimates of population-based mutation carrier prediction performance having greater precision than those obtained from a simple random sample of the same size.
In telephone interviews, participants provided information on date of birth, vital status, date of death or last observation, and diagnosis dates and types of all site-specific cancers for themselves and all first-degree relatives. The questionnaire also elicited the occurrence and age at diagnosis of breast or ovarian cancer in second-degree relatives. When possible, reports of breast or ovarian cancer in relatives were verified by interviewing the relatives themselves, by obtaining medical records, or both. Informed consent was obtained from each study subject; the Institutional Review Boards of the Northern California Cancer Center, Stanford University, and the Dana-Farber Cancer Institute approved the study, in accordance with an assurance filed with and approved by the Department of Health and Human Services. The present analysis was restricted to patients aged <65 years diagnosed with invasive breast cancer between January 1st, 1995 and April 30, 2003, who identified themselves as African American, Hispanic or NHW. The number of BRCA mutation carriers among Asian-American patients was too small for reliable analysis. We restricted NHW patients to those without self-identified Ashkenazi Jewish ancestry given the known elevated prevalence of BRCA mutations among Ashkenazim.
Mutation Testing
Biospecimens from the NC-BCFR were processed by the Coriell Cell Repositories (Coriell Institute for Medical Research, Camden NJ). Testing was performed using Exon Grouping Analysis (EGAN) (28, 29), full sequencing performed by Myriad Genetics (30), or two-dimensional gene scanning (31, 32). The numbers of patients tested by these three methods were 28, 691, and 646, respectively, for BRCA1 and 363, 1002 and 0, respectively, for BRCA2. All coding exons and surrounding intronic sequences were amplified with 34 primer pairs and analyzed on ABI-377 instruments. PCR fragments with aberrant mobility were sequenced. For two-dimensional gene scanning, the entire coding exons and surrounding intronic sequences were amplified in a two-step PCR process involving six individual multiplex reactions (31, 32); these methods permit detection of variants in coding regions and splice site mutations. Mutations were classified according to the Breast Cancer Information Core (BIC) database1 and considered pathogenic as described by Couch et al (33). Regulatory mutations outside of the coding region and splice junctions and large genomic rearrangements are not detected by the methods used here.
Statistical Analysis
We calculated BRCAPRO and BOADICEA probabilities of mutation carriage (hereafter called prediction scores) in patients tested for BRCA1 and BRCA2 mutations. BRCAPRO probabilities were obtained using the CancerGene 4b program (University of Texas Southwestern, Dallas, TX), and BOADICEA probabilities were obtained using the program BOADICEA V3 provided by Antonis Antoniou (19). All available demographic data on all patients’ first- and second-degree relatives aged ≥20 years, whether affected by cancer or not, were included in the analyses. Information on all cases of breast cancer, ovarian cancer, male breast cancer, pancreatic cancer, and prostate cancer among patients and their first- and second-degree relatives was included in the prediction models.
Our goal was to obtain precise, population-based evaluation of the calibration and discrimination of each of the two models applied to each of the three racial/ethnic groups. Here calibration refers to agreement between mean prediction scores and observed carrier prevalences within subgroups of a population. Discrimination refers to the extent of separation between the prediction scores of carriers and noncarriers. We evaluated model calibration by comparing observed prevalences to mean prediction scores visually using attribute diagrams (34). We evaluated model discrimination using the areas under receiver operating characteristic curves (AUCs). Horvitz-Thompson (HT) estimating equations (26, 27) were used to adjust all analyses for the two-stage sampling design of the study.
Calibration
We estimated the prevalence of BRCA mutation carriers in a given racial/ethnic group as a weighted average of the two category-specific prevalences πA and πB. Here, πA is the number of carriers identified in Category A divided by the total number of tested patients in Category A. The overall prevalence estimate (Categories A and B combined) was π = wπA + (1-w) πB, where the weight w is the proportion of all screened patients who were classified in category A. We compared the prevalence π to similarly weighted averages of the category-specific mean prediction scores. In these calculations, we multiplied each prediction score by 90%, assuming that laboratory testing methods were 90% sensitive (35). The overall predicted prevalence was then computed as a weighted average s̄ = ws̄A + (1 – w)s̄B of the two category-specific mean scores s̄A and s̄B. We evaluated statistical significance of the observed/predicted differences via the statistic S = (π – s̄)2 /V where V is the HT variance estimate (26,27). This statistic has approximately a chi-squared distribution on one degree of freedom, under the null hypothesis that the prediction score for each breast cancer patient in a given racial/ethnic population equals her actual probability of mutation carriage.
We also used attribute diagrams to compare observed carrier prevalence to mean prediction scores within subgroups of patients whose scores lie in subintervals of the unit interval. To obtain an even distribution of patients within the subintervals, we grouped the patients according to quantiles of the logits of their scores. Within each subinterval, we computed weighted estimates and HT-based confidence intervals for the proportion of patients with observed mutations. We then plotted these points in attribute diagrams, which are plots of BRCA mutation carrier prevalence against median scores within each score subinterval (1). Results are provided for subgroups according to age and family cancer history, and for the group as a whole.
Discrimination
To evaluate the models’ ability to discriminate between carriers and non-carriers in each racial/ethnic group, we constructed receiver operating characteristic curves and evaluated the areas under these curves (AUCs) (36). We estimated these AUCs as weighted sums of the category-specific AUCs, using weights as described above. We used bootstrap variance estimates to obtain confidence intervals for the AUCs. Results are provided for subgroups according to age and family cancer history, and for the group as a whole.
RESULTS
Participant Characteristics
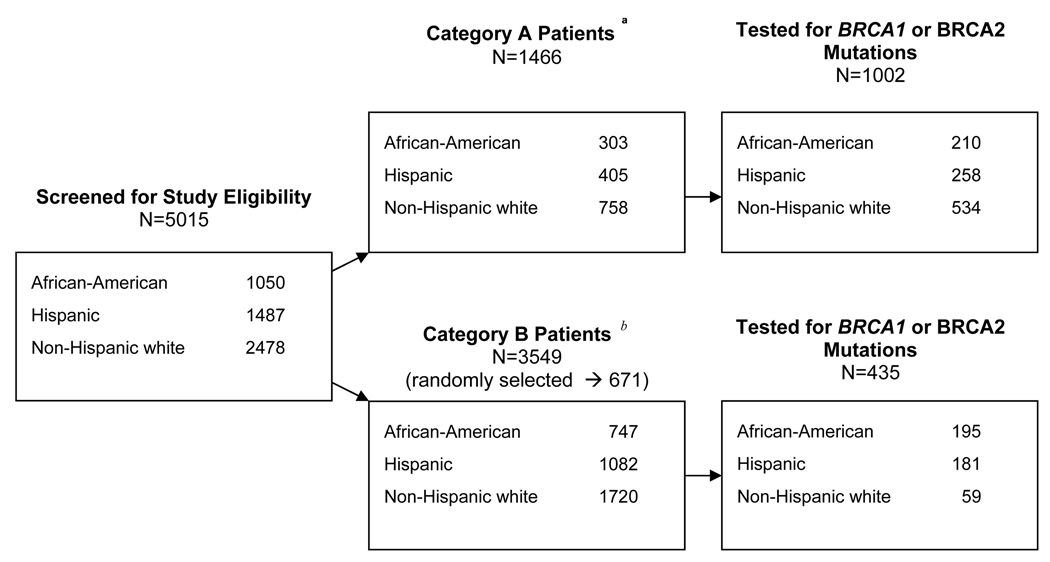
Figure 1 shows the distribution of African-American, Hispanic and NHW patients that were screened according to sampling categories. They include 1,050 (21%) African-Americans, 1,487 (30%) Hispanics, and 2,478 (49%) NHWs; 1,466 (29%) were classified in Category A, and the remaining 3,549 (71%) in Category B. All Category A patients (N=1,466) were invited to participate, and of these, 1002 (68%) provided a blood or buccal sample and were tested for BRCA1 and BRCA2 mutations. Of the 3,549 Category B patients, 671 were randomly selected for participation and of these, 435 (65%) provided a biospecimen sample and were tested. Of the 1,437 patients tested for BRCA1 and BRCA2 mutations, 72 were excluded from analysis: 23 because a family member had already been enrolled in the study through ascertainment of a first-degree relative with breast cancer (7 African Americans, 14 Hispanics, and 2 NHWs), one NHW because no family history information was available, and 48 NHWs because of Ashkenazi Jewish ancestry. A total of 1,365 tested patients were included in the analysis.
Fig. 1.
Flow Chart of Patient Screening, Sampling, and BRCA Mutation Testing
a Breast cancer patients whose cancers are likely to be hereditary.
b All other screened breast cancer patients.
Table 1 shows the distribution of tested patients according to BRCA mutation status, by sampling category (A and B) and race/ethnicity (African-American, Hispanic, and non-AJ NHWs). Of 1,365 patients tested for BRCA mutations, 44 tested positive for BRCA1 mutations (40 in Category A and four in Category B), and 41 tested positive for BRCA2 mutations (32 in Category A and 9 in Category B). Estimates of BRCA1 and BRCA2 mutation prevalence were highest among Hispanic patients (prevalence = 3.2% for each gene) and lowest among African-American patients (1.1% and 1.8% for BRCA1 and BRCA2, respectively), as we previously reported (23).
Table 1.
Distribution of Patients Tested for BRCA1/BRCA2 Mutations Carriage and Carrier Prevalence According to Racial/Ethnic Ancestry, by Type of Mutation.
| BRCA1 |
BRCA2 |
|||||
|---|---|---|---|---|---|---|
| Category A T/P† | Category B T/P | Prevalence (%)* | Category A T/P | Category B T/P | Prevalence(%) | |
| African-American | 203/8 | 195/0 | 1.1 | 203/8 | 195/2 | 1.8 |
| Hispanic | 244/18 | 181/3 | 3.2 | 244/7 | 181/6 | 3.2 |
| Non-Hispanic White | 486/14 | 56/1 | 2.1 | 486/17 | 56/1 | 2.3 |
Horvitz-Thompson weighted estimate
T/P = tested/positive for a mutation
BRCAPRO and BOADICEA Model Calibration
Table 2 shows prediction scores and observed prevalence of BRCA1 and BRCA2 mutations, specific for age and racial/ethnic group. Comparison of prediction scores to observed prevalence indicates generally similar levels of calibration for the two models, with notable differences as follows. The BRCAPRO model under-predicted mutation carriage in Hispanics, with the observed prevalence estimate of 6.4% exceeding the prediction of 3.8% (p=0.04); this under-prediction was statistically significant in the subset of Hispanic patients with no family history (FH) of breast cancer (p=0.01), but not in those with FH (p=0.14) BRCAPRO over-predicted in the subset of patients with FH among African Americans (p<0.01) and NHWs (p<0.01), but did not over-predict significantly in African Americans and NHWs without FH. The BOADICEA model over-predicted in African Americans (3.0% observed vs. 5.0% predicted, p=0.02) and older non-AJ NHWs (2.8% observed vs. 4.0% predicted, p=0.03). BOADICEA’s over-prediction was statistically significant in the subset of patients with FH among African Americans (p<0.01) and NHWs (p<0.001); it under-predicted significantly among Hispanics without FH (p=0.04), but not among those with FH.
Table 2.
Observed and Predicted* Prevalence of BRCA1 and BRCA2 Mutation Carriage in African-American, Hispanic and Non-Hispanic White Breast Cancer Patients, by Age at Diagnosis and Family History of Breast Cancer.
| No. of Patients † |
Carrier Prevalence (%)† |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Category A | Category B | ||||||||
| Tested | Positive | Tested | Positive | Observed | BRCAPRO | p-val‡ | BOADICEA | p-val § | |
| African-American | |||||||||
| All patients | 203 | 16 | 195 | 2 | 3.0 | 4.2 | 0.11 | 5.0 | 0.02 |
| Age (years) | |||||||||
| <50 | 102 | 13 | 79 | 1 | 5.0 | 6.3 | 0.36 | 6.4 | 0.34 |
| 50–64 | 101 | 3 | 116 | 1 | 1.4 | 2.6 | 0.17 | 3.9 | 0.01 |
| Family history (FH)** | |||||||||
| positive | 120 | 8 | 8 | 0 | 5.7 | 12.5 | <0.01 | 12.4 | 0.01 |
| negative | 83 | 8 | 187 | 2 | 2.3 | 2.2 | 0.86 | 3.2 | 0.30 |
| Hispanic | |||||||||
| All patients | 244 | 25 | 181 | 9 | 6.4 | 3.8 | 0.04 | 4.3 | 0.10 |
| Age (years) | |||||||||
| <50 | 146 | 21 | 81 | 4 | 8.0 | 5.0 | 0.09 | 5.4 | 0.16 |
| 50–64 | 98 | 4 | 100 | 5 | 4.8 | 2.8 | 0.24 | 3.2 | 0.34 |
| FH** | |||||||||
| positive | 122 | 14 | 0 | 0 | 11.5 | 15.8 | 0.14 | 14.3 | 0.33 |
| negative | 122 | 11 | 181 | 9 | 5.6 | 2.0 | 0.01 | 2.8 | 0.04 |
| Non-Hispanic White | |||||||||
| All patients | 486 | 31 | 56 | 2 | 5.0 | 4.8 | 0.88 | 5.5 | 0.81 |
| Age (years) | |||||||||
| <50 | 241 | 21 | 17 | 2 | 10.5 | 8.3 | 0.64 | 8.1 | 0.61 |
| 50–64 | 245 | 10 | 39 | 0 | 1.9 | 2.8 | 0.33 | 4.0 | 0.03 |
| FH** | |||||||||
| positive | 328 | 21 | 2 | 0 | 5.7 | 13.9 | <0.01 | 14.2 | <0.001 |
| negative | 158 | 10 | 54 | 2 | 4.8 | 2.1 | 0.23 | 2.9 | 0.40 |
according to BRCAPRO and BOADICEA carrier prediction models
weighted to account for the two-stage sampling, as described in the text
under the null hypothesis that observed and predicted are equal
obtained by adding 0.5 to the zero count for category B
having any first or second-degree relative affected by breast cancer
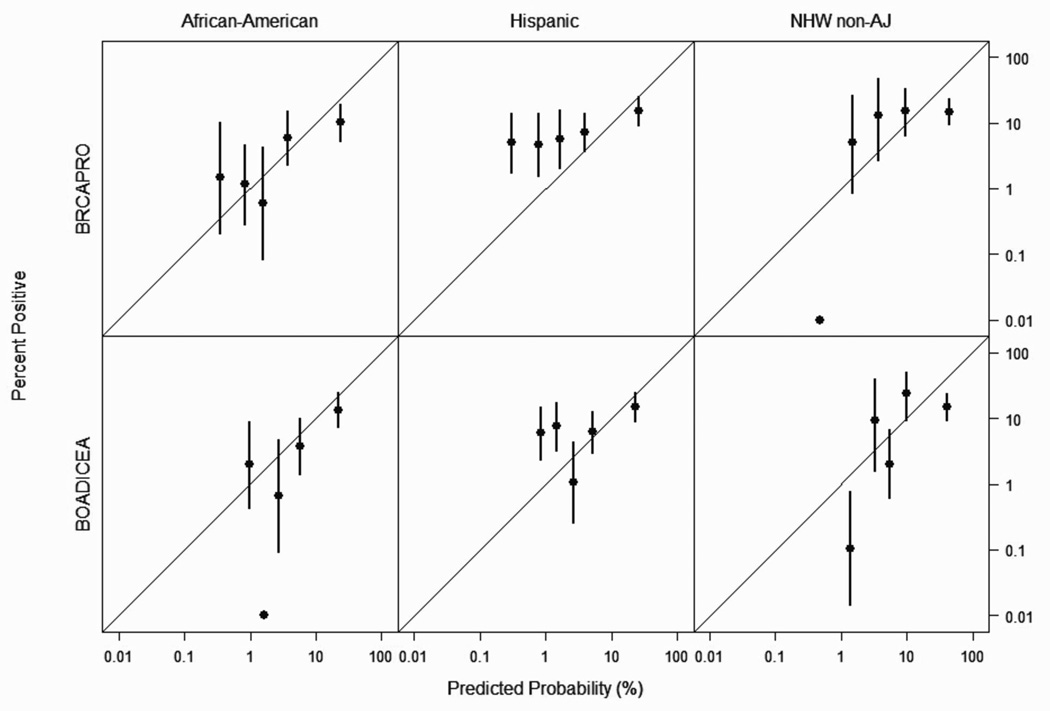
Attribute diagrams for each racial/ethnic group are presented in Figure 2 as a measure of model resolution and reliability, with optimal performance represented by data points on the 45 degree line (1). For both models, data points are clustered on the 45 degree line, with the exception of the BRCAPRO model for Hispanics, in which most points fell above the 45 degree line, consistent with the observed under-prediction of BRCA mutation carriage as reported in Table 2. In all racial/ethnic groups, BRCAPRO shows over-prediction in patients with 80% or greater predicted probability of mutation carriage; BOADICEA shows over-prediction for non-AJ NHWs with 80% or greater predicted probability of mutation carriage (Figure 2).
Fig. 2.
Attribute Diagram (BRCAPRO Model Score versus BRCA Mutation Frequency) for African American, Hispanic and Non-Ashkenazi Jewish Non-Hispanic White (NHW non-AJ) Patients
BRCAPRO and BOADICEA Model Discrimination
Accuracy in discriminating between carriers and non-carriers, as measured by AUC, was similar for both models within each racial/ethnic group (Table 3). The highest AUC values were observed in non-AJ NHWs (BRCAPRO 83%, 95% confidence interval (CI) 63–93%; BOADICEA 83%, CI 63–93%), followed by African Americans (BRCAPRO 74%, CI 59–85%; BOADICEA 75%, CI 60–85%) and Hispanics (BRCAPRO 58%, CI 45–70%; BOADICEA 56%, CI 43–68%). Within subsets defined by age and FH of breast cancer, there was a trend toward worse performance of both models in older (BRCAPRO 49%, CI 22–76%; BOADICEA 44%, CI 17–74%), compared to younger (BRCAPRO 81%, CI 67–90%; BOADICEA 85%, CI 74–92%) African Americans. There also was a trend toward worse discrimination by both models in Hispanics without FH (BRCAPRO 52%, CI 38–65%; BOADICEA 50%, CI 34–66%), compared to Hispanics with FH (BRCAPRO 69%, CI 56–79%; BOADICEA 69%, CI 55–80%).
Table 3.
Area under the Receiver Operating Characteristic Curve (AUC) for BRCAPRO and BOADICEA Models, by Race/Ethnicity, Age, and Family History of Breast Cancer
| BRCAPRO |
BOADICEA |
|||
|---|---|---|---|---|
| AUC (%) | CI* | AUC (%) | CI | |
| African-American | ||||
| All patients | 73.8 | 59.2–84.6 | 74.5 | 60.1–85.0 |
| Age (years) | ||||
| <50 | 81.3 | 67.4–90.2 | 85.1 | 74.0–92.0 |
| 50–64 | 48.7 | 22.0–76.2 | 43.8 | 17.5–74.1 |
| Family History (FH)† | ||||
| positive | 73.1 | 54.7–85.9 | 73.9 | 54.2–87.1 |
| negative | 71.2 | 52.6–84.6 | 70.1 | 53.3–82.8 |
| Hispanic | ||||
| All patients | 58.3 | 45.2–70.4 | 56.2 | 43.3–68.3 |
| Age (years) | ||||
| <50 | 55.8 | 39.8–70.7 | 54.1 | 38.6–68.8 |
| 50–64 | 55.7 | 35.5–74.2 | 54.6 | 33.5–74.1 |
| FH† | ||||
| positive | 68.9 | 56.3–79.2 | 68.5 | 54.6–79.8 |
| negative | 51.8 | 37.9–65.3 | 49.8 | 34.2–65.6 |
| Non-Hispanic White | ||||
| All patients | 82.8 | 62.9–93.2 | 82.6 | 63.1–93.0 |
| Age (yrs) | ||||
| <50 | 68.2 | 39.2–87.7 | 72.8 | 45.2–89.6 |
| 50–64 | 92.7 | 75.4–98.1 | 91.1 | 75.9–97.1 |
| FH | ||||
| positive | 82.3 | 71.7–89.5 | 81.8 | 70.6–89.4 |
| negative | 87.4 | 57.4–97.3 | 87.8 | 57.0–97.5 |
CI = 95% confidence interval
having any first or second-degree relative affected by breast cancer
DISCUSSION
We evaluated the performance of the BRCAPRO and BOADICEA BRCA mutation carrier prediction models in three racial/ethnic groups, consisting of African-American, Hispanic and non-Ashkenazi Jewish NHW breast cancer patients from the San Francisco Bay Area. To our knowledge, this is the first study to compare these BRCA mutation-prediction models across population samples of such racial/ethnic diversity. In general, the models showed similar discrimination within each racial/ethnic group, but differences in calibration: BRCAPRO under-predicted mutation carriage in Hispanics, whereas BOADICEA over-predicted mutations in African Americans and in older NHWs.
The strength of this study is its focus on population-based samples of racial/ethnic minorities, in contrast to most prior evaluations of BRCA mutation prediction models. Some prior studies have found similar accuracy of BRCAPRO in racial/ethnic minorities as in NHWs (13, 17), but we and others reported under-prediction by BRCAPRO and other models among clinic-based minorities including Asian Americans and Hispanics (18, 20). Models may perform less well in racial/ethnic minorities because the prevalence of carriers among breast cancer cases may differ by race/ethnicity. In non-Ashkenazi Jewish NHW cases, our prevalence estimates for BRCA1 (2.1%) and BRCA2 (2.3%) are similar to those used by the BRCAPRO and BOADICEA models (3, 4); by contrast, we found that African-American cases had lower (BRCA1 1.1%; BRCA2 1.8%), and Hispanic cases higher (BRCA1 3.2%; BRCA2 3.2%), carrier prevalence. Notably, the exception to BOADICEA’s general over-prediction occurred in Hispanics, as did a significant under-prediction by BRCAPRO, both consistent with Hispanics’ higher mutation prevalence than observed in non-Ashkenazi Jewish NHWs. Within subsets specific for family history and age, calibration worsened with increasing divergence from the mutation prevalence expected by the models; for example, both models over-predicted significantly only in African Americans with family history of breast cancer, while under-predicting in Hispanics lacking such family history. Recent publications have reported a higher prevalence of the 185delAG BRCA1 founder mutation in Hispanics than was initially appreciated (18, 37, 38), leading some to suggest a common origin for this mutation in Sephardic and Ashkenazi Jewish populations (18, 37, 38). The present results contrast with those from a recent single-center clinic-based study of BRCAPRO in Hispanics, which reported better model performance than we found (17); variations in the use of BRCAPRO between studies may explain some of this difference. Our finding of lesser BRCAPRO model accuracy in Hispanics also prompts questions as to whether BRCA mutation penetrance, or associated cancer risk, might differ between Hispanics and NHW. Given the growing size of the U.S. Hispanic population, further study of this issue has important implications for health policy and resource allocation.
In contrast to prior studies of the calibration of BRCAPRO and similar models in clinic-based settings (1, 2, 6–22), this analysis considered populations having lower BRCA mutation prevalence, with 85 (6%) carriers identified among 1,365 tested patients. This study sample reflects the reality of current clinical BRCA mutation testing, given patient preference and practice guidelines that support more inclusive testing than previously advised2. Our finding that BRCAPRO and BOADICEA over-predicted in a substantial proportion of patients, particularly in patients with family history of breast cancer or with greater than 80% predicted probability of mutation carriage, likely results from lower mutation frequency, and perhaps higher sporadic breast cancer incidence (39, 40), in these groups than the model parameters assume; we anticipate that population-specific corrections may improve model calibration, as others have demonstrated (7).
Prior studies of the BRCAPRO model’s discrimination have reported AUCs in the range of 60–88%. Comparisons of BRCAPRO to other BRCA mutation prediction models, including BOADICEA, Couch, Finnish, National Cancer Institute, Frank/Myriad II, the Manchester Scoring System, the Family History Assessment Tool, and Shattuck-Eidens/Myriad I, have revealed relatively few differences in terms of discrimination (1, 2, 6–16, 18, 19, 41, 42). Exceptions include the slightly superior performance of BOADICEA and the Manchester Scoring System in the United Kingdom, of the Italian IC software modification of BRCAPRO among Italians, and of the LAMBDA model among Ashkenazi Jewish probands; some of these models use population-specific mutation prevalence estimates, which tailored them to the groups under study (7, 9, 19, 43). In the present study, we evaluated model discrimination in three separate racial/ethnic populations, which may differ in their prevalence of BRCA mutations, and in the variance of their carriage probabilities. Variation in the probability of mutation carriage within a racial/ethnic population affects a model’s AUC. For example, if all breast cancer patients in a racial/ethnic group had the same mutation carrier probability, its AUC (which is the likelihood that the carriage probability for a randomly selected carrier exceeds that of a randomly selected noncarrier) would equal its minimum of 50%, indicating that the model is no better at discriminating between carriers and noncarriers than random chance. Given such intra-group variability, it is difficult to compare a model’s discrimination across different racial/ethnic groups. Comparing BRCAPRO’s and BOADICEA’s discriminative abilities within a single population is more straightforward, and we found no difference between models in any of the three racial/ethnic groups under study. Within subsets defined by age and family history of breast cancer, we observed some trends in model performance which did not reach statistical significance (for example, both models discriminated better in younger, compared to older, African Americans). Future research should evaluate race/ethnicity-specific modifications to BRCAPRO’s and BOADICEA’s mutation prevalence assumptions, and compare each model’s discrimination to that of other prediction tools, within the racial/ethnic groups we studied. As understanding of the spectrum of BRCA mutations across race/ethnicity matures, it may prove optimal to develop models specific to each racial/ethnic population.
Although BRCA mutation testing was completed for only 67% of those invited to enroll in the NC-BCFR, the testing rate was similar for patients in Categories A (68%) and B (65%). This similarity suggests that family history was not related to patients’ willingness to participate in the registry and provide biospecimens for research. We assumed that the combination of BRCA testing methods used was 90% sensitive for detection of deleterious mutations (35); however, if testing sensitivity was actually lower than 90%, then the models may over-predict less, and under-predict more, than we report here.
In conclusion, the BRCAPRO and BOADICEA models showed differences in performance across racial/ethnic and age groups in a large population-based series of breast cancer patients. This finding emphasizes the need for further study of BRCA mutations in specific racial/ethnic and age groups, and for development of more accurate mutation prediction methods, with customization for the populations to which they are applied.
Acknowledgments
Notes: This research was supported by the National Cancer Institute, National Institutes of Health, under RFA #CA-95-003 through a cooperative agreement with the Northern California Cancer Center (U01 CA69417), as well as NIH grants CA69417 and CA94069. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast CFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government or the Breast CFR. The authors thank Antonis Antoniou for providing BOADICEA software, and Frederick Li for his contributions to the laboratory work for this analysis.
Financial support: National Cancer Institute, National Institutes of Health, under RFA CA-95-003 through a cooperative agreement with the Northern California Cancer Center (U01 CA69417), and NIH grants CA69417 and CA94069.
Footnotes
Available at: http://research.nhgri.nih.gov/bic/.
Guidelines of the National Comprehensive Cancer Network. Available at: http://www.nccn.org/professionals/physician_gls/PDF/genetics_screening.pdf
References
- 1.Antoniou AC, Durocher F, Smith P, et al. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2006;8:R3. doi: 10.1186/bcr1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry DA, Iversen ES, Jr, Gudbjartsson DF, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20:2701–2712. doi: 10.1200/JCO.2002.05.121. [DOI] [PubMed] [Google Scholar]
- 3.Berry DA, Parmigiani G, Sanchez J, Schildkraut J, Winer E. Probability of carrying a mutation of breast-ovarian cancer gene BRCA1 based on family history. J Natl Cancer Inst. 1997;89:227–238. doi: 10.1093/jnci/89.3.227. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou AC, Cunningham AP, Peto J, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniou AC, Pharoah PP, Smith P, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–1590. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcenas CH, Hosain GM, Arun B, et al. Assessing BRCA carrier probabilities in extended families. J Clin Oncol. 2006;24:354–360. doi: 10.1200/JCO.2005.02.2368. [DOI] [PubMed] [Google Scholar]
- 7.Capalbo C, Ricevuto E, Vestri A, et al. Improving the accuracy of BRCA1/2 mutation prediction: validation of the novel country-customized IC software. Eur J Hum Genet. 2006;14:49–54. doi: 10.1038/sj.ejhg.5201511. [DOI] [PubMed] [Google Scholar]
- 8.Euhus DM, Smith KC, Robinson L, et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. J Natl Cancer Inst. 2002;94:844–851. doi: 10.1093/jnci/94.11.844. [DOI] [PubMed] [Google Scholar]
- 9.Evans DG, Eccles DM, Rahman N, et al. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41:474–480. doi: 10.1136/jmg.2003.017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasching PA, Bani MR, Nestle-Kramling C, et al. Evaluation of mathematical models for breast cancer risk assessment in routine clinical use. Eur J Cancer Prev. 2007;16:216–224. doi: 10.1097/CEJ.0b013e32801023b3. [DOI] [PubMed] [Google Scholar]
- 11.James PA, Doherty R, Harris M, et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006;24:707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- 12.Marroni F, Aretini P, D′Andrea E, et al. Evaluation of widely used models for predicting BRCA1 and BRCA2 mutations. J Med Genet. 2004;41:278–285. doi: 10.1136/jmg.2003.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nanda R, Schumm LP, Cummings S, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–1933. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 14.Oros KK, Ghadirian P, Maugard CM, et al. Application of BRCA1 and BRCA2 mutation carrier prediction models in breast and/or ovarian cancer families of French Canadian descent. Clin Genet. 2006;70:320–329. doi: 10.1111/j.1399-0004.2006.00673.x. [DOI] [PubMed] [Google Scholar]
- 15.Ottini L, Masala G, D′Amico C, et al. BRCA1 and BRCA2 mutation status and tumor characteristics in male breast cancer: a population-based study in Italy. Cancer Res. 2003;63:342–347. [PubMed] [Google Scholar]
- 16.Stuppia L, Di Fulvio P, Aceto G, et al. BRCA1 and BRCA2 mutations in breast/ovarian cancer patients from central Italy. Hum Mutat. 2003;22:178–179. doi: 10.1002/humu.9164. [DOI] [PubMed] [Google Scholar]
- 17.Vogel KJ, Atchley DP, Erlichman J, et al. BRCA1 and BRCA2 genetic testing in Hispanic patients: mutation prevalence and evaluation of the BRCAPRO risk assessment model. J Clin Oncol. 2007;25:4635–4641. doi: 10.1200/JCO.2006.10.4703. [DOI] [PubMed] [Google Scholar]
- 18.Weitzel JN, Lagos V, Blazer KR, et al. Prevalence of BRCA mutations and founder effect in high-risk Hispanic families. Cancer Epidemiol Biomarkers Prev. 2005;14:1666–1671. doi: 10.1158/1055-9965.EPI-05-0072. [DOI] [PubMed] [Google Scholar]
- 19.Antoniou AC, Hardy R, Walker L, et al. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: validation of BOADICEA, BRCAPRO, IBIS, Myriad and the Manchester scoring system using data from UK genetics clinics. J Med Genet. 2008;45:425–431. doi: 10.1136/jmg.2007.056556. [DOI] [PubMed] [Google Scholar]
- 20.Kurian AW, Gong GD, Chun NM, et al. Performance of BRCA1/2 mutation prediction models in Asian Americans. J Clin Oncol. 2008;26:4752–4758. doi: 10.1200/JCO.2008.16.8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao NY, Hu Z, Li WF, et al. Models for predicting BRCA1 and BRCA2 mutations in Han Chinese familial breast and/or ovarian cancer patients. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-9965-9. electronic publication March 15, 2008. [DOI] [PubMed] [Google Scholar]
- 22.Huo D, Senie RT, Daly M, et al. Prediction of BRCA mutations using the BRCAPRO model in clinic-based African American, Hispanic, and other minority families in the United States. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.17.5869. electronic publication February 2, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John EM, Miron A, Gong G, et al. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298:2869–2876. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]
- 24.Noll WW, Reid JE, Burbidge LA, et al. Prevalence of BRCA1 and BRCA2 mutations is similar in non-Ashkenazi women of different ethnic backgrounds undergoing genetic testing. Presented at the American Society of Clinical Oncology Breast Cancer Symposium; San Francisco, CA. 2007. Sep 7–9, abstract 4. [Google Scholar]
- 25.John EM, Hopper JL, Beck JC, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res (Print) 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalbfleisch JD, Lawless JF. Likelihood analysis of multi-state models for disease incidence and mortality. Stat Med. 1988;7:149–160. doi: 10.1002/sim.4780070116. [DOI] [PubMed] [Google Scholar]
- 27.Whittemore AS, Halpern J. Multi-stage sampling in genetic epidemiology. Stat Med. 1997;16:153–167. doi: 10.1002/(sici)1097-0258(19970130)16:2<153::aid-sim477>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Ganguly A, Rock MJ, Prockop DJ. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci U S A. 1993;90:10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korkko J, Annunen S, Pihlajamaa T, Prockop DJ, Ala-Kokko L. Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing. Proc Natl Acad Sci U S A. 1998;95:1681–1685. doi: 10.1073/pnas.95.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WY, Garber JE, Higham S, et al. BRCA1/2 genetic testing in the community setting. J Clin Oncol. 2002;20:4485–4492. doi: 10.1200/JCO.2002.08.147. [DOI] [PubMed] [Google Scholar]
- 31.van Orsouw NJ, Dhanda RK, Elhaji Y, et al. A highly accurate, low cost test for BRCA1 mutations. J Med Genet. 1999;36:747–753. doi: 10.1136/jmg.36.10.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 33.Couch FJ, Weber BL. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Information Core. Hum Mutat. 1996;8:8–18. doi: 10.1002/humu.1380080102. [DOI] [PubMed] [Google Scholar]
- 34.Hsu WRM AH. The attributes diagram: a geometrical framework for assessing the quality of probability forecasts. Int J Forecast. 1986;2:285–293. [Google Scholar]
- 35.Andrulis IL, Anton-Culver H, Beck J, et al. Comparison of DNA-and RNA-based methods for detection of truncating BRCA1 mutations. Hum Mutat. 2002;20:65–73. doi: 10.1002/humu.10097. [DOI] [PubMed] [Google Scholar]
- 36.Faraggi D, Reiser B. Estimation of the area under the ROC curve. Stat Med. 2002;21:3093–3106. doi: 10.1002/sim.1228. [DOI] [PubMed] [Google Scholar]
- 37.Diez O, Osorio A, Robledo M, et al. Prevalence of BRCA1 and BRCA2 Jewish mutations in Spanish breast cancer patients. Br J Cancer. 1999;79:1302–1303. doi: 10.1038/sj.bjc.6690208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullineaux LG, Castellano TM, Shaw J, et al. Identification of germline 185delAG BRCA1 mutations in non-Jewish Americans of Spanish ancestry from the San Luis Valley, Colorado. Cancer. 2003;98:597–602. doi: 10.1002/cncr.11533. [DOI] [PubMed] [Google Scholar]
- 39.Clarke CA, Glaser SL, West DW, et al. Breast cancer incidence and mortality trends in an affluent population: Marin County, California, USA, 1990–1999. Breast Cancer Res. 2002;4:R13. doi: 10.1186/bcr458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbins AS, Brescianini S, Kelsey JL. Regional differences in known risk factors and the higher incidence of breast cancer in San Francisco. J Natl Cancer Inst. 1997;89:960–965. doi: 10.1093/jnci/89.13.960. [DOI] [PubMed] [Google Scholar]
- 41.Kang HH, Williams R, Leary J, et al. Evaluation of models to predict BRCA germline mutations. Br J Cancer. 2006;95:914–920. doi: 10.1038/sj.bjc.6603358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parmigiani G, Chen S, Iversen ES, Jr, et al. Validity of models for predicting BRCA1 and BRCA2 mutations. Ann Intern Med. 2007;147:441–450. doi: 10.7326/0003-4819-147-7-200710020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apicella C, Dowty JG, Dite GS, et al. Validation study of the LAMBDA model for predicting the BRCA1 or BRCA2 mutation carrier status of North American Ashkenazi Jewish women. Clin Genet. 2007;72:87–97. doi: 10.1111/j.1399-0004.2007.00841.x. [DOI] [PubMed] [Google Scholar]