Abstract
Nucleocytoplasmic trafficking of macromolecules, a highly specific and tightly regulated process, occurs exclusively through the Nuclear Pore Complex. This immense structure is assembled from approximately 30 proteins, termed nucleoporins. Here we discuss the four nucleoporins that have been linked to cancers, either through elevated expression in tumors (Nup88) or through involvement in chromosomal translocations that encode chimeric fusion proteins (Tpr, Nup98, Nup214). In each case we consider the normal function of the nucleoporin and its translocation partners, as well as what is known about their mechanistic contributions to carcinogenesis, particularly in leukemias. Studies of nucleoporin-linked cancers have revealed novel mechanisms of oncogenesis and. in the future, should continue to expand our understanding of cancer biology.
Keywords: nuclear pore complex, nucleoporins, cancer, leukemia, chromosomal translocation, oncogene
1. The Nuclear Pore Complex
The nuclear pore complex (NPC) is a massive, multiprotein structure responsible for traffic between the nucleus and cytoplasm. The general structure of the NPC is becoming fairly well defined, but the mechanism of translocation through the pore remains incompletely understood. The NPC is comprised of a central core region consisting of nucleoplasmic and cytoplasmic rings joined by a spoke structure (Figure 1). These are anchored in the nuclear membrane through three transmembrane proteins. Cytoplasmic filaments and the nuclear basket of the pore extend out from this central structure. Pore proteins or nucleoporins (designated Nup followed by their predicted molecular weight) are modular in that a limited number of structural motifs (coiled-coils, α solenoids, β propellers) are used repeatedly to build the symmetrical NPC structure. Approximately one third of nucleoporins contain a domain of phenylalanine-glycine (FG) motifs interspersed with spacer sequences. These repeat domains are natively unstructured and serve as interaction sites for transport receptors (karyopherins) that escort cargo through the pore. For more information on NPC structure and function, a number of excellent reviews are available (1–5).
Figure 1. Structural organization of the Nuclear Pore Complex.
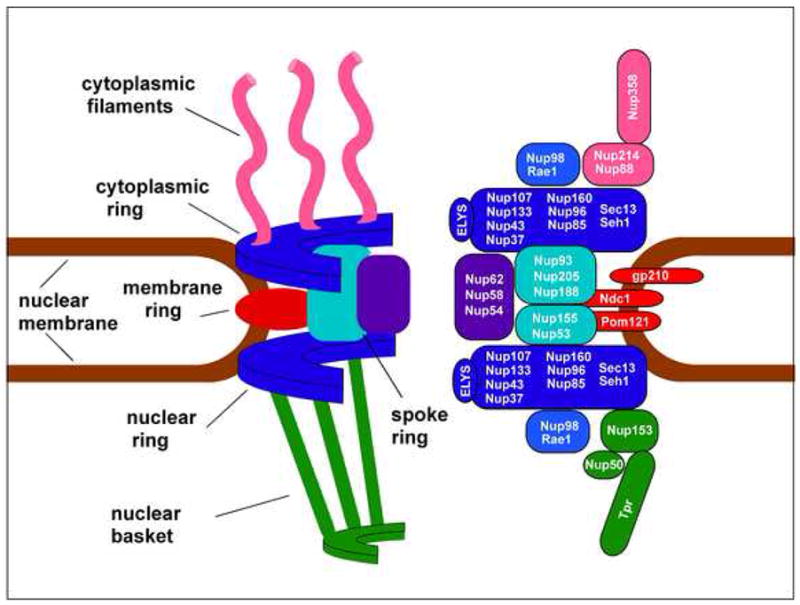
Left: a cutaway representation of the organization of the NPC with the major structural elements indicated. Three of the eight repeating segments that give the NPC 8-fold rotational symmetry are illustrated. Right: Schematic of nucleoporin subcomplexes and approximate position within the NPC. Substructures are color coded to match the left side of the panel.
2. Nup88: a potential marker for high grade tumors
2.1 Nup88 in multiple subcomplexes of the NPC
Nup88 is a non-FG nucleoporin found exclusively on the cytoplasmic face of the NPC (Figure 1). This nucleoporin is comprised of two of the repeating structural motifs of the pore; the N-terminal domain is predicted to form a β-propeller structure and the C-terminal domain contains predicted coiled-coils (Figure 2A). Nup88 is found in a biochemically defined subcomplex of the pore together with the FG repeat nucleoporin Nup214 (6, 7) and, in some systems, Nup62 (8). Formation of this complex is mediated through the N-terminal β-propeller of Nup88 and a central coiled–coil domain of Nup214. Findings differ on whether Nup88 directs Nup214 to the NPC or whether both components of the complex are required simultaneously for NPC association. Additionally, there are some indications that one partner may be unstable in the absence of the other. However, Nup88 can be overexpressed relative to Nup214 and under these conditions it accumulates in the cytoplasm of both cultured human cells and Drosophila melanogaster larvae (9, 10).
Figure 2. Nucleoporin structure and rearrangement following chromosomal translocations.
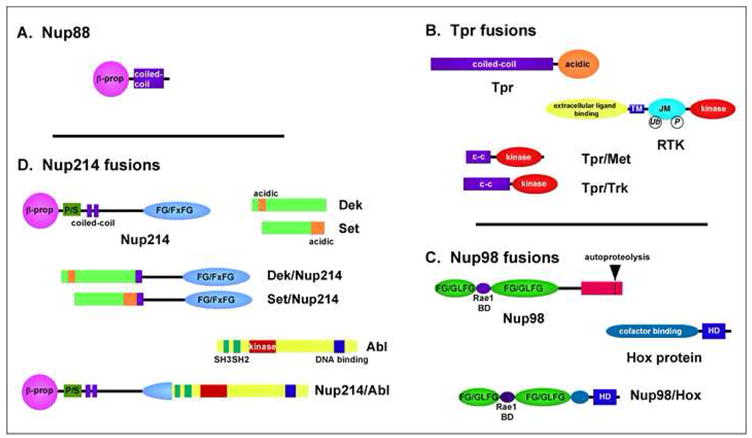
The four nucleoporins discussed are depicted with major structural elements, along with their translocation partners, if applicable, and the structure of the resulting chimeric fusion proteins. Abbreviations: β-prop, β-propeller; c-c, coiled-coil; RTK, receptor tyrosine kinase; P/S, proline, serine rich; TM, transmembrane; JM, juxtamembrane; HD, homeodomain; BD, binding domain; Ub, ubiquitination; P, phosphorylation.
Nup88 can also be found in a second NPC subcomplex with another FG nucleoporin, Nup358 (Figure 1), the major component of cytoplasmic filaments of the pore and the binding site for RanGAP. Again, Nup88 seems to play an important role in targeting this subcomplex to the NPC, although the amount of Nup358 in the cell appears not to be linked to the level of Nup88 (10). Nup88 also interacts with the C-terminus of another FG nucleoporin, Nup98 (11). Rather than forming a separate subcomplex, this interaction likely corresponds to a link between the Nup214/Nup88 complex and Nup98.
2.2 The Nup214/Nup88 subcomplex in nuclear transport
The Nup214/Nup88 subcomplex does not seem to be directly required for nuclear import, but it makes important, if somewhat controversial, contributions to nuclear export. Both the FG repeat domain of Nup214 and the N-terminal β-propeller domain of Nup88 have been reported to bind directly to CRM-1/exportin-1, the receptor for export of most proteins from the nucleus (7, 12). The Nup214/Nup88 subcomplex is crucial for CRM-mediated export of the 60S ribosomal particle from the nucleus although, unexpectedly, this required the Nup88-binding coiled-coils of Nup214 but not the Nup214 FG domain (13). However, the contribution of the Nup214/Nup88 complex in the export of nuclear export signal (NES)-containing proteins varies between systems and substrates. In HeLa cells, depletion of the Nup214/Nup88 complex leads to greatly diminished export of the nuclear transcription factor NFAT (14) but has a minimal affect on export of a classical nuclear localization sequence (cNLS)-GFP-NES reporter (13). In cultured Drosophila S2 cells, depletion of either Nup214 or Nup88 did not alter the localization of a GFP-NES reporter (15). In contrast, Drosophila larvae lacking either Nup214 or Nup88 show a striking cytoplasmic accumulation of the NF-κB homologs, Dorsal and Dif, which contain NESs and normally shuttle between nucleus and cytoplasm. These results led the authors to propose that the normal function of Nup214/Nup88 in larvae is to retain CRM at the NPC and attenuate rather than enhance nuclear export (9). The variation in reported roles for Nup214 and Nup88 in nuclear protein export may reflect either a degree of redundancy in cultured cells that is lacking in embryos, or differences in the specific requirements of individual protein cargoes used in the assays.
A surprising finding came from studies in Drosophila demonstrating that Nup88 (known in D. melanogaster as members only, mbo; Table 1) was expressed in a tissue-specific manner (16). Although some other Nups exhibit tissue-specific expression (17–19), given the role of Nup88 in directing other nucleoporins to the NPC, it was not expected that it would be absent from many developing tissues. In Nup88 null larvae, NF-κB homologs did not relocalize to the nucleus of fat body cells in response to immune stimuli, perhaps due to increased nuclear export in the absence of Nup88. However, in tissues where Nup88 was normally not expressed, NF-κB homologs were correctly relocalized, suggesting that Nup88 is not universally required for regulation of nuclear export. Presumably there are redundant pathways that function in tissues lacking Nup88.
Table 1.
Nomenclature of nucleoporin orthologs
| mammals | S. cerevisae | S. pombe | C. elegans | D. melanogaster |
|---|---|---|---|---|
| Nup88 | Nup82 | Nup82 | – | members only (mbo) |
| Tpr | Mlp1, Mlp2, | – | Npp-21 | Megator |
| Nup98 | Nup145N, Nup100, Nup116 | Nup189 | Npp-10 | Nup98 |
| Nup214, CAN | Nup159 | Nup146 | Npp-14 | Nup214 |
2.3 Nup88 overexpression in tumor tissues
The association of Nup88 with tumors first developed when a monoclonal raised against Candida albicans proved to cross react with a variety of tumor cells and was then found to recognize Nup88 (20). An antibody was then raised specifically against Nup88 and immunohistochemistry revealed overexpression of Nup88 in 75% of ovarian tumors (21). This analysis was extended to a variety of tumor types and Nup88 was consistently found to be overexpressed in a broad spectrum of neoplasias. Staining was especially evident in carcinomas but was also observed in sarcomas, lymphomas and mesotheliomas (22, 23). Immunoblotting of several lung carcinoma samples suggested that overexpression of Nup88 does not correlate with overexpression of the other nucleoporins tested, Nup214 and Nup153 (22). PCR quantitation of Nup88 and Nup107 mRNAs derived from tumors indicated that the level of Nup88, but not Nup107, transcripts correlated with a malignant phenotype (24). Thus, overexpression seems to be unique to Nup88 rather than the result of increased production of NPCs or upregulation of the Nup214/Nup88 subcomplex. In tumors, Nup88 staining is prominent in the cytoplasm, often in granular dots. This correlates well with earlier studies indicating that, when transiently overexpressed in cultured cells, Nup88 accumulates in the cytoplasm (6). Interestingly, in normal fetal tissue samples, strong Nup88 staining was observed only in some tissues, including developing lung epithelia and intestinal crypts (23).
In tumor tissues, the intensity of Nup88 staining has been noted to correlate with tumor grade. Benign tumors and mild hyperplasias showed little to no evidence of overexpression, whereas Nup88 overexpression was routinely detected in more advanced tumors. Highest expression was frequently noted around the edges of tumors suggesting a link to invasivity (22). High Nup88 expression was correlated with tumor aggressiveness in breast, colorectal and hepatocellular carcinomas (24–26). Nup88 expression seems to increase over the progression of carcinogenesis and to be most associated with poorly differentiated tumors. For these reasons, Nup88 has been proposed as a marker of tumor state and a potential indicator of patient prognosis.
The exact nature of the link between Nup88 expression and cancer is uncertain. In Drosophila, active NF-κB accumulates in the cytoplasm due to its increased nuclear export in the absence of Nup88. Conversely, it may be that overexpression of Nup88 in cancer cells leads to decreased NF-κB export, resulting in accumulation in the nucleus. Indeed, in many human tumor types including breast, pancreas and colon carcinomas, NF-κB is predominantly found in the nucleus where it could contribute to constitutive upregulation of target genes (27). Possibly there are additional signaling molecules that also accumulate in the nucleus and alter transcription patterns following overexpression of Nup88, but such a potential mechanism remains to be thoroughly investigated.
3. Tpr: an activator of Receptor Tyrosine Kinases
3.1 Tpr structure and normal function
Tpr is a 265 kDa nucleoporin found exclusively at the nucleoplasmic face of the pore where it is a major component of the nuclear basket (Figure 1) (28). Tpr does not possess nucleoporin FG repeats but it does contain numerous heptad repeat or leucine zipper motifs (Figure 2B). Tpr is comprised of two domains, a large, approximately 190 kDa N-terminal domain which contains the heptad repeats and an unstructured, acidic C-terminal domain (29). The heptad repeat region is predicted to form a coiled-coil structure and indeed Tpr has been characterized as an extended, rod-like homodimer in which the coiled-coils mediate dimerization (30). The C-terminal domain contains an NLS responsible for import of Tpr to the nucleoplasm, whereas the region required for assembly into the NPC is found within the N-terminal coiled-coil domain.
At the NPC, Tpr is a major component of the nuclear basket (Figure 1) where it binds to Nup153; when Nup153 is depleted using siRNA, Tpr is released from the NPC into the nucleoplasm. Tpr has also been reported to form filamentous structures that emanate from the nuclear basket and extend into the nuclear interior of amphibian oocytes (31) although the presence of such filaments in cultured somatic cells is controversial (32, 33). Functional roles for Tpr in both protein and mRNA export have been suggested. Most recently, Tpr has been reported to function in the mitotic spindle checkpoint as a binding partner and regulator of Mad1 and Mad2 (34).
Tpr orthologs have been studied in Drosophila, where it is also known as Megator, and in yeast, where two orthologs have been identified, Mlp1 and Mlp2 (Table 1). The double Mlp1/2 null mutant in yeast remains viable although with increased sensitivity to DNA damage (35). In contrast, deletion of Megator is embryonic lethal in flies. Drosophila Tpr/Megator is found both at the NPC and in the extrachromosomal spaces of the interphase nucleoplasm (36). During mitosis, Megator is reported to colocalize with a spindle matrix structure through association with the Skeletor and Chromator proteins (37). The yeast Mlp1/2 proteins are found at the NPC and at distances into the nuclear interior although it is not certain whether they form into filamentous structures (35). Mlp1/2 have been reported to play multiple roles in functions including protein export, and quality control/export of mRNA (38, 39). Further, Mlp1 and 2 are thought to anchor a variety of functional machinery at the NPC including transcriptional co-activators, some activated genes (40) and, paradoxically, silenced telomeres (41).
3.2 Tpr translocations lead to dimerization of Met and NTrk1
In contrast to the typical Nup nomenclature of nucleoporins, Tpr was named for its initial isolation from a carcinogen-treated osteogenic sarcoma line as part of a chromosomal translocation (1q25:7q31) that fused N-terminal sequences of Tpr (translocated promoter region) to the kinase domain of the proto-oncogene, Met (42). The resulting 65 kDa fusion protein combines two heptad repeats of Tpr with the tyrosine kinase domain of Met (Figure 2B). Dimerization through the heptad repeats leads to constitutive activation of Met kinase activity independent of ligand binding (43). Tpr/Met was the first such activating fusion of a tyrosine kinase receptor to be identified and, served as the prototype for understanding the now more than 25 oncogenes in which translocated tyrosine kinase receptors are activated by dimerization (44).
Met is the cell surface tyrosine kinase receptor for hepatocyte growth factor (HGF), also known as scatter factor (reviewed in (44). Both Met and HGF have essential roles in cell migration during development, as well as in repair of adult tissues and in angiogenesis. In the chromosomal translocation, the extracellular ligand binding and transmembrane domains of Met are lost and the resulting cytosolic fusion protein is no longer subject to downregulation by endocytosis. Also missing is the juxtamembrane domain, which normally functions in downregulation of activated receptor through ubiquitin-mediated recruitment of the proteolytic machinery to internalized receptor (44). Thus, multiple checks on the Met kinase are lost and the cytosolic Tpr/Met fusion protein becomes a constitutive activator of Ras/MAPK and PI3K pathways. Tpr translocations are infrequent in human tumors, but Tpr-Met translocation is associated with gastric carcinomas where it is thought to represent an early step in carcinogenesis (45, 46).
Tpr has also been found translocated with NTrk1 (TrkA), the transmembrane tyrosine kinase receptor for nerve growth factor (reviewed in (47). Tpr/NTrk1 fusion proteins are very similar in structure to the Tpr/Met rearrangements, juxtaposing either two or ten heptad repeats with the NTrk1 kinase domain (Figure 2B). Interestingly, the NPC-targeting site of Tpr is present within the larger of these fusions, although association of the fusion protein with the NPC has not been shown. As in Tpr/Met, the Tpr/NTrk1 translocation leads to constitutively active, unregulated NTrk1 signaling. Tpr-NTrk1 is one of several translocations of the NTrk1 receptor that are associated with papillary thyroid carcinoma, the most common type of thyroid cancer.
4. Nup98: an aberrant regulator of transcription
4.1 Nup98 structure, variants and binding partners
The FG-repeat nucleoporin Nup98 is the only vertebrate nucleoporin with substantial numbers of GLFG-type repeats (48, 49). Nup98 is expressed in two major forms that vary as a result of differential splicing. The first splice variant encodes a 920 amino acid protein (Figure 2C); the N-terminal half contains FG and GLFG repeat motifs and is bisected by a small coiled-coil domain (AA 181–224) that binds to the β-propeller nucleoporin Rae1/Gle2 (hereafter referred to as Rae1; (50). Rae1 is implicated in mRNA export, functions in a cell cycle checkpoint, and, as part of a distinct, RNA-containing complex, has a role in mitotic spindle function (50–52). At the C-terminus of Nup98 is a domain with a unique, β-sandwich structure (53) which possesses autoproteolytic activity (54) and is also important for directing Nup98 to the nuclear pore complex (11, 55). The second major splice variant encodes a 186 kDa Nup98-Nup96 polyprotein which is identical to Nup98 in its first 915 amino acids and goes on to encode a second, non-FG repeat nucleoporin, Nup96. The autoproteolytic activity of Nup98 cleaves both variants after amino acid 863 to produce the 90 kDa mature protein known as Nup98 and either an 8 kDa tail of no known function, or the mature Nup96 nucleoporin (54). Since all chromosomal translocations occur within the Nup98 coding region (see 4.3 below), Nup96 sequences never appear in translocation fusion proteins and will not be further discussed here.
Nup98 and Rae1 comprise a distinct subcomplex of the NPC (Figure 1). Both are dynamic components of the NPC, meaning that they can move on and off the pore, although their dissociation from the NPC is not as rapid as the most dynamic nucleoporins Nup153 and Nup50 (56, 57). Nup98 can also be found within the nucleoplasm, where it is often dynamically associated with intranuclear bodies termed GLFG bodies for the domain required for association with these structures. Additionally, Nup98 can shuttle between nuclear and cytoplasmic compartments. It is not yet known whether Nup98 plays a functional role within the nucleoplasm, but its mobility within that compartment is blocked by inhibitors of transcription (56). Nup98 is found on both nuclear and cytoplasmic faces of the NPC through association of the C-terminal domain with the structural Nup107 complex (Figure 1) (53, 55). The same region of Nup98 also binds to Nup88 and these two interactions appear to be mutually exclusive. Although the C-terminal domain of Nup98 has specific nucleoporin binding partners, it alone is not efficient at directing Nup98 to the NPC but rather requires additional interactions mediated by the GLFG repeats to stabilize it at the NPC (11).
4.2 Nup98 function in transport, viral infection and mitosis
Nup98 has been implicated in a variety of activities and the functional contribution of this nucleoporin is emphasized by the finding that the Nup98 knockout mouse dies early in embryonic development (58). The GLFG repeat region of Nup98 binds nuclear transport receptors including importin β, transportin, and the mRNA export receptor TAP (32, 49, 59). Nuclei assembled in the absence of Nup98 are competent for cNLS-mediated nuclear import, but show structural and replication defects in keeping with a possible role for Nup98 in other import pathways or in NPC assembly (48). Indeed, in cells from the Nup98 knockout mouse, several other nucleoporins were displaced from the NPC in the absence of Nup98 (58). Injection of Nup98 antibodies into oocyte nuclei blocked all RNA export from the nucleus with the exception of tRNA (60). The involvement of Nup98 in mRNA export is supported by exciting work from viral systems wherein components of host pathways are often targeted or redirected in order to favor viral replication. The vesicular stomatitis virus M protein binds the Nup98/Rae1 subcomplex through the Rae1 protein and thus blocks cellular mRNA export (61). Both influenza and polio viruses target Nup98 for degradation early during infection (62, 63). Conversely, IFN-γ, which is induced in cells to combat viral infection, activates transcription of both the Nup98 and Rae1 genes in order to compensate for these viral attacks (64).
Mitotic roles for several nucleoporins have been characterized recently and Nup98 also figures prominently at this stage in the cell cycle. The Nup98/Rae1 complex binds to the cdh1-form of the anaphase promoting complex/cyclosome (APC/C) (65). This form of the APC/C targets securin, the inhibitor of the separase enzyme, for degradation at the metaphase to anaphase transition. Degradation of securin allows separase to cleave the cohesin complex and permit sister chromatid separation. The functional significance of the interaction between Nup98/Rae1 and the APC/C is underscored by the observation of aneupoidy in 32% of splenocytes from Nup98+/−, Rae1+/− double heterozygous mice (66). In an apparently distinct mitotic role, Rae1 was found in a large complex of proteins and RNA that is required for mitotic spindle assembly, although the presence of Nup98 in this complex was not directly demonstrated (52). Additionally, the region of Nup98 between the repeats and the autoproteolytic domain has recently been found to play a role in the regulation of microtubule dynamics during spindle assembly (M. K. Cross and M. A. Powers, in preparation).
4.3 Nup98 translocation partners: homeodomains and more
The first indication of a role for Nup98 in cancer emerged when two groups simultaneously reported Nup98 as part of a rare but recurrent chromosomal translocation in AML patients (67, 68). This translocation (7p15:11p15) fuses the 5′ half of the NUP98 gene with the 3′ portion of the HOXA9 gene (Figure 2C). The resulting 59 kDa fusion protein is expressed under control of the Nup98 promoter and contains nearly all the FG/GLFG repeats of Nup98 together with the Rae1 binding site, and the DNA-binding homeodomain of HoxA9. The Nup98/HoxA9 fusion protein is not found at the NPC but rather is localized to the nucleoplasm in the form of many, finely punctate foci (69). Given this localization it seems likely that the nuclear transport function of Nup98 is not compromised by the fusion protein.
Homeodomain proteins (encoded by the homeobox genes) are transcription factors that function as master regulators of development and differentiation (reviewed in (70). The human genome contains 4 clusters (HOX A–D) of class I homeobox genes that arose from duplications of a single ancestral cluster in Drosophila. Individual clusters contain a representative of 8 to 11 out of the 13 paralog gene groups (designated HOXA4, HOXB4, HOXC4, HOXD4, etc). HOX genes as a group regulate cell proliferation and differentiation along body axes. HOX A, B and C cluster genes are expressed in hematopoietic stem and immature progenitor cell populations, but are downregulated during differentiation to mature hematopoietic cells. Many other, less well conserved, class II homeobox genes are scattered throughout the genome. The class II-encoded proteins, Pbx1 and Meis1, act as Hox co-factors and work in concert with Hox transcription factors during hematopoiesis (Pbx1 works with paralogs 1–9, Meis1 works with paralogs 9–13). Independently, the Hox proteins bind DNA with relatively little selectivity and interaction with these cofactors increases both their affinity and specificity for DNA.
Subsequent to identification of the Nup98/HoxA9 translocation, Nup98 has been found in chromosomal translocations with a remarkably wide array of partners (Table 2). In all cases, the fusion proteins carry the FG/GLFG domains of Nup98, typically the first 469 amino acids as are present in Nup98/HoxA9. Many of these Nup98 fusion partners are homeodomain transcription factors from both class I and class II. Class I Nup98 partners come from the A, C and D clusters and represent genes from the more distal paralogs of each cluster. Nup98/homeodomain translocations are most often found in AML patients but also have been associated with chronic myeloid leukemia, as well as the pre-leukemic myelodysplastic syndrome (MDS).
Table 2.
Nucleoporin translocation partners in cancer
| Nucleoporin | Partner Genes | Partner function | Diseases | Refs | |
|---|---|---|---|---|---|
| Tpr | Met | HGF receptor tyrosine kinase | gastric carcinoma | 42, 45, 46, 123 | |
| NTrk1 | NGF receptor tyrosine kinase | papillary thyroid carcinoma | 124, 125 | ||
|
| |||||
| Nup214 | DEK | chromatin binding/remodeling | AML, MDS | 96 | |
| SET | chromatin binding/remodeling | AML, AUL, T-ALL | 95 | ||
| ABL1 | receptor tyrosine kinase | T-ALL | 120 | ||
|
| |||||
| Nup98 | HD | HoxA9, 11, 13 | transcription factor | AML, MDS, CML | 67, 68, 126, 127 |
| HoxC11, 13 | transcription factor | AML, MDS | 128, 129, 132 | ||
| HoxD11, 13 | transcription factor | AML, MDS, CML | 130, 131 | ||
| PMX1(PRRX1) | transcription factor | AML, MDS, CML | 133 | ||
| PMX2(PRRX2) | transcription factor | AML, MDS | 134 | ||
| HHEX | transcription factor | AML | 76 | ||
|
| |||||
| non-HD | DDX10 | RNA helicase | AML, MDS | 135 | |
| Top1 | type I DNA topoisomerase | AML, t-MDS | 136, 137, 138 | ||
| Top2B | type II DNA topoisomerase | AML | 139 | ||
| LEDGF | transcriptional co-factor | AML, CML | 142 | ||
| NSD1 | histone methyltransferase | AML | 143 | ||
| NSD3 | histone methyltransferase | AML | 144 | ||
| SETBP1 | SET binding protein | T-ALL | 149 | ||
| JARIDIA | RbBP2, histone demethylase | ABL | 147 | ||
| RAP1GDS1 | guanine nucleotide exchange factor | T-ALL | 140, 141 | ||
| Adducin-3 | subunit of actin filament binding complex | AML | 145 | ||
| ANKRD28 | ankyrin domain repeats | AML, MDS | 148 | ||
| PHF23 | a hypothetical PHD finger protein | AML | 150 | ||
| C6ORF80 | unknown | AML, T-ALL | 146 | ||
| LOC348801 | unknown | AML | 151 | ||
| IQCG | unknown | ABL | 89 | ||
AML: acute myleoid leukemia; MDS: myelodysplastic syndrome; CML: chronic myleoid leukemia; T-ALL: T-cell acute lymphoblastic leukemia; ABL: acute biphenotypic leukemia; t-MDS: therapy-related myelodysplastic syndrome; AUL: acute undifferentiated leukemia; NGF: nerve growth factor
Non-homeodomain fusions have been identified in AML, MDS and, albeit infrequently, in other forms of leukemia as well (Table 2). The non-homeodomain Nup98 partners span a wide variety of proteins, most with nuclear functions. These include topoisomerases I and IIB, histone modifying enzymes (NSD1 and NSD3 methyltransferases, and the jumanji domain demethylase, Jarid1A), and the transcriptional co-activator LEDGF. Interestingly, the Nup98 partner protein, SetBp1, interacts with Set, a partner in a Nup214 chromosomal translocation (see 5.2 below). Taken together, these as well as the Hox partners strongly implicate alteration of chromatin structure and resulting changes in transcription as a mechanism for Nup98-induced leukemogenesis. However, other partner genes, encode an RNA helicase, an actin-binding complex subunit, a guanine nucleotide exchange factor, and several proteins of unknown function which raise the possibility of additional mechanistic contributions to leukemogenesis.
4.4 Nup98 fusions and aberrant transcriptional regulation
A major advance in understanding Nup98 leukemias came with the work of Kasper and colleagues (69) who demonstrated that the FG/GLFG repeat domain of Nup98, when directed to DNA, activates transcription through recruitment of the co-activator CBP/p300, a histone acetyl-transferase. This finding strongly implicated altered transcriptional regulation in the development of Nup98 leukemias (Figure 3). Interestingly, this same domain of Nup98 was found to couple the mobility of Nup98 within the nucleus to ongoing transcription, although the basis for this remains unclear (56). Others have subsequently shown that Nup98/PMX, a class II gene fusion protein, can bind both CBP/p300 and HDAC (71).
Figure 3. Model for Nup98 fusions.

Nup98 fusion proteins are proposed to interact with chromatin directly through their homeodomain as depicted here or indirectly through the binding activity of otherfusion partners, e.g. NSD1 as part of a histone methyltransferase complex. The FG/GLFG repeat domain recruits chromatin modifying complexes such as CBP/p300 or HDAC1 which leads to altered transcription of target genes. The basis for target gene selection is not understood.
Microarray studies of Nup98/HoxA9-expressing, CD34+, peripheral blood progenitor cells confirmed that a number of genes are indeed upregulated in these cells, among them HOXA5, A6, A7, and A9, as well as the gene encoding the Meis1 cofactor. A much smaller number of genes are downregulated but among these are genes encoding CCAAT/enhancer binding proteins (C/EBP α and ε), which promote myeloid cell differentiation (72, 73). Cells expressing Nup98/HoxD13, Nup98/PMX1, Nup98/HHEX or the artificial Nup98/HoxA10 fusion, all showed similar gene expression profiles (74–76).
4.5 Mouse models of Nup98 leukemogenesis
Overexpression of endogenous HoxA9 and Meis1 is frequently observed in AML patients (77) where overexpression of HoxA9 is correlated with poor prognosis (78). HoxA9 overexpression in mouse bone marrow transplantation models (BMT; bone marrow cells transduced with retroviral vectors driving HoxA9 expression are transplanted into mice), leads to leukemia in approximately 4 months but when combined with overexpression of Meis, the time required for progression to leukemia is halved (79). In the mouse BMT model, Nup98/HoxA9, Nup98/HoxD13, or Nup98/PMX1 expression resulted in myeloproliferative disease and, in many reports, progression to AML after latency periods approaching a year (summarized in (77). This long latency suggests that, while Nup98 fusions increase proliferation and attenuate differentiation of hematopoietic progenitors, accumulation of additional genetic perturbations is required for full progression to leukemia.
As determined for HoxA9, overexpression of Meis1 can cooperate with the Nup98/homeodomain fusions to facilitate leukemogenesis in mouse models (75, 79, 80). Upregulation of the MEIS1 gene is thus a potential “second hit” although the mechanism by which Meis1 works with Nup98/HoxA9 remains uncertain as the fusion protein does not retain the ability to bind Meis1. Nup98/HoxA9 expression can immortalize HOXA9 −/− cells in vitro, suggesting that HoxA9 is not absolutely required (81). Thus, elevated levels of Meis1 may work with other Hox factors to aberrantly activate additional pathways. Interestingly, the gene encoding Flt3, a receptor tyrosine kinase, is a target of Meis1 transcriptional regulation and elevated Flt3 levels have been observed in AML patients (77). Overexpression of Flt3 can stimulate progression to AML by Nup98 fusions (82). Although not normally expressed in Nup98 AML patients, both the Bcr/Abl and Tel/PDFG activated tyrosine kinases can also cooperate with Nup98/homeodomain fusions (83, 84).
Meis1 does not cooperate with Nup98/TopoI in mouse BMT models, but deficiency of the tumor suppressor gene, ICSBP (IFN consensus sequence binding protein) does accelerate leukemogenesis by Nup98/TopoI. Interestingly, topoisomerase catalytic activity was not required for leukemogenesis by the Nup98/TopoI fusion in a BMT mouse model (85). Consistent with this, catalytic activity was not essential for the previously reported coactivation of transcription by Topoisomerase I (86, 87).
Recently, Wang and colleagues established that Nup98/NSD1, a fusion of Nup98 with a non-HD partner possessing histone methyltransferase activity, caused upregulation of both HOXA9 and MEIS1 genes. Activation of transcription correlated with binding of the fusion protein to the co-activator CBP/p300 (88). Similarly, Nup98/IQCG binds to both CBP/p300 and HDAC (89), and Nup98/PMX1 and Nup98/HoxD13 specifically activate the HoxA9 promoter (75). Thus upregulation of HoxA9 and Meis expression may provide a unifying mechanism to link leukemogenesis induced by members of both classes of Nup98 fusions. However, the means by which Nup98 fusion proteins recognize specific promoters is uncertain and it remains to be seen whether additional non-HD fusions will similarly activate these genes.
5. Nup214: a multi-functional oncogene
5.1 Nup214 structure and function in the NPC
Nup214 is an FG repeat nucleoporin normally confined to the cytoplasmic face of the NPC (Figure 1). Nup214 contains repeats of both the FG and FxFG type and its repeat domain is a high affinity binding site for the nuclear protein export receptor, CRM-1/Exportin-1. The N-terminus of Nup214 is predicted to form a β-propeller structure, as has been demonstrated for the yeast ortholog, Nup159 (90). This is followed by a region rich in proline and serine (17% and 25%, respectively), two stretches of predicted coiled-coil structure, and a structurally uncharacterized region (Figure 2D). The nucleoporin FG repeats are found at the C-terminus of Nup214. As detailed above in 2.1–2.2, Nup214 is found in a subcomplex of the NPC, together with Nup88. Several studies point to a role for Nup214 in mRNA export and indeed, the S. cerevisae Nup214 homolog, Nup159 (Table 1), recruits the RNA helicase Dbp5, a cofactor in mRNA export (90, 91). Nup214 is essential during development; Nup214−/− embryos died once maternal stores of Nup214 were depleted and were impaired in both protein import and mRNA export (92). In contrast, nuclei reconstituted from Xenopus egg extracts depleted of Nup214 showed little to no defect in nuclear protein import (93). Since nuclear export is not monitored in the Xenopus system, it may well be that the critical contribution of Nup214 is in nuclear export.
Nup214 is most frequently described as a component of cytoplasmic fibrils of the pore, however Xenopus nuclei lacking Nup214 appeared by EM to have intact fibrils that were lost in the absence of Nup358 (93). Alternatively, Nup214 has been proposed to form shorter, more central fibers or to be associated with the NPC cytoplasmic ring. An intriguing study using antibodies to individual domains of Nup214 found that whereas the N-terminus remains anchored on the cytoplasmic face, the C-terminal repeat domain is untethered and can reach through the center of the pore, possibly as far as the nuclear basket (94).
5.2 Nup214 fusion with Dek and Set chromatin binding proteins
Like Tpr, the Nup214 gene was first identified as a component of a chromosomal translocation and, presumably due to its proximity to the c-Abl gene, was named Cain (CAN). The protein came to be referred to as Can but has since transitioned to the more standard nomenclature of Nup214. This first chromosomal translocation (6p23:9q34) joined the DEK and CAN genes (95) and was followed shortly thereafter by identification of an intrachromosomal translocation (9q32:9q34) that fused the SET and CAN genes (96). These translocations result in expression of fusion proteins that join virtually the full Dek or Set protein to the C-terminal two-thirds of Nup214, including a portion of the coiled coil domain and the FG repeat domain (Figure 2D).
Dek and Set are abundant nuclear phosphoproteins of 45 and 39 kDa respectively, each with a significant stretch of acidic amino acids. Both are major components of chromatin and are thought to influence transcription through effects on chromatin modification. Set was identified as a member of the INHAT complex (inhibitor of histone acetylation and transcriptional activation; (97). However, paradoxically, Set was found at transcriptionally active sites in Drosophila polytene chromosomes and identified as an activator of transcription from chromatin templates in vitro (98, 99). Set (also known as TAF-1β) was also identified as an inhibitor of protein phosphatase 2A (100),
Dek binds to both naked DNA and chromatin, and, like INHAT, has been described as an inhibitor of the histone acetyltransferase activity of both p300/CBP and PCAF (101). Dek was also identified as a co-repressor of NF-κB-dependent transcription (102). Interestingly, Dek itself is acetylated and this modification appears to drive relocalization of Dek from chromatin to speckles/interchromatin granule clusters (103). Phosphorylation similarly releases Dek from chromatin (104) and phosphorylated Dek can bind to the splicing factor U2AF35 and increase the specificity of 3′ splice site recognition (105). SRm160, a splicing coactivator has also been reported to interact with Dek (106). In that study, Dek remained associated with the spliced RNA, which raises the possibility that Dek might also influence post-splicing events such as proofreading or coupling to the mRNA export system. Dek has also been implicated in protection from p53-dependent apoptosis (107).
An intriguing mechanism by which Dek and Set act as opposing factors to regulate chromatin modification and accessibility to transcription initiation factors has been proposed (108). Addition of Set releases both Dek and PARP from chromatin templates in vitro and leaves the chromatin more accessible for transcriptional activation. Subsequently there may be a second contribution from Set in recruitment of the Mediator co-activating complex to the chromatin.
The Dek/Nup214 fusion was identified in a subtype of AML characterized by early age of onset and generally poor response to therapy (109). Set/Nup214 was reported in one patient with acute undifferentiated leukemia (AUL) (96). In a few patients with T-cell acute lymphoblastic leukemia (T-ALL), the same SET/CAN fusion was generated by deletion of the intervening region of chromosome 9 (110, 111).
Given the many reports of Dek and Set as modifiers of chromatin structure, a reasonable expectation is that, like Nup98 fusions, Nup214 fusions might similarly alter transcription patterns during hematopoiesis. Surprisingly, there are relatively few mechanistic studies of leukemogenesis by the Dek/Nup214 or Set/Nup214 fusion proteins. Early on, Fornerod et al observed that Dek/Nup214 or Set/Nup214 fusion proteins are localized in a punctate fashion in the nucleoplasm and no longer associate with the NPC (112). Some binding of the fusion protein to Nup88 may persist, despite the loss of half of the coiled-coil region involved in Nup88 binding (Figure 2D) (113). Nup214 fusion proteins do retain the ability to bind to and relocalize Crm-1, the NES receptor, raising the possibility of altered nuclear transport in leukemic cells (113, 114).
Set was found to be overexpressed in cells from a variety of solid tumors including uterine, stomach and rectal tumors (115). However, neither fusion increased proliferation of a myeloid precursor cell line, although Set-Nup214 did block differentiation of these cells (116, 117). Following BMT of Set/Nup214 expressing cells into mice, expansion of an early progenitor cell pool and reduction in peripheral lymphocytes were observed, but the mice showed no tendency to develop leukemia (115). ChIP experiments with Set/Nup214-expressing T-ALL patient cells indicated that Set/Nup214 bound to a subset of HoxA gene promoters. HoxA expression is normally seen in only the earliest T-cell precursors, thus expression of these genes could enforce a block in T-cell maturation (111). It may be that the role of Set/Nup214 in leukemogenesis is limited to this block to differentiation.
Like Set, Dek overexpression has been reported in solid tumors including hepatocellular carcinoma, glioblastoma and melanoma, as well as in AML (118). Surprisingly, Dek/Nup214 was found to activate global protein translation only in myeloid-derived cell lines, a property that required both Dek and Nup214 sequences and thus may be specific to the fusion protein (119). Whether the contribution of the Dek/Nup214 and Set/Nup214 fusions to leukemogenesis is simply through blocks to differentiation or whether any of the diverse functions ascribed to Dek and Set will have significant contributions remains to be seen.
5.3 Nup214 translocation activates abl kinase activity
A third and unique Nup214 translocation was identified in a screen for novel Abl translocations associated with T-cell ALL (120). Approximately 6% of T-ALL patients examined exhibited a marked, extrachromosomal amplification of the ABL1 gene. This resulted from amplification of an episome derived from circularization of a small region of chromosome region 9q34. The episome contains the 3′ half of the ABL1 gene encoding the Abl kinase, the entire LAMC3 gene, which encodes a protein found on the apical surface of ciliated epithelial cells, and the majority of the CAN gene. Circularization generates an in-frame fusion of the CAN and ABL gene fragments and leads to expression of a Nup214/Abl fusion protein (Figure 2D). Analogous to BCR/ABL translocations, the breakpoint in the ABL1 gene is always within the first intron, leading to inclusion of exon 2 which encodes most of the Abl protein including the SH3, SH2, and tyrosine kinase domains. Nup214 contributes its β-propeller, P/S-rich, coiled-coil domains as well as a variable amount of the FG repeat domain. In one patient, the Nup214 breakpoint occurred just downstream of the coiled-coil domain suggesting that the nucleoporin FG repeats may not be essential for leukemogenesis.
The Nup214/Abl protein is an active tyrosine kinase (120), but the means of constitutive kinase activation was not readily apparent until a recent study characterized a novel potential mechanism for activation of a tyrosine kinase (121). The Nup214-Abl fusion protein retains the ability to associate with the NPC through binding of the Nup214 coiled-coils to Nup88. Since Nup214/Abl is significantly overexpressed from multicopy episomes (5–50 copies per cell;(120), it may compete effectively with endogenous Nup214 for sites at the NPC. The resulting close association of Nup214-Abl proteins at the NPC would allow for some activation of kinase activity, leading to phosphorylation and constitutive activation of adjacent fusion proteins. This model is supported by experiments in which constructs encoding the Nup214 coiled-coil domain compete for Nup214-Abl association with the NPC and for downstream indicators of kinase activity. A direct test of this mechanism of kinase activation would be the ability of a minimal Nup214 (coiled-coil domain)/Abl fusion to induce T-ALL in mice.
Given the cytoplasmic orientation of Nup214 at the NPC, it is possible that Nup214/Abl would be in sufficient contact with the cytoplasmic substrates to allow for activation of signaling. Alternatively, the Nup214/Abl kinase may be ideally positioned to contact substrates as they translocate through the NPC. Nup214/Abl phosphorylates other nucleoporins such as Nup358, although whether this plays a part in oncogenic transformation is unknown. Nup214/Abl is a relatively weak oncogenic protein, requiring 70–200 days after BMT of cells expressing Nup214/Abl to induce T-cell disease in mice. This is compared to the short 20 day latency of Bcr/Abl-induced myeloproliferative disorder in the same system (121).
Conclusions
The study of nucleoporins and their roles in cancer has been a fruitful one, revealing novel mechanisms by which cells are advanced along the path to transformation. From our current level of understanding, each nucleoporin contributing to in carcinognesis seems to do so in a unique manner. It is somewhat surprising that, in the case of nucleoporin translocations, defects in nuclear transport have not been reported. Patient cells are heterozygous for these translocations and thus the level of the nucleoporin involved should be halved. This has not been thoroughly investigated, but perhaps the NPC is remarkably able to compensate for reduction in the level of many of its components thus circumventing selection against nucleoporin-based transformation. A recent report indicated that knockout of Nup358 also leads to increased aneuploidy in mice (122) although this nucleoporin has not yet been implicated in any form of cancer. It may be that additional Nups will prove to contribute to cancer in novel ways and the ongoing study of nucleoporin-induced cancers may continue to uncover surprising new mechanisms in cancer biology.
Acknowledgments
The authors regret the many publications we were unable to cite due to lack of space. We thank the members of the Powers laboratory, especially Marie Cross and Dr. Amy Pierce for their very helpful discussions and comments on the manuscript, and Dr. Katharine Ullman and Dr. Paula Vertino for comments on the manuscript. Work in the authors’ laboratory is supported by National Institutes of Health grant GM-059975 to M. A. P.
Abbreviations
- NPC
nuclear pore complex
- cNLS
classical nuclear localization signal
- NES
nuclear export signal
- FG
phenylalanine-glycine
- FxFG
phenylalanine-any amino acid-phenylalanine-glycine
- GLFG
glycine-leucine-phenylalanine-glycine
- AA
amino acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125(6):1041–53. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 2.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18(10):456–66. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15(2):221–6. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–6. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 5.Lim RY, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol. 2008;129(2):105–16. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137(5):989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fornerod M, Deursen Jv, Baal Sv, Reynolds A, Davis D, Murti KG, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–16. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270(1):254–62. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- 9.Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila. J Cell Sci. 2006;119(Pt 21):4409–19. doi: 10.1242/jcs.03201. [DOI] [PubMed] [Google Scholar]
- 10.Bernad R, van der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol. 2004;24(6):2373–84. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell. 2003;14(2):600–10. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth P, Xylourgidis N, Sabri N, Uv A, Fornerod M, Samakovlis C. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J Cell Biol. 2003;163(4):701–6. doi: 10.1083/jcb.200304046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernad R, Engelsma D, Sanderson H, Pickersgill H, Fornerod M. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J Biol Chem. 2006;281(28):19378–86. doi: 10.1074/jbc.M512585200. [DOI] [PubMed] [Google Scholar]
- 14.Hutten S, Kehlenbach RH. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol. 2006;26(18):6772–85. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabri N, Roth P, Xylourgidis N, Sadeghifar F, Adler J, Samakovlis C. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol. 2007;178(4):557–65. doi: 10.1083/jcb.200612135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uv AE, Roth P, Xylourgidis N, Wickberg A, Cantera R, Samakovlis C. members only encodes a Drosophila nucleoporin required for rel protein import and immune response activation. Genes Dev. 2000;14(15):1945–57. [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson C, Rustum C, Hallberg E. Dynamic properties of nuclear pore complex proteins in gp210 deficient cells. FEBS Lett. 2004;572 (1–3):261–5. doi: 10.1016/j.febslet.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 18.Olsson M, Scheele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res. 2004;292(2):359–70. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Fan F, Liu CP, Korobova O, Heyting C, Offenberg HH, Trump G, et al. cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics. 1997;40(3):444–53. doi: 10.1006/geno.1996.4557. [DOI] [PubMed] [Google Scholar]
- 20.Schneider J, Moragues D, Martinez N, Romero H, Jimenez E, Ponton J. Cross-reactivity between Candida albicans and human ovarian carcinoma as revealed by monoclonal antibodies PA10F and C6. Br J Cancer. 1998;77(6):1015–20. doi: 10.1038/bjc.1998.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez N, Alonso A, Moragues MD, Ponton J, Schneider J. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer Res. 1999;59(21):5408–11. [PubMed] [Google Scholar]
- 22.Gould VE, Orucevic A, Zentgraf H, Gattuso P, Martinez N, Alonso A. Nup88 (karyoporin) in human malignant neoplasms and dysplasias: correlations of immunostaining of tissue sections, cytologic smears, and immunoblot analysis. Hum Pathol. 2002;33(5):536–44. doi: 10.1053/hupa.2002.124785. [DOI] [PubMed] [Google Scholar]
- 23.Gould VE, Martinez N, Orucevic A, Schneider J, Alonso A. A novel, nuclear pore-associated, widely distributed molecule overexpressed in oncogenesis and development. Am J Pathol. 2000;157(5):1605–13. doi: 10.1016/S0002-9440(10)64798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agudo D, Gomez-Esquer F, Martinez-Arribas F, Nunez-Villar MJ, Pollan M, Schneider J. Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int J Cancer. 2004;109(5):717–20. doi: 10.1002/ijc.20034. [DOI] [PubMed] [Google Scholar]
- 25.Knoess M, Kurz AK, Goreva O, Bektas N, Breuhahn K, Odenthal M, et al. Nucleoporin 88 expression in hepatitis B and C virus-related liver diseases. World J Gastroenterol. 2006;12(36):5870–4. doi: 10.3748/wjg.v12.i36.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang ZY, Zhao ZR, Jiang L, Li JC, Gao YM, Cui DS, et al. Nup88 expression in normal mucosa, adenoma, primary adenocarcinoma and lymph node metastasis in the colorectum. Tumour Biol. 2007;28(2):93–9. doi: 10.1159/000099154. [DOI] [PubMed] [Google Scholar]
- 27.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4(2):106–17. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 28.Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as Components of the Nuclear Pore Complex Core Structure and Tpr as the Architectural Element of the Nuclear Basket. Mol Biol Cell. 2004 doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrd DA, Sweet DJ, Pante N, Konstantinov KN, Guan T, Saphire AC, et al. Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. JCell Biol. 1994;127:1515–26. doi: 10.1083/jcb.127.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hase ME, Kuznetsov NV, Cordes VC. Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol Biol Cell. 2001;12(8):2433–52. doi: 10.1091/mbc.12.8.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136(3):515–29. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontoura BM, Dales S, Blobel G, Zhong H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci U S A. 2001;98(6):3208–13. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol. 2002;156(4):617–30. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22(21):2926–31. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144(5):839–55. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci. 1997;110(Pt 8):927–44. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
- 37.Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15(11):4854–65. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116(1):63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 39.Green DM, Johnson CP, Hagan H, Corbett AH. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci U S A. 2003;100(3):1010–5. doi: 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, et al. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282(5):3042–9. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 41.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403(6765):108–12. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 42.Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311(5981):29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 43.Park M, Dean M, Cooper CS, Schmidt M, O’Brien SJ, Blair DG, et al. Mechanism of met oncogene activation. Cell. 1986;45(6):895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 44.Peschard P, Park M. From Tpr-Met to Met, tumorigenesis and tubes. Oncogene. 2007;26(9):1276–85. doi: 10.1038/sj.onc.1210201. [DOI] [PubMed] [Google Scholar]
- 45.Soman NR, Correa P, Ruiz BA, Wogan GN. The TPR-MET oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc Natl Acad Sci U S A. 1991;88(11):4892–6. doi: 10.1073/pnas.88.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu J, Miehlke S, Ebert MP, Hoffmann J, Breidert M, Alpen B, et al. Frequency of TPR-MET rearrangement in patients with gastric carcinoma and in first-degree relatives. Cancer. 2000;88(8):1801–6. [PubMed] [Google Scholar]
- 47.Pierotti MA, Greco A. Oncogenic rearrangements of the NTRK1/NGF receptor. Cancer Lett. 2006;232(1):90–8. doi: 10.1016/j.canlet.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 48.Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128(5):721–36. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–22. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 50.Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145(2):237–54. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, Van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160(3):341–53. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121(2):223–34. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 53.Hodel A, Hodel M, Griffis E, Hennig K, Ratner G, Xu S, et al. The three-dimensional structure of the autoproteolytic, nuclear pore- targeting domain of the human nucleoporin nup98. Mol Cell. 2002;10(2):347–58. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 54.Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144(6):1097–112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155(3):339–54. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 Is a Mobile Nucleoporin with Transcription-dependent Dynamics. Mol Biol Cell. 2002;13(4):1282–97. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154(1):71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci U S A. 2001;98(6):3191–6. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278(23):20979–88. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- 60.Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136(2):241–50. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 62.Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104(6):1853–8. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park N, Katikaneni P, Skern T, Gustin KE. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol. 2008;82(4):1647–55. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enninga J, Levy DE, Blobel G, Fontoura BM. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 2002;295(5559):1523–5. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- 65.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438(7070):1036–9. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 66.Jeganathan KB, Baker DJ, van Deursen JM. Securin associates with APCCdh1 in prometaphase but its destruction is delayed by Rae1 and Nup98 until the metaphase/anaphase transition. Cell Cycle. 2006;5(4):366–70. doi: 10.4161/cc.5.4.2483. [DOI] [PubMed] [Google Scholar]
- 67.Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, et al. Fusion of the nucleoporin gene Nup98 to HoxA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nature Genetics. 1996;12:154–14. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 68.Borrow J, Shearman AM, Stanton VP, Becher R, Collins T, Williams AJ, et al. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin Nup98 and class1 homeoprotein HoxA9. Nature Genetics. 1996;12:159–67. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 69.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19(1):764–76. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharkey M, Graba Y, Scott MP. Hox genes in evolution: protein surfaces and paralog groups. Trends Genet. 1997;13(4):145–51. doi: 10.1016/s0168-9525(97)01096-2. [DOI] [PubMed] [Google Scholar]
- 71.Bai XT, Gu BW, Yin T, Niu C, Xi XD, Zhang J, et al. Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 2006;66(9):4584–90. doi: 10.1158/0008-5472.CAN-05-3101. [DOI] [PubMed] [Google Scholar]
- 72.Chung KY, Morrone G, Schuringa JJ, Plasilova M, Shieh JH, Zhang Y, et al. Enforced expression of NUP98-HOXA9 in human CD34(+) cells enhances stem cell proliferation. Cancer Res. 2006;66(24):11781–91. doi: 10.1158/0008-5472.CAN-06-0706. [DOI] [PubMed] [Google Scholar]
- 73.Takeda A, Goolsby C, Yaseen NR. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006;66(13):6628–37. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- 74.Palmqvist L, Pineault N, Wasslavik C, Humphries RK. Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PLoS ONE. 2007;2 (1):e768. doi: 10.1371/journal.pone.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hirose K, Abramovich C, Argiropoulos B, Humphries RK. Leukemogenic properties of NUP98-PMX1 are linked to NUP98 and homeodomain sequence functions but not to binding properties of PMX1 to serum response factor. Oncogene. 2008;27(46):6056–67. doi: 10.1038/onc.2008.210. [DOI] [PubMed] [Google Scholar]
- 76.Jankovic D, Gorello P, Liu T, Ehret S, La Starza R, Desjobert C, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111(12):5672–82. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
- 77.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 78.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 79.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. Embo J. 2001;20(3):350–61. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pineault N, Buske C, Feuring-Buske M, Abramovich C, Rosten P, Hogge DE, et al. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003;101(11):4529–38. doi: 10.1182/blood-2002-08-2484. [DOI] [PubMed] [Google Scholar]
- 81.Calvo KR, Sykes DB, Pasillas MP, Kamps MP. Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1. Oncogene. 2002;21(27):4247–56. doi: 10.1038/sj.onc.1205516. [DOI] [PubMed] [Google Scholar]
- 82.Palmqvist L, Argiropoulos B, Pineault N, Abramovich C, Sly LM, Krystal G, et al. The Flt3 receptor tyrosine kinase collaborates with NUP98-HOX fusions in acute myeloid leukemia. Blood. 2006;108(3):1030–6. doi: 10.1182/blood-2005-12-007005. [DOI] [PubMed] [Google Scholar]
- 83.Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99(11):7622–7. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mayotte N, Roy DC, Yao J, Kroon E, Sauvageau G. Oncogenic interaction between BCR-ABL and NUP98-HOXA9 demonstrated by the use of an in vitro purging culture system. Blood. 2002;100(12):4177–84. doi: 10.1182/blood-2002-04-1244. [DOI] [PubMed] [Google Scholar]
- 85.Gurevich RM, Aplan PD, Humphries RK. The NUP98-Topoisomerase I AML-associated fusion gene has potent leukemogenic activities independent of an engineered catalytic site mutation. Blood. 2004 doi: 10.1182/blood-2003-10-3550. [DOI] [PubMed] [Google Scholar]
- 86.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365(6443):227–32. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 87.Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11(3):397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 88.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9(7):804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 89.Pan Q, Zhu YJ, Gu BW, Cai X, Bai XT, Yun HY, et al. A new fusion gene NUP98-IQCG identified in an acute T-lymphoid/myeloid leukemia with a t(3;11)(q29q13;p15)del(3)(q29) translocation. Oncogene. 2008;27(24):3414–23. doi: 10.1038/sj.onc.1210999. [DOI] [PubMed] [Google Scholar]
- 90.Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell. 2004;16(5):749–60. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 91.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8(7):711–6. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 92.van Deursen J, Boer J, Kasper L, Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. Embo J. 1996;15(20):5574–83. [PMC free article] [PubMed] [Google Scholar]
- 93.Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, et al. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158(1):63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Paulillo SM, Phillips EM, Koser J, Sauder U, Ullman KS, Powers MA, et al. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol. 2005;351(4):784–98. doi: 10.1016/j.jmb.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 95.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12(4):1687–97. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12(8):3346–55. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104(1):119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 98.Gamble MJ, Erdjument-Bromage H, Tempst P, Freedman LP, Fisher RP. The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol Cell Biol. 2005;25(2):797–807. doi: 10.1128/MCB.25.2.797-807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nowak SJ, Pai CY, Corces VG. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol Cell Biol. 2003;23(17):6129–38. doi: 10.1128/MCB.23.17.6129-6138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271(19):11059–62. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 101.Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, et al. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580(13):3217–22. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 102.Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281(37):26802–12. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- 103.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, et al. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280(36):31760–7. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 104.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24(13):6011–20. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312(5782):1961–5. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 106.McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150(2):309–20. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26(20):7506–19. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14(6):548–55. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 109.von Lindern M, Fornerod M, Soekarman N, van Baal S, Jaegle M, Hagemeijer A, et al. Translocation t(6;9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. Baillieres Clin Haematol. 1992;5(4):857–79. doi: 10.1016/s0950-3536(11)80049-1. [DOI] [PubMed] [Google Scholar]
- 110.Boer J, Bonten-Surtel J, Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol. 1998;18(3):1236–47. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111(9):4668–80. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fornerod M, Boer J, van Baal S, Jaeglé M, von Lindern M, Murti M, et al. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–48. [PubMed] [Google Scholar]
- 113.Saito S, Miyaji-Yamaguchi M, Nagata K. Aberrant intracellular localization of SET-CAN fusion protein, associated with a leukemia, disorganizes nuclear export. Int J Cancer. 2004;111(4):501–7. doi: 10.1002/ijc.20296. [DOI] [PubMed] [Google Scholar]
- 114.Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13(8):1801–8. [PubMed] [Google Scholar]
- 115.Cervoni N, Detich N, Seo SB, Chakravarti D, Szyf M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277(28):25026–31. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- 116.Boer J, Bonten-Surtel J, Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol. 1998;18(3):1236–47. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kandilci A, Mientjes E, Grosveld G. Effects of SET and SET-CAN on the differentiation of the human promonocytic cell line U937. Leukemia. 2004;18(2):337–40. doi: 10.1038/sj.leu.2403227. [DOI] [PubMed] [Google Scholar]
- 118.Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5(11):1202–7. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 119.Ageberg M, Drott K, Olofsson T, Gullberg U, Lindmark A. Identification of a novel and myeloid specific role of the leukemia-associated fusion protein DEK-NUP214 leading to increased protein synthesis. Genes Chromosomes Cancer. 2008;47(4):276–87. doi: 10.1002/gcc.20531. [DOI] [PubMed] [Google Scholar]
- 120.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36(10):1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 121.De Keersmaecker K, Rocnik JL, Bernad R, Lee BH, Leeman D, Gielen O, et al. Kinase activation and transformation by NUP214-ABL1 is dependent on the context of the nuclear pore. Mol Cell. 2008;31(1):134–42. doi: 10.1016/j.molcel.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 122.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133(1):103–15. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dean M, Park M, Le Beau MM, Robins TS, Diaz MO, Rowley JD, et al. The human met oncogene is related to the tyrosine kinase oncogenes. Nature. 1985;318(6044):385–8. doi: 10.1038/318385a0. [DOI] [PubMed] [Google Scholar]
- 124.Greco A, Pierotti MA, Bongarzone I, Pagliardini S, Lanzi C, Della Porta G. TRK-T1 is a novel oncogene formed by the fusion of TPR and TRK genes in human papillary thyroid carcinomas. Oncogene. 1992;7(2):237–42. [PubMed] [Google Scholar]
- 125.Greco A, Miranda C, Pagliardini S, Fusetti L, Bongarzone I, Pierotti MA. Chromosome 1 rearrangements involving the genes TPR and NTRK1 produce structurally different thyroid-specific TRK oncogenes. Genes Chromosomes Cancer. 1997;19(2):112–23. [PubMed] [Google Scholar]
- 126.Fujino T, Suzuki A, Ito Y, Ohyashiki K, Hatano Y, Miura I, et al. Single-translocation and double-chimeric transcripts: detection of NUP98-HOXA9 in myeloid leukemias with HOXA11 or HOXA13 breaks of the chromosomal translocation t(7;11)(p15;p15) Blood. 2002;99(4):1428–33. doi: 10.1182/blood.v99.4.1428. [DOI] [PubMed] [Google Scholar]
- 127.Suzuki A, Ito Y, Sashida G, Honda S, Katagiri T, Fujino T, et al. t(7;11)(p15;p15) Chronic myeloid leukaemia developed into blastic transformation showing a novel NUP98/HOXA11 fusion. Br J Haematol. 2002;116(1):170–2. doi: 10.1046/j.1365-2141.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- 128.Taketani T, Taki T, Shibuya N, Kikuchi A, Hanada R, Hayashi Y. Novel NUP98-HOXC11 fusion gene resulted from a chromosomal break within exon 1 of HOXC11 in acute myeloid leukemia with t(11;12)(p15;q13) Cancer Res. 2002;62(16):4571–4. [PubMed] [Google Scholar]
- 129.Gu BW, Wang Q, Wang JM, Xue YQ, Fang J, Wong KF, et al. Major form of NUP98/HOXC11 fusion in adult AML with t(11;12)(p15;q13) translocation exhibits aberrant trans-regulatory activity. Leukemia. 2003;17(9):1858–64. doi: 10.1038/sj.leu.2403036. [DOI] [PubMed] [Google Scholar]
- 130.Taketani T, Taki T, Shibuya N, Ito E, Kitazawa J, Terui K, et al. The HOXD11 gene is fused to the NUP98 gene in acute myeloid leukemia with t(2;11)(q31;p15) Cancer Res. 2002;62(1):33–7. [PubMed] [Google Scholar]
- 131.Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998;58(19):4269–73. [PubMed] [Google Scholar]
- 132.La Starza R, Trubia M, Crescenzi B, Matteucci C, Negrini M, Martelli MF, et al. Human homeobox gene HOXC13 is the partner of NUP98 in adult acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36(4):420–3. doi: 10.1002/gcc.10182. [DOI] [PubMed] [Google Scholar]
- 133.Nakamura T, Yamazaki Y, Hatano Y, Miura I. NUP98 is fused to PMX1 homeobox gene in human acute myelogenous leukemia with chromosome translocation t(1;11)(q23;p15) Blood. 1999;94(2):741–7. [PubMed] [Google Scholar]
- 134.Gervais C, Mauvieux L, Perrusson N, Helias C, Struski S, Leymarie V, et al. A new translocation t(9;11)(q34;p15) fuses NUP98 to a novel homeobox partner gene, PRRX2, in a therapy-related acute myeloid leukemia. Leukemia. 2005;19(1):145–8. doi: 10.1038/sj.leu.2403565. [DOI] [PubMed] [Google Scholar]
- 135.Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, et al. The inv (11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89(11):3936–44. [PubMed] [Google Scholar]
- 136.Ahuja HG, Felix CA, Aplan PD. The t(11;20)(p15;q11) chromosomal translocation associated with therapy- related myelodysplastic syndrome results in an NUP98-TOP1 fusion. Blood. 1999;94(9):3258–61. [PubMed] [Google Scholar]
- 137.Chen S, Xue Y, Chen Z, Guo Y, Wu Y, Pan J. Generation of the NUP98-TOP1 fusion transcript by the t(11;20) (p15;q11) in a case of acute monocytic leukemia. Cancer Genet Cytogenet. 2003;140(2):153–6. doi: 10.1016/s0165-4608(02)00642-8. [DOI] [PubMed] [Google Scholar]
- 138.Iwase S, Akiyama N, Sekikawa T, Saito S, Arakawa Y, Horiguchi-Yamada J, et al. Both NUP98/TOP1 and TOP1/NUP98 transcripts are detected in a de novo AML with t(11;20)(p15;q11) Genes Chromosomes Cancer. 2003;38(1):102–5. doi: 10.1002/gcc.10239. [DOI] [PubMed] [Google Scholar]
- 139.Nebral K, Schmidt HH, Haas OA, Strehl S. NUP98 is fused to topoisomerase (DNA) IIbeta 180 kDa (TOP2B) in a patient with acute myeloid leukemia with a new t(3;11)(p24;p15) Clin Cancer Res. 2005;11(18):6489–94. doi: 10.1158/1078-0432.CCR-05-0150. [DOI] [PubMed] [Google Scholar]
- 140.Hussey DJ, Nicola M, Moore S, Peters GB, Dobrovic A. The (4;11)(q21;p15) translocation fuses the NUP98 and RAP1GDS1 genes and is recurrent in T-cell acute lymphocytic leukemia. Blood. 1999;94(6):2072–9. [PubMed] [Google Scholar]
- 141.Mecucci C, La Starza R, Negrini M, Sabbioni S, Crescenzi B, Leoni P, et al. t(4;11)(q21;p15) translocation involving NUP98 and RAP1GDS1 genes: characterization of a new subset of T acute lymphoblastic leukaemia. Br J Haematol. 2000;109(4):788–93. doi: 10.1046/j.1365-2141.2000.02106.x. [DOI] [PubMed] [Google Scholar]
- 142.Ahuja HG, Hong J, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75- lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60(22):6227–9. [PubMed] [Google Scholar]
- 143.Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98(4):1264–7. doi: 10.1182/blood.v98.4.1264. [DOI] [PubMed] [Google Scholar]
- 144.Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99(10):3857–60. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 145.Lahortiga I, Vizmanos JL, Agirre X, Vazquez I, Cigudosa JC, Larrayoz MJ, et al. NUP98 is fused to adducin 3 in a patient with T-cell acute lymphoblastic leukemia and myeloid markers, with a new translocation t(10;11)(q25;p15) Cancer Res. 2003;63(12):3079–83. [PubMed] [Google Scholar]
- 146.Tosi S, Ballabio E, Teigler-Schlegel A, Boultwood J, Bruch J, Harbott J. Characterization of 6q abnormalities in childhood acute myeloid leukemia and identification of a novel t(6;11)(q24.1;p15.5) resulting in a NUP98-C6orf80 fusion in a case of acute megakaryoblastic leukemia. Genes Chromosomes Cancer. 2005;44(3):225–32. doi: 10.1002/gcc.20233. [DOI] [PubMed] [Google Scholar]
- 147.van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45(5):437–46. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 148.Ishikawa M, Yagasaki F, Okamura D, Maeda T, Sugahara Y, Jinnai I, et al. A novel gene, ANKRD28 on 3p25, is fused with NUP98 on 11p15 in a cryptic 3-way translocation of t(3;5;11)(p25;q35;p15) in an adult patient with myelodysplastic syndrome/acute myelogenous leukemia. Int J Hematol. 2007;86(3):238–45. doi: 10.1532/IJH97.07054. [DOI] [PubMed] [Google Scholar]
- 149.Panagopoulos I, Kerndrup G, Carlsen N, Strombeck B, Isaksson M, Johansson B. Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12) Br J Haematol. 2007;136(2):294–6. doi: 10.1111/j.1365-2141.2006.06410.x. [DOI] [PubMed] [Google Scholar]
- 150.Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21(4):842–4. doi: 10.1038/sj.leu.2404579. [DOI] [PubMed] [Google Scholar]
- 151.Gorello P, Brandimarte L, La Starza R, Pierini V, Bury L, Rosati R, et al. t(3;11)(q12;p15)/NUP98-LOC348801 fusion transcript in acute myeloid leukemia. Haematologica. 2008;93(9):1398–401. doi: 10.3324/haematol.12945. [DOI] [PubMed] [Google Scholar]


