Abstract
Background and Aims
Gagea is a Eurasian genus of petaloid monocots, with a few species in North Africa, comprising between 70 and approximately 275 species depending on the author. Lloydia (thought to be the closest relative of Gagea) consists of 12–20 species that have a mostly eastern Asian distribution. Delimitation of these genera and their subdivisions are unresolved questions in Liliaceae taxonomy. The objective of this study is to evaluate generic and infrageneric circumscription of Gagea and Lloydia using DNA sequence data.
Methods
A phylogenetic study of Gagea and Lloydia (Liliaceae) was conducted using sequences of nuclear ribosomal internal transcribed spacer (ITS) and plastid (rpl16 intron, trnL intron, trnL-F spacer, matK and the psbA-trnH spacer) DNA regions. This included 149 accessions (seven as outgroups), with multiple accessions of some taxa; 552 sequences were included, of which 393 were generated as part of this research.
Key Results
A close relationship of Gagea and Lloydia was confirmed in analyses using different datasets, but neither Gagea nor Lloydia forms a monophyletic group as currently circumscribed; however, the ITS and plastid analyses did not produce congruent results for the placement of Lloydia relative to the major groups within Gagea. Gagea accessions formed five moderately to strongly supported clades in all trees, with most Lloydia taxa positioned at the basal nodes; in the strict consensus trees from the combined data a basal polytomy occurs. There is limited congruence between the classical, morphology-derived infrageneric taxonomy in Gagea (including Lloydia) and clades in the present phylogenetic analyses.
Conclusions
The analyses support monophyly of Gagea/Lloydia collectively, and they clearly comprise a single lineage, as some previous authors have hypothesized. The results provide the basis for a new classification of Gagea that has support from some morphological features. Incongruence between plastid and nuclear ITS results is interpreted as potentially due to ancient hybridization and/or paralogy of ITS rDNA.
Key words: Gagea, Lloydia, Liliaceae, ITS ribosomal DNA sequences, plastid DNA sequences
INTRODUCTION
Gagea (Liliaceae) is a geophytic, perennial, largely Eurasian genus with a few species in North Africa; it comprises somewhere between 70 and approx. 275 species, depending on the author (Stroh, 1937; Uphof, 1958–1960; Melchior, 1964; Willis, 1980; Hyam and Pankhurst, 1995; Mabberley, 1997; Levichev, 1999; Peruzzi, 2003; Govaerts, 2006; Zarrei et al., 2007; Peterson et al., 2008). At the time of original publication (Salisbury, 1806), the genus contained only seven species; these had long been placed in Ornithogalum L. (Hyacinthaceae; e.g. Gerard, 1663; Linnaeus, 1753, 1762; Pallas, 1773, 1776). Salisbury did not take into consideration priority of the names he used, and this was a source of confusion for later authors (Heyn and Dafni, 1971). Many novel species have been added, particularly in the last two decades (Levichev, 1981, 1988, 1991, 2000, 2001, 2006a; Dasgupta and Deb, 1983; Rechinger, 1986; Levichev and Navruzshoev, 1997; Tison, 2004; Zhao and Zhao, 2004; Henker, 2005; Zarrei and Zarre, 2005a; Ali, 2006; Levichev and Ali, 2006; Zhao and Yang, 2006; Peruzzi et al., 2007; Hamzaoğlu et al., 2008), but a comparative systematic study is needed to elucidate relationships of species and species groups and to clarify taxonomic boundaries.
The first attempt to classify species within Gagea was carried out by Koch (1849), who divided the genus into two sections, Holobolbos K.Koch and Didymobolbos K.Koch, and recognized 17 species; this was followed by the addition of sections Tribolbos Boiss. and Platyspermum Boiss. (Koch, 1882; Table 1). At the beginning of the 20th Century, Terracciano (1905a, b, 1906) and Pascher (1904, 1907) made major contributions by revising the Asiatic species. Both authors erected independent classifications nearly simultaneously. Pascher (1904; Table 1) classified the species of Gagea into two subgenera, Gagea (Eugagea Pascher, which is properly subgenus Gagea; McNeill et al., 2006) with four sections and Hornungia (Bernh.) Pascher with two sections, based mainly on seed shape and bulb characters. Three years later, Pascher (1907) published a more extensive treatment with the addition of new species and complete Latin descriptions, but he maintained his previous infrageneric taxa with some additional subsections (Table 1). Terracciano (1905a, b, 1906) also adopted two subgenera, Gagea and Gageastrum, with two sections under each (Table 1). Neither author produced a complete revision. However, Pascher's treatment was favoured by several authors, including Stroh (1937), Uphof (1958–1960) and Rechinger (1986). Two major monographic classifications of Gagea were produced by Stroh (1937) and Uphof (1958–1960), with 124 and 106 species names, respectively, each representing an updated version of Pascher's (1907) classification. Following the classification of Pascher (1904, 1907), Stroh and Uphof divided Gagea into his subgenera, sections and subsections. Because many new descriptions have since appeared, particularly from the Middle East and Central Asia, the species treatments of Stroh (1937) and Uphof (1958–1960) are now substantially out of date. Although there has been no attempt to revise them completely, there are some more recent regional classifications. For example, after studying morphology and karyology, Davlianidze (1976) treated 26 species present in the Caucasus and accepted the two subgenera previously followed by Uphof (1958) and Stroh (1937); he also established six new sections within each of those subgenera (Table 1). Levichev (1990) published a new classification of the western Tien Shan species using general morphological features as well as cross-sections of radical leaves and stem-base characters. Levichev (1990) did not use the subgeneric rank and divided the genus into ten sections, some of which were the same as those of Davlianidze (1976). Apart from Davlianidze (1976) and Levichev (1990), who published new classifications, several authors (Heyn and Dafni, 1971, 1977; Dasgupta and Deb, 1983; Feinburn-Dotham, 1986; Wendelbo and Rechinger, 1990; Federov, 2001; Grubov and Egorova, 2003) have accepted the general outline of Pascher's infrageneric classification (1904, 1907) with few modifications. However, recent papers such as Zarrei and Zarre (2005a, b), Peruzzi et al. (2008a, b) and Peterson et al. (2008) have instead referred to Levichev's classification (Levichev, 1990).
Table 1.
Overview of previous infrageneric classifications of Gagea
| Boissier (1882) | Terracciano (1905a, b) | Pascher (1904, 1907)* | Grossheim (1935) | Davlianidze (1976) | Levichev (1990, 1999b)† |
|---|---|---|---|---|---|
| Gagea | Gagea | Gagea | Gagea | Gagea | Gagea |
| Subgenus | Subgenus | Subgenus | Subgenus | ||
| Gagea | Gagea (= Eugagea) | Gagea | Gagea | ||
| Section | Section | Section | Section | Section | Section |
| Gagea | Gagea | Gagea | Gagea | Gagea | Gagea |
| ( = Holobolbos) | (=Nudiscaposae) | (Holobolbos) | (=Nudiscaposae) | ||
| Tribolbos | Tribolbos | ||||
| Monophyllos | |||||
| Subsection | |||||
| Minimae | Minimae | Minimae | |||
| Spathaceae | |||||
| Fistulosae | Fistulosae | Fistulosae | |||
| Didymobolbos | Foliatae | Didymobolbos | Foliatae | Didymobolbos | Didymobolbos |
| Gageastrum | Hornungia | ||||
| Platyspermum | Verticillatae | Platyspermum | Platyspermum | Platyspermum | Platyspermum |
| Graminifoliae | |||||
| Incrustatae | |||||
| Bulbiferae | |||||
| Stipitatae | Stipitatae | Stipitatae | |||
| Dschungaricae | |||||
| Plecostigma | Plecostigma | Plecostigma | |||
| Anthericoides | Anthericoides |
The classifications are complete only at subgeneric and sectional levels. The only subsections listed are those later raised to section level by Davlianidze (1976).
* Stroh (1937) and Uphof (1958–1960) followed the same classification as Pascher (1904, 1907). See text for details.
† Levichev's classification (1990, 1999b, 2006b) has been updated by Levichev in Peterson et al. (2008).
Lloydia Salisb., a small bulbiferous herb from the temperate Northern Hemisphere, has always been considered the closest relative of Gagea. Lloydia consists of 12–20 species (Willis 1980; Hyam and Pankhurst, 1995; Mabberley, 1997; Govaerts, 2006) that have a mostly eastern Asian distribution. The only species that occurs in Europe is L. serotina (L.) Rchb., which is also distributed in western North America (Phillips and Rix, 1989). It is a protected species in Britain and has some ornamental use, unlike most species of Gagea. Lloydia replaces Gagea in the Himalayas and adjoining areas, although some species of Gagea reach Japan [e.g. G. lutea (L.) Ker-Gawl]. The taxonomic status of Lloydia has been problematic since its description by Salisbury and validation by Reichenbach in 1830 (Dasgupta and Deb, 1986). Many species have been moved between Gagea and Lloydia in the last two centuries. For example, L. libanotica Hochst. and L. graeca (L.) Endl. ex. Kunth are now known as G. libanotica (Hochst) Greuter and G. graeca (L.) Irmisch, respectively (Greuter, 1970).
The study of core Liliales conducted by Patterson and Givnish (2002) using a combined sequence matrix of plastid rbcL and ndhF genes (with only one accession each from Gagea and Lloydia) showed that these two are sister taxa in a highly supported clade [bootstrap percentage (BP) 100]. Rønsted et al. (2005) produced the same result for Gagea wilczekii (= G. algeriensis) and Lloydia serotina using a different plastid dataset (matK, trnK intron). Another analysis of psbA-trnH and trnL-F sequence data (Peterson et al., 2004) confirmed the monophyly of seven species of Gagea and Lloydia serotina from Germany, with BPs of 99 and 100 using different datasets.
Analysis based on phenotypic (morphological) data (Patterson and Givnish, 2002) resulted in a weakly supported clade (BP 56) containing Gagea and Lloydia. This low support may be because they included all genera of Liliales and did not include additional characters that are specific to tribe Tulipeae, in which both Gagea and Lloydia are placed.
A detailed examination of pollen morphology of Iranian representatives of Gagea (Zarrei and Zarre, 2005b) revealed that sculpturing of the exine provides valuable characters for separation of species, sometimes even closely related ones, and delimitation of natural groups within the genus. Zarrei and Zarre (2005b) distinguished four basic pollen types within Gagea.
Chromosome counts for 100 taxa have been reported (Peruzzi, 2003, 2008; Peruzzi and Aquaro, 2005). The base chromosome number is x = 12 among species of known chromosome number, and 37·8 % of the studied species have this number and are diploid (Peruzzi, 2003). Chromosome studies suggested that asymmetric karyotypes are an ancestral feature, whereas more balanced ones are derived (Peruzzi and Aquaro, 2005), but this hypothesis needs reconsideration within a phylogenetic framework.
Molecular phylogenetic studies (mostly of plastid DNA) that have included single exemplars of Gagea [Gagea wilczekii Braun-Blanq. & Maire (= Gagea algeriensis Chabert)] and Lloydia (L. serotina) support a close relationship of the genera, but provide no insight into generic circumscription (Kosenko and Levichev, 1988; Kosenko, 1999; Fay and Chase, 2000; Patterson and Givnish, 2002; Rønsted et al., 2005). A molecular phylogenetic study of seven Gagea species from Germany was undertaken by Peterson et al. (2004), using plastid DNA sequences (trnL intron, trnL-F spacer and the psbA-trnH spacer) and the nuclear the internal transcribed spacer (ITS) ribosomal region. In this analysis, L. serotina was used initially as outgroup, but it was placed among the Gagea species in all analyses of plastid data. Subsequent analyses that included morphological data also cast doubt on the validity of maintaining Lloydia and Gagea as distinct genera (Peterson and Peterson, 2005, 2006). Combined analyses of plastid and ITS DNA demonstrated that G. section Didymobolbos forms a clade with G. section Monophyllos sensu Pascher (in particular with G. section Minimae and G. section Euspathaceae sensu Levichev) and that G. section Gagea (Holobolbos sensu Pascher) forms a clade with G. section Tribolbos. Indeed, the last section has been merged by recent authors with G. section Gagea (Levichev, 1990; Peruzzi & Aquaro, 2005; Peruzzi et al., 2007). Molecular and morphological study of Gagea and Lloydia has been conducted by Peterson et al. (2008), revealing a close relationship between these two genera and further undermining the concept of Lloydia as a distinct genus. Moreover, these studies broadly supported Levichev's classification (Levichev, 1990). Peruzzi et al. (2008a) proposed the genus Lloydia as a section within Gagea. A detailed phylogenetic study of Gagea species in Italy has also been conducted by Peruzzi et al. (2008b).
To further evaluate phylogenetic relationships of Lloydia and Gagea and the infrageneric classification of Gagea, nuclear (ITS) and plastid (rpl16 intron, trnL intron, trnL-F spacer, matK and the psbA-trnH spacer) DNA data are used here. We have included species representing as many morphologically based species-groups of Lloydia and Gagea as possible and also aimed for broad geographical sampling of species. The phylogenetic analyses include previously published DNA data and address relationships between and within the two genera; the results are also compared with previous classifications of Gagea.
MATERIALS AND METHODS
Plant material
Silica gel-preserved samples of leaf tissue from field collections and, in a few cases, herbarium specimens were used for DNA extraction (see Appendix for source information). The ingroup comprised 142 accessions. In all analyses, Tulipa clusiana DC., T. lehmanniana Merckl., T. uniflora (L.) Besser ex Baker, Amana erythronioides (Baker) D.Y.Tan & D.Y.Hong, Erythronium japonicum Decne., Fritillaria persica L. and Lilium ledebourii (Baker) Boiss. served as outgroups, based on the results of Rønsted et al. (2005).
DNA extraction, marker amplification and sequencing
Genomic DNA extractions were performed using 0·01–0·23 g of silica-dried leaves or 0·01–0·08 g of leaf tissue from herbarium sheets and a modified version of the 2× CTAB method of Doyle and Doyle (1987). Before precipitation, an aliquot of 150 µL was purified using the NucleoSpin Extract II PCR purification kit (Machery-Nagel, GmbH & Co. KG, Düren, Germany) following the manufacturer's protocols; this provided a small amount of DNA that was able to be used the same day for amplification. The remainder of the DNA was precipitated in 2·5 volumes ethanol (for herbarium specimens, 2/3 volume isopropanol was used instead of ethanol). DNA samples were then purified using a caesium chloride/ethidium bromide gradient (1·55 g mL−1) followed by removal of the ethidium bromide with butanol, dialysis and storage at −80 °C in the DNA Bank at the Royal Botanic Gardens, Kew (http://www.data.kew.org/dnabank/homepage.html).
Amplification of the psbA-trnH spacer was undertaken using previously published primers for psbA (Sang et al., 1997) and trnH (Tate and Simpson, 2003). Owing to the small size of this fragment (339–438 bp), only the psbA primer was used for sequencing, unless there were ambiguities that needed resolving in the single electropherogram produced. Amplification of the rpl16 intron was carried out using the primers 71F and 1661R of Jordan et al. (1996). In many cases the internal primer 158F, designed originally for palms (5′-AAGAAACAGTCACTATATGA-3′; C. Asmussen, University of Copenhagen, unpubl. res.), was used to avoid a long region of T/A, which interfered with sequencing at the beginning of the rpl16 intron. For degraded DNA from herbarium material, two internal primers were designed for this project, 576F (5′-GATGGCGGAATGAACCAAGA-3′) and 657R (5′-GTTTCGCGGGCGAATAT TGACT-3′), and both were used to amplify the rpl16 intron in two pieces, in this case 71F + 657R and 576F + 1661R. These primers were also used for sequencing.
The trnL-F region (including trnL intron and trnL-F spacer) was amplified with primers c and f of Taberlet et al. (1991). In some older herbarium material, the trnL-F region was amplified in two pieces using primers d and e designed by Taberlet et al. (1991; c + d and e + f).
In a similar way, matK was amplified in two pieces using primers 19F (Molvray et al., 2000) and 1326R (Sun et al., 2001) for the first piece and 390F (Sun et al., 2001) and 1565R (5′-TCACCAGGTCATTGACACGAA-3′), which we designed for this study. In three cases, 2R (Johnson and Soltis, 1994) was used instead for the reverse primer. For sequencing, 19F and 1326R were used for the first fragment, but only 1565R was used for the second piece because of the large degree of overlap of the two fragments.
Amplification of the ITS region of 18S–26S nuclear ribosomal DNA was carried out using primers 17SE and 26SE of Sun et al. (1994). Primers ITS2, ITS3, ITS4 and ITS5 (White et al., 1990) were used for herbarium material to amplify ITS in two pieces (ITS5 + ITS2, ITS3 + ITS4). DMSO (2 %; dimethylsulfoxide) was added to reduce secondary structure problems common in ITS (Winship, 1989; Baldwin et al., 1995; Chase et al., 2003). In all cases, amplified products were purified using NucleoSpin PCR purification columns in accordance with the manufacturer's protocols. Cycle sequencing reactions were performed using the BigDye Terminator Kit ver. 3.1 (Applied Biosystems, Inc., ABI, Warrington, UK). Cycle sequencing products were cleaned using Magnesil (Promega product, Southampton, UK) on a Beckman Coulter robot (Biomek NX S8, Buckinghamshire, UK) following the manufacturer's protocols. Cleaned products were then sequenced on an ABI 3730 following the manufacturer's protocols.
Sequence alignment and phylogenetic analysis
For this paper, 393 new sequences were generated; 159 sequences of ITS, psbA-trnH and trnL-F intergenic spacer (IGS) of some taxa were downloaded from GenBank (http://www.ncbi.nlm.nih.gov). GenBank accession numbers for all sequences are listed in the Appendix. New DNA sequences were edited and assembled using Sequence Navigator ver. 1.0 and Autoassembler ver. 1.4.0 (ABI), respectively. All sequences were easily aligned by eye using PAUP v. 4.0b10 for Macintosh (Swofford, 2002), following the guidelines of Kelchner (2000). The matrices are available as NEXUS files upon email request from M.W.C. or M.Z. Parsimony analyses were undertaken using PAUP v. 4.0b10 for Macintosh (Swofford, 2002). All changes were assessed as unordered and were equally weighted (Fitch parsimony; Fitch, 1971).
The data were analysed in three steps. First (analysis I; results not shown), all data were analysed as separate plastid regions. Species for which sequences were taken from GenBank had significant missing data for these plastid regions (e.g. no sequences were available for matK and rpl16 intron in GenBank). BPs were low, which is why these are not shown; these analyses were performed to determine the degree to which our plastid sequences and those previously published were in agreement, which they generally were. Then, we ran analyses of newly generated sequences only as separate plastid and ITS matrices (i.e. no separate analyses of the individual plastid regions; analysis II). Thirdly, combined analyses were run for all data (ITS plus plastid regions) generated in this paper plus the sequences downloaded from GenBank (analysis III). We were worried that the amount of missing data would make evaluation of incongruence and internal support difficult; missing data might reduce bootstrap support and thus might conceal hard incongruence (Cameron et al., 2001). Thus, we analysed all data produced only by us as combined plastid and ITS matrices on only the sequences generated for this study, which are with few exceptions (Appendix) complete for each of our accessions (analysis IV).
All searches were conducted using 1000 random taxon-addition replicates, tree-bisection-reconnection (TBR) branch swapping and MulTrees on (i.e. keeping multiple, equally parsimonious trees). Ten trees only were saved from each replicate to reduce search time on potentially thousands of trees. All trees collected were then used as starting trees in another search without a tree limit. Support for clades was estimated using 1000 bootstrap replicates (Felsenstein, 1985), with simple taxon addition, and TBR swapping but permitting only ten trees per replicate to be held. Groups were retained with BP ≥ 50. Summary data for all analyses are presented in Table 2.
Table 2.
Tree and matrix statistics related to the various datasets and analysis
| Analyses | No. of positions | No. of variable positions | No. of parsimony-informative positions | No. of trees | Length | CI | RI |
|---|---|---|---|---|---|---|---|
| ITS | 706 | 312 (44 %) | 253 (36 %) | 8152 | 850 | 0·62 | 0·89 |
| Plastid | 3474 | 727 (21 %) | 507 (15 %) | 7620 | 1115 | 0·75 | 0·93 |
| Combined total* | 4180 | 1039 (25 %) | 760 (18 %) | 7276 | 1993 | 0·68 | 0·91 |
| Combined total† | 4180 | 1088 (26 %) | 810 (19 %) | 6430 | 2388 | 0·62 | 0·90 |
* Only newly generated datasets.
† Whole datasets including sequences from GenBank.
RESULTS
Analysis I
The trees obtained from separate analyses of the plastid regions of all available datasets show limited resolution, and therefore these trees are not presented. Statistics from these separate plastid analyses are shown in Table 2.
Analysis II, ITS
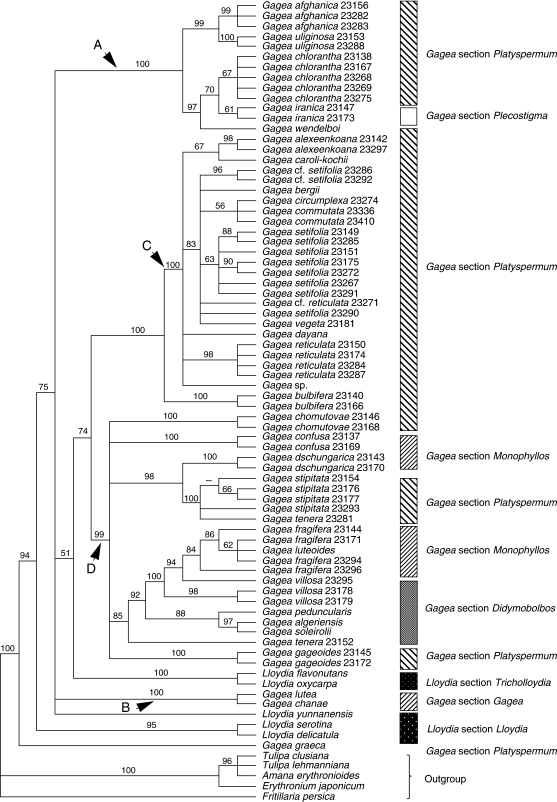
Analysis of 78 ITS sequences yielded 8152 equally most-parsimonious trees, each of length (L) = 850 steps, consistency index (CI) = 0·62 and retention index (RI) = 0·89. Tree and matrix statistics are presented in Table 2. One of the most-parsimonious trees was randomly selected and is shown in Fig. 1. Gagea and Lloydia collectively are supported as monophyletic within Liliaceae (BP 100), but neither is monophyletic (Fig. 1). Species of Lloydia are dispersed throughout the tree. Lloydia serotina and L. delicatula Noltie comprise a well-supported clade (BP 95). Lloydia flavonutans and L. oxycarpa also form a pair (BP 100). Apart from Gagea graeca, which is sister to the rest of the ingroup (BP 94), all other members of Gagea form four well-supported clades (clades A–D; BPs = 99–100). These five Gagea clades (including G. graeca) were recovered in all the different analyses presented here. Bootstrap support for each clade is high in every tree. However, there are a small number of soft incongruences regarding the placement of some constituent taxa of those clades, and these are discussed below.
Fig. 1.
Bootstrap percentages (≥50) are indicated above branches on the strict consensus tree of 8152 trees obtained from analysis of nuclear ribosomal ITS (sequences from GenBank are excluded). Branches with a hyphen have BP < 50.
Analysis II, plastid regions
Analyses of the newly generated data for plastid regions included 87 accessions, of which seven were outgroups (tree and matrix statistics are presented in Table 2). The strict consensus tree with BPs is presented in Fig. 2. The ingroup is strongly supported (BP 100). Neither Gagea nor Lloydia accessions form monophyletic groups. The topology of the tree differs from that obtained from analysis of the combined matrix (see Fig. 4) regarding the relative positions of Lloydia taxa. All Lloydia accessions and G. graeca form a grade within a larger, moderately supported clade (BP 81; Fig. 2) that also includes clade A. Although the major clades A–D are strongly supported (BP = 90–100), they form a polytomy in the strict consensus tree.
Fig. 2.
Bootstrap percentages (≥50) are indicated above branches on the strict consensus tree of 7620 trees obtained from analysis of the combined plastid data matrix (rpl16 intron, trnL intron, trnL-F spacer, matK and the psbA-trnH spacer; sequences from GenBank are excluded). Branches with a hyphen have BP < 50. Pascher's classification (1904, 1907) is shown next to the tree.
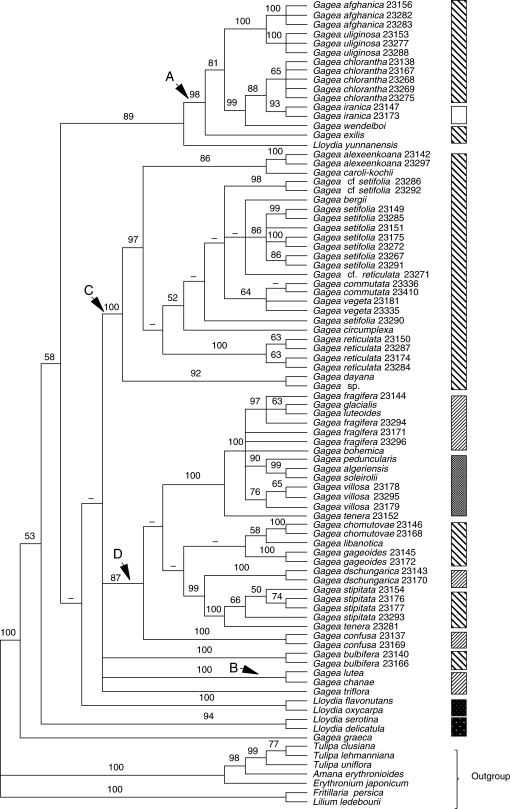
Fig. 4.
The strict consensus of 7276 trees obtained from analysis of the combined data matrix (nuclear ITS rDNA and plastid sequences; sequences from GenBank are excluded). Bootstrap percentages (≥50) are indicated above branches. Branches with a hyphen have BP < 50.
Combined matrix of all datasets including sequences from GenBank (analysis III)
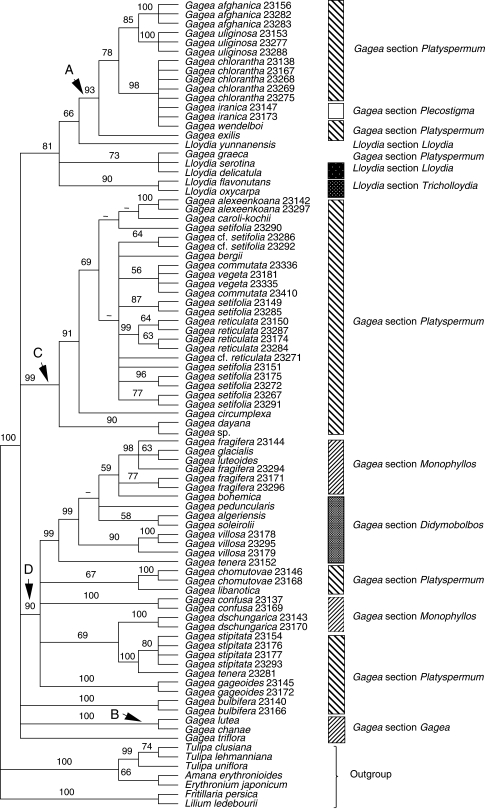
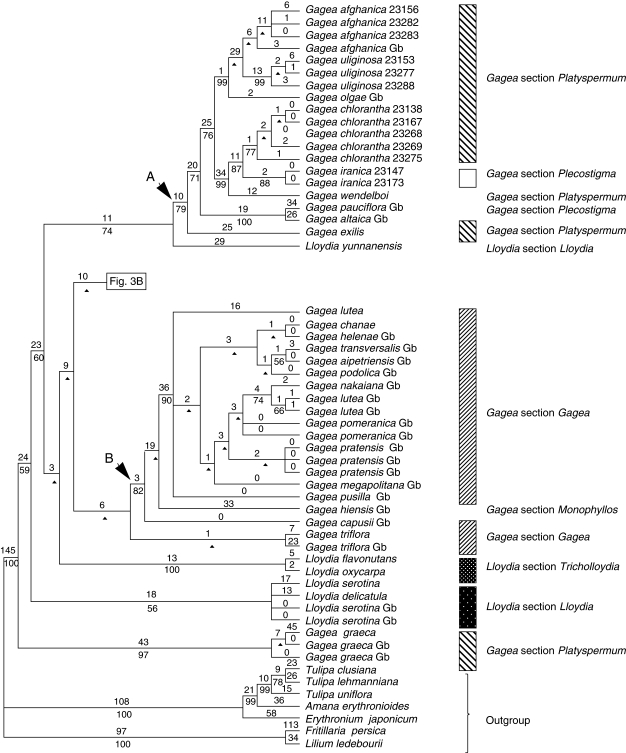
The combined matrix included 135 accessions of Gagea, seven of Lloydia and seven outgroups species. Tree and matrix statistics are presented in Table 2. Figure 3A and B show one of the most-parsimonious trees selected randomly from 6430 trees. In this analysis, Lloydia and Gagea together comprise a strongly supported clade (BP 100; Fig. 3A, B). All G. graeca accessions are collectively sister to all other accessions of Lloydia and Gagea, but with low support (BP 59).
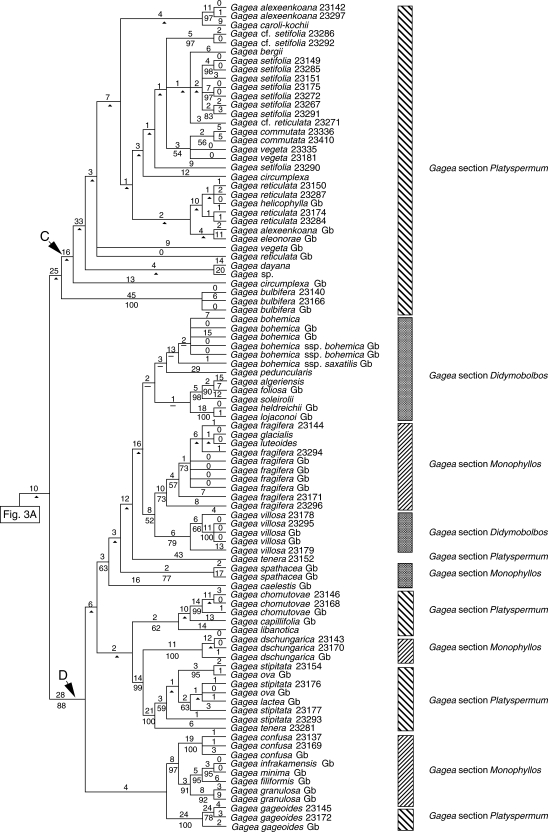
Fig. 3.
One of the most-parsimonious trees, randomly selected from 6430 trees, obtained from analysis of the combined data matrix (nuclear ribosomal ITS, and plastid rpl16 intron, trnL intron, trnL-F spacer, matK and the psbA-trnH spacer; sequences from GenBank are included). Tree length = 2388, CI = 0·62, RI = 0·90. Branch lengths (DELTRAN optimization) are indicated above branches and bootstrap percentages below. An arrowhead indicates nodes collapsing in the strict consensus of all most-parsimonious trees. Branches with a hyphen have BP < 50. Gb after the species names indicates those taken from GenBank. For cases in which more than one accession of a species were analysed, numbers after the species names are RBG, Kew, DNA Bank accession numbers (see Appendix).
The spine of the tree is poorly resolved, but multiple accessions of the same species and groups of closely related species form clades, in some cases with moderate to strong bootstrap support. Clades A, B and D are well supported, but apart from within clade A, taxa within these clades mainly form polytomies. These polytomies usually comprise species that morphology indicates are closely related.
Lloydia species occur in three distinct parts of the trees. Lloydia serotina and L. delicatula are among a weakly supported polytomy towards the basal nodes of the tree. However, L. yunnanensis Franch. is sister to other accessions in clade A (BP 79; Fig. 3A). Lloydia oxycarpa Franch. and L. flavonutans H.Hara form a clade (BP 100).
Combined matrix of newly generated sequences (analysis IV)
Owing to the amount of missing data for taxa obtained from GenBank, separate analyses were performed using only newly generated sequences (tree and matrix statistics presented in Table 2). Bootstrap percentages (BP ≥ 50) are shown on the strict consensus tree (Fig. 4). The tree generally has the same topology as that obtained from analysis of the ITS matrix (Fig. 1). However, there is incongruence between them in the positions of G. bulbifera (Pall.) Salisb. and L. yunnanensis. Clade support is relatively high in comparison with those in Fig. 3A and B; for example, clade C, which is completely unresolved in Fig. 3B, has a BP of 100 in Fig. 4. Moreover, clade membership is not identical in clade C in all the analyses. However, there is weak support (BP 53) for G. graeca being sister to all other members of the ingroup.
DISCUSSION
In the present survey, sequence data were generated from both biparentally (nuclear; Álvarez and Wendel, 2003) and maternally (plastid; Bohdanowicz and Lewandowska, 1999) inherited genomes to reconstruct a phylogenetic tree for Gagea and Lloydia, controversial and difficult taxa within Liliaceae. For several species, multiple accessions were used to assess intraspecific genetic variation and species delimitation. Consideration was also given to morphological variation within species and geographical distribution when developing the taxon sampling strategy. In many cases, multiple accessions of a single species form clades, usually with high bootstrap support. However, in some species such as G. setifolia Baker, accessions are nested in separate clades or interdigitated or unresolved in groupings with accessions of other taxa that have previously been considered to be closely related.
Incongruence of plastid and ITS matrices
Owing to the unreliability of the partition homogeneity test in assessing combinability (Farris et al., 1995) as shown by several authors (Reeves et al., 2001; Yoder et al., 2001; Rønsted et al., 2005), incongruence between the plastid and ITS data was investigated by comparing the combined results (Figs 3 and 4) with the those of the separate analyses (Figs 1 and 2) with respect to level of resolution and bootstrap support. Although G. bulbifera (Pall.) Salisb. accessions form a strongly supported group with clade C (BP 100) using ITS sequence data (Fig. 1), it does not have support as sister to clade C in analyses of the plastid sequences (Fig. 2). In the combined analysis of all data, this species forms a polytomy with clades B–D (Fig. 4). Thus. its position in the plastid tree is less resolved. We interpreted this as soft rather than a hard incongruence between the ITS rDNA and plastid DNA data, which could be resolved by incorporating more data, particularly more rapidly evolving DNA regions than those used in this research.
Although all Lloydia species are part of a moderately supported group (BP 81) with clade A in the tree resulting from analyses of combined plastid data (Fig. 2), there are three distinct clades that include Lloydia species based on combined ITS and plastid sequences and also the ITS dataset alone (Figs 1, 3 and 4).
We do not have any clear hypotheses regarding the causes of the incongruence. Both ancient hybridization (with Lloydia species perhaps sharing a maternal lineage) and paralogy of ITS sequences (Doyle, 1992; Baldwin et al., 1995; Wendel et al., 1995) are possibilities. Differences in tree topologies may also be due to sampling error (too few data, in the case of Lloydia species) (see Hulsenbeck et al., 1996; Whitten et al., 2000; Rønsted et al., 2005), but these hypotheses are difficult to distinguish. We are currently sequencing several low-copy, protein-coding nuclear regions and hope that the results of these analyses can shed some light on the phenomena that may have generated the incongruence detected in our analyses. It is clear in all cases that Lloydia and Gagea are a single entity and cannot be separately recognized. Incongruence within this clade did not prevent the necessary taxonomic transfers. Peruzzi et al. (2008a) suggested the transfer of L. serotina and Lloydia delicatula to G. section Lloydia.
Monophyly of Gagea plus Lloydia
A close relationship of Gagea and Lloydia was confirmed in all of the analyses derived from the independent datasets presented here. All six species of Lloydia and all Gagea accessions form a highly supported clade (BP 100; Figs 1–4). These results are consistent with those from previous phylogenetic analyses (Patterson and Givnish, 2002; Peterson et al., 2004, 2008; Rønsted et al., 2005; Peruzzi et al., 2008).
Gagea and subgenera
The widely accepted subgeneric classification first published by Pascher (1904, 1907) and subsequently used by other botanists, e.g. Stroh (1937) and Uphof (1958–1960), is superimposed on all figures presented here with the difference that Gagea section Tribolbos has been merged with G. section Gagea in this study as in Davlianidze (1976) and Levichev (1990). According to Pascher (1904, 1907), there are two subgenera, i.e. Gagea subgenus Gagea and Hornungia (Table 1). The former is characterized by having globose, angular or edged seeds whereas the latter possesses flat, thin seeds. Neither of these subgenera is monophyletic in the present study, and sections belonging to these taxa are dispersed throughout the tree (Figs 1–4). This indicates that either or both of the two seed forms, which may have specific adaptive roles, have arisen several times during the evolution of the genus, and that these characters are not suitable for classification within Gagea due to parallelism. Most species belonging to G. subgenus Gagea, which possess thick seeds, are adapted to relatively more humid areas. They are generally found in the Euro-Siberian floristic region (Takhtajan, 1986) and less in the Irano-Turanian region; these species are restricted to higher elevations where there is more precipitation, particularly in the form of snow. The other subgenus, G. subgenus Hornungia, with flattened seeds, is adapted to drier conditions and can usually be found in the Irano-Turanian and Saharo-Arabian regions. There are a few species of this group distributed in Europe, but they are mainly restricted to southern and southeastern areas where the climate is drier.
The placement of species in clades A–D described below indicates that these subgenera are polyphyletic.
Phylogenetic relationships within Gagea and Lloydia and infrageneric classification
Although the monophyly of Lloydia/Gagea is strongly supported, with few exceptions relationships within this clade are not well resolved. Lloydia species are mostly positioned towards the base of trees and in strict consensus trees form a polytomy. This may be due to missing data in the various accessions of Lloydia (mostly obtained from herbarium material).
As was the case for Gagea, there is no consistency between classical infrageneric taxa in Lloydia and the results of our phylogenetic analyses. Lloydia section Lloydia (L. yunnanensis, L. delicatula and L. serotina included in the present study) and Lloydia section Tricholloydia Engl. (with L. oxycarpa and L. flavonutans in the present study) are the two recognized infrageneric taxa of Lloydia (Dasgupta and Deb, 1986). These species are dispersed throughout the trees without a recognizable pattern, although closely related species such as L. oxycarpa and L. flavonutans form well-supported clades (BPs 90–100; Figs 1–4). Lloydia yunnanensis is usually sister to clade A in trees from analyses of plastid sequences alone and those from combined plastid and nuclear datasets (Figs 2, 3A and 4). However, nuclear data (ITS) did not resolve it in this position.
In some analyses, L. serotina and L. delicatula are in a clade with high support (BPs 94–95; Figs 1 and 4). In analysis of the combined plastid dataset (Fig. 2), five species, i.e. G. graeca, L. serotina, L. delicatula, L. flavonutans and L. oxycarpa, form two clades that are moderately to highly supported but for which relationships to each other and clade A are not resolved. These belong to the two sections of Lloydia.
Five major monophyletic, moderately to strongly supported groups are revealed for Gagea accessions (Figs 1–4); the first clade includes only G. graeca, and the other four are referred to as clades A–D. Gagea graeca has an eastern Mediterranean distribution and is one of the few Gagea species possessing white to pale pink flowers, which resemble those of some species of Lloydia. Gagea graeca was placed under G. section Anthericoides by Terracciano (1905b) and Peterson et al. (2008). Stroh (1937) classified this species under G. section Platyspermum. The present results do not support Stroh's (1937) treatment of the species, and it reveals an isolated position for the species as suggested by Terracciano (1905b), Peruzzi et al. (2008a) and Peterson et al. (2008).
The second group comprises clade A (Figs 1–4), which includes species of G. subgenus Hornungia [excluding G. pauciflora (Turcz. ex Trautv.) Turcz. ex Ledeb., G. altaica Schischk. & Sumnev.]. All species included in clade A belong to G. section Platyspermum except G. iranica and G. pauciflora, which belong to G. section Plecostigma [according to Pascher's (1904, 1907) classification]. According to Levichev (1990), all species in this clade, apart from G. iranica and G. pauciflora, G. bulbifera, G. chlorantha and G. exilis (Figs 2–4), belong to G. section Plecostigma. Possession of a cymose inflorescence is a potential morphological synapomorphy for clade A.
In the analysis of the combined matrix of all datasets (Fig 3A), in addition to the grouping of G. pauciflora (Turcz. ex Trautv.) Turcz. ex Ledeb. and G. altaica Schischk. & Sumnev., which form a well-supported clade (BP 100; Fig. 3A), there are two well-defined subclades (BP 99) within clade A. The first subclade includes G. uliginosa, G. afghanica and G. olgae. All species of this subclade belong to G. section Platyspermum (sensu Pascher, 1907; Stroh, 1937; Uphof, 1958–1960) and G. section Plecostigma Pascher (sensu Levichev, 1990). There is little morphological similarity between the first two species; G. uliginosa has a single-flowered inflorescence (rarely with two flowers) and grows in moist meadows in alpine areas of north-western Iran, eastern Turkey and north-western Iraq; G. afghanica is a multi-flowered, cymose plant, mostly growing throughout the eastern part of Iran to Central Asia and preferring a drier habitat than G. uliginosa. Gagea olgae is morphologically similar to G. afghanica, with smaller tepals (6–9 mm). Cord-like roots around the bulb are a synapomorphy for G. afghanica and G. olgae. However, there is no resolution between G. olgae and G. afghanica and relatives in analyses using both nuclear and plastid sequence data, and the four accessions of G. afghanica and the single accession of G. olgae are unresolved in the strict consensus (Fig. 3A).
The second subclade comprises G. chlorantha, G. iranica and G. wendelboi. These species are morphologically similar. Gagea iranica and G. wendelboi are endemic species to northern and north-eastern Iran, whereas G. chlorantha is widely distributed through western Iran and other countries of the Middle East. Fewer taxa are included in the other analyses, but to the extent that they overlap in sampling, the same subclades are recovered in all analyses of clade A (Figs 1–4).
The next clade, B (BP 82, in Fig. 3A and BP 100, in Figs 1, 2 and 4), is moderately to strongly supported, although only two taxa are included in Figs 1, 2 and 4. An umbellate inflorescence plus a leathery bulb tunic are potential morphological synapomorphies for this clade. The species forming this clade all grow in humid areas. Relationships between members of clade B are poorly resolved in the analysis conducted using all datasets including sequences from GenBank (Fig. 3A). Clade B accessions are members of two sections, G. sections Gagea and Monophyllos, sensu Pascher (1907) Stroh (1937) and Uphof (1958–1960), compared with only one section, G. section Gagea of Davlianidze (1972), Levichev (1990) and Peterson et al. (2008).
Clade C is well supported in all but the combined analysis of all datasets (BPs 99–100 in Figs 1, 2 and 4). All taxa included in clade C are characterized by having a multi-flowered, umbellate inflorescence. Possession of acute to long-acuminate tepal apices is another potential synapomorphy for this group. All taxa in clade C belong to G. section Platyspermum subsection Reticulatae Pascher of G. subgenus Hornungia (sensu Pascher, 1907; Stroh, 1937; Uphof, 1958–1960). Grossheim (1935) and Davlianidze (1976) also treated taxa of clade C as belonging to G. section Platyspermum. In contrast, Levichev (1990) classified taxa of clade C in three sections, G. section Platyspermum, G. section Graminifoliae Levichev and G. section Incrustatae Levichev, but support for this in the combined analysis of all data is weak (BP < 50), and all taxa collapse in the strict consensus tree (Fig. 3B). Gagea section Platyspermum (sensu Pascher, 1907) is characterized by having a weakly trilobed stigma and flattened seeds.
Gagea bulbifera, sister to the rest of clade C in the analysis of the ITS alone (BP 100, Fig. 1), is usually a single-flowered species. However, in some cases multi-flowered stems have been observed in the field (M. Zarrei, pers. obs.). It was placed in Gagea section Plecostigma by Levichev (1990). Peterson et al. (2008) referred G. bulbifera to a new unpublished G. section Bulbiferae based on possession of a few-flowered, paniculate inflorescence.
Although the accessions are mostly unresolved in the combined analysis of all data (Fig 3B), some well-supported monophyletic groups, usually including multiple accessions of one species, are recognizable within clade C. As shown in Fig. 4, G. alexeenkoana Miscz. and G. caroli-kochii Grossh. form a moderately supported clade (BP 86), and they are morphologically similar taxa. Gagea caroli-kochii is more slender and smaller than G. alexeenkoana and possesses a narrower basal leaf and shorter, narrower tepals. In contrast to our accessions of G. alexeenkoana, G. alexeenkoana from GenBank formed a clade with accessions of G. reticulata sensu lato (s.l.) (Fig. 3B), and this accession might be just a robust form of G. reticulata (Pall.) Schult. & Schult.f., a morphologically polymorphic species. Gagea reticulata s.l. accessions form a clade with strong BP support (Figs 1, 2 and 4; BP > 97). This species can be recognized by its long, reticulate, multi-layered neck, single to multi-flowered umbellate inflorescence and tepals with long-acuminate apices. However, it is a polymorphic species with regards to morphological features, and different forms have been designated as distinct species by many authors. Clumped forms with a circinate, narrowly linear basal leaf are recognized as G. tenuifolia (Boiss.) Fomin (our accessions 23287, 23174 and 23284), solitary plants with a straight, linear basal leaf as G. reticulata sensu stricto (s.s.) (23150), and forms with a shorter bulb neck, broader basal leaf and tepals are recognized as G. tehranica Gand. (not included in the present study). Although lacking support, an accession of Gagea helicophylla Levichev from GenBank is also positioned in the G. reticulata group (Fig. 3B). Although G. reticulata s.l. accessions fall into two weakly supported groups (BP approx. 64; Figs 2 and 4), these clades do not appear to us to be referable to any named taxa.
Gagea setifolia s.l. accessions are dispersed throughout clade C. This species, like G. reticulata, shows considerable morphological variation (Peruzzi and Zarrei, 2007), and many forms have been designated as species by some authors (e.g. G. anonyma Rech f. and G. perpusilla Pascher). In analyses of the G. setifolia complex, there is thus support for recognition of some forms that grow in similar ecological conditions. Gagea setifolia 23272 (from the locus classicus of G. anonyma) and G. setifolia 23175 collected from fine sand habitats form a strongly supported group in all analyses (BPs 90–100); this group corresponds to G. anonyma. Gagea setifolia 23291 and 23267 share the same habit as 23272, but they are slightly more robust and grow in soils of coarse sand. These two samples, determined as G. setifolia s.s., also form a monophyletic group in analyses including new plastid data and combined datasets (BPs 77 and 86 in Figs 2 and 4, respectively). However, there is no support for this group in the ITS analysis (Fig. 1). The two accessions of G. cf. setifolia (23286 and 23292) that are morphologically similar to G. setifolia s.s. but differ from it in having broader bracts and a longer-necked tunic fall outside the main G. setifolia clade (Fig. 4). Gagea bergii Litv. also groups with G. setifolia s.l., but it is morphologically distinct from it. It has a short peduncle and long pedicels with a long-villous indumentum. Gagea vegeta and G. commutata K.Koch are morphologically similar and form a clade in the plastid and plastid plus ITS analyses (BPs 56 and 64 in Figs 2 and 4, respectively). Gagea vegeta is the only species of G. section Graminifoliae (sensu Levichev, 1990) included in this analysis. The next species nested in clade C is G. circumplexa Vved. Levichev (1990) placed this species in a separate section, G. section Incrustatae, the only species of this section included in the present analyses, but other authors have classified it in G. section Platyspermum of G. subgenus Hornungia. Gagea circumplexa falls towards the basal nodes of clade C (Figs 2 and 4), but its position is not well supported.
The last monophyletic group within Gagea is clade D (BPs 87–99; Figs 1–4). The presence of a leathery, dark-grey to dark-brown tunic is a potential morphological synapomorphy for this clade. Although clade D includes several moderately to well-supported clades, the deeper nodes are unresolved. Species included in clade D belong to G. sections Didymobolbos and Monophyllos of G. subgenus Gagea and G. section Platyspermum of G. subgenus Hornungia (Fig. 3B). Three subclades include members of G. subgenus Hornungia (labelled as section Platyspermum in Figs 1–5), i.e. G. gageoides (Zucc.) Vved., the clade from G. libanotica to G. chomutovae and the grouping from G. tenera 23281 to G. stipitata 23154. Clade D also includes a well-supported grouping of G. granulosa Gb to G. confusa Gb 23137, which are morphologically similar, sharing a flattened basal leaf and umbellate to subumbellate inflorescences (Fig. 3B). All of these species belong to G. section Monophyllos subsection Minimae Pascher, later promoted to sectional rank by Davlianidze (1976; G. section Minimae (Pascher) Davlianidze).
The grouping from Gagea tenera 23281 to G. dschungarica 23143 is a well-supported group (BP 99) within clade D based on combined analysis of all datasets (Fig. 3B). To the extent that there is overlap in sampling, this group is recovered in all analyses (Figs 1–4). Gagea dschungarica Regel (G. section Monophyllos subsection Minimae) is morphologically distinct relative to the rest of the members of this group, which are all members of G. section Platyspermum subsection Stipitatae Pascher (G. section Stipitatae sensu Davlianidze, 1976; Levichev, 1990). Gagea dschungarica has recently been placed in a new G. section Dschungaricae Levichev (Peterson et al., 2008). However, we do not believe that creating a new section for a taxon that is sister to the rest of the clade enhances systematic understanding. The next group within clade D includes Gagea caelestis GB–Gagea bohemica, which is weakly supported (BP 63; Fig. 3B). This group includes representatives from three sections, G. section Platyspermum, Monophyllos and Didymobolbos, and is recovered in all analyses. The production of bulbils in the axil of the lower cauline leaf is a potential synapomorphy for a group comprising G. tenera and species such as G. villosa and G. fragifera (Vill.) E. Bayer & G. López.
All accessions of G. bohemica form a group with <50 % bootstrap support. All accessions of G. villosa form a moderately supported clade (BP 79; Fig. 3B). Gagea fragifera, G. luteoides Stapf and G. glacialis K.Koch (all belong to G. section Monophyllos) form a clade with moderate support (BP 73; Fig. 3B).
In the analysis conducted using only newly generated sequences, G. fragifera, G. luteoides and G. glacialis, together with G. villosa accessions and other morphologically similar species, form a strongly supported group (BPs 85–100; see Figs 1, 2 and 4). These species share a hollow basal leaf and umbellate to sub-umbellate inflorescence; they usually grow in heavy clay soils. The differences between them are so slight that it is difficult to separate them, particularly in the case of herbarium material. Gagea glacialis and G. fragifera, for example, are distinguished on the basis of tepal length (less than or more than 12 mm, respectively) and number of flowers per inflorescence. Gagea glacialis usually has fewer flowers (mostly one and rarely up to three), whereas G. fragifera has more than three flowers. The monotypic subsect. Luteoides (G. luteoides) of Pascher, which was not recognized by later authors, needs to be included in G. section Monophyllos. The species composition of clade D is compatible with neither Pascher's (1907) nor Levichev's (1990 and in Peterson et al., 2008) classification.
CONCLUSIONS
The present analyses support the collective monophyly of Gagea and Lloydia – they are clearly a single taxon. They provide a basis for a new classification of Gagea that is supported by some previously unused morphological features. Incongruence between the plastid and nuclear ITS results is interpreted as potentially due to ancient hybridization and/or paralogy of ITS rDNA. To resolve the trees, particularly along the spine of the tree and also within closely related species complexes, we will need to conduct additional analyses using more variable, low-copy nuclear genes. Such genes are not subject to concerted evolution and generally show higher evolutionary rates, which make them better tools to understand species relationships when levels of variation in plastid markers and nuclear ribosomal ITS are too low to resolve relationships and hybridization/paralogy prevent clear assessments of patterns of species evolution.
SUPPLEMENTARY DATA
The original version of this manuscript as submitted by the authors to Annals of Botany on 3 January 2007 is available as Supplementary Data online at www.aob.oxfordjournals.org.
ACKNOWLEDGMENTS
We would like to thank the Stanley Smith Horticultural Trust, Norman Scarfe Trust, Faith Raven and Paul Miles for their financial support. Thanks also to Martyn Rix for his support. Thanks to Laura Kelly for critical reading of the manuscript. At RBG, Kew, the work could not have been completed without the help of Martyn Powell, Ben Davis, Imalka Kahandawala, Maria Vorontsova, Laszlo Csiba, Dion Devey, Jim Clarkson, Rhian Smith, Ilia Leitch, Félix Forest and Lola Lledó. At the Shahed University of Tehran, we thank Iraj Rasooli, Dean of the Faculty of Science, for his valuable support. Asghar Kamrani, Taiebeh Radjabian and Amir Habibi helped in collecting material. We also thank Amots Dafni for providing additional plant materials. We wish to thank Lorenzo Peruzzi and other anonymous reviewers for their helpful comments on the manuscript. The constructive comments of Jeannette Whitton are gratefully acknowledged.
APPENDIX
Sources of DNA for taxa included in this study (RBG Kew DNA Bank numbers are in parentheses after voucher information) and GenBank accession numbers for sequences generated in this research and by others.
| Taxa | Voucher information | ITS | matK | psbA-trnH | rpl16 | trnL-trnF region |
|---|---|---|---|---|---|---|
| Gagea subgenus Gagea:Gagea sect. Didymobolbos K. Koch | ||||||
| Gagea bohemica (Zauschn.) Schult. & Schult. f. | Andy Jones s.n. (Kew 7952) | – | EU912103 | – | EU912175 | EU912253 |
| Gagea bohemica (Zauschn.) Schult. & Schult. f. | Germany: Saxony-Anhalt | – | – | – | – | AJ437197 |
| Gagea bohemica (Zauschn.) Schult. & Schult. f. | Levichev 50 (LE) | AM162672 | – | AM085142 | – | AJ969117 |
| Gagea bohemica subsp. bohemica | Czech Republic: Moravia | AJ427549 | – | AJ416370 | – | AJ419161 |
| Gagea bohemica subsp. bohemica | Germany: Saxony-Anhalt | AJ427548 | – | – | – | AJ419160 |
| Gagea bohemica subsp. saxatilis (Mert. & W.D.J.Koch) Asch. & Graebn | Germany: Saxony-Anhalt | AJ427547 | – | AJ416371 | – | AJ419159 |
| Gagea algeriensis Chabert | Chase 748 (K)(Kew 748) | EU912088 | AY624470 | EU939280 | EU912232 | EU912311 |
| Gagea foliosa (J.Presl & C.Presl) Schult. & Schult.f | Italy: Sardegna 34697 (Z) | AM162676 | – | AM049258 | – | AJ969124 |
| Gagea heldreichii (A.Terracc.) Stroh | Levichev 8 (LE) | AM265534 | – | AM161464 | – | AM180467 |
| Gagea lojaconoi Peruzzi | Italy 9256 (CLU) | AM287272 | – | AM282997 | – | AM283106 |
| Gagea peduncularis (J.Presl & C.Presl) Pascher | Davis 40349 (K)(Kew 23333) | EU912054 | EU912127 | EU939252 | EU912204 | EU912283 |
| Gagea soleirolii F.W.Schultz | Montserrat et al. s.n. (Kew 20651) | – | EU912166 | EU939297 | EU912244 | EU912330 |
| Gagea villosa (M.Bieb.) Sweet | Zarrei & Kamrani 35273 (TUH)(Kew 23178) | EU912084 | EU912151 | EU939276 | EU912228 | EU912307 |
| Gagea villosa (M.Bieb.) Sweet | Zarrei & Golzarian 35247 (TUH)(Kew 23179) | EU912085 | EU912152 | EU939277 | EU912229 | EU912308 |
| Gagea villosa (M.Bieb.) Sweet | Zarrei & Golzarian 35253 (TUH)(Kew 23295) | EU912087 | EU912154 | EU939279 | EU912231 | EU912310 |
| Gagea villosa (M.Bieb.) Sweet | Levichev 7 (LE) | AM180453 | – | AJ973170 | – | AM238538 |
| Gagea villosa (M.Bieb.) Sweet | Germany: Saxony-Anhalt | AJ427545 | – | AJ416373 | – | AJ419163 |
| Gagea sect. Monophyllos Pascher | ||||||
| Gagea caelestis Levichev | Levichev 44 (LE) | AM180456 | – | AJ973165 | – | AJ969118 |
| Gagea confusa A.Terracc. | TUH-E BOT.EXP. 35712 (TUH)(Kew 23169) | EU912041 | EU912117 | EU939239 | EU912189 | EU912268 |
| Gagea confusa A.Terracc. | Zarrei & Zarrei 35266 (TUH)(Kew 23137) | EU912040 | EU912116 | EU939238 | EU912188 | EU912267 |
| Gagea confusa A.Terracc. | Levichev 13 (LE) | AM087949 | – | AJ973173 | – | AJ890369 |
| Gagea dschungarica Regel | Zarrei 35815 (TUH)(Kew 23143) | EU912043 | EU912118 | EU939240 | EU912191 | EU912270 |
| Gagea dschungarica Regel | Zarrei 35290 (TUH)(Kew 23170) | EU912044 | EU912119 | EU939241 | EU912192 | EU912271 |
| Gagea dschungarica Regel | Levichev 14 (LE) | AM087952 | – | AJ973164 | – | AJ970175 |
| Gagea filiformis (Ledeb.) Kunth | Levichev 12 (LE) | AM180457 | – | AM161459 | – | AM084904 |
| Gagea fragifera (Vill.) E.Bayer & G.López | Zarrei 35820 (TUH)(Kew 23144) | EU912045 | EU912120 | EU939243 | EU912194 | EU912273 |
| Gagea fragifera (Vill.) E.Bayer & G.López | TUH-E BOT.EXP. 35711 (TUH)(Kew 23171) | EU912046 | EU912121 | EU939244 | EU912195 | EU912274 |
| Gagea fragifera (Vill.) E.Bayer & G.López | TUH-E BOT.EXP. 35307 (TUH)(Kew 23294) | EU912086 | EU912153 | EU939278 | EU912230 | EU912309 |
| Gagea fragifera (Vill.) E.Bayer & G.López | Zarrei & Zarrei 35265 (K, TUH)(Kew 23296) | EU912047 | EU912122 | EU939245 | EU912196 | EU912275 |
| Gagea fragifera (Vill.) E.Bayer & G.López | Italy: 12692 (CLU) | AM287285 | – | AM282995 | – | AM283102 |
| Gagea fragifera (Vill.) E.Bayer & G.López | Switzerland: Canton Graubuenden 10726 (ZT) | – | – | AM238531 | – | AJ890375 |
| Gagea fragifera (Vill.) E.Bayer & G.López | Bulgaria: Pirin-mountains 070407 (HAL) | AM162677 | – | AJ973158 | – | AJ890368 |
| Gagea fragifera (Vill.) E.Bayer & G.López | Levichev 29b (LE) | AM180455 | – | AM238521 | – | AM161467 |
| Gagea glacialis K.Koch | Marais 1565 (K)(Kew 23279) | – | – | – | EU912199 | EU912278 |
| Gagea granulosa Turcz. | Levichev 11b (LE) | AM287278 | – | AM238517 | – | AM180463 |
| Gagea granulosa Turcz. | Levichev 11a (LE) | AM265533 | – | AM238518 | – | AM180462 |
| Gagea hiensis Pascher | Mongolia: Bogd-Ul Mountains 070426 (HAL) | AM287279 | – | AJ973169 | – | AJ890367 |
| Gagea infrakamensis Levichev | Levichev 10 (LE) | AM180459 | – | AM238519 | – | AM180471 |
| Gagea luteoides Stapf | Baytor, T. ISTE 44270 (K)(Kew 23280) | EU912053 | EU912126 | EU939251 | EU912203 | EU912282 |
| Gagea minima (L.) Ker-Gawl. | Germany: Saxony-Anhalt | AJ427546 | – | AJ416374 | – | AJ419164 |
| Gagea spathacea (Hayne) Salisb. | Levichev 37 (LE) | – | AJ973174 | AJ969126 | ||
| Gagea spathacea (Hayne) Salisb. | Germany: Saxony-Anhalt 095844 (Hal) | AJ427541 | – | AJ416369 | – | AJ419166 |
| Gagea sect. Gagea (Holobolbos K.Koch) | ||||||
| Gagea aipetriensis Levichev | Levichev 15 (LE) | AM087955 | – | AM049259 | – | AJ970178 |
| Gagea capusii A.Terracc. | Levichev 24 (LE) | – | – | AM085143 | – | AJ969123 |
| Gagea chanae Grossh. | Zarrei 867 (K)(Kew 23270) | EU912082 | EU912167 | EU939298 | EU912245 | – |
| Gagea helenae Grossh. | Levichev 22 (LE) | AM265531 | – | AM161461 | – | AJ969120 |
| Gagea lutea (L.) Ker-Gawl. | Zarrei 35285 (TUH)(Kew 23148) | EU912052 | EU912125 | EU939250 | EU912202 | EU912281 |
| Gagea lutea (L.) Ker-Gawl. | Germany: Saxony-Anhalt, Rothenschirmbach | AJ488569 | – | AJ416368 | – | AJ488279 |
| Gagea lutea (L.) Ker-Gawl. | Levichev 16 (LE) | AM265530 | – | AM161456 | – | AM110255 |
| Gagea megapolitana Henker | Henker (HAL) | – | – | AM161455 | – | AM084902 |
| Gagea nakaiana Kitag. | Levichev 17 (LE) | AM180454 | – | AM161457 | – | AM110256 |
| Gagea pratensis (Pers.) Dumort. | Germany: Saxony-Anhalt | AJ437203 | – | AJ416372 | – | AJ437196 |
| Gagea pratensis (Pers.) Dumort. | Germany: Brandenburg | AJ437202 | – | – | – | AJ437195 |
| Gagea pratensis (Pers.) Dumort. | Germany: Saxony-Anhalt | AJ437201 | – | – | – | AJ419162 |
| Gagea podolica Schult. & Schult.f. | Levichev 21 (LE) | AM409334 | – | AM238525 | – | AM084903 |
| Gagea pomeranica R.Ruthe | Germany: Mecklenburg-Western Pomeranica 095846 (HAL) | AJ429193 | – | AJ429194 | – | – |
| Gagea pomeranica R.Ruthe | Germany: Saxony-Anhalt 095842 (HAL) | AJ427543 | – | AJ416375 | – | AJ419167 |
| Gagea pusilla (F.W.Schmidt) Sweet | Levichev 18 (LE) | – | – | AM161458 | – | AM180464 |
| Gagea triflora Schult. fl. | Furse & Miyoshi 26159 (K)(Kew 23409) | – | – | – | EU912246 | EU912331 |
| Gagea triflora Schult. fl. | Levichev 46 (LE) | AM162674 | – | AM049261 | – | AJ890377 |
| Gagea transversalis (Pall.) Steven | Levichev 56 (LE) | AM162671 | – | AJ973167 | – | AJ890370 |
| Gagea subgenus Hornungia (Bernh.) Pascher: Gagea sect. Platyspermum Boiss. | ||||||
| Gagea afghanica A.Terracc. | Zarrei & Golzarian 35257 (TUH)(Kew 23156) | EU912021 | EU912097 | EU939221 | EU912171 | EU912247 |
| Gagea afghanica A.Terracc. | Zarrei & Golzarian 35223 (K, TUH)(Kew 23282) | EU912022 | EU912098 | EU939222 | – | EU912248 |
| Gagea afghanica A.Terracc. | Zarrei & Golzarian 35207 (K, TUH)(Kew 23283) | EU912023 | EU912099 | EU939223 | – | EU912249 |
| Gagea afghanica A.Terracc. | Levichev 52 (LE) | AM087953 | – | AJ973160 | – | AJ890373 |
| Gagea alexeenkoana Miscz. | TUH-E BOT.EXP. 35305 (TUH)(Kew 23142) | EU912024 | EU912100 | EU939224 | EU912172 | EU912250 |
| Gagea alexeenkoana Miscz. | TUH-E BOT.EXP. 35306 (TUH)(Kew 23297) | EU912030 | EU912106 | EU939229 | EU912179 | EU912257 |
| Gagea alexeenkoana Miscz. | Levichev 34 (LE) | AM180458 | – | AM161460 | – | AM110257 |
| Gagea altaica Schischk. & Sumnev. | Levichev 51 (LE) | AM162670 | – | AJ973159 | – | AJ890374 |
| Gagea bergii Litv. | Zarrei & Golzarian 35222 (TUH)(Kew 23141) | EU912026 | EU912102 | – | EU912174 | EU912252 |
| Gagea bulbifera (Pall.) Salisb. | TUH-E BOT.EXP. 35713 (TUH)(Kew 23140) | EU912027 | EU912104 | EU939226 | EU912176 | EU912254 |
| Gagea bulbifera (Pall.) Salisb. | TUH-E BOT.EXP. 35709 (TUH)(Kew 23166) | EU912028 | EU912105 | EU939227 | EU912177 | EU912255 |
| Gagea bulbifera (Pall.) Salisb. | Levichev 2 (LE) | AM162669 | – | AM049260 | – | AJ969119 |
| Gagea capillifolia Vved. | Levichev 42 (LE) | AM087951 | – | AJ973171 | – | AJ970177 |
| Gagea caroli-kochii Grossh. | TUH-E BOT.EXP. 35715 (TUH)(Kew 23139) | EU912029 | EU912170 | EU939228 | EU912178 | EU912256 |
| Gagea chlorantha (M.Bieb.) Schult. & Schult. f. | Zarrei & Kamrani 35192 (TUH)(Kew 23138) | EU912031 | EU912107 | EU939230 | EU912180 | EU912258 |
| Gagea chlorantha (M.Bieb.) Schult. & Schult. f. | Zarrei & Kamrani 35195 (TUH)(Kew 23167) | EU912032 | EU912108 | EU939231 | EU912181 | EU912259 |
| Gagea chlorantha (M.Bieb.) Schult. & Schult. f. | Zarrei & Zarre 778 (K, TUH)(Kew 23268) | EU912033 | EU912109 | EU939232 | EU912182 | EU912260 |
| Gagea chlorantha (M.Bieb.) Schult. & Schult. f. | Zarrei 872 (K, TUH)(Kew 23269) | EU912034 | EU912110 | EU939233 | EU912183 | EU912261 |
| Gagea chlorantha (M.Bieb.) Schult. & Schult. f. | Hikmat Abbas Al-Ani & Danail & Danail Aoraha 9354 (K)(Kew 23275) | EU912035 | EU912111 | – | – | EU912262 |
| Gagea chomutovae (Pascher) Pascher | Zarrei & Golzarian 35214 (TUH)(Kew 23146) | EU912036 | EU912112 | EU939234 | EU912184 | EU912263 |
| Gagea chomutovae (Pascher) Pascher | Zarrei 35814 (TUH)(Kew 23168) | EU912037 | EU912113 | EU939235 | EU912185 | EU912264 |
| Gagea chomutovae (Pascher) Pascher | Levichev 37 (LE) | AM087950 | – | AM049262 | – | AJ970176 |
| Gagea circumplexa Vved. | Carter 721 (K)(Kew 23274) | EU912038 | EU912114 | EU939236 | EU912186 | EU912265 |
| Gagea circumplexa Vved. | Levichev 30 (LE) | AM265529 | – | AJ973172 | – | AJ969122 |
| Gagea commutata K.Koch | Zarrei 876 (K, LE, TUH)(Kew 23336) | EU912039 | EU912115 | EU939237 | EU912187 | EU912266 |
| Gagea commutata K.Koch | Dafni s.n. (Kew 23410) | EU912096 | – | EU939296 | EU912243 | EU912329 |
| Gagea dayana Chodat & Beauverd | Davis 8235 (K)(Kew 23273) | EU912042 | – | – | EU912190 | EU912269 |
| Gagea eleonorae Levichev | Levichev 57 (LE) | AM287274 | – | AJ973163 | – | AJ970179 |
| Gagea exilis Vved. | Moussavi & Tehrani 29971 (IRAN)(Kew 23182) | – | – | EU939242 | EU912193 | EU912272 |
| Gagea gageoides (Zucc.) Vved. | Zarrei & Kamrani 35274 (TUH)(Kew 23172) | EU912049 | EU912169 | EU939247 | EU912198 | EU912277 |
| Gagea gageoides (Zucc.) Vved. | TUH-E BOT.EXP. 35714 (TUH)(Kew 23145) | EU912048 | EU912168 | EU939246 | EU912197 | EU912276 |
| Gagea gageoides (Zucc.) Vved. | Levichev 41 (LE) | AM162673 | – | AM161462 | – | AM084905 |
| Gagea graeca (L.) Irmisch. | Davis 40591 (K)(Kew 23339) | EU912077 | EU912159 | EU939285 | EU912235 | EU912316 |
| Gagea graeca (L.) Irmisch. | Greece: Crete, Lassithi plateau 099962 (HAL) | AJ810089 | – | AM049263 | – | AJ810090 |
| Gagea graeca (L.) Irmisch. | Greece: Lakonia | AJ810088 | – | – | – | – |
| Gagea helicophylla Levichev” ined. | Levichev 35a (LE) | – | – | AM085145 | – | AM084901 |
| Gagea lactea Levichev | Levichev 53 (LE) | AM180452 | – | AJ973166 | – | AJ969125 |
| Gagea libanotica (Hochst.) Greuter | Townsend 74/38 (K)(Kew 23338) | – | EU912160 | EU939286 | EU912236 | EU912317 |
| Gagea olgae Regel | Levichev 3 (LE) | – | – | AM085144 | – | AM161465 |
| Gagea ova Stapf | Levichev 39b (LE) | AM287277 | – | AM265588 | – | AM180466 |
| Gagea ova Stapf | Levichev 39a (LE) | AM287276 | – | AM238526 | – | AM180465 |
| Gagea reticulata (Pall.) Schult. & Schult. f. | Zarrei & Golzarian 35260 (TUH)(Kew 23150) | EU912056 | EU912129 | EU939254 | EU912206 | EU912285 |
| Gagea reticulata (Pall.) Schult. & Schult. f. | Zarrei & Kamrani 35196 (TUH)(Kew 23174) | EU912058 | EU912131 | EU939256 | EU912208 | EU912287 |
| Gagea reticulata (Pall.) Schult. & Schult. f. | Zarrei & Kamrani 35186 (K, TUH)(Kew 23284) | EU912060 | EU912133 | EU939258 | EU912210 | EU912289 |
| Gagea reticulata (Pall.) Schult. & Schult. f. | Zarrei & Ajani 832 (IRAN, K, M, TUH)(Kew 23287) | EU912061 | EU912134 | EU939259 | EU912211 | EU912290 |
| Gagea reticulata (Pall.) Schult. & Schult. f. | Levichev 56 (LE) | – | – | AJ973162 | – | – |
| Gagea cf. reticulata (Pall.) Schult. & Schult. f. | Zarrei & Zarre 1032 (K)(Kew 23271) | EU912059 | EU912132 | EU939257 | EU912209 | EU912288 |
| Gagea setifolia Baker | Zarrei 35289 (TUH)(Kew 23149) | EU912055 | EU912128 | EU939253 | EU912205 | EU912284 |
| Gagea setifolia Baker | Zarrei & Golzarian 35246 (TUH)(Kew 23151) | EU912063 | EU912136 | EU939261 | EU912213 | EU912292 |
| Gagea setifolia Baker | Zarrei & Golzarian 35254 (TUH)(Kew 23175) | EU912064 | EU912137 | EU939262 | EU912214 | EU912293 |
| Gagea setifolia Baker | Heydari 30547 (K)(Kew 23267) | EU912065 | EU912138 | EU939263 | EU912215 | EU912294 |
| Gagea setifolia Baker | Zarrei 1017 (K)(Kew 23272) | EU912066 | EU912139 | EU939264 | EU912216 | EU912295 |
| Gagea setifolia Baker | Mohammadi 35198 (K, TUH)(Kew 23285) | EU912067 | EU912140 | EU939265 | EU912217 | EU912296 |
| Gagea setifolia Baker | Zarrei & Zarrei 35268 (K, TUH)(Kew 23290) | EU912068 | EU912141 | EU939266 | EU912218 | EU912297 |
| Gagea setifolia Baker | Zarrei & Golzarian 35213 (K, TUH)(Kew 23291) | EU912069 | EU912142 | EU939267 | EU912219 | EU912298 |
| Gagea cf. setifolia Baker | Zarre 1009 (K, TUH)(Kew 23286) | EU912025 | EU912101 | EU939225 | EU912173 | EU912251 |
| Gagea cf. setifolia Baker | Zarrei & Golzarian 35252 (K, TUH)(Kew 23292) | EU912062 | EU912135 | EU939260 | EU912212 | EU912291 |
| Gagea stipitata Merckl. ex Bunge | Zarre & Zarrei 35297 (TUH)(Kew 23154) | EU912070 | EU912143 | EU939268 | EU912220 | EU912299 |
| Gagea stipitata Merckl. ex Bunge | Zarrei & Kamrani 35275 (TUH)(Kew 23176) | EU912071 | EU912144 | EU939269 | EU912221 | EU912300 |
| Gagea stipitata Merckl. ex Bunge | Zarrei & Kamrani 35197 (TUH)(Kew 23177) | EU912072 | EU912145 | EU939270 | EU912222 | EU912301 |
| Gagea stipitata Merckl. ex Bunge | Zarrei & Golzarian 35215 (K, TUH)(Kew 23293) | EU912073 | EU912146 | EU939271 | EU912223 | EU912302 |
| Gagea tenera Pascher | Zarrei & Golzarian 35256 (TUH)(Kew 23152) | EU912074 | EU912147 | EU939272 | EU912224 | EU912303 |
| Gagea tenera Pascher | Zarrei & Golzarian 35219 (K, TUH)(Kew 23281) | EU912075 | EU912148 | EU939273 | EU912225 | EU912304 |
| Gagea uliginosa Siehe & Pascher | TUH-E BOT.EXP. 35304 (TUH)(Kew 23153) | EU912089 | EU912155 | EU939281 | EU912233 | EU912312 |
| Gagea uliginosa Siehe & Pascher | Moussavi et al. 30018 (IRAN)(Kew 23288) | EU912090 | EU912157 | EU939283 | – | EU912314 |
| Gagea uliginosa Siehe & Pascher | Rawi & Serhang 18286 (K)(Kew 23277) | – | EU912156 | EU939282 | EU912234 | EU912313 |
| Gagea vegeta Vved. | Shafii 475 (Shahed University Herbarium)(Kew 23181) | EU912076 | EU912149 | EU939274 | EU912226 | EU912305 |
| Gagea vegeta Vved. | Zarrei and Zarre, 1033 (K, TUH)(Kew 23335) | – | EU912150 | EU939275 | EU912227 | EU912306 |
| Gagea vegeta Vved. | Levichev 32 (LE) | AM287275 | – | AM238520 | – | AM180468 |
| Gagea wendelboi Rech.f. | Matin 35605 (IRAN)(Kew 23183) | EU912091 | EU912158 | EU939284 | – | EU912315 |
| Gagea sp. | Zarrei & Kamrani 35194 (TUH)(Kew 23155) | EU912057 | EU912130 | EU939255 | EU912207 | EU912286 |
| Gagea sect. Plaecostigma (Turcz.) Pascher | ||||||
| Gagea iranica Zarrei & Zarre | Zarrei & Golzarian 35210 (TUH)(Kew 23147) | EU912050 | EU912123 | EU939248 | EU912200 | EU912279 |
| Gagea iranica Zarrei & Zarre | Zarrei & Golzarian 35251 (TUH)(Kew 23173) | EU912051 | EU912124 | EU939249 | EU912201 | EU912280 |
| Gagea pauciflora (Turcz. ex Trautv.) Turcz. ex Ledeb. | Mongolia: Ulan Bator 070423 (HAL) | AM409330 | – | AJ973168 | – | AJ890372 |
| Lloydia delicatula Noltie | AGSES 212 (K)(Kew 23340) | EU912079 | – | – | – | EU912320 |
| Lloydia flavonutans H.Hara | AGSES 77 (K)(Kew 23341) | EU912080 | – | – | EU912238 | EU912321 |
| Lloydia oxycarpa Franch. | ACE 137 (K)(Kew 23342) | EU912081 | – | EU939289 | – | EU912322 |
| Lloydia serotina (L.) Rchb. | Jones s.n. (K)(Kew 1004) | EU912092 | AY624471 | EU939288 | – | EU912319 |
| Lloydia serotina (L.) Rchb. | Levichev 45a (LE) | AM087956 | – | AM238530 | – | AJ890376 |
| Lloydia serotina (L.) Rchb. | Bulgaria: Ovtscharez 074806 (HAL) | – | – | AJ585048 | – | AJ585049 |
| Lloydia yunnanensis Franch. | Luo, Yi-bo 64 (K)(Kew 23337) | EU912078 | EU912161 | EU939287 | EU912237 | EU912318 |
| Outgroup members | ||||||
| Tulipa clusiana DC. | Zarrei 35183 (TUH)(Kew 23348) | EU912093 | EU912162 | EU939290 | EU912239 | EU912323 |
| Tulipa lehmanniana Merckl. | Zarrei & Golzarian 35228A (TUH)(Kew 23349) | EU912094 | EU912163 | EU939291 | EU912240 | EU912324 |
| Tulipa uniflora (L.) Besser ex Baker | Chase 751 (K) | – | EU912164 | EU939292 | EU912241 | EU912325 |
| Amana erythronioides (Baker) D. Y. Tan & D. Y. Hong | Chase 742 (K) | EU912095 | AY624472 | EU939293 | EU912020 | EU912326 |
| Erythronium japonicum Decne. | Chase 780 (K) | EU912083 | AF485323 | EU939295 | AF485323* | EU912332 |
| Fritillaria persica L. | Chase 3496 (K) | AY616736 | AY624451 | AY624399 | AY624399 | EU912327 |
| Lilium ledebourii (Baker) Boiss. | Zarrei s.n. (TUH)(Kew 23346) | – | EU912165 | EU939299 | EU912242 | EU912328 |
* Voucher differs from those on the voucher list; it has been taken from NCBI.
LITERATURE CITED
- Ali SI. Two new species of Gagea Salisb. (Liliaceae) from Pakistan. Pakistan Journal of Botany. 2006;38:43–46. [Google Scholar]
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- Bohdanowicz J, Lewandowska B. Participation of endoplasmic reticulum in the unequal distribution of plastids during generative cell formation in Gagea lutea (L.) Ker-Gawl. (Liliaceae) Acta Biologica Cracoviensia Series Botanica. 1999;41:77–183. [Google Scholar]
- Boissier E. Gagea. Flora Orientalis. 1882;5:203–211. Geneva and Basel. [Google Scholar]
- Cameron KM, Chase MW, Anderson WR, Hills HG. Molecular systematics of Malpighiaceae: evidence from plastid rbcL and matK sequences. American Journal of Botany. 2001;88:1847–1862. [PubMed] [Google Scholar]
- Chase MW, Knapp S, Cox AV, et al. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae) Annals of Botany. 2003;92:107–127. doi: 10.1093/aob/mcg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Deb DB. Taxonomic revision of the genus Gagea Salisb. (Liliaceae) in India and adjoining regions. The Journal of the Bombay Natural History Society. 1983;83:78–97. [Google Scholar]
- Dasgupta S, Deb DB. Taxonomic revision of the genus Lloydia (Liliaceae) in India and adjoining region. Indian Journal of Forestry. 1986;9:104–112. [Google Scholar]
- Davlianidze MT. Conspectus of the Caucasian representatives of the genus Gagea Salisb. I. Notulae Systematicae ac Geographicae. Instituti Botanici Thbilissiensis. 1972;29:69–75. [Google Scholar]
- Davlianidze MT. Kavkazaskie predstaviteli roda Gagea Salisb. 1976 Tbilisi: Metsniereba, 1–160 [In English translation entitled ‘A review of the systematics and the taxonomy of the Caucasian representatives of the genus Gagea Salisb.’]. [Google Scholar]
- Doyle JJ. Gene trees and species trees: molecular systematics as one-character taxonomy. Systematic Botany. 1992;71:144–163. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Farris JS, Kjällersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- Fay MF, Chase MW. Modern concepts of Liliaceae with a focus on the relationship of Fritillaria. Curtis's Botanical Magazine. 2000;17:146–149. [Google Scholar]
- Federov A. Flora of Russia, the European Part and Bordering Regions. Vol. 4. CRC Press Inc.: 2001. Magnoliophyta (=Angiospermae), Magnoliopsida (=Dicotyledones), Liliopsida (=Monocotyledones) [Google Scholar]
- Feinburn-Dotham N. Gagea. In: Zohary M., editor. Flora Paleastina. Vol. 4. Jerusalem: The Israeil Academy of Science and Humanities; 1986. pp. 32–39. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Towards defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–426. [Google Scholar]
- Gerard J. The herbal of general history of plants. New York: Dover Publications (Reprint); 1663. [Google Scholar]
- Govaerts R. World checklist of Liliaceae. Kew: The Board of Trustees of the Royal Botanic Gardens; 2006. http://www.kew.org/wcsp/ accessed October 2008. [Google Scholar]
- Greuter W. The taxonomic position of Lloydia graeca (Liliaceae) and related species. Israel Journal of Botany. 1970;19:155–160. [Google Scholar]
- Grossheim AA. Gagea. In: Komarov VL, editor. Fl. USSR. Jerusalem: Israel Program for Scientific Translation Ltd; 1935. pp. 61–112. 4. Leningrad, English ed. 1968. [Google Scholar]
- Grubov VI, Egorova TV. Gagea. In: Grubor VI, editor. Plants of Central Asia. Vol. 7. Liliaceae–Orchidaceae: Science Publishers, Inc; 2003. pp. 10–17. [Google Scholar]
- Hamzaoğlu E, Budak Ü, Aksoy A. A new species of Gagea Salisb. (Liliaceae) from Sivas (Central Anatolia, Turkey) Turkish Journal of Botany. 2008;32:261–264. [Google Scholar]
- Heyn CH, Dafni A. Studies in the genus Gagea (Liliaceae) I. The platyspermous species in Israel and neighbouring areas. Israel Journal of Botany. 1971;20:214–233. [Google Scholar]
- Heyn CH, Dafni A. Studies in the genus Gagea (Liliaceae) II, The non-platyspermous species from the Galilee, the Golan Heights and Mt. Hermon. Israel Journal of Botany. 1977;26:11–22. [Google Scholar]
- Henker H. Goldsterne und Stinsenpflanzen in Mecklenburg-Vorpommern. Teil 1. Die Goldsterne von Mecklenburg-Vorpommern unter besonderer Berücksichtigung kritisher und neuer Sippen. Botanischer Rundbrief für Mecklenburg-Vorpommern. 2005;39:3–90. [Google Scholar]
- Hulsenbeck JF, Bull JJ, Cunningham CW. Combining data in phylogenetic analysis. Trends in Ecology and Evolution. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- Hyam R, Pankhurst R. Plants and their names, a concise dictionary. Oxford University Press; 1995. [Google Scholar]
- Kelchner SA. The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden. 2000;87:482–498. [Google Scholar]
- Koch K. Beiträge zu einer Flora des Orientes (Fortezung): Gagea. Linnaea. 1849;22:226–231. [Google Scholar]
- Johnson LA, Soltis DE. matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s.str. Systematic Botany. 1994;19:143–156. [Google Scholar]
- Jordan WC, Courtney MW, Neigle JE. Low levels of intraspecific genetic variation at a rapidly evolving chloroplast DNA locus in North American duckweeds (Lemnaceae) American Journal of Botany. 1996;83:430–439. [Google Scholar]
- Kosenko VN. Contribution to the pollen morphology and taxonomy of Liliaceae. Grana. 1999;38:20–30. [Google Scholar]
- Kosenko VN, Levichev IG. Pollen morphology in the genera Gagea and Lloydia (Liliaceae) Botanicheskii Zhurnal. 1988;73:965–976. [Google Scholar]
- Levichev IG. The new species of the genus Gagea (Liliaceae) from the western Tian-Shan (in Russian) Botanicheskii Zhurnal. 1981;66:1635–1645. [Google Scholar]
- Levichev IG. New species of the genus Gagea (Liliaceae) from western part of Tian-Shan (in Russian) Botanicheskii Zhurnal. 1988;73:1617–1623. [Google Scholar]
- Levichev IG. The synopsis of the genus Gagea (Liliaceae) from the western Tien-Shan. (in Russian) Botanicheskii Zhurnal. 1990;75:225–234. [Google Scholar]
- Levichev IG. The new species of the genus Gagea (Liliaceae) from the western Tian-Shan. Botanicheskii Zhurnal. 1991;76:999–1004. (in Russian) [Google Scholar]
- Levichev IG. Zur Morphologie in der Gattung Gagea Salisb. (Liliaceae). I. Die unterirdischen Organe. Flora. 1999;194:379–392. [Google Scholar]
- Levichev IG. A new species of the genus Gagea (Liliaceae) Botanicheskii Zhurnal. 2000;85:125–127. [Google Scholar]
- Levichev IG. New species of the genus Gagea (Liliaceae) from western district of Asia. Turczaninowia. 2001;4:5–35. [Google Scholar]
- Levichev IG. Four new species of the genus Gagea Salisb. (Liliaceae) from Western Himalayas and the adjoining regions. Pakistan Journal of Botany. 2006a;38:47–54. [Google Scholar]
- Levichev IG. A review of the Gagea (Liliaceae) species in the flora of Caucasus. Botanicheskii Zhurnal. 2006b;91:917–951. [Google Scholar]
- Levichev IG, Ali SI. Seven new species of the genus Gagea Salisb. (Liliaceae) from Western Himalayas and the adjoining regions. Pakistan Journal of Botany. 2006;38:55–62. [Google Scholar]
- Levichev IG, Navruzshoev D. A new species of the genus Gagea (Liliaceae) from the Pamiro-Alayi. Botanicheskii Zhurnal. 1997;82:91–92. [Google Scholar]
- Linnaeus C. Species Plantarum. Vol. 2. Stockholm: 1753. [Google Scholar]
- Linnaeus C. Species Plantarum. 7. Vol. 2. Stockholm: 1762. [Google Scholar]
- Mabberley DJ. The plant book. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- McNeill J, Barrie FR, Burdet HM, et al. International Code of Botanical Nomenclature (Vienna Code) adopted by the Seventeenth International Botanical Congress Vienna; Austria, July 2005; Ruggell, Liechtenstein: Gantner Verla; 2006. [Google Scholar]
- Melchior H. Reihe Liliiflorae. In: Melchior H, editor. A. Engler's Syllabus der Pflanzenfamilien. Band. Berlin: Nikolassee; 1964. pp. 513–542. 12. Aufl., 2. [Google Scholar]
- Molvray MP, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influence on floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melborne: CSRIO; 2000. pp. 441–448. [Google Scholar]
- Pallas PS. Reise Durch verich Ledene provinzen des Russischen. Vol. 2. St. Petersburg: 1773. [Google Scholar]
- Pallas PS. Reise des Russischen Reichs: Reise aus Sibirien zuruch an die Wolga. Vol. 3. St. Petersburg: 1776. [Google Scholar]
- Pascher A. Übersicht über die Arten der Gattung Gagea. Sitzungberichtedes deutschen naturwissenschaflich-medicischen Vereins für Böhmen ‘Lotos’. 1904:109–131. [Google Scholar]
- Pascher A. Conspectus Gagearum Asiae. Bulletin of the de la Societe Imperiale des Naturalistes de Moscou. 1907;19:353–375. [Google Scholar]
- Patterson TB, Givnish TJ. Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: insights from rbcL and ndhF sequence data. Evolution. 2002;56:233–252. doi: 10.1111/j.0014-3820.2002.tb01334.x. [DOI] [PubMed] [Google Scholar]
- Peruzzi L. Contribution to cytotaxonomical knowledge of Gagea Salisb. (Liliaceae). sect. Foliatae A.Terrac. and synthesis of karyological data. Caryologia. 2003;56:115–128. [Google Scholar]
- Peruzzi L. Contribution to the cytotaxonomical knowledge of the genus Gagea Salisb. (Liliaceae). III. New karyological data from the central Mediterranean area. Caryologia. 2008;61:92–106. [Google Scholar]
- Peruzzi L, Aquaro G. Contribution to cytotaxonomical knowledge of Gagea Salisb. (Liliaceae). II. Further karyological studies on Italian populations. Candollea. 2005;60:237–253. [Google Scholar]
- Peruzzi L, Zarrei M. Typification some critical taxa of Gagea Salisb. (Liliaceae) from W Asia. Candollea. 2007;62:237–244. [Google Scholar]
- Peruzzi L, Bartolucci F, Frignani F, Minutillo F. G. tisoniana, a new species of Gagea Salisb. sect. Gagea (Liliaceae) from Central Italy. Botanical Journal of the Linnean Society. 2007;153:337–347. [Google Scholar]
- Peruzzi L, Tison J-M, Peterson A, Peterson J. On the phylogenetic position and taxonomic value of Gagea trinervia (Viv.) Greuter and the whole G. sect. Anthericoides A. Terracc. (Liliaceae) Taxon. 2008a;57:1201–1214. [Google Scholar]
- Peruzzi L, Peterson A, Tison J-M, Peterson J. Phylogenetic relationships of Gagea Salisb. (Liliaceae) in Italy, inferred from molecular and morphological data matrices. Plant Systematics and Evolution. 2008b;276:219–234. [Google Scholar]
- Peterson A, Peterson J. Phylogenetic relationships among Gagea and Lloydia (Liliaceae). Abstract, in 17th International Botanical Congress; Vienna, Austria. 2005. p. 389. [Google Scholar]
- Peterson A, Peterson J. Molecular analysis of phylogenetic relationships among Gagea and Lloydia (Liliaceae). Abstract, in International Botanical Conference; California State University. 2006. p. 248. [Google Scholar]
- Peterson A, John H, Koch E, Peterson J. A molecular phylogeny of the genus Gagea (Liliaceae) in Germany inferred from non-coding chloroplast and nuclear DNA sequences. Plant Systematics and Evolution. 2004;245:145–162. [Google Scholar]
- Peterson A, Levichev IG, Peterson J. Systematics of Gagea and Lloydia (Liliaceae) and infrageneric classification of Gagea based on molecular and morphological data. Molecular Phylogenetics and Evolution. 2008;46:446–465. doi: 10.1016/j.ympev.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Phillips R, Rix M. The Pan garden plants series. London: Pan; 1989. Bulbs. [Google Scholar]
- Reeves G, Chase MW, Goldblatt P, et al. Molecular systematics of Iridaceae: evidence from four plastid DNA regions. American Journal of Botany. 2001;88:2074–2087. [PubMed] [Google Scholar]
- Rechinger KH. Six new species of Gagea (Liliaceae) from Flora. Iranica area. Plant Systematics and Evolution. 1986;153:287–292. [Google Scholar]
- Rønsted N, Law S, Thornton H, Fay MF, Chase MW. Molecular phylogenetic evidence for the monophyly of Fritillaria and Lilium (Liliaceae, Liliales) and the infrageneric classification of Fritillaria. Molecular Phylogenetics and Evolution. 2005;35:509–527. doi: 10.1016/j.ympev.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Salisbury RA. On the characters of a distinct genus hitherto confounded with Ornithogalum, and called Gagea; with some remarks on the importance of the inflorescence in distinguishing genera. Annals of Botany. 1806;2:553–557. [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) American Journal of Botany. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Stroh G. Die Gattung Gagea Salisb. Beihefte zum Botanischen Centralblatt. 1937;57:485–520. [Google Scholar]
- Sun H, McLewin W, Fay MF. Molecular phylogeny of Helleborus (Ranunculaceae), with an emphasis on the East Asian-Mediterranean disjunction. Taxon. 2001;50:1001–1018. [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacer of nuclear ribosomal DNA. Theoretical and Applied Genetics. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 2002. Verson 4. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Takhtajan A. Florestic regions of the world. Berkeley: University of California Press; 1986. [Google Scholar]
- Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Systematic Botany. 2003;28:723–737. [Google Scholar]
- Terracciano A. Gagearum species Florae Orientalis. Bulletin de I'herbier Boissier. 1905a;2:1061–1076. 1113–1128. [Google Scholar]
- Terracciano A. Les espèces du genre Gagea dans la flore del l'Afrique boréale. Bulletin de la Societe Botanique de France. 1905b;52:1–26. [Google Scholar]
- Terracciano A. Gagearum species Florae Orientalis. Bulletin de I'herbier Boissier. 1906;2:105–120. [Google Scholar]
- Tison JM. Gagea polidorii J.-M. Tison, espèce méconnue du sud-ouest des Alpes et des Apennins. Acta Botanica Gallica. 2004;15:319–326. [Google Scholar]
- Uphof JC. A review of the genus Gagea Salisb. I-III. Plant Life. 1958–1960;14:124–132. 151–161; 15: 163–176. [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. An unusual ribosomal DNA sequence from Gossypium gossypioides reveals ancient, cryptic, intergenomic introgression. Molcular Phylogenetics and Evolution. 1995;4:298–313. doi: 10.1006/mpev.1995.1027. [DOI] [PubMed] [Google Scholar]
- Wendelbo P, Rechinger KH. Gagea. In: Rechinger KH, editor. Flora Iranica. Vol. 165. Graz, Austria: Akademische Druck- und Verlagsanstalt; 1990. pp. 13–57. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Whitten WM, Williams NH, Chase MW. Subtribal and genetic relationships of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: combined molecular evidence. American Journal of Botany. 2000;87:1842–1856. [PubMed] [Google Scholar]
- Willis JC. A dictionary of the flowering plants and ferns. Cambridge: Cambridge University Press; 1980. [Google Scholar]
- Winship PR. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Research. 1989;17:1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Irwin JA, Payseur BA. Failure of the ILD to determine data combinability for slow loris phylogeny. Systematic Biology. 2001;50:408–424. [PubMed] [Google Scholar]
- Zarrei M, Zarre Sh. A new species of Gagea (Liliaceae) from Iran. Nordic Journal of Botany. 2005a;23:269–274. [Google Scholar]
- Zarrei M, Zarre Sh. Pollen morphology of the genus Gagea (Liliaceae) in Iran. Flora. 2005b;200:96–108. [Google Scholar]
- Zarrei M, Zarre Sh, Wilkin P, Rix M. Systematic revision of the genus Gagea Salisb. (Liliaceae) in Iran. Botanical Journal of the Linnean Society. 2007;154:559–588. [Google Scholar]
- Zhao LQ, Yang J. Gagea daqingshanensis (Liliaceae), a new species from inner Mongolia, China. Annales Botanici Fennici. 2006;43:223–224. [Google Scholar]
- Zhao YZ, Zhao LQ. Gagea chinensis (Liliaceae), a new species from inner Mongolia, China. Annales Botanici Fennici. 2004;41:297–298. [Google Scholar]