Abstract
Background and Aims
In clonal plants producing vegetative offspring, performance at the genet level as well as at the ramet level should be investigated in order to understand the entire picture of the population dynamics and the life history characteristics. In this study, demography, including reproduction and survival, the growth patterns and the spatial distributions of ramets within genets of the clonal herb Convallaria keiskei were explored.
Methods
Vegetative growth, flowering and survival of shoots whose genets were identified using microsatellite markers were monitored in four study plots for 3 years (2003–2005). The size structures of ramets in genets and their temporal shifts were then analysed. Their spatial distributions were also examined.
Key Results
During the census, 274 and 149 ramets were mapped in two 1 × 2 m plots, and 83 and 94 ramets in two 2 × 2 m quadrats. Thirty-eight genotypes were identified from 580 samples. Each plot included 5–18 genets, and most ramets belonged to the predominant genet(s) in each plot. Shoots foliated yearly for several years, but flowering ramets did not have an inflorescence the next year. A considerable number of new clonal offspring persistently appeared, forming a bell-shaped curve of the size structure of ramets in each genet. Comparing the structures modelled by the normal distributions suggested variation among ramets belonging to a single genet and variation among genets. Furthermore, spatial analyses revealed clumped and distant distributions of ramet pairs in a genet, in which the distant patterns corresponded to the linearly elongating clonal growth pattern of this species.
Conclusion
Characteristics of ramet performances such as flowering and recruitment of clonal offspring, in addition to growth, played a large part in the regulation of genet dynamics and distribution, which were different among the studied genets. These might be characteristics particularly relevant to clonal life histories.
Key words: Clonal plant, Convallaria keiskei, demography, genet, genetic identification, growth pattern, life history, ramet, spatial distribution
INTRODUCTION
Clonality is a widespread characteristic of herbaceous plants. One striking feature of clonal plants is hierarchical organization, in which each genetic individual (genet), i.e. the developmental product of a single zygote, consists of genetically identical and semi-autonomous construction units (ramets; Tuomi and Vuorisalo, 1989; Eriksson and Jerling, 1990). As a consequence, the life history characteristics of clonal plants, as related to population structures and dynamics, are quite different from those of non-clonal plants (Eriksson and Bremer, 1993; Fischer and van Kleunen, 2002; Honnay et al., 2005). In terms of reproduction, for example, vegetative offspring ramets develop faster and become adults more quickly than seedlings; in contrast to seed dispersal, mobility via clonal growth is limited, and potentially independent offspring sometimes remain connected to the parent (Araki and Ohara, 2008). Moreover, such features should be addressed along the hierarchy of ramet, genet and population (Stone and Ezrati, 1996; de Kroon and van Groenendael, 1997) because the capacity for horizontal spreading by propagating ramets is likely to affect the spatial distribution at the genet level as well as the population level (Abrahamson, 1980; Tuomi and Vuorisalo, 1989). In order to understand the life history of clonal plants, it is ultimately important to keep precise track of genotypic individuals, i.e. genets, and to evaluate the phenomenon at the ramet and population level as dynamic features related to the genet level, taking into consideration spatio-temporal variations (Eriksson, 1993; Tanner, 2001).
The results of a number of demographic studies of clonal species without genotypic identifications (e.g. Cook, 1985; Navas and Garnier, 1990; Sampaio et al., 2005) cannot be compared with those of non-clonal genets, although ramets not genets behave independently, like non-clonal genets. Genet-level approaches based on morphological features such as rhizomatous connections, clustering units or marked differences have also been attempted (e.g. Hartnett and Bazzaz, 1985; Geber et al., 1992; Falińska, 1995; Fair et al., 1999; Guárdia et al., 2000), but these approaches could not discriminate individual genets because of the complex clonal structure of intermingled genets and isolated ramets. Molecular genetic studies of population structures have provided demographic information such as numbers, recruitment and mortality of genets (Steinger et al., 1996; Persson and Gustavsson, 2001) in addition to estimating the contributions of the two reproductive modes (sexual and asexual; Watkinson and Powell, 1993; Harada et al., 1997). However, these assessments were indirect and general, and did not consider fine-scale variations or temporal fluctuations of recent years. In particular, it is of interest to determine how clonal features found in not only ramets but genets influence their demography and the population dynamics in natural populations. If the genets of the analysed ramets could be identified, demographic observations of them in the populations would characterize the population dynamics and the life history characteristics of clonal plant species; it may be quite different with non-clonal species.
Our previous study of a rhizomatous clonal herb, Convallaria keiskei, investigating the clonal structure based on allozyme variation in a 90 × 100 m population, demonstrated that not only were several large clones widely distributed, but also many small clones were scattered there (Araki et al., 2007). Therefore, this might indicate that habitat conditions were heterogeneous at the fine-scale and each genet in the population had adapted to its microhabitat conditions (Stuefer, 1996; Price and Marshall, 1999; Prati and Schmid, 2000). Because the detailed distributions are still unclear at the ramet level, it should be clarified whether clones converge in one genet or intermingle with other clones. We would also like to know whether genet diversity increases at the ramet level (Stehlik and Holderegger, 2000; Ruggiero et al., 2005) and whether genetic diversity may also differ according to ramet density. Linearly elongating rhizomes of C. keiskei connect distant ramets, which causes intermingled structure for different genets (Araki and Ohara, 2008). The results also suggest significant contributions of both sexual reproduction and clonal growth, as large genets have been maintained by clonal growth after seedling establishment (Soane and Watkinson, 1979; Eriksson, 1993; Watkinson and Powell, 1993). However, it is still unclear whether such a variety of clones have been produced continuously and to what extent the two reproductive modes contribute to maintaining the clones. Detailed studies using molecular techniques and field observations to examine the demography of growth, reproduction and spatial expansion at the ramet level could reveal the spatio-temporal patterns at the genet level.
In this study, we explored the population dynamics to clarify the life history characteristics of a clonal herb, C. keiskei, which has two reproductive systems, by monitoring genetically identified shoots of seedlings through to reproductive stages in natural populations. First, we obtained basic demographic information, such as survival, flowering, and recruitment, about the ramets in each genet. Secondly, we analysed genet-based size structures and their temporal changes (growth patterns). Thirdly, we examined the spatial distributions of ramets in a single genet or between two genets. The overall goal of this study was to employ a demographic–genetic approach in order to understand the life history of C. keiskei and the complicated population structures of clonal plants.
MATERIALS AND METHODS
Study species
Convallaria keiskei (Convallariaceae) is a perennial herb distributed in Japan (Hokkaido, Honshu and Kyushu), Sakhalin Island, Korea, China and eastern Siberia. New aerial shoots (sheath leaves) elongate and appear above ground in late April to May, and flowering takes place in late May to June. On the forest floor, there are two distinct types of vegetative aerial shoots (one-leaf shoots and two-leaf shoots), as well as a two-leaf shoot that bears an inflorescence. An inflorescence typically has 3–15 bell-shaped flowers with a mild fragrance, and produces fruits containing 3–17 seeds (Ohara et al., 2006). Our previous pollination experiments revealed that C. keiskei is highly self-incompatible, and outcrossing is mediated by insect pollinators of the orders Diptera, Coleoptera and Hymenoptera (Araki et al., 2005). No effective seed dispersers have been identified, although wild mice eat both the fruits and seeds of this plant (K. Araki et al., unpubl. res.). The aerial shoots die back during September and October, leaving only belowground structures to overwinter. New shoot and flower buds pre-formed at the basal area by the next spring elongate at the same position every year. Convallaria keiskei also propagates clonally by growing linear rhizomes that extend from the parent to a new offspring ramet. A clonal fragment usually comprises an average of 1·86 (s.d. ± 1·60) and up to nine ramets, and isolated ramets are also scattered throughout the genet (Araki and Ohara, 2008).
Study sites and field measurements
The present study was carried out at two forest sites in Nakasatsunai (143°10′N, 42°40′E) in eastern Hokkaido, northern Japan. In 2003, two permanent plots (A and B; 1 × 2 m) were established in a deciduous forest dominated by Quercus dentata, where highly dense patches of C. keiskei were found, and two other plots (C and D; 2 × 2 m) in a windbreak forest of Larix leptolepis, with relatively low densities of C. keiskei.
The field study was performed between 2003 and 2005. In 2003, all aerial shoots observed within the plots were marked and mapped. The leaf size (length) of each individual shoot was measured and recorded, as were the ontogenetic stages, i.e. single-leaf or two-leaf sterile or two-leaf fertile stages. For the two-leaf sterile and fertile stages, the interior leaf length of a sheath when foliage leaves were fully expanded was measured every season. Because a new aerial shoot of ramets grows from the same underground organ every year, we can adequately discern individual shoots. Thus, the fate and successive change of growth stages and leaf size of marked shoots were re-measured and newly emerged shoots within the plots were also marked and measured until 2005.
DNA analysis
Leaf tissues of all ramets (those present in 2003 and newly emerged in 2004 and 2005) existing in the plots were collected when the ramets were marked, and then stored in a freezer until analysis. To minimize the damage to plant growth, only about 1 cm2 was collected from each leaf. DNA was extracted using the CTAB (cetyltrimethylammonim bromide) method, and 5 ng of the extracted DNA was then amplified using a set of four labelled primer pairs targeting highly polymorphic DNA microsatellite loci according to previously reported protocols (GenBank accession nos AB251398, AB251399, AB251400 and AB251401; see Araki et al., 2006). Size separation of the PCR products was carried out using capillary electrophoresis on an ABI 3100 genetic analyser (Applied Biosystems). Size scoring of banding patterns and genotyping were performed in a semi-automated manner using the software program GENESCAN (Applied Biosystems). The likelihoods of errors falsely ascribing genotypes to the same genet, Pgen (Parks and Werth, 1993), of the primers used were all <0·001 when calculated for the whole population of A and B plots (Araki et al., 2006). Thus, all of the same genotypes identified by using the primers were considered to be members of the same clone. Moreover, in order to distinguish clearly between nearby genets in small plots, several samples of each genotype were additionally analysed using further primers (GenBank accession nos AB251396, AB251397, AB25142 and AB25143). For ramets showing unclear banding patterns, the genotypes of connected bands were determined by analysis with other primers described above. By this analysis, no additional genotypes were identified. Clonal diversity in each plot was assessed by Simpson's index, D = 1 – Σqg2, where qg is the frequency of the g-th genotype.
Data analysis
Size structure of genets
Based on field measurements and molecular data, ramet size class structures of the predominant genets for the four plots were determined. For convenience, ramets of the predominant genets were classified into six categories according to the leaf length: <9, 9–12, 12–15, 15–18, 18–21 and >21 (cm).
The size class histograms were bell-shaped, like a normal distribution (Fig. 2). To compare size distributions across genets, their changes in each genet over 3 years and differences between offspring and parental ramets, the normal distribution model was applied (Sakamoto et al., 1986) to the size class structures. When leaf sizes are l1, l2, … , ln, the likelihood of the normal distribution model of mean μ and variance σ2 is expressed as
and the maximum likelihood method leads us to estimates μ = Σli/n and σ = Σ(li – μ)2/n. The maximum likelihood estimates were calculated using all possible combinations of equal and different μ and σ, and the results were compared by calculating the Akaike information criterion (AIC) = −2 (maximum log-likelihood) + (number of parameters). The combination that exhibited the smallest AIC was selected. Unlike the popular t-test, this method enables differences of both means and variances to be testede at the same time. Because of the rarity of being able to compare ramets in different growth stages, i.e. ramets shifted from one leaf to two leaves and vice versa during the 3 years, we used only two-leaf ramets in this analysis.
Fig. 2.
Size class structure of Convallaria keiskei ramets of a predominant genet(s) in each plot.
Spatial pattern analysis
The spatial patterns of ramet distribution were also analysed for the four plots by using the pair correlation function, g(r), which estimates how many pairs of ramets are present with interdistance r and is normalized so as to be equal to 1 when the distribution is random (Stoyan and Stoyan, 1994). g(r) > 1 for small r suggests the clustering of ramets, while g(r) < 1 means that ramets are regularly spaced. If g(r) » 0 for r » 0 and increases toward 1 for r » r0, this scale (r0) can be interpreted as a general interval of ramet spacing. Otherwise, if g(r) increases past 1 and reaches a peak at r0 satisfying g(r0) > 1, ramets are clustered while maintaining this distance. The analysed data were those of predominant genets, as described above.
For each genet, mapped shoots {Xi} (i = 1, 2, … , n) in a rectangular plot a × b in 2005, g(r) was estimated by:
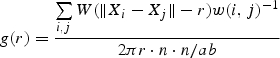 |
where w (i, j) is Ripley's edge correction factor, the proportion of a circumference centred at Xi with a radius of ‖Xi – Xj‖ that belong to the inside of the plot, W(t) = ¾δ(1 – t2/δ2) if |t| <δ and 0 otherwise (the Epanecnikov kernel function) where δ is an arbitrarily fixed constant. We tested δ (=1, 2, … , 10), and settled on δ = 5 cm as the most appropriate value for illustrating the spatial patterns of ramets.
For three plots (i.e. plots A, B and D), the spatial pattern of all ramets and flowering ramets of one genet dominating in 2005 was explored. For plot C, where two major clones were detected, the patterns of ramets of the two genets and the largest clone (c-1) were each calculated and compared.
RESULTS
Population structure of C. keiskei
A total of 274 and 149 ramets were found in plots A and B, respectively, during 3 years, and 83 and 94 in plots C and D (Table 1). In the total of 580 samples analysed, 38 different genets were detected. The number of genets in each plot ranged from five (B) to 18 (C). In plots A, B and D, one genet was predominant and consisted of >90 % of the ramets. In plot C, there were two predominant genets, which included 58 and 21 % of the ramets. The other genotypes detected consisted of only 1–10 ramets in all plots. Thus, clonal diversity (Simpson's index) in each plot was 0·13 (A), 0·08 (B), 0·62 (C) and 0·17 (D), respectively. Small one-leaf ramets (<6 cm in leaf length) were distinguished by unique genotypes, and were accordingly identified as seedlings germinated in relatively recent seasons. The proportion of ramets having one leaf (including seedlings) was the highest in plot C in 2005 (22·2 % of ramets), and seedlings sprouting in dense clumps were also observed every year in plot C. On the other hand, there were no seedlings in plot B during the 3 years.
Table 1.
Summary of census of genets and ramets in each plot
| Ramets of predominant genets |
Ramets of the other genets |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plot | Area (m2) | No. of ramets | No. of samples analysed | No. of genets identified | Year | Genet ID | Total | FL | NEW* | UEM | Total | FL | NEW* | UEM | Seedling† |
| A | 2 × 1 | 274 | 265 | 9 | 2003 | a-1 | 209 | 24 | – | – | 15 | 1 | – | – | (1) |
| 2004 | 207 | 64 | 12 | 14 | 16 | 2 | 3 | 2 | 0 | ||||||
| 2005 | 235 | 55 | 24 (8) | 4 | 17 | 2 | 0 (1) | 0 | 0 | ||||||
| B | 2 × 1 | 149 | 146 | 5 | 2003 | b-1 | 123 | 25 | – | – | 5 | 1 | – | – | 0 |
| 2004 | 122 | 25 | 7 | 8 | 5 | 0 | 0 | 0 | 0 | ||||||
| 2005 | 135 | 17 | 10 (7) | 4 | 6 | 1 | 1 | 0 | 0 | ||||||
| C | 2 × 2 | 83 | 81 | 18 | 2003 | c-1 | 45 | 1 | – | – | 3 | 0 | – | – | (3) |
| 2004 | 47 | 3 | 2 | 0 | 4 | 0 | 1 | 0 | 6 | ||||||
| 2005 | 47 | 5 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | ||||||
| 2003 | c-2 | 14 | 0 | – | – | ||||||||||
| 2004 | 16 | 0 | 2 | 0 | |||||||||||
| 2005 | 17 | 2 | 1 | 0 | |||||||||||
| D | 2 × 2 | 94 | 88 | 6 | 2003 | d-1 | 59 | 13 | – | – | 3 | 1 | – | – | 0 |
| 2004 | 67 | 8 | 8 | 0 | 5 | 0 | 2 | 0 | 3 | ||||||
| 2005 | 80 | 23 | 13 | 0 | 4 | 3 | 0 | 1 | 0 | ||||||
The number of total, flowering (FL), newly emerged (NEW) and unemerged (UEM) ramets of predominant genets and the other genets and seedlings in 2003, 2004 and 2005 in each plot were integrated according to genotypes. Dashes indicate data neither calculated nor estimated.
*The number in parentheses represents recovering ramets, which did not emerge in the previous year.
†The number in parentheses indicates the ramets that were <6 cm and detected by unique genotypes even though their birth year could not be detected.
Figure 1 illustrates the distribution of individual ramets and its yearly changes in plot A. It can be recognized that one genet exclusively expanded in this plot. Ramets of rare genets were scattered among the ramets of the predominant genet. In plot A, most ramets had two leaves, and the frequency of flowering ramets (genotypically identified ramets) varied from 10·9 % (25/224) in 2003 to 29·6 % (66/223) in 2004 (cf. Table 1). On the other hand, the frequency of flowering ramets was low in plot C, ranging from 1·6 % (1/62) in 2003 to 10·3 % (7/68) in 2005 (data not shown). It is interesting to note that the position of flowering shoots in the plots also changed from year to year, and ramets having an inflorescence rarely produced flowers the next year even though most developed new aerial shoots (Fig. 1). After 2 years, some had another inflorescence. New ramets produced by clonal growth, belonging to both one-leaf and two-leaf stages, were found in all the plots every year. The number of unemerged ramets was a few or none in all study plots. Even in plots A and B with highly dense ramets, only 7·0 % (16 ramets) and 6·3 % (eight ramets) vanished in 2004 at most, and nearby half recovered the next year; thus, ramet mortality seems quite low.
Fig. 1.
Distributions and seasonal changes of Convallaria keiskei ramets in plot A over 3 years (2003–2005).
Size structure of ramets of a genet
Although the plot sizes and number of ramets differed among the plots, overall the size class structures of (a) predominant genet(s) in each plot were consistent, the intermediate size always being the best represented. Ramets in the 15–18 cm leaf size class were the most dominant in genets a-1, b-1 and d-1 (Fig. 2). In plot C, where two major genets (c-1 and c-2) were detected, the largest number of ramets was found in the 12–15 cm class for c-1, whereas all the ramets were larger than the 15–18 cm class for c-2 (Fig. 2), showing that these two co-existing clones were morphologically quite different.
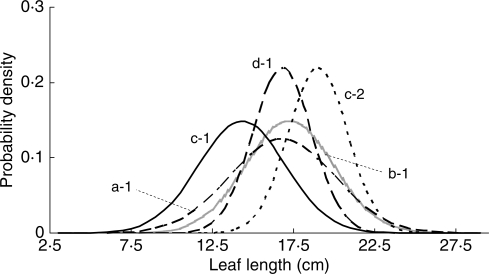
Comparing and optimizing the normal distribution model of each genet suggested that ramets in different genets comprised different size class structures with discrete means and variances (Fig. 3). Of the predominant genets that were compared, a-1, b-1 and d-1 showed similar mean values (16·3, 16·7 and 16·3 cm, respectively), while c-1 (13·8 cm) was smaller and c-2 (18·5 cm) was larger, which was consistent with the results from the size class histograms (Fig. 2). The variance of c-2 and d-1 (3·3), on the other hand, was smaller than that of b-1 (10·2), a-1 and c-1 (7·2). These values revealed the morphological characteristics of genets, regardless of differences in ramet densities and/or their mean ramet sizes. For instance, the curve for a-1 showed higher within-genet variation in ramet size than that for b-1 (Fig. 3) despite their similar densities (Table 1) and average sizes. The normalized model of c-1, in contrast, clearly showed the characteristic that this clone was composed of larger ramets.
Fig. 3.
The selected normal distribution model of the size structures for five genets.
Analysis with normal distribution models also revealed that for new ramets, seedlings (with unique genotypes) were significantly smaller than the offspring ramets produced by clonal growth (Table 2). Offspring ramets seemed to be as large as parents persisting for 3 years, although their sizes differed depending on the genet according to the parental characteristics.
Table 2.
The mean and s.d. of the selected normal distribution model for the size structure of new ramets
| Ramet | Mean | s.d. |
|---|---|---|
| Seedling | 4·07 | 0·58 |
| a-1 clonal | 14·85 | 3·45 |
| b-2 clonal | 14·85 | 2·93 |
| c-1 clonal | 10·90 | 2·10 |
| c-2 clonal | 16·50 | 0·80 |
| d-1 clonal | 12·54 | 2·97 |
| *Parent | 15·58 | 3·29 |
The model was selected by AIC values.
*Parent ramets were emerging over 3 years in all study plots.
Temporal dynamics of ramets and genets
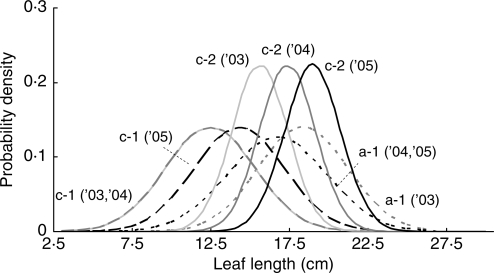
Census data revealed that most ramets foliated the same number of leaves in succeeding years, while 18·0 % of shoots shifted from one-leaf to two-leaf ramets or vice versa. As for the five major genets detected in the present study, the size class structure of the same genet varied over the years, and the variation seemed to reflect the growth at the genet level (Fig. 4). For example, in genet a-1, the average leaf length of 17·9 cm in 2003 changed to 16·2 cm in succeeding years. Genet c-1 grew from 12·1 cm (2003 and 2004) to 13·8 cm (2005), while genet c-2 shifted toward a larger size every year (13·8, 16·9 and 18·5 cm, respectively). Thus, the temporal patterns of the structures also differed among genets, in addition to the differences of the structures themselves even for genets in the same place. No different variances were detected for any of the genets over the years.
Fig. 4.
The selected normal distribution models of the transition patterns of the size structures of three genets (a-1, c-1 and c-2) over 3 years (2003–2005).
Spatial distribution of ramets
Spatial distribution analysis of three dense genets, i.e. a-1, b-1 and d-1, revealed the clustered distribution of all ramets in one genet (Fig. 5). Flowering ramets of each genet, on the other hand, showed a weaker clumping (a-1), or distant distribution of 5, 20 and 30 cm (b-1), and 3, 15 and 30 cm (d-1), respectively. In plot C, where two dominant clones (c-1 and c-2) were sparsely scattered, if both clones were included, some ramets were slightly clustered and others were 10 and 28 cm apart from each other. In contrast, in a single genet, c-1, ramets tended not to be close to each other, and were instead located at least 7–8 cm away (Fig. 5).
Fig. 5.
Spatial patterns of ramets in a genet analysed in Convallaria keiskei by using the pair correlation function. Spatial patterns of all ramets and flowering ramets (as indicated) of genets a-1, b-1 and d-1 are shown. For genet c-1 and c-2, the patterns of ramets of two genets and that of a single genet (c-1) were analysed in 2005.
DISCUSSION
Clonal distribution at the fine-scale
Despite a total of 94 genotypes found in the population of C. keiskei (Araki et al., 2007), in the current study focusing on all ramets in 1 × 2 m and 2 × 2 m quadrats, much smaller numbers of genets to which most ramets belonged were discovered (Fig. 1 and Table 1). This distribution indicated patchy structures which were usually formed by a single genet, although long rhizomes are often related to intermingled distributions of multiple genets in other species. In other plants such as Uvularia perfoliata L. (Kudoh et al., 1999), Anemone nemorosa (Stehlik and Holderegger, 2000), Cymodocea nodosa (Ruggiero et al., 2005) and Stenocereus eruca (Clark-Tapia et al., 2005), a larger number of genets discovered on a small scale suggested local genotypic diversity in clonal populations. In our investigation, only plot C included two predominant genets and unique genotypes which formed an intermingled distribution. Because clones of C. keiskei expand to several metres (Araki and Ohara, 2008), the degree of clonal intermingling may vary even in the same genet, depending on the position such as the middle and edge of the genet.
Seedling recruitment
Seedlings were discovered in plot C every year, and the occasional seedling was found in the other plots (Table 1), which provided direct evidence for sustained recruitment by sexual reproduction in the current C. keiskei population. Many studies of the genetic structure and genotypic diversity of clonal plant populations have indicated a significant contribution of sexual reproduction to the populations (e.g. Ellstrand and Roose, 1987; Honnay et al., 2005; Sherman and Ayre, 2008), but there has been little evidence of seedling recruitment (Eriksson, 1993; Damman and Cain, 1998). The emergence of rare genets at the fine-scale (Fig. 1; Kudoh et al., 1999; Stehlik and Holderegger, 2000; Ruggiero et al., 2005) is also indirect evidence for the repeated establishment of seedlings (Eriksson, 1989). Low and irregular sexual recruitment has so far been described as a trend in clonal rhizomatous woodland species (Bierzychudek, 1982; Whigham, 2004) and its frequency seems to be sufficient to maintain genetic variation in populations (Shirreffs, 1985; Eriksson, 1989; Watkinson and Powell, 1993). Further monitoring of the germinated seedlings must throw light on the their fates and their contribution to the population dynamics of the clonal herb C. keiskei.
Demography of ramets and genets
It should be possible to evaluate the genet dynamics by detecting the number of fertile ramets that are capable of producing offspring ramets in genotypically identified genets. Our demographic data revealed that a significant number of clonal offspring were persistently produced in the studied genets every year (Table 1). Such sustained clonal recruitment formed the bell-shaped size class structure of ramets, which has been reported in some populations of pseudo-annual clonal species (e.g. Disporum sessile and D. smilacinum, Kawano et al., 1987). Clonal growth maintaining and expanding genets has a great impact on the population dynamics (Eriksson, 1989; Watkinson and Powell, 1993; Tanner, 2001). Then, in our case of C. keiskei, their temporal patterns differed even among co-existing genets (Fig. 4). This might reflect genetic differences among genets and/or variations of the ramet generation of genets, and thus might indicate different responses of genets to microhabitat conditions (Argyres and Schmitt, 1991; Stratton, 1994; Falińska, 1995; Prati and Schmid, 2000).
Wijesinghe and Whigham (1997) reported genet growth patterns and concluded that population dynamics can be influenced by ramet size, because the number of clonal progeny and resource allocation to them can increase with increasing ramet size. For C. keiskei, the ramets that flowered but produced no fruits showed a marginal tendency to grow larger, while the ramets which had produced fruits became smaller than sterile ramets (data not shown). Hence, the trade-offs between ramet growth and sexual reproduction might also negatively affect the genet growth pattern (Cain and Damman, 1997; Lambrecht-McDowell and Radosevich, 2005). Finally, low mortality of ramets and fluctuating frequency of flowering (Table 1) also have an impact on such genet growth.
Clonal growth patterns
Analysis of the spatial distribution revealed that the placements of ramets in a given genet ranged from regularly spaced to clustering patterns (Fig. 5). Clonal distributions are generally constrained by the growth forms or clonal architecture (Lovett Doust, 1981; Navas and Garnier, 1990; Klimeš et al., 1997). Thus, the distant patterns found in less dense areas (e.g. c-1) in this study corresponded to linearly elongating rhizomes of C. keiskei. We previously observed that the length of rhizomes connecting two ramets was up to 22·72 (s.d. ± 9·17) cm long (Araki and Ohara, 2008). In general, long spacers (called guerrillas; sensu Lovett Doust, 1981) usually form an intermingled distribution (Humphrey and Pyke, 1998; Ruggiero et al., 2005), but we found such a distribution in just one plot (plot C in Fig. 5).
At the same time, clustering patterns, which usually correspond to relatively compact growth forms (called a phalanx) accompanied by branching (de Kroon and Schieving, 1990; Humphrey and Pyke, 1998; Ye et al., 2006; Thomas and Hay, 2008), were also observed in this study (Figs 1 and 5). However, our excavating investigation discovered that shoots in a dense area were connected with distant ramets rather than with neighbouring ramets (Araki and Ohara, 2008). Thus, dense patches seem to result from the co-mingling of many disconnected fragments and independent ramets in a single genet. Otherwise, in areas where ramets were scarce, ramets of different genets tended to be located closer to each other than ramets in the same genet (Figs 1 and 5). Therefore, local conditions substantially affect at least the way of clonal spreading (Navas and Garnier, 1990; Wijesinghe and Hutchings, 1997; Lenssen et al., 2004). Moreover, growth forms could change plastically depending on habitat conditions such as the amount of surrounding vegetation (Fischer et al., 2004; Macek and Lepš, 2008) or available nutrients (Niva et al., 2006; Ye et al., 2006). In natural populations, furthermore, clonal distributions must reflect not only the growth form, but also the historical background of the populations.
On the other hand, the regular space patterns of flowering ramets indicated that flowering occurred in ramets not in a certain area but throughout the genet even in dense genets (Fig. 5). This may imply that sexual reproduction of each ramet is less influenced by the microhabitat condition than clonal growth. Although the mechanisms of flowering onset in C. keiskei's ramets producing no inflorescence the year after flowering remain unclear, many factors other than habitat conditions may have more impact on determining reproduction (Huber et al., 2004; Lambrecht-McSowell and Radosevich, 2005).
Although earlier studies which detected clustering distributions suggested that two neighbouring ramets were often genetically identical (e.g. Harada and Iwasa, 1994; Harada et al., 1997; Holderegger et al., 1998; Suzuki et al., 2006), in the present study, by performing individual-based analysis using a method of point process and pair correlation function (Stoyan and Stoyan, 1994), we examined whether neighbouring ramets exist at a given distance, in addition to comparing genetic identity between the ramets.
Demographic–genetic studies of clonal plants
In this study, we performed a series of field observations in combination with genet identification, using a molecular marker in a clonal plant, C. keiskei. In contrast to previous demographic studies in clonal species that were based on either ramet behaviours or genetic analysis of population structures alone, our study emphasized particularly the importance of continuously monitoring ramets that were genotypically identified, not just combining the information of both field observations and genetic analysis; we call this approach ‘demographic–genetic analysis’. This approach is essential for differentiating the fates of clonal ramets from those of sexual seedlings. Our longitudinal data then successfully tracked the spatio-temporal transitions of both ramets and genets. Our results revealed that not only the processes of the two types of reproduction (sexual and clonal), but also horizontal spreading by clonal growth, may contribute importantly to the demographic features of clonal plants. Furthermore, because the results of studies using molecular markers are strongly dependent on the sampling strategy, the combination of different scales of sampling from large (Araki et al., 2007) to small (as in this study) is an effective approach for achieving a more precise understanding of the life history of clonal plants.
ACKNOWLEDGEMENTS
We would like to thank the village officers of Nakasatsunai for providing access to the study areas. We also thank Hiroko Tanaka for helping us with field measurements. This study was supported by Grants-in Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (18405010 and 1957001108), and a research fellowship of the JSPS for Young Scientists (19-256); the Ministry of Education, Culture, Sports, Science and Technology for the 21st COE Program (E-01) and Global COE Program (AO6), Japan; ISM Cooperative Research Program (17-2063, 18-2034, 19-2008); and the Hokkaido University Clark Commemoration Foundation support.
LITERATURE CITED
- Abrahamson WG. Demography and vegetative reproduction. In: Solbrig OT, editor. Demography and evolution in plant populations. Oxford: Blackwell; 1980. pp. 89–106. [Google Scholar]
- Araki K, Ohara M. Reproductive demography of ramets and genets in a rhizomatous clonal plant Convallaria keiskei. Journal of Plant Research. 2008;121:147–154. doi: 10.1007/s10265-007-0141-9. [DOI] [PubMed] [Google Scholar]
- Araki K, Yamada E, Ohara M. Breeding system and floral visitor of Convallaria keiskei. Plant Species Biology. 2005;20:151–155. [Google Scholar]
- Araki K, Lian CL, Shimatani K, Ohara M. Development of microsatellite markers in a clonal perennial herb, Convallaria keiskei. Molecular Ecology Notes. 2006;6:1144–1146. [Google Scholar]
- Araki K, Shimatani K, Ohara M. Floral distribution, clonal structure, and their effects on pollination success in a self-incompatible Convallaria keiskei population in northern Japan. Plant Ecology. 2007;189:175–186. [Google Scholar]
- Argyres AZ, Schmitt J. Microgeographic genetic structure of morphological and life-history traits in a natural population of Impatiens capensis. Evolution. 1991;45:178–189. doi: 10.1111/j.1558-5646.1991.tb05276.x. [DOI] [PubMed] [Google Scholar]
- Bierzychudek P. Life histories and demography of shade-tolerant temperate forest herbs: a review. New Phytologist. 1982;90:757–776. [Google Scholar]
- Cain ML, Damman H. Clonal growth and ramet performance in the woodland herb, Asarum canadense. Journal of Ecology. 1997;85:883–897. [Google Scholar]
- Clark-Tapia R, Alfonso-Corrado C, Eguiarte LE, Molina-Freaner F. Clonal diversity and distribution in Stenocereus eruca (Cactaceae), a narrow endemic cactus of the Sonoran desert. American Journal of Botany. 2005;92:272–278. doi: 10.3732/ajb.92.2.272. [DOI] [PubMed] [Google Scholar]
- Cook RE. Growth and development in clonal plant populations. In: Jackson JBC, Buss LW, Cook RE, editors. Population biology and evolution of clonal organisms. New Haven: Yale University Press; 1985. pp. 259–296. [Google Scholar]
- Damman H, Cain ML. Population growth and viability analyses of the clonal woodland herb, Asarum canadense. Journal of Ecology. 1998;86:13–26. [Google Scholar]
- Ellstrand NC, Roose ML. Patterns of genotypic diversity in clonal plant species. American Journal of Botany. 1987;74:123–131. [Google Scholar]
- Eriksson O. Seedling dynamics and life histories in clonal plants. Oikos. 1989;55:231–238. [Google Scholar]
- Eriksson O. Dynamics of genets in clonal plants. Trends in Ecology and Evolution. 1993;8:313–316. doi: 10.1016/0169-5347(93)90237-J. [DOI] [PubMed] [Google Scholar]
- Eriksson O, Bremer B. Genet dynamics of the clonal plant Rubus saxatilis. Journal of Ecology. 1993;81:533–542. [Google Scholar]
- Eriksson O, Jerling L. Hierarchical selection and risk spreading in clonal plants. In: van Groenendael J, de Croon H, editors. Clonal growth in plants: regulation and function. The Hague: SPB Academic Publishing; 1990. pp. 79–94. [Google Scholar]
- Fair J, Lauenroth WK, Coffin DP. Demography of Bouteloua gracilis in a mixed prairie: analysis of genets and individuals. Journal of Ecology. 1999;87:233–243. [Google Scholar]
- Falińska K. Genet disintegration in Filipendula ulmaria: consequences for population dynamics and vegetation succession. Journal of Ecology. 1995;83:9–21. [Google Scholar]
- Fischer M, van Kleunen M. On the evolution of clonal plant life histories. Evolutionary Ecology. 2002;15:565–582. [Google Scholar]
- Fischer M, van Kleunen M, Schmid B. Experimental life-history evolution: selection on growth form and its plasticity in a clonal plant. Journal of Evolutionary Biology. 2004;17:331–341. doi: 10.1046/j.1420-9101.2003.00677.x. [DOI] [PubMed] [Google Scholar]
- Geber A, Watson MA, Furnish R. Genetic differences in clonal demography in Eichhornia crassipes. Journal of Ecology. 1992;80:329–341. [Google Scholar]
- Guárdia R, Raventós J, Caswell H. Spatial growth and population dynamics of a perennial tussock grass (Achnatherum calamagrostis) in a badland area. Journal of Ecology. 2000;88:950–963. [Google Scholar]
- Harada Y, Iwasa Y. Lattice population dynamics for plants with dispersing seeds and vegetative propagation. Research on Population Ecology. 1994;36:237–249. [Google Scholar]
- Harada Y, Kawano S, Iwasa Y. Probability of clonal identity: inferring the relative success of sexual versus clonal reproduction from spatial genetic patterns. Journal of Ecology. 1997;85:591–600. [Google Scholar]
- Hartnett DC, Bazzaz FA. The genet and ramet population dynamics of Solidago Canadensis in an abandoned field. Journal of Ecology. 1985;73:407–413. [Google Scholar]
- Holderegger R, Stehlik I, Schneller JJ. Estimation of the relative importance of sexual and vegetative reproduction in the clonal woodland herb Anemone nemorosa. Oecologia. 1998;117:105–107. doi: 10.1007/s004420050637. [DOI] [PubMed] [Google Scholar]
- Honnay O, Jacquemyn H, Bossuyt B, Hermy M. Forest fragmentation effects on patch occupancy and population viability of herbaceous plant species. New Phytologist. 2005;166:723–734. doi: 10.1111/j.1469-8137.2005.01352.x. [DOI] [PubMed] [Google Scholar]
- Huber H, Whigham DF, O'Neill J. Timing of disturbance changes the balance between growth and survival of parent and offspring ramets in the clonal forest understory herb Uvularia perfoliata. Evolutionary Ecology. 2004;18:521–539. [Google Scholar]
- Humphrey LD, Pyke DA. Demographic and growth responses of a guerrilla and a phalanx perennial grass in competitive mixtures. Journal of Ecology. 1998;86:854–865. [Google Scholar]
- Kawano S, Takada T, Nakayama S, Hiratsuka A. Demographic differentiation and life-history evolution in temperate woodland plants. In: Urbanska KM, editor. Differentiation patterns in higher plants. London: Academic Press; 1987. pp. 153–181. [Google Scholar]
- Klimeš L, Klimesova J, Hendriks R, van Groenendael J. Clonal plant architecture: a comparative analysis. In: de Kroon H, van Groenendael J, editors. The ecology and evolution of clonal plants. Leiden: Backhuys Publishers; 1997. pp. 1–29. [Google Scholar]
- de Kroon H, Schieving F. Resource partition in relation to clonal growth strategy. In: van Groenendael J, de Kroon H, editors. Clonal growth in plants: regulation and function. The Hague: SPB Academic Publishing; 1990. pp. 113–130. [Google Scholar]
- de Kroon H, van Groenendael J. The ecology and evolution of clonal plants. Leiden: Backhuys Publishers; 1997. [Google Scholar]
- Kudoh H, Shibaike H, Takasu H, Whigham DF, Kawano S. Genet structure and determinants of clonal structure in a temperate deciduous woodland herb, Uvularia perfoliata. Journal of Ecology. 1999;87:244–257. [Google Scholar]
- Lambrecht-McDowell SC, Radosevich SR. Population demographics and trade-offs to reproduction of an invasive and noninvasive species of Rubus. Biological Invasions. 2005;7:281–295. [Google Scholar]
- Lenssen JPM, van Kleunen M, Fischer M, de Kroon H. Local adaptation of the clonal plant Ranunculus reptans to flooding along a small-scale gradient. Journal of Ecology. 2004;92:696–706. [Google Scholar]
- Lovett Doust L. Population dynamics and local specialization in a clonal perennial (Ranunculus repens). I. The dynamics of ramets in contrasting habitats. Journal of Ecology. 1981;69:743–755. [Google Scholar]
- Macek P, Lepš J. Environmental correlates of growth traits of the stoloniferous plant Potentilla palustris. Evolutionary Ecology. 2008;22:419–435. [Google Scholar]
- Navas M-L, Garnier E. Demography and growth forms of the clonal perennial Rubia peregrina in Mediterranean vineyard and unmanaged habitats. Journal of Ecology. 1990;78:691–712. [Google Scholar]
- Niva M, Svensson BM, Karlsson PS. Effects of light and water availability on shoot dynamics of the stoloniferous plant Linnaea borealis. Ecoscience. 2006;13:318–323. [Google Scholar]
- Ohara M, Araki K, Yamada E, Kawano S. Life-history monographs of Japanese plants. 6: Convallaria keiskei Miq. (Convallariaceae) Plant Species Biology. 2006;21:119–126. [Google Scholar]
- Parks JC, Werth CR. A study of spatial features of clones in a population of bracken fern, Pteridium aquilium (Dennstaedtiaceae) American Journal of Botany. 1993;80:537–544. doi: 10.1002/j.1537-2197.1993.tb13837.x. [DOI] [PubMed] [Google Scholar]
- Persson HA, Gustavsson BA. The extent of clonality and genetic diversity in lingonberry (Vaccinium vitis-idaea L.) revealed by RAPDs and leaf-shape analysis. Molecular Ecology. 2001;10:1385–1397. doi: 10.1046/j.1365-294x.2001.01280.x. [DOI] [PubMed] [Google Scholar]
- Prati D, Schmid B. Genetic differentiation of life-history traits within populations of the clonal plant Ranunculus reptans. Oikos. 2000;90:442–456. [Google Scholar]
- Price EAC, Marshall C. Clonal plants and environmental heterogeneity. Plant Ecology. 1999;141:3–7. [Google Scholar]
- Ruggiero MV, Reusch TBH, Procaccini G. Local genetic structure in a clonal dioecious angiosperm. Molecular Ecology. 2005;14:957–967. doi: 10.1111/j.1365-294X.2005.02477.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Ishiguro M, Kitagawa G. Akaike information criterion statistics. Tokyo: Kluwer Academic Publishers; 1986. [Google Scholar]
- Sampaio MC, Picó FX, Scarano FR. Ramet demography of a nurse bromeliad in Brazilian restingas. American Journal of Botany. 2005;92:674–681. doi: 10.3732/ajb.92.4.674. [DOI] [PubMed] [Google Scholar]
- Sherman CDH, Ayre DJ. Fine-scale adaptation in a clonal sea anemone. Evolution. 2008;62:1373–1380. doi: 10.1111/j.1558-5646.2008.00375.x. [DOI] [PubMed] [Google Scholar]
- Shirreffs DA. Biological flora of the British Isles: Anemone nemorosa L. Journal of Ecology. 1985;73:1005–1020. [Google Scholar]
- Soane ID, Watkinson AR. Clonal variation in populations of Ranunculus repens. New Phytologist. 1979;82:557–573. [Google Scholar]
- Stehlik I, Holderegger R. Spatial genetic structure and clonal diversity of Anemone nemorosa in late successional deciduous woodlands of central Europe. Journal of Ecology. 2000;88:424–435. [Google Scholar]
- Steinger T, Körner C, Schmid B. Long-term persistence in a changing climate: DNA analysis suggests very old ages of clones of alpine Carex curvula. Oecologia. 1996;105:94–99. doi: 10.1007/BF00328796. [DOI] [PubMed] [Google Scholar]
- Stone L, Ezrati S. Chaos, cycles and spatiotemporal dynamics in plant ecology. Journal of Ecology. 1996;84:279–291. [Google Scholar]
- Stoyan D, Stoyan H. Fractals, random shapes and point fields. Chichester: Wiley and Sons; 1994. [Google Scholar]
- Stratton DA. Genotype-by-environment interactions for fitness of Erigeron annuus show fine-scale selective heterogeneity. Evolution. 1994;48:1607–1618. doi: 10.1111/j.1558-5646.1994.tb02199.x. [DOI] [PubMed] [Google Scholar]
- Stuefer JF. Potential and limitations of current concepts regarding the response of clonal plants to environmental heterogeneity. Vegetatio. 1996;127:245–271. [Google Scholar]
- Suzuki J, Herben T, Krahulec F, Storchová H, Hara T. Effects of neighbourhood structure and tussock dynamics on genet demography of Festuca rubra in a mountain meadow. Journal of Ecology. 2006;94:66–76. [Google Scholar]
- Tanner JE. The influence of clonality on demography: patterns in expected longevity and survivorship. Ecology. 2001;82:1971–1981. [Google Scholar]
- Thomas RG, Hay MJM. Adaptive variation in physiological traits underpinning stem elongation responses among nodally-rooting stoloniferous herbs. Evolutionary Ecology. 2008;22:369–381. [Google Scholar]
- Tuomi J, Vuorisalo T. Hierarchical selection in modular organisms. Trends in Ecology and Evolution. 1989;4:209–213. doi: 10.1016/0169-5347(89)90075-X. [DOI] [PubMed] [Google Scholar]
- Watkinson AR, Powell JC. Seedling recruitment and the maintenance of clonal diversity in plant populations – a computer simulation of Ranunculus repens. Journal of Ecology. 1993;81:707–717. [Google Scholar]
- Whigham DF. Ecology of woodland herbs in temperate deciduous forests. Annual Review of Ecology, Evolution and Systematics. 2004;35:583–621. [Google Scholar]
- Wijesinghe DK, Hutchings MJ. The effects of spatial scale of environmental heterogeneity on the growth of a clonal plant: an experimental study with Glechoma hederacea. Journal of Ecology. 1997;85:17–28. [Google Scholar]
- Wijesinghe DK, Whigham DF. Costs of producing clonal offspring and the effects of plant size on population dynamics of the woodland herb Uvularia perfoliata (Liliaceae) Journal of Ecology. 1997;85:907–919. [Google Scholar]
- Ye X-H, Yu F-H, Dong M. A trade-off between guerrilla and phalanx growth forms in Leymus secalinus under different nutrient supplies. Annals of Botany. 2006;98:187–191. doi: 10.1093/aob/mcl086. [DOI] [PMC free article] [PubMed] [Google Scholar]