Abstract
Background and Aims
Hieracium subgenus Hieracium is one of the taxonomically most intricate groups of vascular plants, due to polyploidy and a diversity of breeeding systems (sexuality vs. apomixis). The aim of the present study was to analyse nuclear genome size in a phylogenetic framework and to assess relationships between genome size and ploidy, breeding system and selected ecogeographic features.
Methods
Holoploid and monoploid genome sizes (C- and Cx-values) of 215 cultivated plants from 89 field populations of 42 so-called ‘basic’ Hieracium species were determined using propidium iodide flow cytometry. Chromosome counts were available for all analysed plants, and all plants were tested experimentally for their mode of reproduction (sexuality vs. apomixis). For constructing molecular phylogenetic trees, the external transcribed spacer region of nuclear ribosomal DNA was used.
Key Results
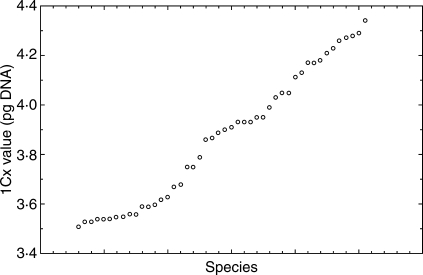
The mean 2C values differed up to 2·37-fold among different species (from 7·03 pg in diploid to 16·67 in tetraploid accessions). The 1Cx values varied 1·22-fold (between 3·51 and 4·34 pg). Variation in 1Cx values between conspecific (species in a broad sense) accessions ranged from 0·24% to 7·2%. Little variation (not exceeding the approximate measurement inaccurracy threshold of 3·5%) was found in 33 species, whereas variation higher than 3·5% was detected in seven species. Most of the latter may have a polytopic origin. Mean 1Cx values of the three cytotypes (2n, 3n and 4n) differed significantly (average of 3·93 pg in diploids, 3·82 pg in triploids and 3·78 pg in tetraploids) indicating downsizing of genomes in polyploids. The pattern of genome size variation correlated well with two major phylogenetic clades which were composed of species with western or eastern European origin. The monoploid genome size in the ‘western’ species was significantly lower than in the ‘eastern’ ones. Correlation of genome size with latitude, altitude and selected ecological characters (light and temperature) was not significant. A longitudinal component was only apparent for the whole data set, but absent within the major lineages.
Conclusions
Phylogeny was the most important factor explaining the pattern of genome size variation in Hieracium sensu stricto, species of western European origin having significantly lower genome size in comparison with those of eastern European origin. Any correlation with ecogeographic variables, including longitude, was outweighed by the divergence of the genus into two major phylogenetic lineages.
Key words: Apomixis, chromosome numbers, Compositae, genome size, hawkweeds, Hieracium subgenus Hieracium, mode of reproduction, nuclear DNA content, phylogeny, polyploidy
INTRODUCTION
Genome size has become a widely studied phenomenon, beginning in the 1950s when large differences in the nuclear content of different organisms were detected (e.g. Swift, 1950; Laurie and Bennett, 1985; Bennett and Leitch, 1995). Large differences in DNA content can be caused by several mechanisms. It has been found that nuclear DNA content is primarily influenced by the proportion of non-genic repetitive DNA, much of which is generated by transposable elements (Flavell et al., 1977; Barakat et al., 1997). In particular, it has been found that retrotransposon copy number can vary among genomes (Arumuganathan and Earle, 1991; Vicient et al., 1999; Kalendar et al., 2000; Piegu et al., 2006; Wicker and Keller, 2007; Hawkins et al., 2008). Decrease in genome size can result from a higher overall rate of deletions than insertions, selection against transposable elements, unequal crossing over and illegitimate recombination (Morgan, 2001; Petrov, 2002; Wendel et al., 2002; Ma et al., 2004; Bennetzen et al., 2005).
Correlations between genome size and specific life traits, most importantly life history and breeding systems, have been documented. Selfers were found to have smaller Cx-values than related outcrossers (Labani and Elkington, 1987; Govindaraju and Cullis, 1991; Albach and Greilhuber, 2004). Annuals, especially weedy species, tend to have lower genome size in comparison with related perennials (Bennett, 1972; Rejmanek and Richardson, 1996; Bennett et al., 1998; Garnatje et al., 2004; Grotkopp et al., 2004), probably due to an association of annual life history with selfing (e.g. Albach and Greilhuber, 2004). Relationships between genome size and ecological factors are less clear (see, for example, Knight et al., 2005). Correlations between genome size and frost resistance in the British flora (MacGillivray and Grime, 1995), elevation in some groups of Centaurea (Bancheva and Greilhuber, 2006), Veronica (Albach and Greilhuber, 2004), Dactylis (Reeves et al., 1998) and Berberis (Bottini et al., 2000) and continentality and habitat conditions (moisture) in Cirsium (Bureš et al., 2004) have already been documented. There are also more or less close associations between genome size and cell size and leaf anatomical traits (Bennett, 1972; Edwards and Endrizzi, 1975; Knight et al., 2005; Sugiyama, 2005; Beaulieu et al., 2008), cell cycle duration (e.g. Rees et al., 1966; Bennett et al., 1983; Lawrence, 1985), seed mass (e.g. Knight et al., 2005; Beaulieu et al., 2007) and photosynthetic rate (e.g. Knight et al., 2005).
Polyploids often have smaller Cx-values than their diploid relatives (e.g. Leitch and Bennett, 2004; Weiss-Schneeweiss et al., 2006). These decreases correlate with a mutational bias towards deletion over insertions (Petrov, 2002), and illegitimate recombination has been shown to eliminate retrotransposon sequences (Bennetzen, 2002; Devos et al., 2002; Ma et al., 2004). However, exceptions of this downsizing pattern have been found, e.g. in the genus Orobanche (tetraploid O. transcaucasica). It was hypothesized that such polyploids are relatively young and that there was not enough time for a substantial reduction in nuclear DNA content (Weiss-Schneeweiss et al., 2006).
Genome size alone is of little value as a phylogenetic indicator at higher taxonomic levels, but can be helpful in infrageneric classification assessments, species delimitation or hybrid identification (Keller et al., 1996; Buitendijk et al., 1997; Morgan et al., 1998; Thalmann et al., 2000; Zonneveld, 2001; Šiško et al., 2003; Bureš et al., 2004; Baack et al., 2005; Záveský et al., 2005; Suda et al., 2007). An important issue that is still largely neglected in the literature, mostly due to a lack of comparative analyses between DNA sequence and genome size data sets, is the understanding of how genome size variation is linked with species evolution (but see Wendel et al., 2002; Albach and Greilhuber, 2004; Grotkopp et al., 2004; Jakob et al., 2004; Weiss-Schneeweiss et al., 2006). Species relationships were therefore assessed based on the external transcribed spacer (ETS) of nuclear ribosomal DNA in order to relate genome size variation to their evolutionary history.
Hieracium subgenus Hieracium is distributed in temperate regions of Europe, Asia, Mediterranean Africa and North America and has been introduced to several other regions, e.g. New Zealand. The genus is suitable for the study of genome size variation due its remarkable diversity in ploidy (coupled with breeding systems), habitat preferences and distribution of particular species. Polyploid (triploid, tetraploid and rarely pentaploid, x = 9) taxa with asexual reproduction through parthenogenetic development of the unreduced egg cell (Antennaria-type diplospory) prevail in this group, i.e. they are (near-)obligate apomicts (e.g. Nogler, 1984). Sexual reproduction is rather rare and restricted to diploid species (Schuhwerk, 1996; Chrtek et al., 2004). The species occupy forests, forest margins, various grasslands and rocks from the lowlands to the alpine belt.
Species concepts in Hieracium have long been a matter of discussion (e.g. Schuhwerk, 2003). The Central European school of ‘hieraciology’ (founded by Nägeli and Peter in the 19th century) accepts a broad species definition (species are then divided into subspecies, varieties, etc.), whereas Scandinavian, British and Russian botanists follow a narrow species concept, i.e. nearly all morphologically recognizable forms are treated at species rank (‘microspecies’). We follow the Central European concept because, in our opinion, it better reflects the situation across the whole distribution area, especially in central and southern Europe where most diploids occur, from which the apomictic polyploids are thought to be derived. According to this concept, approx. 500 species (in the broad sense) are accepted (Zahn, 1921–1923; and species described since that time), being either so-called ‘basic’ or ‘intermediate’ taxa. The latter share morphological characters of two or more basic species and are supposed to be of hybridogenous origin (hybrids stabilized by agamospermy). Basic species (about 45, including diploids and polyploids) are tentatively considered as main units of species evolution in Hieracium.
Here, a nuclear DNA content analysis of 42 basic species of Hieracium subgenus Hieracium (sensu Zahn, 1921–1923, with a few exceptions, see Materials and methods) is reported. The following questions were addressed: (a) how does the level of intraspecific variation in holoploid and monoploid genome sizes relate to the circumscription of species sensu Zahn? (b) how does monoploid genome size (Cx) relate to ploidy (diploids, triploids and tetraploids), i.e. is there evidence for downsizing of genomes in polyploids? (c) is there any congruence between the phylogenetic structure and the pattern of genome size variation? and (d) how does nuclear genome size relate to selected ecogeographic features (latitude, longitude, altitude, temperature and light)?
MATERIALS AND METHODS
Plant material
Two hundred and fifteen samples from 89 populations of 42 Hieracium species were collected in the field (or grown from seeds in a few cases) throughout Europe and transferred to the experimental garden of the Institute of Botany in Průhonice (Table 1; for details of sample localities see Supplementary Data, available online). Taxon sampling was restricted to so-called ‘basic’, supposedly non-hybridogenous species, generally following Zahn (1921–1923) with a few exceptions. The species concept of section Cerinthoidea followed Mateo (2005), H. plumulosum (H. waldsteinii sensu lato) was treated as a separate species, and two newly described diploid Balkan species (H. kittanae and H. petrovae; Vladimirov, 2003; Vladimirov and Szeląg, 2006) were included. Complete analysis covering all recognized and mostly (allo)polyploid hybridogenous species (approx. 500 ‘broad’ species) was not feasible, and interpretation of estimated genome sizes would be extremely complicated due to the often unknown origin of polyploids and the reticulate patterns of variation.
Table 1.
Accession origin and genome size
| Species | Locality (no. of plants) (S): plant cultivated from seed | 2n | 2C (pg) mean ± s.e. | 2C (pg) range | 1Cx (pg) | 1Cx (pg) species mean ± s.e. | 1Cx (pg) species range | 1Cx species variation (%) | ETS clade† |
|---|---|---|---|---|---|---|---|---|---|
| H. alpinum L. | Ukraine: Chornohora (4) | 18 | 7·90 ± 0·01 | 7·88–7·92 | 3·95 | 3·95 ± 0·01 | 3·94–3·97 | 0·76 | E |
| France: Hautes Alpes (1) | 27 | 11·87 | – | 3·96 | n.d. | ||||
| H. amplexicaule L. | Austria: Hohe Tauern, Fragant (2) | 27* | 10·70 ± 0·01 | 10·7–10·71 | 3·57 | 3·60 ± 0·02 | 3·54–3·68 | 3·95 | X(W) |
| Austria: Hohe Tauern, Mallnitz (1) | 27* | 10·74 | – | 3·58 | n.d. | ||||
| Spain: prov. Gerona (1) | 27* | 10·8 | – | 3·6 | n.d. | ||||
| France: Hautes Alpes (1) | 27 | 10·62 | – | 3·54 | n.d. | ||||
| Italy: Rhaetian Alps (2) | 36* | 14·66 ± 0·07 | 14·59–14·72 | 3·67 | n.d. | ||||
| H. bifidum Kit. | Slovakia: Roháče (1) | 27 | 10·67 | – | 3·56 | 3·53 ± 0·01 | 3·51–3·56 | 1·42 | W |
| Czech Rep.: distr. Beroun | 27 | 10·52 ± 0·01 | 10·52–10·53 | 3·51 | n.d. | ||||
| Czech Rep.: Krkonoše Mts. | 27 | 10·62 ± 0·01 | 10·61–10·63 | 3·54 | n.d. | ||||
| H. bracteolatum Sibth. & Sm. | Greece: Pilion (2) (S) | 27 | 12·39 ± 0·08 | 12·31–12·47 | 4·13 | 4·13 ± 0·03 | 4·10–4·16 | 1·46 | X |
| H. bupleuroides C.C.Gmel. I. | Slovakia: Biele Karpaty (2) | 27* | 11·95 ± 0·05 | 11·91–12·01 | 3·99 | E(H) | |||
| H. bupleuroides C.C.Gmel. II. | Slovakia: Chočské vrchy (2) | 27* | 12·00 ± 0·02 | 11·96–12·00 | 3·99 | n.d. | |||
| Slovakia: Roháče (2) | 27 | 11·73 | – | 3·91 | E | ||||
| Austria: Dachstein massif (1) | 27* | 11·61 ± 0·05 | 11·56–11·66 | 3·87 | n.d. | ||||
| Slovakia: Slovenský raj (1) | 27* | 12·03 | – | 4·01 | n.d. | ||||
| Austria: Allgäuer Alpen (1) | 27 | 11·63 | – | 3·88 | n.d. | ||||
| H. bupleuroides s.l. mean | 3·95 ± 0·06 | 3·85–4·01 | 4·20 | ||||||
| H. caesium (Fr.) Fr. | Sweden: prov. Gotland (3) | 36* | 14·66 ± 0·12 | 14·52–14·89 | 3·67 | 3·68 ± 0·01 | 3·64–3·72 | 2·20 | X(W) |
| Sweden: prov. Gästrikland (3) | 36* | 14·75 ± 0·05 | 14·66–14·83 | 3·69 | n.d. | ||||
| H. cerinthoides L. | Spain: Pyrenees (2) | 27* | 10·68 ± 0·03 | 10·66–10·71 | 3·56 | 3·56 ± 0·01 | 3·55–3·57 | 0·56 | W(H) |
| H. cordifolium Lapeyr. | Andorra: Pyrenees (6) | 18* | 7·18 ± 0·02 | 7·11–7·22 | 3·59 | 3·59 ± 0·01 | 3·56–3·61 | 1·40 | W(H) |
| H. eriophorum St.-Amans | France: Landes, Labenne (6) (S) | 18* | 8·51 ± 0·03 | 8·45–8·61 | 4·25 | 4·27 ± 0·01 | 4·22–4·31 | 2·13 | E |
| France: Landes, Vieux-Boucau-les-Bains (6) (S) | 18* | 8·55 ± 0·03 | 8·44–8·61 | 4·27 | n.d. | ||||
| H. glaucum All. | Slovenia: Julijske Alpe, Podklanec (2) | 27* | 11·31 ± 0·03 | 11·29–11·34 | 3·77 | 3·79 ± 0·01 | 3·76–3·81 | 1·33 | n.d. |
| Slovenia: Julijske Alpe, Zadnjica (2) | 27* | 11·42 ± 0·00 | 11·42–11·42 | 3·81 | n.d. | ||||
| Slovenia: Julijske Alpe, izvir Soče (2) | 27 | 11·39 ± 0·05 | 11·34–11·44 | 3·8 | X | ||||
| H. gouani Arv.-Touv. | Spain: prov. Gerona (8) | 18* | 7·10 ± 0·01 | 7·07–7·12 | 3·55 | 3·55 ± 0·00 | 3·54–3·56 | 0·56 | X(W) |
| H. gymnocephalum Griseb. ex Pant. | Albania: Jezerce (2) | 18 | 8·44 ± 0·01 | 8·43–8·45 | 4·22 | 4·23 ± 0·01 | 4·22–4·23 | 0·24 | X |
| H. gymnocerinthe Arv.-Touv. & G.Gaut. | Spain: Serra del Cadí (2) | 27* | 10·6 ± 0·02 | 10·58–10·61 | 3·53 | 3·54 ± 0·01 | 3·53–3·54 | 0·28 | W(H) |
| H. heterogynum (Froel.) Guterm. | Montenegro: Lovčen (2) (S) | 27 | 12·52 ± 0·02 | 12·52–12·55 | 4·17 | 4·18 ± 0·01 | 4·17–4·18 | 0·24 | X |
| H. humile Jacq. | Austria: Dachstein massif (2) | 36* | 14·25 ± 0·01 | 14·24–14·26 | 3·56 | 3·55 ± 0·01 | 3·54–3·57 | 0·85 | W |
| France: Corbières (2) | 27 | 10·64 ± 0·01 | 10·61–10·66 | 3·55 | W | ||||
| H. kittanae Vladimirov | Bulgaria: Rodopi (3) | 18* | 8·41 ± 0·04 | 8·38–8·46 | 4·21 | 4·21 ± 0·01 | 4·19–4·23 | 0·95 | E |
| H. lachenalii Suter | Czech Rep.: Křivoklátsko (2) | 27* | 11·25 ± 0·01 | 11·27–11·29 | 3·76 | 3·75 ± 0·01 | 3·73–3·76 | 0·80 | n.d. |
| Czech Rep.: distr. Znojmo (2) | 27* | 11·22 ± 0·03 | 11·19–11·24 | 3·74 | X(W) | ||||
| Czech Rep.: distr. Praha-east (2) | 27 | 11·24 ± 0·06 | 11·20–11·32 | 3·75 | n.d. | ||||
| H. laevigatum Willd. | Czech Rep.: Brdy (2) | 27* | 12·05 ± 0·02 | 12·01–12·08 | 4·02 | 4·05 ± 0·02 | 3·97–4·14 | 4·28 | X |
| Czech Rep.: Hradec Králové (2) | 27* | 12·00 ± 0·08 | 11·94–12·10 | 4 | n.d. | ||||
| Germany: Kamenz (2) (S) | 27 | 12·41 ± 0·01 | 12·40–12·42 | 4·14 | n.d. | ||||
| H. lawsonii Vill. | France: Corbières (2) | 27 | 10·76 ± 0·02 | 10·74–10·78 | 3·59 | 3·62 ± 0·03 | 3·58–3·68 | 2·79 | W |
| France: Briançon (1) | 36 | 14·71 | – | 3·68 | n.d. | ||||
| H. murorum L. | Czech Rep.: Plzeň (2) | 27* | 10·62 ± 0·06 | 10·55–10·68 | 3·54 | 3·56 ± 0·01 | 3·52–3·59 | 1·99 | W |
| Czech Rep.: Doupovské hory (2) | 27* | 10·67 ± 0·05 | 10·63–10·72 | 3·56 | n.d. | ||||
| Czech Rep.: Český kras (2) | 27* | 10·74 ± 0·02 | 10·72–10·76 | 3·58 | n.d. | ||||
| H. naegelianum Pančić | Montenegro: Durmitor Mts (2) | 27* | 10·89 ± 0·08 | 10·81–10·97 | 3·63 | 3·63 ± 0·03 | 3·60–3·66 | 1·67 | E |
| H. olympicum Boiss. | Bulgaria: Kaloferska Planina (2) | 27* | 12·27 ± 0·05 | 12·22–12·33 | 4·09 | 4·05 ± 0·01 | 4·04–4·06 | 0·50 | X |
| H. pannosum Boiss. I | Bulgaria: Trojanska Planina (2) | 27* | 11·71 ± 0·05 | 11·66–11·77 | 3·9 | E | |||
| H. pannosum Boiss. II | Greece: Peloponnesos (2) (S) | 36 | 16·67 ± 0·01 | 16·66–16·67 | 4·17 | n.d. | |||
| H. pannosum s.l. mean | 4·04 ± 0·08 | 3·89–4·17 | 7·20 | ||||||
| H. petrovae Vladimirov & Szeląg | Bulgaria: Rodopi (1) | 18* | 7·9 | – | 3·95 | E | |||
| H. pictum Pers. | France: Montegenèvre (2) | 27 | 10·78 ± 0·27 | 10·5–11·05 | 3·59 | 3·59 ± 0·04 | 3·50–3·68 | 5·14 | n.d. |
| France: Briançon (2) | 27 | 10·75 ± 0·13 | 10·62–10·88 | 3·58 | W | ||||
| H. piliferum Hoppe | Austria: Gurktaler Alpen (2) | 36 | 15·57 ± 0·04 | 15·54–15·60 | 3·89 | 3·91 ± 0·02 | 3·86–4·01 | 3·89 | E-int. |
| Austria: Reisseck-Gruppe (1) | 27 | 11·58 | – | 3·86 | |||||
| Italy: Alps, Spluga (1) | 27 | 11·58 | – | 3·86 | |||||
| France: Hautes Alpes (2) | 27 | 11·95 ± 0·07 | 11·89–12·02 | 3·99 | |||||
| H. pilosum Schleich. ex Froel. I. | Slovenia: Julijske Alpe (1) | 27 | 11·57 | 3·86 | E | ||||
| H. pilosum Schleich. ex Froel. II. | Slovenia: Julijske Alpe (1) | 27 | 11·8 | 3·93 | X | ||||
| H. pilosum s.l. mean | 3·90 ± 0·04 | 3·86–3·93 | 1·81 | ||||||
| H. plumulosum A.Kern. | Montenegro: Mrtvica canyon (1) | 18* | 8·59 | – | 4·29 | X(E) | |||
| H. porrifolium L. | Austria: Karawanken (6) | 18* | 7·76 ± 0·01 | 7·7–7·79 | 3·88 | 3·89 ± 0·01 | 3·85–3·93 | 2·08 | E |
| Austria: Karawanken (1) | 18 | 7·7 | – | 3·85 | n.d. | ||||
| Slovenia: Julijske Alpe (6) | 18* | 7·82 ± 0·02 | 7·74–7·85 | 3·91 | n.d. | ||||
| H. prenanthoides Vill. I. | Poland: Karkonosze (2) | 27 | 10·82 ± 0·03 | 10·78–10·85 | 3·61 | X(W) | |||
| France: Hautes Alpes, La Grave (1) | 18 | 7·11 | – | 3·55 | X(W) | ||||
| France: Hautes Alpes, Briançon (2) | 18 | 7·29 ± 0·01 | 7·24–7·24 | 3·64 | n.d. | ||||
| H. prenanthoides Vill. II. | Andorra: Canillo (2) | 27* | 11·41 ± 0·02 | 11·40–11·43 | 3·8 | X(W) | |||
| H. prenanthoides s.l. mean | 3·67 ± 0·04 | 3·56–3·81 | 7·02 | ||||||
| H. racemosum Waldst. & Kit. ex Willd. | Czech Rep.: distr. Znojmo (2) | 27 | 12·41 ± 0·02 | 12·39–12·44 | 4·14 | 4·11 ± 0·06 | 4·08–4·15 | 1·72 | X |
| Czech Rep.: Ústí nad Orlicí (2) | 27 | 12·26 ± 0·01 | 12·26–12·26 | 4·09 | n.d. | ||||
| Slovakia: Gemer (1) (S) | 27 | 12·24 | - | 4·08 | n.d. | ||||
| H. ramondii Griseb. | Andorra: Encamp (2) | 27* | 10·63 ± 0·07 | 10·56–10·7 | 3·54 | 3·54 ± 0·03 | 3·51–3·57 | 1·71 | W |
| H. recoderi De Retz | Spain: prov. Barcelona (8) | 18* | 7·09 ± 0·01 | 7·00–7·09 | 3·53 | 3·53 ± 0·01 | 3·50–3·68 | 1·43 | W |
| H. sabaudum L. | Czech Rep.: Praha (2) | 27 | 12·51 ± 0·05 | 12·47–12·56 | 4·17 | 4·17 ± 0·02 | 4·12–4·23 | 2·67 | n.d. |
| Germany: Oberlausitz (2) | 27 | 12·65 ± 0·03 | 12·62–12·68 | 4·22 | X | ||||
| Czech Rep.: České středohoří (2) | 27 | 12·36 ± 0·01 | 12·35–12·37 | 4·12 | n.d. | ||||
| H. schmidtii Tausch | Czech Rep.: České středohoří (2) | 27* | 10·64 ± 0·05 | 10·59–10·69 | 3·55 | 3·54 ± 0·01 | 3·52–3·56 | 1·14 | n.d. |
| Czech Rep.: České středohoří (2) | 27* | 10·58 ± 0·00 | 10·61–10·61 | 3·54 | W | ||||
| Czech Rep.: Křivoklátsko (2) | 27 | 10·60 ± 0·01 | 10·60–10·61 | 3·53 | n.d. | ||||
| H. sparsum Friv. | Bulgaria: Vitoša (1) | 18 | 8·15 | - | 4·08 | 4·03 ± 0·03 | 3·99–4·08 | 2·56 | E(H) |
| Bulgaria: Pirin (2) | 18 | 8·01 ± 0·03 | 7·98–8·04 | 4·01 | |||||
| H. stelligerum Froel. | France: Ardèche (3) | 18* | 7·03 ± 0·06 | 6·91–7·14 | 3·51 | 3·51 ± 0·03 | 3·47–3·57 | 2·89 | W |
| H. tomentosum L. | France: Alpes Maritimes (8) | 18* | 7·48 ± 0·02 | 7·41–7·58 | 3·74 | 3·75 ± 0·01 | 3·71–3·79 | 2·16 | W |
| France: Briançon (1) | 27 | 11·27 | – | 3·76 | n.d. | ||||
| France: Hautes Alpes (2) | 27 | 11·25 ± 0·08 | 11·17–11·33 | 3·78 | n.d. | ||||
| H. transylvanicum Heuff. | Ukraine: Marmaros'ki Al'py (8) | 18* | 8·56 ± 0·01 | 8·52–8·59 | 4·28 | 4·28 ± 0·00 | 4·26–4·30 | 0·94 | W |
| H. umbellatum L. | Poland: Baltic coast, Jantar (1) | 18 | 8·34 | – | 4·17 | 4·26 ± 0·01 | 4·17–4·30 | 3·12 | E |
| Czech Rep.: Praha (8) | 18* | 8·54 ± 0·01 | 8·48–8·59 | 4·27 | n.d. | ||||
| Germany: Nordfriesland (2) (S) | 18 | 8·48 ± 0·03 | 8·45–8·52 | 4·24 | n.d. | ||||
| H. villosum Jacq. I. | Slovakia: Strážovské vrchy (2) | 36 | 15·71 ± 0·04 | 15·66–15·75 | 3·93 | E | |||
| H. villosum Jacq. II. | France: Hautes Alpes (1) | 27 | 11·6 | – | 3·87 | X(E) | |||
| H. villosum s.l. mean | 3·91 ± 0·02 | 3·87–3·94 | 1·81 | ||||||
| H. virosum Pall. | Russia: Rostov-na-Donu (2) (S) | 27 | 13·06 ± 0·03 | 13·02–13·09 | 4·35 | 4·34 ± 0·00 | 4·33–4·36 | 0·69 | E |
| Russia: Altajskij kraj (3) (S) | 27 | 13·00 ± 0·01 | 12·98–13·03 | 4·34 | E | ||||
| ‘Hieracium’ intybaceum All. | Italy: Rhaetian Alps (1) | 18 | 7·5 | – | 3·75 | 3·76 ± 0·01 | 3·74–3·79 | 1·34 | |
| Italy: Alpi Oróbie (2) | 18 | 7·53 ± 0·07 | 7·48–7·58 | 3·77 | |||||
| Austria: Ötztaler Alpen (2) | 18 | 7·53 ± 0·07 | 7·52–7·54 | 3·77 | |||||
| Pilosella lactucella (Wallr.) P.D.Sell & C.West | Germany: Erzgebirge (1) | 18 | 4·07‡ | 2·04 | |||||
| Andryala integrifolia L. | Spain: Andalusia (1) | n.d. | n.d. | ||||||
| Andryala levitomentosa (Nyár.) P.D.Sell | Romania: Pietrosul Bogolini (1) | 18 | 5·31§ | 2·66 | |||||
| Hispidella hispanica Barnades ex Lam. | Spain: Sierra de Guadarrama (1) | 18# | ∼4·00 | ∼2·00 |
* From Chrtek et al. (2007).
† E, ‘eastern’ clade; W: ‘western’ clade; X, interclade hybrid; X(E) and X(W), interclade hybrids with predominant ‘eastern’ or ‘western’ sequence variant; E(H) and W(H), hybrids within ‘eastern’ or ‘western’ clade; hybrid origin of H. sparsum and H. lachenalii inferred from plastid DNA (J. Fehrer et al., unpubl. res.). E-int. indicates ETS character additivity between the ‘eastern’ clade and ‘Hieracium’ intybaceum indicative of hybrid origin.
‡ See also Suda et al. (2007).
§ With Pilosella lactucella standard.
For diploid, sexually reproducing species and for agamospermous polyploids with a rather small distribution area, one or two populations were chosen. For sexual diploids with large geographic areas and for more widely distributed agamospermous polyploids, two to six populations were selected. The number of plants analysed per population varied from two in agamospermous species with likely clonal population structure (e.g. Shi et al., 1996; Mráz et al., 2001; Štorchová et al., 2002) to eight in supposedly genetically variable populations of sexual diploids. Two species, H. petrovae and H. plumulosum, were represented by a single plant due to their rarity in the field or because of cultivation problems. When two or more ploidies have been reported for a species, this diversity was covered as far as possible. Voucher specimens of all samples are deposited in the herbarium PRA.
Chromosome numbers and breeding system
At least two plants per population were checked for their chromosome number using the method described in Chrtek et al. (2007); counts for selected accessions have been published (Chrtek et al., 2007). The mode of reproduction was also tested, generally following Gadella (1987) and Krahulcová and Krahulec (1999). In diploids (in which sexual reproduction was expected), randomly selected capitula were bagged at the bud stage and tested for late-acting autogamy (in the absence of active pollination); results were compared with control capitula from the same plant in open pollination treatments. In polyploids (in which agamospermy was expected), the upper part of the capitulum was cut off at the bud stage (emasculation) and the number of ‘full’ achenes was counted as a measure of seed set and compared with the number of achenes from untreated capitula of the same plant. Percentages of ‘full’ achenes after emasculation in particular plants/species are available upon request.
Genome size estimation
Genome size was determined by flow cytometry using a Partec CyFlow cytometer equipped with a green (532 nm) solid-state laser. Zea mays ‘CE–777’ (2C = 5·48 pg; Lysák and Doležel, 1998) and Pisum sativum ‘Ctirad’ (2C = 8·85 pg; Doležel et al., 1994; Suda et al., 2007) were used as internal standards for diploid and polyploid species. The modified two step-procedure described by Otto (1990) was employed for sample preparation. Intact leaf tissue (approx. 1 cm2) of the analysed species and an appropriate quantity of the internal standard were co-chopped with a sharp razor blade in a plastic Petri dish with 1 mL of ice-cold Otto I buffer (0·1 m citric acid, 0·5% Tween 20) as the nuclear isolating solution. The suspension was filtered through a 42-μm nylon filter and centrifuged at 15 g for 5 min. The supernatant was discarded, and the pellet was resuspended in 100 µL fresh Otto I buffer. Samples were incubated for at least 10 min at room temperature and mixed with 1 mL Otto II buffer (0·4 m Na2HPO4) supplemented with propidium iodide as the fluorochrome, RNase IIA (both at a concentration of 50 µg mL–1) and β-mercaptoethanol (2 µL mL–1). Samples were stained for 5 min at room temperature before measurement. Usually, 5000 nuclei were analysed for each sample. Nuclear genome size was calculated as a linear relationship between the ratio of 2C peaks of sample and standard. Each plant was measured at least three times on different days by the same operator to eliminate potential artefacts. If the difference between the three measurements exceeded 2%, the value was discarded, and the sample was re-analysed. The coefficients of variation (CVs) of G0/G1 peaks did not exceed 5% (with two exceptions in Hieracium samples).
Molecular methods
A representative subset of 49 Hieracium accessions was selected for phylogenetic analysis. As the outgroup, species of the most closely related genera Andryala, Hispidella and Pilosella (sometimes treated as a subgenus of Hieracium) and ‘Hieracium’ intybaceum were chosen according to previous results (Fehrer et al., 2007). Although the latter species was traditionally placed in Hieracium subgenus Hieracium, molecular data (ITS sequences) suggested it belongs to an older isolated lineage clearly separated from a cluster formed by Hieracium s.l. and its closely related genera Andryala and Hispidella (Fehrer et al., 2007), which is also supported by the present data based on the ETS region.
Total genomic DNA was extracted from fresh or CTAB-preserved material using a sorbitol extraction method (Štorchová et al., 2000). The ETS region of the nuclear ribosomal DNA was amplified using the primers Ast-8 and 18 S (Noyes, 2006). PCR amplifications were done in 25-μL reactions containing 2·5 mm MgCl2, 0·2 mm of each dNTP, 0·5 µm of each primer, 0·5 unit of Taq DNA polymerase (Fermentas, Ontario, Canada), 1 × Taq buffer with KCl (Fermentas) and a few nanograms of genomic DNA. An initial denaturation step at 94°C for 3 min was followed by 35 cycles of denaturation (94°C for 30 s), annealing (55°C for 30 s) and extension (72°C for 40 s) steps, and a final extension at 72°C for 10 min. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced (GATC Biotech, Konstanz, Germany). Both strands were sequenced using the PCR primers. For one accession, Pilosella lactucella, direct sequencing was not successful and, therefore, the amplified fragment was subcloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions, but downscaled to half reactions. Three clones of this sample were sequenced (GATC Biotech), using the primer Ast-8.
Molecular data analyses
Sequence chromatograms were inspected by eye. In many accessions intra-individual polymorphism, i.e. more than one allele of the ETS region, was detected. Polymorphic sites were represented by the NC-IUB ambiguity symbols (e.g. R for A or G).
Initial sequence alignment was done with Clustal X (Thompson et al., 1997) and further edited manually in Bioedit (Hall, 1999). It was unambiguous due to low overall variation. Visual inspection of the alignment revealed the existence of two major groups within Hieracium s.s., and a proportion of accessions were identified as hybrids among these groups according to the additive pattern of polymorphic sites (these are referred to as ‘interclade hybrids’). Furthermore, the Hieracium piliferum accession analysed was identified as a hybrid between one of the major groups and ‘Hieracium’ intybaceum. Sequences were submitted to GenBank (EU821362–EU821419).
Bayesian (MrBayes V3·1·2; Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) and maximum likelihood (RAxML V7·0·3; Stamatakis, 2006) analyses were performed (a) on the complete data set and (b) on a modified data set excluding interclade hybrids and H. piliferum. Bayesian analyses were run with two nucleotide substitution rates and gamma distribution. This corresponds to a HKY + G model found in hierarchical Likelihood Ratio Tests as the model of molecular evolution best fitting to the data as implemented in Modeltest V3·5 (Posada and Crandall, 1998). Two parallel runs with four chains each were used for both analyses, sampling every 1000th tree. The analysis of the complete data set was computed for 2 million generations until convergence. The first 500 trees per run were discarded as burn-in and the remaining 1501 trees per run were summarized. For the modified data set, 1 million generations were sufficient for reaching convergence, the first 250 trees per run were discarded as burn-in and the remaining 751 trees per run were summarized. Maximum likelihood analyses were done using the rapid BS algorithm in combination with a maximum likelihood search, using the GTR model of nucleotide substitutions with a gamma model of rate heterogeneity and 1000 bootstrap replicates.
Statistical analyses of genome size
Intraspecific variation in morphologically defined species sensu Zahn was assessed and species with variation exceeding the approximate inaccuracy threshold of 3·5% [following Suda et al. (2007) in Hieracium subgenus Pilosella] were marked (H. amplexicaule, H. bupleuroides, H. laevigatum, H. pannosum, H. pictum, H. piliferum and H. prenanthoides). In H. bupleuroides and H. prenanthoides, differences in ETS sequences among accessions corresponding with the differences in genome size were found (J. Fehrer et al., unpubl. res.) which were caused by hybridization with other species. These heterogeneous species were therefore split into more natural units (accessions or groups of accessions) according to genome size and treated separately in the following analyses. The same was done for H. pannosum, in which the accessions differed distinctly in genome size (and in ploidy), and for H. pilosum and H. villosum, in which differences in ETS sequences due to introgression were found between accessions (although genome size variation was rather low in these cases). On the other hand, morphologically homogeneous species with high intrapopulation variation in genome size (H. pictum) and species with unclear patterns of genome size variation (H. amplexicaule, H. laevigatum and H. piliferum) were not split. The units after splitting held the name of the broad species and were numbered (I, II) (Table 1). For convenience, they are referred to as species in the following paragraphs. Forty six taxa were recognized after the split (Table 1). To test the correlation between monoploid DNA amount (1Cx) and chromosome number, the Spearman rank order correlation coefficient and the one-way ANOVA were used with a matrix of all samples (2n = 18, 27 and 36).
Evolution of genome size was investigated on a sample of trees using the generalized least-squares method implemented in BayesTraits (Pagel and Meade, 2007). For this purpose, the last 751 trees from each run of the Bayesian analysis of the modified data set (without hybrids) were sampled and merged into one file. All 1502 trees were rooted manually with the outgroup (Andryala integrifolia, the taxon used as outgroup in the Bayesian analysis), using the program Dendroscope (Huson et al., 2007). Analyses were conducted on three different data sets: (1) on a complete set of these species; (2) separately for species of the western clade; and (3) separately for species of the eastern clade. All genome size data were log10 transformed prior to analyses. Two models of trait evolution were compared, using likelihood ratio statistics (Huelsenbeck and Rannala, 1997) or BayesFactor. Model A (drift model) corresponds to the standard constant-variance random-walk model, and model B is a directional random-walk model (Pagel, 1999, 2004). The scaling parameters lambda (λ), kappa (κ) and delta (δ) were optimized for 1Cx-values. λ assesses the contribution of the phylogeny to a character, κ scales branch lengths and can be used to test punctual vs. gradual modes of trait evolution, and δ scales the overall path length in the phylogeny. Values of 1·0 correspond to the null model (tree topology and branch lengths accurately describe models A and B). To test whether a model with estimated scaling parameters is a better fit than the null model where all scaling parameters are set to 1 (i.e. a strict Brownian motion model) the likelihood ratio test or BayesFactor was used. Two different methods of analysis were used, namely maximum likelihood and Monte Carlo Markov chain (MCMC).
How the variation in genome size matched the two major lineages of Hieracium (named ‘western’ and ‘eastern’) suggested by molecular phylogenetic analyses of the ETS region was tested further. In addition, genome sizes of interclade hybrid accessions were analysed. Five informal groups were recognized and used for these analyses, restricting the genome size data set to the accessions for which sequence data were available (Table 1): (a) ‘western’, corresponding to the phylogenetically distinguished ‘western’ group and containing ‘pure’ accessions as well as hybrid/hybridogenous accessions within the ‘western’ group [W and W(H)]; (b) ‘eastern’, corresponding to the phylogenetically distinguished ‘eastern’ group and containing ‘pure’ accessions and hybrid/hybridogenous accessions within the ‘eastern’ group [E and E(H)]; (c) hybrid/hybridogenous accessions between ‘western’ and ‘eastern’ clade species with about equal contribution of ETS variants from each parent (X); (d) interclade hybrid accessions with strongly dominating ‘western’ ETS sequence type [X(W)]; and (e) interclade hybrid accessions with strongly dominating ‘eastern’ group ETS [X(E)] (Table 1). For the purpose of this analysis, no distinction was made between ‘pure’ and intraclade hybrids (a, b) because of the low sequence variation within each clade and because their genome sizes did not differ. Two comparisons were performed: (1) the ‘western’ and ‘eastern’ groups with a group comprising all E–W hybrids; and (2) the ‘western’ and ‘eastern’ groups with the three different groups of hybrids (c–e) specified above. More details about ETS sequence features and the identification of particular hybrids will be presented elsewhere (J. Fehrer et al., unpubl. res.). Both comparisons were conducted separately with and without H. transylvanicum (which fell into the phylogenetically defined western lineage, but has a genome size and geographic range congruent with the ‘eastern’ group; see Discussion). The correlation between genome size and phylogenetic pattern (five groups, see above) was tested.
The Spearman non-parametric rank order correlation coefficient was used in testing whether DNA amounts correlated with selected Ellenberg's indicator values, namely for light and temperature (Ellenberg et al., 1992). Mean 1Cx values for species sensu Zahn were used for this analysis; only a subset of central European species (for which these values are available) was chosen. Genome size variation was also tested against altitudinal and geographical position (longitude and latitude) for (a) the complete set of accessions (mean accessions' 1Cx values were used), and (b) excluding accessions of widely distributed species (H. bifidum, H. lachenalii, H. laevigatum, H. murorum, H. sabaudum and H. umbellatum) for which the results are strongly affected by the collection site of the samples.
The only significant correlations of genome size variation with other parameters concerned phylogeny and longitude. In order to distinguish between these two factors, three correlation tests concerning longitude were performed for each pair-wise comparison, constrained to accessions for which sequence data were available: (1) across all accessions from the ‘western’ and ‘eastern’ clades; (2) within the ‘western’ clade only; and (3) within the ‘eastern’ clade only. If the correlation is significant across all species, but not significant within either of the two clades analysed separately, this would argue for a connection between phylogenetic relationships and genome size variation. If, however, significant correlations are found for each of these tests, a relationship of genome size to longitude independent of species relationships would be supported. Data were analysed using the statistical package ‘Statistica for Windows 6·0’ (StatSoft, 1984–2002).
RESULTS
Chromosome counts and mode of reproduction
Chromosome numbers for plants of 43 populations analysed in the present paper belonging to 28 species were published elsewhere (Chrtek et al., 2007), and counts for the remaining 46 populations are presented here (Table 1). A new ploidy is reported for H. gymnocephalum (2n = 18). Other counts confirmed previously published chromosome numbers. All diploids studied were found to be sexual and allogamous, and all polyploids (3x, 4x) were agamospermous (data not provided).
Flow cytometry
Flow cytometric analyses yielded high-resolution histograms with CVs of G0/G1 peaks for Hieracium samples ranging from 0·83 to 5·76% (mean 2·28%), the values for internal reference standards were 0·97 to 5·0% (mean 2·19%). Generally, CVs of Pisum sativum were lower than those of Zea mays.
Nuclear DNA content: within-species variation
Intraspecific variation was assessed in 40 of 42 species sensu Zahn. Variation within accessions (populations) was generally low (Table 1). Mean values with standard errors (ranges for 2C and means for 1Cx genome sizes) for each population and mean values with standard errors and ranges for 1Cx for each species are summarized in Table 1. Variation in 1Cx values between conspecific accessions (in both homo- and multiploid species) ranged from 0·24% in H. gymnocephalum and H. heterogynum to 7·2% in H. pannosum. Variation exceeding the approximate measurement inaccuracy threshold of 3·5% was detected in seven species, namely H. amplexicaule, H. bupleuroides, H. laevigatum, H. pannosum, H. pictum, H. piliferum and H. prenanthoides.
However, variation in the more naturally delimited (without heterogeneity in ETS sequences and inter-population genome size; see Materials and methods) H. bupleuroides was below the threshold of 3·5% variation. Variation within the separately treated populations of morphologically heterogeneous H. pannosum was also below 3·5% (Table 1). In further paragraphs, these ‘narrower’ taxa (a total of 46 taxa) are used.
C-values in the total set of ‘basic’ species
The mean 2C values differed up to 2·37-fold among different species (from 7·03 pg in diploid H. stelligerum to 16·67 in a tetraploid accession of H. pannosum). The 1Cx values varied 1·22-fold between 3·51 pg in H. stelligerum and 4·34 pg in H. virosum (mean 1Cx value of 3·87, s.d. 0·27; Fig. 1). The 1Cx values of diploids (including diploid accessions of multiploid species, means for species/cytodemes) varied 1·22-fold between 3·51 pg in H. stelligerum and 4·29 pg in H. plumulosum (mean 1Cx value 3·92 pg, s.d. 0·30), in triploids [including triploid accessions of multiploid species (means for species/cytodemes)] 1·23-fold between 3·53 pg in H. bifidum and 4·35 pg in H. virosum (mean 1Cx value of 3·81 pg, s.d. 0·25), and in tetraploids 1·17-fold between 3·56 pg in H. humile and 4·17 pg in H. pannosum II (mean 1Cx value of 3·79, s.d. 0·19).
Fig. 1.
1Cx-value variation in 46 taxa of Hieracium subgenus Hieracium.
Correlation between genome size, ploidy and breeding system
Diploids differed significantly in their 1Cx values from both triploids (t = 2·71, d.f. = 196, P = 0·007) and tetraploids (t = 2·01, d.f. = 109, P = 0·047), but triploids did not differ significantly from tetraploids (t = 0·72, d.f. = 119, P = 0·476) (Fig. 2). The value of the Spearman non-parametric rank order correlation coefficient was r = –0·179, P = 0·009. The mean 1Cx value was 3·93 pg in diploids, 3·82 pg in triploids and 3·78 pg in tetraploids, suggesting a trend towards smaller genome size with increasing ploidy.
Fig. 2.
Variation of genome size among diploids, triploids and tetraploids (all samples). 1Cx values of all accessions are shown. Differences between diploids and triploids, and between diploids and tetraploids are significant. The box indicates the interquartile (25–75%) range, the small square within the box is the median. The whiskers indicate minimum and maximum values.
In multiploid species, there was no general trend to either genome downsizing or upsizing. In H. prenanthoides 2x/3x there was 2·47% upsizing, in H. villosum 3x/4x 0·06% downsizing, in H. tomentosum 2x/3x 0·36% upsizing, in H. humile 3x/4x 0·36% upsizing and in H. alpinum 2x/3x 0·17% upsizing.
Comparison between 1Cx values of sexually reproducing plants (i.e. all diploids; polyploids were exclusively apomictic) and apomicts (triploids and tetraploids) revealed significant differences at α = 0·01 (t-test, t = 3·04, d.f. = 213, P = 0·003); the mean 1Cx value in sexuals was 3·93 pg, whereas in apomicts it was 3·82 pg, corresponding to the value for triploids due to the low number of tetraploid accessions.
Molecular phylogenetics of Hieracium
Analysis of ETS data including all sequenced accessions resulted in the same tree with both methods. It indicates monophyly of Hieracium subgenus Hieracium, but species relationships remained completely unresolved as reflected by a large polytomy with only two small subclusters that received low support (Fig. 3). However, as two major species groups could be identified by visual inspection of the alignment and many sequences showed additive patterns indicative of hybridization involving both groups, these accessions were deleted from subsequent analysis, because reticulation is known to collapse branches (Feliner et al., 2001; Soltis et al., 2008). With the reduced data set, a clear separation into two major clades with strong statistical support was found with both methods (Fig. 4). These lineages were designated ‘eastern’ and ‘western’ clade because they contained species of predominantly eastern or western European origin. A large number of accessions (18) showed ETS variants of both clades in either equal proportion or with the ‘eastern’ or ‘western’ sequence type dominating as indicated in Fig. 4. Details of these analyses will be given in a parallel paper (J. Fehrer et al., unpubl. res.).
Fig. 3.
Phylogenetic analysis of ETS sequences based on all accessions. A Bayesian consensus tree of 3002 saved trees is shown with posterior probabilities above branches. The maximum likelihood tree has the same topology; bootstrap values are indicated below branches. Hieracium subgenus Hieracium (=Hieracium sensu stricto) is monophyletic, but species relationships are completely unresolved when hybrids are included in the analysis. Support for the two subclusters is low.
Fig. 4.
Phylogenetic analysis of ETS sequences excluding interclade hybrid accessions. A Bayesian consensus tree of 1502 saved trees is shown with posterior probabilities above branches. The maximum likelihood tree has the same topology; bootstrap values are indicated below branches. After exclusion of hybrids based on character additivity, two major groups are resolved (referred to as ‘eastern’ and ‘western’ clades). Hybrid accessions composed of parents from both clades (interclade hybrids) are listed to the right. (W), Interclade hybrids with predominant ‘western’ ETS type; (E), interclade hybrids with predominant ‘eastern’ ETS type. Hieracium piliferum is a hybrid between ‘Hieracium’ intybaceum and an ‘eastern’ clade taxon. For details about accessions, see Table 1 and Supplementary Data, available online.
Correlation of genome size with phylogenetic signal
The ‘western’ clade included 15 accessions: 2C values ranged from 7·03 pg in diploid H. stelligerum to 14·25 pg in a tetraploid accession of H. humile; 1Cx values ranged from 3·51 pg in H. stelligerum to 4·28 pg in H. transylvanicum (mean ± s.d.: 3·61 ± 0·19 pg; with H. transylvanicum excluded: up to 3·74 pg in H. tomentosum, 3·57 ± 0·06 pg). The ‘eastern’ clade also comprised 15 accessions: 2C values ranged from 7·78 pg in diploid H. porrifolium to 15·71 pg in a tetraploid accession of H. villosum; 1Cx values ranged from 3·63 pg in H. naegelianum to 4·35 pg in H. virosum (4·02 ± 0·20 pg). Significant differences in 1Cx values were found between the clades at α = 0·001 (Student's t-test), with (t = –5·71, d.f. = 28, P < 0·001) and without (t = –8·23, d.f. = 27, P < 0·001) H. transylvanicum.
Differences in 1Cx values between accessions of the ‘western’ (W) and ‘eastern’ (E) clades and of interclade hybrid accessions (X) are significant, independent of the inclusion of H. transylvanicum (F = 13·79, d.f. = 45, P < 0·001 with H. transylvanicum, F = 20·87, d.f. = 44, P < 0·001 without H. transylvanicum; Fig. 5A). However, post hoc comparison (Scheffé test) revealed only two groups at α = 0·05, the first comprising all ‘western’ accessions, and the second embracing ‘eastern’ and ‘hybrid’ accessions. Thus, ‘eastern’ and ‘hybrid’ accessions do not differ significantly from each other. Significant differences were also found between five groups, i.e. after splitting the bulk of hybrids into three groups, namely hybrids with intermediate position (X) and hybrids with strongly dominating ‘western’ [X(W)] or ‘eastern’ [X(E)] ETS sequences (F = 17·07, d.f. = 43, P < 0·001 with H. transylvanicum, F = 28·86, d.f. = 42, P < 0·001 without H. transylvanicum). The Scheffé test revealed only two groups at α = 0·05, the first including W and X(W) accessions, the second X, X(E) and E accessions (Fig. 5B). A significant correlation (Spearman rank coefficient r = 0·705, P < 0·001) between phylogenetic signal and hybrid origin [all five groups – W, E, X, X(W) and X(E)] and the pattern of genome size variation was found. Hieracium piliferum (1Cx = 3·9) occupies an isolated position, and it was identified as a hybrid between an ‘eastern’ clade taxon and ‘Hieracium’ intybaceum (1Cx = 3·76).
Fig. 5.
Correlation of 1Cx values with phylogeny. Only accessions for which sequence data were available are included. (A) W1, ‘western’ clade accessions without H. transylvanicum; W2, ‘western’ clade accessions including H. transylvanicum; X, interclade hybrid accessions; E, accessions of the ‘eastern’ clade. (B) W1, W2 and E as before, hybrids divided into those with predominant ‘western’ [X(W)], equal (X) and predominant ‘eastern’ [X(E)] ETS sequence composition (see also Table 1). The box indicates the interquartile (25–75%) range, the small square within the box is the median. The whiskers indicate minimum and maximum values.
Evolution of genome size
Maximum likelihood method
For the complete data set (all species), a directional model of evolution (model B) did not result in significantly higher likelihood scores than the drift model of evolution (model A; 74·856 vs. 75·746), indicating that there is no general trend to either genome size increase or decrease. Scaling parameters leading to the highest likelihood for 1Cx values were λ = 0·908, δ = 0·819 and κ = 1·035 in model A and λ = 0·701, δ = 0·637 and κ = 1·201 in model B, respectively. For both models, the values of scaling parameters did not differ significantly from 1 (the null expectation, LR test), indicating that the phylogenetic tree correctly predicts the pattern of covariance among species on the trait (1Cx) and that there is no evidence of accelerated evolution.
For the western clade, likelihood scores of models A and B did not differ significantly (44·061 vs. 44·790). The maximum likelihood values for λ (<1; 0·550 in model A, 0·523 in model B) show a role of adaptive response to some external factors. Values of δ and κ are >1 in both models (not shown) indicating that longer paths contribute more to 1Cx evolution (accelerated evolution as time progresses) and that longer branches contribute more to the trait evolution. Likelihoods of the null model (with scaling paramaters set to 1·0) are significantly lower in both models, indicating that scaling parameters improve the fit of the data to the models.
For the eastern clade, likelihood scores of models A and B also did not differ significantly (40·915 vs. 42·141). Scaling parameters are not significantly different from 1 (the null expectation, Brownian motion, data not shown) indicating that the phylogenetic tree correctly predicts the pattern of covariance among species and that there is no evidence of accelerated evolution.
MCMC method
For the complete data set, comparison of harmonic means of log maximum likelihoods of models A and B showed a somewhat higher value in the latter (82·544 vs. 84·670). The model with estimated scaling parameters is a better fit than the null model (with scaling parameters set to 1) for both models A and B, showing that the scaling parameters improve the fit of the data to the model. The values of λ did not differ significantly from 1, and relatively high values of κ (3·379 and 3·606, respectively) indicate that longer branches contribute more to genome size evolution.
For the western clade only, the harmonic mean of model B is also higher than that of model A (49·888 vs. 45·494) and the harmonic mean of the null model is significantly lower than that for the model with estimated scaling parameters. Scaling parameters are similar to those found with the maximum likelihood method, i.e. λ and δ < 1, κ > 1 (for interpretation see above).
For the eastern clade, harmonic means of models A and B do not differ significantly. Values of λ and δ do not differ from 1, higher values of κ (1·988 in model A and 2·212 in model B) again indicate accelerated rates of evolution within long branches.
Correlation between genome size and ecogeographic features
The genome size of particular accessions was significantly correlated with their geographic position (longitude) in a west–east direction, both in the complete set of accessions (Spearman rank coefficient r = 0·562, P < 0·001; Fig. 6A) and after exclusion of widely distributed species (r = 0·617, P < 0·001; i.e. without H. bifidum, H. lachenalii, H. laevigatum, H. murorum, H. sabaudum and H. umbellatum; Fig. 6B). The correlation was stronger in the second case due to the strong dependence on the part of the geographic area from which the target plants of widespread species were sampled.
Fig. 6.
Distribution of 1Cx values versus longitudinal position of collection sites: (A) based on the complete set of accessions/populations (Spearman rank coefficient r = 0·562, P < 0·001); (B) based on a subset after excluding accessions of widely distributed species (H. bifidum, H. lachenalii, H. laevigatum, H. murorum, H. sabaudum and H. umbellatum; r = 0·617, P < 0·001).
No correlation between genome size and latitude (r = 0·049, P = 0·646) or genome size and altitude (r = –0·224, P = 0·034) was found (complete set of accessions, results not shown). Also, no significant correlation was found between genome size and selected ecological parameters (Ellenberg's indicator values), namely temperature (r = 0·194, P = 0·427) and light (r = –0·236, P = 0·331) in a subset of species occurring in central Europe (results not shown).
Distinction between longitudinal and phylogenetic correlation
In order to determine whether the increase in genome size towards the east/‘eastern’ clade is based on geographic distribution or on species relationships, the longitudinal correlation was re-analysed for those accessions for which molecular data were available. Even stronger correlation was found between longitude and genome size in a set of accessions with known ETS sequences independent of including (r = 0·656, P < 0·001) or excluding (r = 0·688, P < 0·001) accessions of widely distributed species (Fig. 7A, B). In contrast, no significant correlation was found either among species of the ‘western’ (r = 0·161, P = 0·567) or among species of the ‘eastern’ (r = 0·394, P = 0·245) clade when tested separately (Fig. 7C and D). These results reveal that the evolutionary history due to eastern or western origin of the species is the dominant parameter affecting genome size in Hieracium rather than longitude.
Fig. 7.
Longitudinal component of genome size variation for accessions of known phylogenetic origin: (A) based on a complete set of ‘western’ and ‘eastern’ accessions/populations analysed by molecular methods (excluding interclade hybrid accessions; Spearman rank coefficient r = 0·656, P < 0·001); (B) based on a subset after excluding accessions of widely distributed species (H. bifidum, H. murorum and H. umbellatum; r = 0·688, P < 0·001); (C) based on a subset of accessions with ‘western’ ETS type (r = 0·161, P = 0·567); (D) based on subset of accessions with ‘eastern’ ETS type (r = 0·394, P = 0·245).
DISCUSSION
Chromosome numbers and mode of reproduction
Chromosome numbers for plants from 46 populations belonging to 26 species are published here for the first time, and counts for the remaining accessions have been published elsewhere (Chrtek et al., 2007). Among the new data, a new ploidy (diploid) is reported for H. gymnocephalum (s.l.); previously reported counts (Niketić et al., 2006) refer only to triploids (2n = 3x = 27). After H. petrovae, this is the second diploid count within section Pannosa. Also worth mentioning is the diploid (2n = 2x = 18) count for H. prenanthoides from the western Alps. Although this number had been reported from the same area in the 1960s (Favarger, 1969; Löve, 1969), it has not been confirmed until now. The remaining chromosome numbers correspond to previously published counts for the target species [cf. Schuhwerk (1996) and other standard reference manuals]. Analysis of the mode of reproduction confirmed the pattern already published for selected species and suggested it to be generally valid throughout the genus: diploid species reproduce sexually and are allogamous whereas polyploids are agamospermous.
Intraspecific genome size variation
Variation beyond arbitrary fluctuation (3·5%) was found in seven species sensu Zahn, namely H. piliferum (3·89%), H. amplexicaule (3·95%), H. bupleuroides (4·2%), H. laevigatum (4·28%), H. pictum (5·14%), H. prenanthoides (7·02%) and H. pannosum (7·2%). All are agamospermous polyploids, two of them including two cytodemes (H. amplexicaule, 3x /4x; H. prenanthoides, 2x /3x). As only a subset of populations was used for phylogenetic analysis, these results need to be interpreted with caution. In several cases in which sequence data were obtained for more than one accession, multiple origins of a given taxon, namely in H. bupleuroides, H. pilosum, H. prenanthoides and H. villosum, were found, which allows a plausible explanation of genome size variation at least for H. prenanthoides. In this species, sequences of three accessions, one being diploid and two triploid, were analysed. All three showed identical signatures of an ancient interclade hybridization already affecting the diploid (J. Fehrer et al., unpubl. res.). The triploids resulted from different subsequent hybridizations, one involving a ‘western’ and one an ‘eastern’ lineage. This fits well with the 1Cx values: whereas values of the diploid and ‘western’ introgressed triploid populations ranged from 3·56 pg to 3·67 pg, the 1Cx value of the ‘eastern’ introgressed triploid was higher (3·81 pg). Hieracium laevigatum, H. amplexicaule and H. piliferum are, according to the ETS sequences, hybrids/hybridogenous types (at least the analysed accessions), and higher intraspecific variation might reflect recurrent polytopic origins. High inter-population variation in H. pannosum could also be related to multiple origins as suggested by the different ploidies of the analysed accessions. The high variation in H. pictum cannot be explained by the present data. Broader sampling for molecular analysis in these species might reveal hybrid accessions that have not yet been discovered. Thus, unequivocal evidence for intraspecific genome size variation was not found in ‘good’ species of subgenus Hieracium.
Similar results were obtained for Hieracium subgenus Pilosella in which the majority of wild species/cytotypes possess constant nuclear DNA amounts (variation in fluorescence intensity lower than 3·5%). Nevertheless, higher divergence was observed in six cytotypes belonging to three ‘intermediate’ species (the same terms as in subgenus Hieracium, i.e. hybrids/hybridogenous species) and among genetically variable F1 offspring of experimental crosses between hexaploid H. rubrum and tetraploid H. pilosella (Suda et al., 2007).
Intraspecific genome size variation in ‘non-hybridogenous’ species recently became a matter of debate (Murray, 2005). Whereas many of the examples of variation have been shown to be artefacts of measurement methods (e.g. Teoh and Rees, 1976; Greilhuber, 1998, 2005), there are some reports documenting C-value variation where appropriate controls and standards have been used (Reeves et al., 1998; Hall et al., 2000; Moscone et al., 2003; Pecinka et al., 2006).
Interspecific genome size variation
The present estimates of nuclear DNA content in Hieracium subgenus Hieracium are the first to be published for this group; all other data on Hieracium available so far refer to species of (subgenus) Pilosella (Bennett and Leitch, 2005), genomes of which are considerably smaller than those of subgenus Hieracium (see below). Holoploid (2C) genome sizes in Hieracium species included in the present study ranged 2·37-fold from 7·03 pg to 16·67 pg (mean 2C value 10·16 pg, median 10·61 pg). As almost all so-called ‘basic’ species were investigated, the present results should cover the genome size variation within the subgenus. A few rare pentaploid hybridogenous (i.e. not ‘basic’) taxa exist which were not analysed, and thus the upper limit could be higher. Variation in Cx values among species is relatively high (up to about 20%), but more or less continuous.
In Hieracium subgenus Pilosella with the same basic chromosome number (n = 9), holoploid (2C) genome size differs 4·33-fold and ranges from 3·53 pg to 15·30 pg (Suda et al., 2007). However, Pilosella has more extensive variation in ploidy, ranging from diploid to octoploid. Monoploid genome sizes (1Cx values) in subgenus Hieracium ranged 1·22-fold from 3·51 pg to 4·29 pg (mean 2C value 3·86 pg, median 3·85 pg), whereas genome size in Pilosella is distinctly lower (it varies 1·23-fold from 1·72 pg to 2·16 pg). Thus, Pilosella has consistently about half the DNA content compared with Hieracium. The reasons for these large differences among closely related groups (Fehrer et al., 2007) are unclear at the moment. Chromosomes of subgenus Hieracium are distinctly larger than those of Pilosella (no quantitative assessments available). Accumulation of repetitive sequence elements as in other plant groups might be one of the causes, but insights into Hieracium genomes are still lacking.
Genome size and ploidy
Diploid hawkweeds differ significantly in their 1Cx values from both triploids and tetraploids, but the latter do not differ from each other. This might indicate general downsizing of genomes in polyploid hawkweeds. However, there is no general trend to either downsizing or upsizing within multiploid species. Their origin remains to be elucidated in many cases and could involve autopolyploid origin as well as participation of another taxon (introgression which cannot always be detected by morphology). In Asteraceae, a similar situation was documented in the genus Centaurea s.l. in four multiploid (consisting of diploid and tetraploid cytotypes) species: downsizing was found in two species, upsizing in one species and equal monoploid genome size in one species (Bancheva and Greilhuber, 2006). However, downsizing of the genome after polyploidization is widely supposed to be a general trend in angiosperms (Kellogg and Bennetzen, 2004; Leitch and Bennett, 2004; Weiss-Schneeweiss et al., 2006), as seems to be the case for Hieracium. The present data also suggest that species of autopolyploid origin might have more uniform genome size (and morphology) than allopolyploids of multiple (hybrid) origin.
Genome size and phylogeny
Genome size distribution basically matches two phylogenetically defined major lineages, i.e. a ‘western’ and an ‘eastern’ group (Figs 4 and 5). It thus reveals a strong correlation of nuclear DNA content with the basal evolutionary divergence of Hieracium subgenus Hieracium. Both clades include sexual diploids, agamospermous triploids and rare tetraploid apomicts. A similar pattern has also been observed for the geographic ranges. Both groups comprise local endemics (e.g. H. stelligerum and species of section Cerinthoidea in the ‘western’ group and H. kittanae, H. petrovae and H. eriophorum in the ‘eastern’ group) and widely distributed species (e.g. H. murorum and H. bifidum in the ‘western’ and H. umbellatum in the ‘eastern’ group). Differences of genome size between the two clades were also significant when only diploid or triploid accessions were compared (despite indication for some genome downsizing in polyploids), and thus all cytodemes were analysed together.
As mentioned above, H. transylvanicum falls into the western lineage but has a genome size and geographic range congruent with the ‘eastern’ group. Two alternative scenarios for its origin can be proposed: (1) the species has an eastern origin as suggested by its current distribution and DNA content and shows some ancient introgression from western species, some of which are widespread. Its ETS sequence then became completely homogenized towards the western type by concerted evolution (Arnheim, 1983); and (2) the species originated in western Europe, spread towards the east, the original populations became extinct probably during the Ice Ages and only the eastern populations survived in an eastern glacial refuge like the Carpathian basin. In this case, the high DNA content may be due to other reasons than phylogenetic relationships. Another accession of a ‘western’ clade species, H. lachenalii, has plastid DNA matching some ‘eastern’ clade species which suggests ancient introgression (J. Fehrer et al., unpubl. res.). Correspondingly, its DNA content is also slightly higher than that of most other species of the ‘western’ clade. Hieracium eriophorum, a local endemic of the Atlantic coast near Arcachon in western France, is another species with an incongruent geographic range and position in the phylogenetic tree. Despite its western European distribution, it is most likely derived from widespread H. umbellatum, a species belonging to the same ‘eastern’ clade (Fig. 4). The morphology of H. eriophorum could therefore be interpreted as a local adaptation to sand dunes along the sea coast. The lowest genome size within the ‘eastern’ clade (2C = 10·89 pg, 1Cx = 3·63 pg) was detected in triploid H. naegelianum. The distribution of this species fits well with other ‘eastern’ species as it occurs in the Balkan Peninsula and in the Abruzzi Mountains in central Italy, mostly in refugial areas. With respect to morphology, it is the only species of Hieracium subgenus Hieracium with long underground stolons, which enable the plant to spread vegetatively. Although no evidence of introgression from a ‘western’ species is apparent from the present molecular data, its occurrence in Italian glacial refuges could be indicative of past contacts and introgression from which only an unusually small genome size is left. Its plastid DNA is unique, and its relationships with other ‘eastern’ clade species are unresolved.
Hybrid (hybridogenous) ‘basic’ species
Fourteen ‘basic’ species were found to be of hybrid origin between ‘eastern’ and ‘western’ clade species (Fig. 4): H. amplexicaule, H. bracteolatum, H. caesium, H. glaucum, H. gouanii, H. gymnocephalum, H. heterogynum, H. lachenalii, H. laevigatum, H. olympicum, H. plumulosum, H. prenanthoides, H. racemosum and H. sabaudum. In addition, individual accessions of H. pilosum and H. villosum also had this type of hybrid origin. Based on the known mean 1Cx values for the ‘western’ and ‘eastern’ groups (3·61 and 4·02 pg, respectively), intermediate genome sizes of the previously mentioned species might be expected. However, hybrid accessions were more similar to the ‘eastern’ species group and significantly different from the ‘western’ group. The median 1Cx values of intermediate hybrid taxa and those with dominating ‘eastern’ ETS sequence were even higher than those for both clades (Fig. 5B). Potential interpretations could be to assume extinct parents with higher genome sizes or, more likely, an increase in DNA content in species of hybrid origin in comparison to their parents, as has been documented in Helianthus by Baack et al. (2005). The present results also show that hybrids/hybridogenous types with strongly dominating ‘western’ type ETS (e.g. H. amplexicaule, H. caesium, H. gouani, H. lachenalii and H. prenanthoides) have similar DNA content in comparison with ‘western’ species, which is significantly different from hybrids with equal contribution of ‘eastern’ and ‘western’ parents. This might suggest repeated backcrossing towards ‘western’ species at the diploid level before genomes became fixed by apomixis. Intermediate hybrids with dominant ‘eastern’ ETS, as expected, did not differ significantly from ‘intermediate’ hybrids or ‘eastern’ species.
One accession of H. piliferum, a basic species distributed in European mountains was found to be a hybrid between an unidentified ‘eastern’ taxon and ‘H.’ intybaceum by character additivity in ETS sequences. Comparing 1Cx values of ‘H.’ intybaceum (3·76 pg, Table 1), H. piliferum (3·91 pg) and members of the two (‘western’ and ‘eastern’) major clades, past hybridization between ‘H.’ intybaceum and an ‘eastern’ species might be expected, which is also congruent with the results of the molecular analyses. Intraspecific genome size variation beyond the arbitrary fluctuation in H. piliferum could be indicative of multiple origins.
Evolution of genome size
Two different approaches implemented in BayesTraits for continuous data were applied, namely the maximum likelihood method and the MCMC approach, and two models of evolution (A, drift model; B, directional model) were compared. In most comparisons, our data fit (higher log-likelihoods and harmonic means) model B better, indicating that there is some but no strong trend to genome size increase.
For the complete data set and the maximum likelihood method, the values of scaling parameters did not depart significantly from the default values (1·0) suggesting that tree topology and branch lengths accurately describe the constant variance random-walk model A or B. Thus, genome size is evolving, as expected, given the tree topology, fitting well with the basal split into the two clades. Using the MCMC method, more changes on longer branches were indicated (longer branches contribute more to genome size evolution).
The tempo and mode of evolution differ between the western and eastern clade. In the western clade, values of scaling parameters depart from the default value (1·0) indicating that genome size evolution has not followed the topology or the branch lengths. Phylogenetic history has lower impact (λ < 1, presumed adaptive response to some external pressures) in this case which may also be reflected by the almost complete lack of resolution of species within the western clade (Fig. 4). Nevertheless, longer paths and branches contribute more to 1Cx evolution (accelerated evolution as time progresses). In contrast, for the eastern clade, the maximum likelihood method revealed that topology and branch lengths accurately describe the constant variance random-walk model A or B. However, using the MCMC method, accelerated rate of evolution in long branches is indicated.
Studies using a phylogenetic approach to evaluate the directionality of genome size evolution revealed both DNA decrease and increase and often different tendencies in genome size diversification in different phylogenetic lineages (e.g. Wendel et al., 2002; Jakob et al., 2004; Caetano-Annolés, 2005; Price et al., 2005). Ancient genome size enlargement followed by more or less drastic parallel reduction in the main phylogenetic lineages was found in Festuca (Šmarda et al., 2008).
Genome size and ecogeographic features
In order to identify further components of genome size variation for Hieracium, correlations were tested with a number of other factors. A significant, positive correlation was found between 1Cx value and longitude of sampling sites, both in the complete set of accessions and in a restricted set without species with large distribution areas for which the results depend strongly on sampling (Fig. 6). Restriction of these analyses to accessions analysed by molecular data showed that these correlations were even stronger when accessions of ambiguous origin were excluded (Fig. 7A, B). As no significant correlation was found within either of the two clades (‘western’ and ‘eastern’; Fig. 7C, D), it can be concluded that the basal divergence into two phylogenetic lineages is most likely the determining factor of genome size variation in Hieracium (or vice versa) rather than longitudinal distribution.
In Hieracium subgenus Pilosella (Suda et al., 2007), a longitudinal component of genome size distribution was also found: the highest 1Cx values were detected in H. echioides, a species distributed mainly in steppic habitats in Asia and eastern Europe (and well differentiated from the remaining species by the absence of a basal leaf rosette at flowering time). However, no comparison with species relationships is available. An opposite relationship between genome size distribution and geographic ranges has been observed in the genus Cirsium (Bureš et al., 2004). At the intraspecific level, a geographically correlated variation in DNA content with an increase towards the east has been documented, e.g. in several taxa of Koeleria (Pecinka et al., 2006), but no correlation was found in Sesleria albicans (Lysák et al., 2000). Thus, there does not seem to be a general trend in genome size variation in relation to longitude. The same holds for a relationship between genome size and latitude, where positive, negative, or non-significant correlations with genome size have been found (reviewed in Knight et al., 2005).
Altitude was also studied. The correlation between this parameter and genome size has been a matter of debate in the past years, and divergent results have been obtained. In subgenus Hieracium, genome size variation does not depend on altitude. However, the present data had to be based on altitudes of the sampling sites and are therefore biased by this selection, especially in species occurring across a large range of different altitudes (e.g. H. bifidum). Similarly, no correlation between genome size and altitude in Asteraceae was found in Cirsium (Bureš et al., 2004) or Artemisia and Tripleurospermum (García et al., 2004, 2005). In other families, no or even a negative correlation between genome size and altitude was found by Creber et al. (1994), Reeves et al. (1998) and Vilhar et al. (2002) (all on intraspecific variation in Dactylis glomerata). On the other hand, an increase in genome size with higher altitute was found in Centaurea s.s. (Bancheva and Greilhuber, 2006) and in some groups of grasses (Bennett, 1976; Laurie and Bennett, 1985; Rayburn and Auger, 1990). Thus, altitudinal genome size variation also seems to be dependent on the particular plant group analysed (Knight et al., 2005). In Hieracium, there are specifically montane or alpine taxa, but they are distributed in the Pyrenees, the Alps or the Balkan mountains, i.e. in western, central and eastern European regions, and therefore, the geographic origin of the species strongly dominates any altitudinal genome size variation that might be found.
Also no correlation was found between Cx values and two selected approximate ecological parameters published for German plant species by Ellenberg et al. (1992), namely light and temperature. However, the use of Ellenberg's indicator values for Hieracium is ambiguous. Many Hieracium species have large ecological amplitudes and therefore these approximate values could indeed be useful indicators, but these values are only available for central European species, and therefore species confined to either western or eastern Europe had to be excluded.
Many published correlations between ecogeographical factors (and others such as life form, etc.) and genome size must be interpreted with caution, as phylogenetic information is lacking. Albach and Greilhuber (2004) showed quite different correlations between selected factors (habitat, life history and breeding system) and genome size and DNA C-values in the genus Veronica if using a simple statistical test without phylogenetic information or more sophisticated methods incorporating phylogeny (independent contrast, GLSM). Furthermore, the use of linear regression analysis could obscure patterns in relationships if they are not linear. Knight and Ackerly (2002), Knight et al. (2005) and Beaulieu et al. (2007) used quantile regression analysis and showed that, although the relationship between genome size and a particular parameter was poor for species with small genomes, as genome size increased, the relationship became increasingly significant.
Conclusions
Genome size variation in Hieracium subgenus Hieracium is congruent with the phylogenetic pattern, with species of putative western European origin having significantly lower genome size than those of eastern European origin. Consequently, a significant longitudinal correlation can also be inferred. Separate analyses of closely related species (i.e. within each phylogenetic clade) clearly show that despite considerably overlapping scales, no significant geographic component is apparent. Thus, in Hieracium, any correlation of genome size with longitude, and with other ecogeographic variables such as latitude, altitude, light and temperature, is outweighed by the basal phylogenetic divergence into species of eastern or western European origin.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank S. Bräutigam, R. Douzet, E. Forey, J. Hadinec, P. Ignatova, A. Krahulcová, F. Krahulec, M. Král, K. Marhold, P. Mráz, M. Niketić, A. N. Sennikov, Z. Szeląg, T. Tyler, V. Vladimirov and V. Zavadil for providing us with plant material of several species; G. Mateo, J. A. Rosselló and B. Vreš for their generous help with the field sampling and G. Mateo, F. Schuhwerk and Z. Szeląg for the determination of some plants. We are grateful to P. Trávníček for expert help in the flow cytometry laboratory, P. Caklová for most of the molecular laboratory work, J. Loureiro and J. Suda for critically reading the manuscript and particularly S. Bräutigam and P. Mráz for discussions and many valuable comments. Three anonymous reviewers greatly helped to improve the manuscript. The work was supported by the Czech Science Foundation (grant no 206/05/0657) and partly also by the Academy of Sciences of the Czech Republic (AV0Z60050516) and Ministry of Education, Youth and Sports of the Czech Republic (grant no 0021620828).
LITERATURE CITED
- Albach DC, Greilhuber J. Genome size variation and evolution in Veronica. Annals of Botany. 2004;94:897–911. doi: 10.1093/aob/mch219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N. Concerted evolution of multigene families. In: Nei M, Koehn R, editors. Evolution of genes and proteins. Sunderland, MA: Sinauer; 1983. pp. 38–61. [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Molecular Biology Reports. 1991;9:208–218. [Google Scholar]
- Baack EJ, Whitney KD, Rieseberg LH. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytologist. 2005;167:623–630. doi: 10.1111/j.1469-8137.2005.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancheva S, Greilhuber J. Genome size in Bulgarian Centaurea s.l. (Asteraceae) Plant Systematics and Evolution. 2006;257:95–117. [Google Scholar]
- Barakat A, Carels N, Bernardi G. The distribution of genes in the genomes of Gramineae. Proceedings of the National Academy of Sciences of the USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Moles AT, Leitch IJ, Bennett MD, Dickie JB, Knight CA. Correlated evolution of genome size and seed mass. New Phytologist. 2007;173:422–437. doi: 10.1111/j.1469-8137.2006.01919.x. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist. 2008;179:975–986. doi: 10.1111/j.1469-8137.2008.02528.x. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society London, Series B, Biological Sciences. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD. DNA amount, latitude and crop plant distribution. In: Jones K, Brandham PE., editors. Current chromosome research. Amsterdam: North-Holland; 1976. pp. 151–158. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Annals of Botany. 1995;76:113–176. [Google Scholar]
- Bennett MD, Leitch IJ. Plant DNA C-values Database. 2005. (release 5·0, December 2004). http://www.kew.org/cval/homepage.html. (last accessed 16 September 2008) [DOI] [PubMed]
- Bennett MD, Heslop-Harrison JS, Smith JB, Ward JP. DNA density in mitotic and meiotic metaphase chromosomes of plants and animals. Journal of Cell Science. 1983;63:173–179. doi: 10.1242/jcs.63.1.173. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Annals of Botany. 1998;82(Suppl. A):121–134. [Google Scholar]
- Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. doi: 10.1023/a:1016015913350. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ma JX, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini M, Greizerstein E, Aulicino M, Poggio L. Relationships among genome size, environmental conditions and geographical distribution in natural populations of NW Patagonian species of Berberis L. (Berberidaceae) Annals of Botany. 2000;86:565–573. [Google Scholar]
- Buitendijk JH, Boon EJ, Ramanna MS. Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Annals of Botany. 1997;79:343–353. [Google Scholar]
- Bureš P, Yi-Feng Wang, Horová L, Suda J. Genome size variation in central European species of Cirsium (Compositae) and their natural hybrids. Annals of Botany. 2004;94:353–363. doi: 10.1093/aob/mch151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Annolés G. Evolution of genome size in the grasses. Crop Science. 2005;45:1809–1816. [Google Scholar]
- Chrtek J, Jr, Mráz P, Severa M. Chromosome numbers in selected species of Hieracium s.str. (Hieracium subgen. Hieracium) in the Western Carpathians. Preslia. 2004;76:119–139. [Google Scholar]
- Chrtek J, Jr, Mráz P, Zahradníček J, Mateo G, Szeląg Z. Chromosome numbers and DNA-ploidy levels of selected species of Hieracium s.str. (Asteraceae) Folia Geobotanica. 2007;42:411–430. [Google Scholar]
- Creber HMC, Davies MS, Francis D, Walker HD. Variation in DNA C value in natural populations of Dactylis glomerata L. New Phytologist. 1994;128:555–561. doi: 10.1111/j.1469-8137.1994.tb03001.x. [DOI] [PubMed] [Google Scholar]
- Devos K, Brown J, Bennetzen J. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Doleželová M, Novák F. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana) Biologia Plantarum. 1994;36:351–357. [Google Scholar]
- Edwards GA, Endrizzi JL. Cell size nuclear size and DNA content relationships in Gossypium. Canadian Journal of Genetics and Cytology. 1975;17:181–186. [Google Scholar]
- Elena-Rosselló JA, González-Zapatero MA, Navarro F. Notas cariológicas sobre algunos oroendemismos ibéricos. Lazaroa. 1985;8:91–96. [Google Scholar]
- Ellenberg H, Weber HE, Düll R, Wirth V, Werner W, Paulissen D. Indicator values of plants in Central Europe. Scripta Botanica. 1992;18:1–258. [Google Scholar]
- Favarger C. Notes de caryologie alpine V. Bulletin de la Société Neuchâteloise des Sciences Naturelles. 1969;92:13–30. [Google Scholar]
- Fehrer J, Gemeinholzer B, Chrtek J, Jr, Bräutigam S. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae) Molecular Phylogenetics and Evolution. 2007;42:347–361. doi: 10.1016/j.ympev.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Feliner NG, Aguilar JF, Rosselló JA. Can extensive reticulation and concerted evolution result in cladistically structured molecular dataset? Cladistics. 2001;17:301–312. [Google Scholar]
- Flavell RB, Rimpau J, Smith DB. Repeated sequence DNA relationships in four cereal genomes. Chromosoma. 1977;63:205–222. [Google Scholar]
- Gadella TWJ. Sexual tetraploid and apomictic pentaploid populations of Hieracium pilosella (Compositae) Plant Systematics and Evolution. 1987;157:219–246. [Google Scholar]
- García S, Sanz M, Garnatje T, Kreitschitz A, McArthur ED, Vallès J. Variation of DNA amount in 47 populations of the subtribe Artemisiinae and related taxa (Asteraceae, Anthemideae): karyological, ecological, and systematic implications. Genome. 2004;47:1004–1014. doi: 10.1139/g04-061. [DOI] [PubMed] [Google Scholar]
- García S, Inceer H, Garnatje T, Vallès J. Genome size variation in some representatives of the genus Tripleurospermum. Biologia Plantarum. 2005;49:381–387. [Google Scholar]
- Garnatje T, Vallès J, García S, et al. Genome size in Echinops L. and related genera (Asteraceae, Cardueae): karyological, ecological and phylogenetic implication. Biology of the Cell. 2004;96:117–124. doi: 10.1016/j.biolcel.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Govindaraju DR, Cullis CA. Modulation of genome size in plants: the influence of breeding systems and neighbourhood size. Evolutionary Trends in Plants. 1991;5:43–51. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size: a critical reassessment. Annals of Botany. 1998;82(Suppl. A):27–35. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkopp E, Rejmanek M, Sanderson MJ, Rost TL. Evolution of genome size in pines (Pinus) and its life-history correlates: supertree analyses. Evolution. 2004;58:1705–1729. doi: 10.1111/j.0014-3820.2004.tb00456.x. [DOI] [PubMed] [Google Scholar]
- Hall SE, Dvorak WS, Johnston JS, Price HJ, Williams CG. Flow cytometric analysis of DNA content for tropical and temperate new world pines. Annals of Botany. 2000;86:1081–1086. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis suite. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hawkins JS, HU G, Rapp RA, Grafenberg JL, Wendel JF. Phylogenetic determination of the pace of transposable element proliferation in plants: copia and LINE-like elements in Gossypium. Genome. 2008;51:11–18. doi: 10.1139/g07-099. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob SS, Meister A, Blattner FR. The considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Molecular Biology and Evolution. 2004;21:860–869. doi: 10.1093/molbev/msh092. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences of the USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller ERJ, Schubert I, Fuchs J, Meister A. Interspecific crosses of onion with distant Allium species and characterization of the presumed hybrids by means of flow cytometry, karyotype analysis and genomic in situ hybridization. Theoretical and Applied Genetics. 1996;92:417–424. doi: 10.1007/BF00223688. [DOI] [PubMed] [Google Scholar]
- Kellogg EA, Bennetzen JL. The evolution of nuclear genome structure in seed plants. American Journal of Botany. 2004;91:1709–1725. doi: 10.3732/ajb.91.10.1709. [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. Genome size variation across environmental gradients: a quantile regression analysis. Ecology Letters. 2002;5:66–76. [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome size constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahulcová A, Krahulec F. Chromosome numbers and reproductive systems in selected representatives of Hieracium subgen. Pilosella in the Krkonoše Mts (the Sudeten Mts) Preslia. 1999;71:217–234. [Google Scholar]
- Labani RM, Elkington TT. Nuclear DNA variation in the genus Allium L. (Liliaceae) Heredity. 1987;59:119–128. [Google Scholar]
- Laurie DA, Bennett MD. Nuclear DNA content in the genera Zea and Sorghum: intergeneric, interspecific and intraspecific variation. Heredity. 1985;55:307–313. [Google Scholar]
- Lawrence ME. Senecio L. (Asteraceae) in Australia: nuclear DNA amounts. Australian Journal of Botany. 1985;33:221–232. [Google Scholar]
- Leitch I, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Löve Á. IOPB Chromosome number reports XXII. Taxon. 1969;18:433–442. [Google Scholar]
- Luque T. Números cromosómicos para la flora española. Lagascalia. 1981;10:225–256. [Google Scholar]
- Lysák MA, Doležel J. Estimation of nuclear DNA content in Sesleria (Poaceae) Caryologia. 1998;51:123–132. [Google Scholar]
- Lysák MA, Rostková A, Dixon JM, Rossi G, Doležel J. Limited genome size variation in Sesleria albicans. Annals of Botany. 2000;86::399–403. [Google Scholar]
- MacGillivray CW, Grime JP. Genome size predicts frost-resistance in British herbaceous plants – implication for rates of vegetation response to global warming. Functional Ecology. 1995;9:320–325. [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genomic Research. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo G. Aportaciones al conocimiento del género Hieracium en España. X. Novedades para el Pirineo catalán. Flora Montiberica. 2005;31:62–69. [Google Scholar]
- Morgan ER, Burge GK, Seelye JF, Hopping ME, Grant JE. Production of inter-specific hybrids Limonium perezii (Stapf) Hubb. and Limonium sinuatum (L.) Mill. Euphytica. 1998;102:109–115. [Google Scholar]
- Morgan MT. Transposable element number in mixed mating populations. Genetical Research. 2001;77:261–275. doi: 10.1017/s0016672301005067. [DOI] [PubMed] [Google Scholar]
- Moscone EA, Baryani M, Ebert I, Greilhuber J, Ehrendorfer F, Hunziger AT. Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and Feulgen densitometry. Annals of Botany. 2003;92:21–29. doi: 10.1093/aob/mcg105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mráz P, Chrtek J, Jr, Kirschner J. Genetic variation in the Hieracium rohacsense group (Hieracium sect. Alpina) Phyton (Horn) 2001;41:269–276. [Google Scholar]
- Murray BG. When does intraspecific C-value variation become taxonomically significant? Annals of Botany. 2005;95:119–125. doi: 10.1093/aob/mci007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niketić M, Vladimirov V, Mráz P. Chromosome numbers and taxonomic-chorological notes on selected species of Hieracium s.str. (Asteraceae) from Montenegro. Phytologia Balcanica. 2006;12:85–97. [Google Scholar]
- Nogler GA. Gametophytic apomixis. In: Johri BM, editor. Embryology of angiosperms. Berlin: Springer; 1984. pp. 475–518. [Google Scholar]
- Noyes RD. Intraspecific nuclear ribosomal DNA divergence and reticulation in sexual diploid Erigeron strigosus (Asteraceae) American Journal of Botany. 2006;93:470–479. doi: 10.3732/ajb.93.3.470. [DOI] [PubMed] [Google Scholar]
- Otto F. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods in Cell Biology. 1990;33:105–110. doi: 10.1016/s0091-679x(08)60516-6. [DOI] [PubMed] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pagel M. Continuous. Manual. 2004. Available at http://www.evolution.rdg.ac.uk/BayesTraits.html .
- Pagel M, Meade A. BayesTraits. 2007. Computer program and documentation available at http://www.evolution.rdg.ac.uk/BayesTraits.html .
- Pecinka A, Suchánková P, Lysak MA, Trávníček B, Doležel J. Nuclear DNA content variation among Central European Koeleria taxa. Annals of Botany. 2006;98:117–122. doi: 10.1093/aob/mcl077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. DNA loss and evolution of genome size in Drosophila. Genetica. 2002;115:81–91. doi: 10.1023/a:1016076215168. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L, Johnston JS. Genome evolution in the genus Sorghum (Poaceae) Annals of Botany. 2005;95:219–227. doi: 10.1093/aob/mci015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn AL, Auger JA. Genome size variation in Zea mays ssp. mays adapted to different altitudes. Theoretical and Applied Genetics. 1990;79:470–474. doi: 10.1007/BF00226155. [DOI] [PubMed] [Google Scholar]
- Rees H, Cameron FM, Hararika MH, Jones GH. Nuclear variation between diploid angiosperms. Nature. 1966;211:828–830. doi: 10.1038/211828a0. [DOI] [PubMed] [Google Scholar]
- Reeves G, Francis D, Davies MS, Rogers HJ, Hodkinson TR. Genome size is negatively correlated with altitude in natural populations of Dactylis glomerata. Annals of Botany. 1998;82:99–105. [Google Scholar]
- Rejmanek M, Richardson DM. What attributes make some plant species more invasive? Ecology. 1996;77:1655–1661. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schuhwerk F. Published chromosome counts in Hieracium. 1996. http://www.botanik.biologie.uni-muenchen.de/botsamml/projects/chrzlit.html. (last accessed 16 September 2008)
- Schuhwerk F. Some thoughts on the taxonomy of Hieracium. Berichte der Bayerischen botanischen Gesellschaft. 2003;72:193–198. [Google Scholar]
- Shi Y, Gornall RJ, Draper J, Stace CA. Intraspecific molecular variation in Hieracium sect. Alpina (Asteraceae), an apomictic group. Folia Geobotanica Phytotaxonomica. 1996;31:305–313. [Google Scholar]
- Soltis DE, Mavrodiev EV, Doyle JJ, Rauscher J, Soltis PS. ITS and ETS sequence data and phylogeny reconstruction in allopolyploids and hybrids. Systematic Botany. 2008;33:7–20. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Statsoft. Statistica for Windows. Tulsa: Statsoft Inc; 1984–2002. [Google Scholar]
- Suda J, Krahulcová A, Trávníček P, Rosenbaumová R, Peckert T, Krahulec F. Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Annals of Botany. 2007;100:1323–1335. doi: 10.1093/aob/mcm218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S. Developmental basis of interspecific differences in leaf size and specific leaf area among C3 grass species. Functional Ecology. 2005;19:916–924. [Google Scholar]
- Swift H. The constancy of deoxyribose nucleic acid in plant nuclei. Proceedings of the National Academy of Sciences of the USA. 1950;36:643–654. doi: 10.1073/pnas.36.11.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šiško M, Ivanič A, Bohanec B. Genome size analysis in the genus Cucurbita and its use for determination of interspecific hybrids obtained using the embryo-rescue technique. Plant Science. 2003;165:663–669. [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. Genome size and CG content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany. 2008;101:421–433. doi: 10.1093/aob/mcm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štorchová H, Hrdličková R, Chrtek J, Jr, Tetera M, Fitze D, Fehrer J. An improved method of DNA isolation from plants collected in the field and conserved in saturated NaCl/CTAB solution. Taxon. 2000;49:79–84. [Google Scholar]
- Štorchová H, Chrtek J, Jr, Bartish IV, Tetera M, Kirschner J, Štěpánek J. Genetic variation in agamospermous taxa of Hieracium sect. Alpina (Compositae) in the Tatry Mts (Slovakia) Plant Systematics and Evolution. 2002;235:1–17. [Google Scholar]
- Teoh SB, Rees H. Nuclear DNA amounts in populations of Picea and Pinus species. Heredity. 1976;36:123–137. [Google Scholar]
- Thalmann C, Guadagnuolo R, Felber F. Search for spontaneous hybridization between oilseed rape (Brassica napus L.) and wild radish (Raphanus raphanistrum L.) in agricultural zones and evaluation of the genetic diversity of the wild species. Botanica Helvetica. 2000;111:107–119. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthawat-Jónsson K, et al. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. The Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhar B, Vidic T, Jogan N, Dermastia M. Genome size and the nucleolar number as estimators of ploidy level in Dactylis glomerata in the Slovenian Alps. Plant Systematics and Evolution. 2002;234:1–13. [Google Scholar]
- Vladimirov V. A new diploid Hieracium (Asteraceae: Lactuceae) from Bulgaria. Botanical Journal of the Linnean Society. 2003;143:213–218. [Google Scholar]
- Vladimirov V, Szeląg Z. A new diploid species of Hieracium sect. Pannosa (Asteraceae) from Bulgaria. Botanical Journal of the Linnean Society. 2006;150:261–265. [Google Scholar]
- Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany. 2006;93:148–156. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC, Johnston S, Price HJ. Feast and famine in plant genomes. Genetica. 2002;115:37–47. doi: 10.1023/a:1016020030189. [DOI] [PubMed] [Google Scholar]
- Wicker T, Keller B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Research. 2007;17:1072–1081. doi: 10.1101/gr.6214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KH. In: Hieracium. Engler A, editor. Leipzig: Wilhelm Engelmann; 1921–1923 Das Pflanzenreich 75, 76, 77, 80, 82 (IV/280) [Google Scholar]
- Záveský L, Jarolímová V, Štěpánek J. Nuclear DNA content variation within the genus Taraxacum (Asteraceae) Folia Geobotanica. 2005;40:91–104. [Google Scholar]
- Zonneveld BJM. Nuclear DNA content of all species of Helleborus (Ranunculaceae) discriminate between species and sectional division. Plant Systematics and Evolution. 2001;229:125–130. [Google Scholar]