Abstract
Chemokine receptors are a specific class of G protein-coupled receptors (GPCRs) that control cell migration associated with routine immune surveillance, inflammation and development. In addition to their roles in normal physiology, these receptors and their ligands are involved in a large number of inflammatory diseases, cancer and AIDS, making them prime therapeutic targets in the pharmaceutical industry. Like other GPCRs, a significant obstacle in determining structures and characterizing mechanisms of activation has been the difficulty in obtaining high levels of pure, functional receptor. Here we describe a systematic effort to express the chemokine receptor CCR1 in mammalian cells, and to purify and reconstitute it in functional form. The highest expression levels were obtained using an inducible HEK293 system. The receptor was purified using a combination of N- (StrepII or Hemagglutinin) and C-terminal (His8) affinity tags. Function was assessed by ligand binding using a novel fluorescence polarization assay with fluorescein-labeled chemokine. A strict dependence of function on the detergent composition was observed, as solubilization of CCR1 in n-dodecyl-β-D-maltopyranoside/cholesteryl hemisuccinate yielded functional receptor with a Kd of 21 nM for the chemokine CCL14, whereas it was non-functional in phosphocholine detergents. Differences in function were observed despite the fact that both these detergent types maintained the receptor in a state characterized by monomers and small oligomers, but not large aggregates. While optimization is still warranted, yields of ~ 0.1–0.2mgs of pure functional receptor per 109 cells will permit biophysical studies of this medically important receptor.
INTRODUCTION
GPCRs are seven-transmembrane helical receptors that form the largest family of cell-surface signal transduction receptors. Following ligand binding at their extracellular surface, GPCRs elicit diverse physiological responses, mediated in large part by receptor-associated heterotrimeric G-proteins at the intracellular surface [1]. It has recently become apparent that many GPCRs exist as homo- or heterodimers in vivo, and that this also plays a role in their function and downstream signaling pathways [2]. The importance of GPCRs in health and disease is highlighted by the fact that drugs that target GPCRs constitute ~30% of all known-marketed medicines [3, 4]. However, GPCRs are notoriously difficult to express and purify in sufficient quantities for structural and biochemical studies [5]. The reasons for these difficulties are well reported, and include low levels of expression, cellular toxicity, post-translational modifications that may not be faithfully reproduced in heterologous expression systems (but may be necessary for function), and difficulties with mimicking the native membrane environment upon reconstitution [6]. As a consequence, only a few high-resolution structures have been published to date, despite the fact that there are ~850 GPCRs in the human genome alone [7–13].
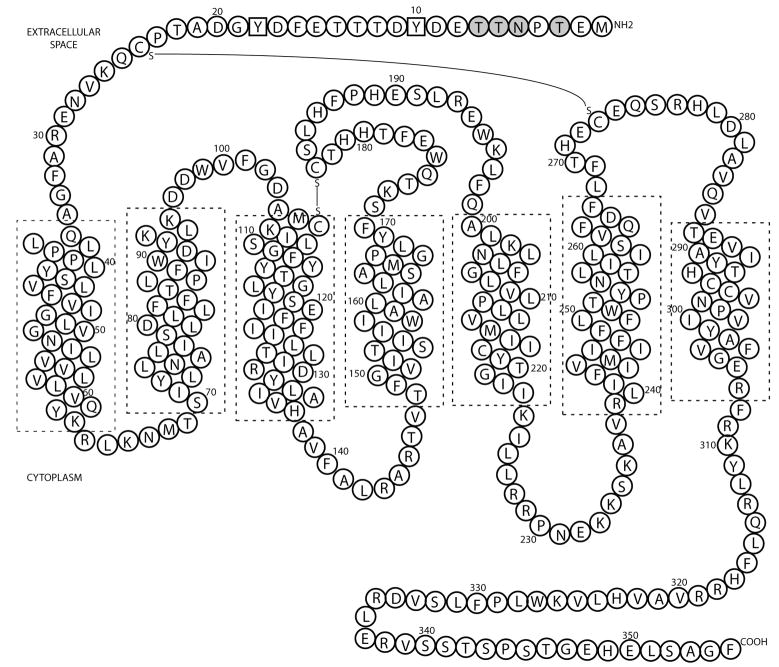
Chemokine receptors represent a subfamily of ~20 GPCRs that were originally identified by their roles in immune cell trafficking. They function by binding to chemokines (chemotactic cytokines) and activating signaling pathways that are linked to cell migration in many processes, including lymphocyte development, cardiogenesis, development in the CNS, and angiogenesis [Rev. in [14]]. Due to their multiple functions, it is not surprising that chemokines and their receptors are also associated with many diseases characterized by inappropriate cell migration and activation, ranging from auto-immune disorders (e.g. multiple sclerosis, rheumatoid arthritis, psoriasis) to pulmonary diseases (e.g. asthma), atherosclerosis and cancer. CCR1, the focus of this study (Figure 1) has been the target of a number of small-molecule drug development programs to treat auto-inflammatory disorders [Rev. in [15]], and therefore learning more about the structure and biochemistry of CCR1 is both scientifically and medically important. However, no high-level expression systems of CCR1 have been published and no chemokine receptor structures have been solved to date.
Figure 1. Sequence and predicted topology of the chemokine receptor CCR1.
Potential N-linked and O-linked glycosylation sites (gray circles) and tyrosine sulfation sites (squares) are highlighted. Chemokines bind to the extracellular surface, whilst G-proteins bind to the cytoplasmic surface, in a region encompassing the second and third cytoplasmic loops, and C-terminal tail of CCR1. Transmembrane helices were positioned according to Vaidehi et al. [21].
Here we describe a systematic study to obtain milligram quantities of CCR1 for biochemical and biophysical studies. Mammalian expression systems represent the best chance to facilitate correct folding of GPCRs and also faithfully reproduce post-translational modifications, and are therefore desirable for both cell-based assays and solubilization studies. In this study, four mammalian (HEK293) systems were tested for the expression of CCR1. Three of these systems (FlpIn, T-REx and FlpIn/T-REx) were commercially available from Invitrogen, whilst the fourth (inducible HEK293) was based upon published reports of high-level expression of two other GPCRs (rhodopsin and β2-adrenergic receptor) [16, 17]. The Invitrogen systems contain either (i) the Flp recombinase (FlpIn), whereby the gene of interest is specifically integrated into the genome at a single site behind an active promoter, (ii) tetracycline regulation (T-REx), whereby protein expression is induced by the addition of tetracycline, or (iii) a combination of the two (FlpIn/T-REx). FlpIn/CHO cells have been previously shown to be amenable to high-level expression of the chemokine receptors CCR2 and CCR3 [18].
The HEK293 system features three main advantages over common mammalian expression systems. Firstly, like the Invitrogen T-REx system, protein expression can be regulated by the presence of tetracycline. This means that cells can be grown to near-confluence before induction, which can be advantageous for the expression of toxic proteins. Secondly, a glycosylation deficient cell line is available, which lacks N-acetylglucosaminyltransferase I (GNTI) activity. GNTI− cells are unable to synthesize complex glycans, thereby making it possible to produce more uniformly glycosylated proteins for crystallographic studies [19]. Finally, although this property was not fully utilized in this study, this HEK293 cell line can also be grown in suspension, making it considerably easier and cheaper to scale-up expression to larger volumes. CCR1 was successfully expressed in HEK293 cells, purified and functionally reconstituted into a mixed artificial lipid/detergent system. High affinity binding of CCR1 to CCL14 was inhibited in a dose-dependent manner by the small-molecule specific CCR1 antagonist BX-471 [20].
MATERIALS AND METHODS
Materials
Flp-In, Flp-In/T-REx and T-REx HEK293 cells, their corresponding vectors (pcDNA5/FRT, pcDNA5/FRT/TO and pcDNA4/TO respectively) and the helper vector pOG44 were purchased from Invitrogen (Carlsbad, CA). HEK293 cells containing pcDNA6/TR and the pACMV-TetO vector were generous gifts from H.G. Khorana (MIT, Cambridge, MA). 125I-CCL3 was obtained from Perkin Elmer (Waltham, MA) and wild-type CCL3 from R&D Systems (Minneapolis, MN). Codon-optimized CCR1 and CCR5 were purchased from GenScript (Piscataway, NJ) and detergents obtained from Anatrace (Maumee, OH). Unless stated otherwise, all tissue culture reagents were from Invitrogen.
Cloning of expression plasmids
Nucleotides encoding HA (Hemagglutinin), His8 and/or StrepII (IBA, Germany) tags were cloned onto the N- and/or C-termini of human CCR1, CCR2, CCR3 and CCR5 constructs (see Supplementary Figure S1 for DNA sequences). The resulting protein sequences were as follows: StrepII-CCRx-His8 (MASWSHPQFEKGA-CCRx-HHHHHHHH), HA-CCRx-His8 (MYPYDVPDYAGPG-CCRx-HHHHHHHH) and CCRx-StrepII (CCRx-PSAWSHPQFEK), where x corresponds to the identity of the receptor. Receptor constructs were cloned into the appropriate mammalian vectors containing an extended Kozak sequence (GCCGCCGCCACCATG) (initiator Met underlined).
Transfections and generation of stable cell lines
Prior to transfection, HEK293 cell lines were grown at 37°C, 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM)+Glutamax, supplemented with 10% tetracycline-screened fetal bovine serum (FBS) (Hyclone, Logan, UT) and the appropriate drugs (5μg/mL Blasticidin for HEK239s and T-REx 293 cells, 5μg/mL Blasticidin + 100μg/mL Zeocin for Flp-In/T-REx 293 cells and 100μg/mL Zeocin for Flp-In 293 cells). Vectors were transfected into all systems using Fugene 6 (Roche, Indianapolis, IN) according to manufacturer instructions and pOG44 was co-transfected at ratios ranging from 2:1 to 1:2 pOG44:pcDNA5 (w/w) in the Flp-In systems. After recovery, transformed cells were selected by their ability to grow in the presence of 100μg/mL Hygromycin B (Flp-In), 5μg/mL Blasticidin + 100μg/mL Hygromycin B (Flp-In-T-REx), 5μg/mL Blasticidin + 200μg/mL Zeocin (T-Rex) or 5μg/mL Blasticidin + 700μg/mL Geneticin (HEK293). Stable cell lines were obtained in 2–4 weeks. A combination of FACS sorting (FACS Aria, BD Biosciences) and single colony selection by limiting dilution were used to select for the highest expressing cells.
Expression tests
HEK293 cells were grown to ~70–80% confluency and induced with 1–2μg/mL doxycycline and 10mM sodium butyrate (Sigma) for 24 hrs. Cells were harvested by incubation with PBS containing 0.5mM EDTA and washed with PBS/0.5% bovine serum albumin (BSA). For cell-surface expression tests, non-specific binding was blocked by incubating the cells with IgG (Sigma) for 15 mins at 4°C before addition of anti-CCRx-PE antibody (R&D Systems) for 45 mins. Cells were washed and analyzed by flow cytometry (FACS Scan, BD Biosciences), using FlowJo software or in a plate reader format using a SpectraMax M5 plate reader (MDS Analytical Technologies). In all cases un-transfected or un-induced cells were used as controls. For fluorescence microscopy assays, cells were harvested and spun down using a cytofuge (Statspin, Westwood, MA), fixed using formaldehyde and antibodies added as above. For large-scale expression tests, HEK293 cells were induced as above in cellSTACK chambers (Corning, NY), or in spinner flasks, as described previously [17]. Cells were harvested, washed and flash frozen in the presence of protease inhibitors (Roche). To determine the rates of CCR1 degradation, CCR1 expression was induced as above. After 24 hours the induction media was replaced with DMEM/5% BSA in the presence or absence of 100μg/mL cycloheximide for various times. Upon harvesting, cells were lysed in the following buffer: 20mM Tris, pH 8.0, 150mM NaCl, 2mM EDTA, 1% IGEPAL and protease inhibitors (Roche). Samples were run on SDS-PAGE gels and visualized using western blotting with an anti-CCR1 antibody (ABR, Golden, CO).
Radioligand binding and calcium mobilization assays
Binding assays were performed using SPA (scintillation proximity assay) technology (GE Healthcare, Piscataway, NJ) as previously described [21]. Briefly, 2×104 cells per well were harvested, rinsed with PBS and re-suspended in buffer containing 0.2mg of WGA-PVT-SPA beads, 0.05nM 125I-CCL3 and increasing concentrations of unlabeled competitor. Each data point was assayed in quadruplicate using a Microbeta (Perkin Elmer) and data are presented as a percentage of counts obtained in the absence of competing ligand. Calcium flux assays were performed using a FLIPR Calcium 4 assay kit (Molecular Devices), using 1.3×105 cells per well in a 96-well assay format. Chemokine-dependent increases in cytosolic Ca2+ were measured using a FlexStation 3 microplate reader (Molecular Devices).
Expression and purification of chemokines
CCL7 and CCL14 (full length (1–74) and active (9–74) variants) were expressed using a ubiquitin (ub) fusion system as previously described [22]. His6-ub-chemokine constructs were purified using a combination of Ni-affinity and reverse phase C18 HPLC chromatography (Crown et al, in preparation). Labeled variants were obtained by the addition of a C-terminal cysteine using Quikchange (Stratagene, LA Jolla, CA), followed by purification and derivatization using Alexa Fluor 488/647-maleimide or fluorescein-5-maleimide (Invitrogen) (Allen and Handel, in preparation). Masses and purities were confirmed using electrospray mass spectrometry.
Purification of CCR1
Cells expressing CCR1 were resuspended in buffer A (50mM Tris, pH7.4, 0.5M NaCl, 10% (v/v) glycerol) and disrupted using an Emulsiflex (Avestin, Ottawa, Canada) (2 runs, 1000psi). Cell debris was removed by centrifugation (5000g, 5mins, 4°C). Membranes were pelleted by ultracentrifugation (125,000g, 1hr, 4°C), resuspended in buffer A containing one of the following detergents or detergent/lipid combinations: 2% n-octyl-β-D-glucopyranoside (OG), 0.2% n-decyl-β-D-glucopyranoside (DG), 2% 5-cyclohexyl-1-pentyl-β-D-maltoside (Cymal-5), 1% 7-cyclohexyl-1-heptyl-β-D-maltoside (Cymal-7), 1% n-dodecylphosphocholine (FC-12), 1% n-tetradecylphosphocholine (FC-14), 1% n-hexadecylphosphocholine (FC-16), 2% 5-cyclohexyl-1-2-pentylphosphocholine (Cyclofos-5), 2% n-decyl-β-D-maltopyranoside (DM), 1% n-dodecyl-β-D-maltopyranoside (DDM), 1% n-tetradecyl-β-D-maltopyranoside (TDM), 1% DDM + 0.2% cholesteryl hemisuccinate (CHS), 1% DDM + 0.02% CHS or 2% OG + 0.02% CHS. After detergent/lipid addition, membranes were gently homogenized using a Dounce homogenizer and incubated on a shaking platform for 2 hrs at 4°C. After a second ultracentrifugation step, the supernatant containing solubilized CCR1 was loaded onto a Ni-Sepharose column (GE Healthcare), washed and eluted using 250mM imidazole. Fractions were pooled and flash frozen in liquid N2 before being placed at −80°C. For further purification, samples were diluted to decrease the concentration of NaCl, and mixed in batch with either Streptavidin (IBA) or anti-HA (Roche) affinity resins. In both cases the buffer used (buffer B) was as follows: 50mM Tris, pH7.4, 0.1–0.15M NaCl, 10% glycerol, 1mM EDTA + detergents. Columns were poured and washed with >5 column volumes (cv) of buffer B before eluting CCR1. Due to relatively weak affinity between CCR1 and Streptavidin resin, the solution was incubated at 4°C for 30 mins before pouring the column. StrepII tagged CCR1 was eluted by the addition of 4cv of buffer B containing 2.5mM desthiobiotin. HA-tagged CCR1 was eluted by 3 consecutive rounds of incubation with 2cv of buffer B containing 0.5M NaCl and 2mg/mL HA peptide (GenScript) at 25°C/37°C for 15 minutes before elution. The elevated temperature was necessary to elute all CCR1 from the column. In both cases, samples were pooled and flash frozen. For gel filtration, samples were concentrated to 100uL and loaded onto a Superdex 200 gel filtration column with a 24mL bed volume (GE Healthcare). All procedures except for those stated were undertaken at 4°C and protease inhibitor tablets (Roche) were used throughout.
Coomassie and western blotting
Protein samples containing CCR1 were mixed with gel loading buffer and separated on 12% Tris Glycine gels according to standard protocols. Following electrophoresis, gels were either stained using Coomassie Blue, or transferred to nitrocellulose membranes for immunodetection. Antibodies used were as follows: anti-StrepII (IBA), anti-His (R&D Systems), anti-HA (Roche), anti-CCR1 (ABR, Golden, CO), anti-mouse IgG-HRP (Promega, Madison, WI), anti rat IgG-HRP (Chemicon, Billerica, MA) and anti-goat IgG-HRP (Calbiochem, Gibbstown, NJ). Blots were developed using a chemiluminescence detection kit (GE Healthcare) and His and StrepII tag protein ladders were used to aid in molecular weight determination. CCR1 samples were quantified by comparing to BSA standards on Coomassie gels using ImageQuant (GE Healthcare).
Fluorescence anisotropy assays
5–10nM CCL7/CCL14-fluorescein was incubated with increasing concentrations of CCR1 at 25°C for 10 minutes, before measuring fluorescence anisotropy (FA) using a SpectraMax M5 plate reader. Final buffer conditions were as follows: 10mM Tris, pH7.4, 0.1M NaCl, 2% (v/v) glycerol + detergents. For competition assays, 0.17μM CCR1 was incubated with CCL14-fluorescein in the presence of increasing concentrations of the CCR1 inhibitor BX-471 [23]. Experiments were undertaken in quadruplicate and data was normalized to FA in the absence of CCR1. KD values were determined using Origin 7 (Microcal, Northampton, MA). To demonstrate stability of CCR1 over time, 0.3μM samples were incubated for various times at 4°C, 25°C or 37°C before diluting 5-fold and measuring FA as above. Control samples containing no CCR1 were also included and values were plotted as a percentage of initial anisotropy.
RESULTS
Expression of chemokine receptors in different cell lines
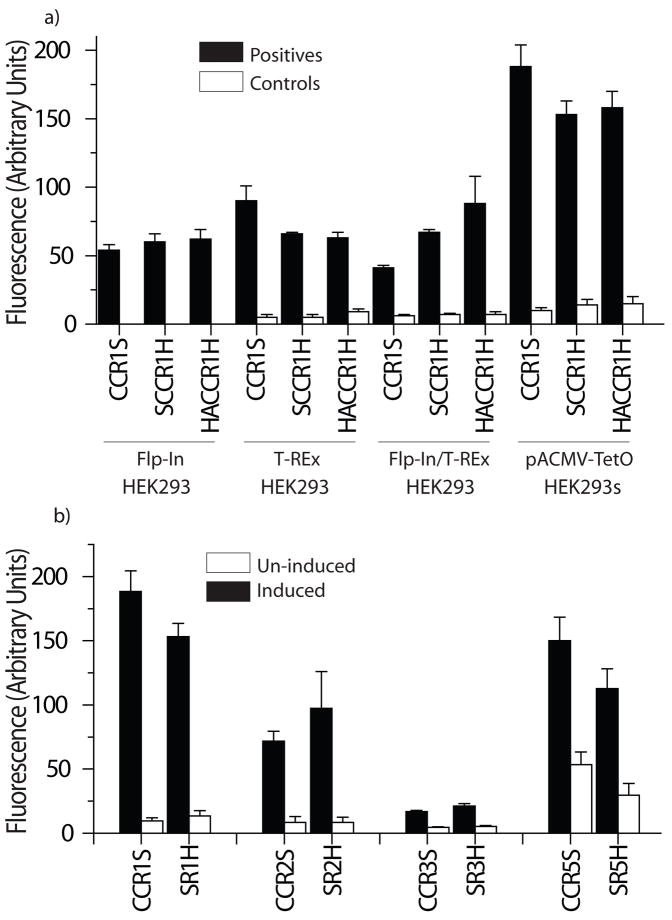
Four different HEK293 mammalian expression systems were studied to identify the best expressing system for CCR1 (Figure 2a). Overall, the presence of N-terminal and/or C-terminal affinity tags designed to aid purification had little effect upon the surface expression levels of CCR1, but the actual levels varied depending upon the vector/cell line used. Of the systems tested, the inducible pACMV-TetO/HEK293 vector/cell system developed by Khorana et al for the high level expression of rhodopsin showed the highest levels of CCR1 cell surface expression [17]. The average level of expression observed in unsorted HEK293 cell populations following transfection was about 2-fold higher than the other HEK293 cell lines, and >5-fold higher than that of the highest expressing insect cell/vector combination tested. HEK293 cells also expressed a number of other chemokine receptors (Figure 2b), indicating general applicability, and in all cases, expression results were consistent regardless of whether a fluorescence plate reader assay or flow cytometry was used (Figure 2b versus Figure S2). In addition to higher expression levels, HEK293 cells had some over advantages over the other systems used, including faster growth rates, higher viability than the other cells tested, the ability to sort for the highest expressing cells, (which was not possible for Flp-In variants, as all transfected Flp-In cells should be identical), and the ability to grow the cells in suspension. For these reasons the HEK293 system was used for all further experiments.
Figure 2. Expression of chemokine receptors in different systems.
a) CCR1 variants containing different N- and/or C-terminal tags were stably transfected into mammalian cell systems (FlpIn/HEK293, T-Rex/HEK293, FlpIn/T-Rex/HEK293 and pACMV-TetO/HEK293). Where appropriate, cells were induced and cell surface receptor levels detected as described in the experimental procedures. b) Chemokine receptor variants CCR1, CCR2, CCR3 and CCR5 were transfected into HEK293 cells and expression levels of induced and un-induced cells were measured (n=4, shown as mean ± S.D.). Key: CCRXS: CCRX-StrepII, SCCRXH: StrepII-CCRX-His8, HACCRXH: Hemagglutinin-CCRX-His8.
Cell surface expression levels of CCR1 and CCR5 variants synthesized using optimal codons for mammalian cells were similar to those observed for the wild-type (non-codon optimized) versions (Figure S3a). This observation remained the same regardless of whether pools of cells, or sorted cells containing only the highest expressing populations were tested (results not shown), indicating that with the present receptor constructs, codon optimization does not make a major difference to expression levels in this system, whether the expression levels are low or high.
Localization and functional analyses of CCR1 at the cell surface
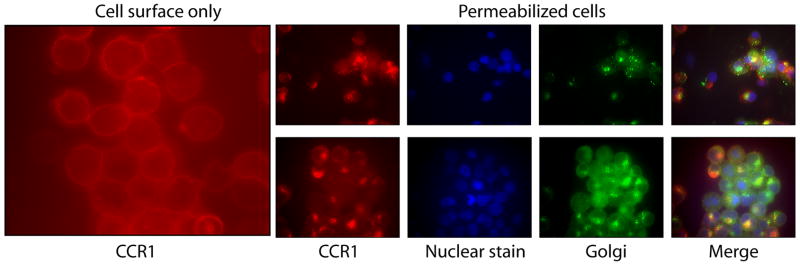
Cell surface expression was confirmed using immunofluorescence staining, which indicated that CCR1 is located both on the cell surface as well as intracellularly, co-localizing with cis and trans Golgi proteins (Figure 3). However, a significant fraction of the protein was present at the cell surface, in stark contrast to the non-signaling chemokine receptor D6, which is constitutively recycled and is almost all intracellular in HEK293 cells [24, 25].
Figure 3. Immunofluorescence staining of CCR1.
Cells expressing CCR1 were harvested, permeabilized where indicated, and stained with the following antibodies: anti-CCR1-phycoerythrin antibody (red), nuclear stain (blue) and either the cis Golgi antibody anti-GM130 (green, top panel) or the trans Golgi antibody 58K (green, bottom panel).
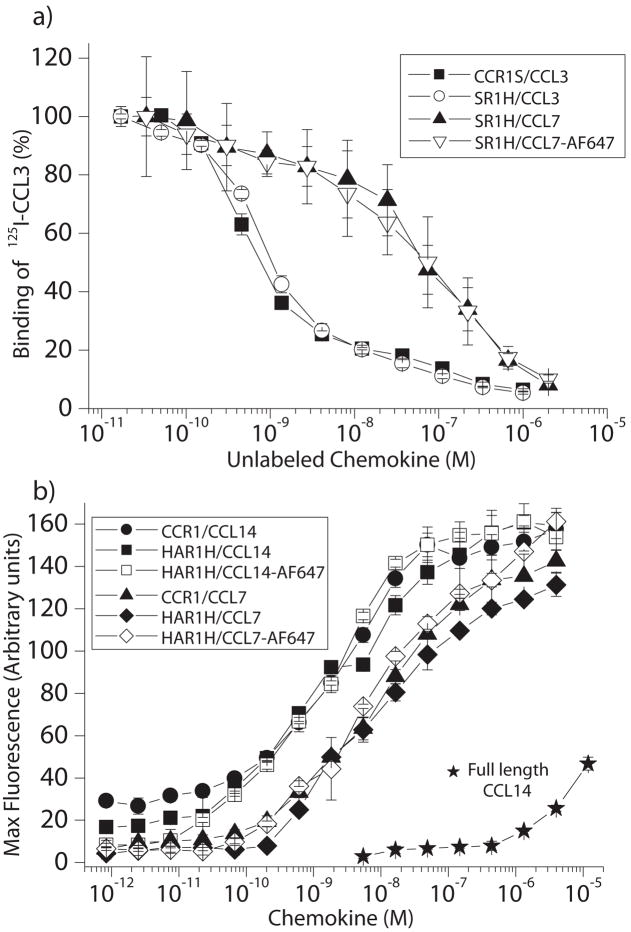
Radiolabeled competition and calcium mobilization assays were undertaken to determine the effects of over-expression and the presence of affinity tags at the N- and/or C-termini upon the activity of CCR1. All constructs tested were as functional as wild-type CCR1 (IC50 for displacement of 125I-CCL3 by CCL3 =0.9nM and EC50 values for calcium mobilization by CCL14 and CCL7 were 2nM and 8nM respectively) (Figure 4). These values are comparable to those previously published [26–28], and indicate that both chemokine binding on the extracellular surface and G-protein coupling on the intracellular surface are not hindered by over-expression, or by the presence of affinity tags. In addition, chemokine variants with C-terminal Alexa-Fluor 647 moieties behaved identically to wild type variants, with respect to their ability to bind to CCR1 and induce a transient calcium flux. Consistent with previous studies [26], N-terminal truncation of residues 1–8 from CCL14 is necessary for high affinity binding and signaling of CCR1 through this chemokine ligand (Figure 4b), as the EC50 for calcium flux by full length CCL14 was >1μM.
Figure 4. Functional studies of CCR1 in HEK293 cells.
a) Chemokine binding to CCR1 was studied by the ability of CCL3, CCL7 or CCL7 containing a C-terminal AlexaFluor 647 moiety (CCL7-AF647) to displace radiolabeled CCL3. b) Chemokine-dependent calcium mobilization in CCR1 transfected HEK293 cells. Data were expressed as chemokine concentration versus maximum fluorescence of a calcium-specific dye (n≥3, shown as mean ± S.D.). Except where indicated, CCL14 (9-74) was used.
Solubilization and purification of CCR1
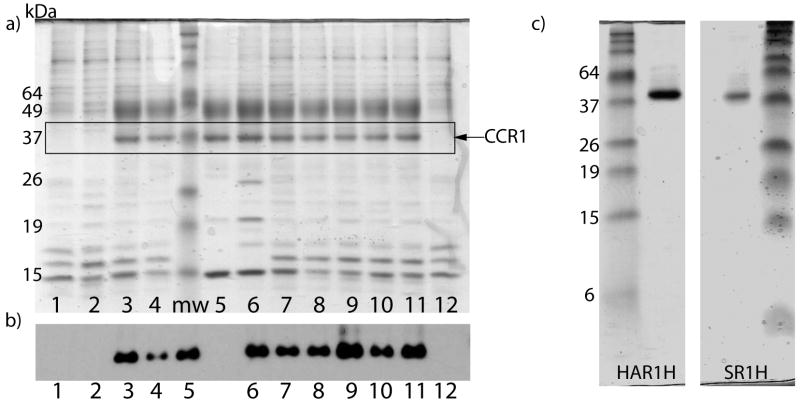
Biophysical studies of chemokine receptors require the protein to be solubilized into artificial detergent/lipid systems, such as micelles, bicelles, vesicles or nanodiscs. However, this is a challenging problem and few studies to date have shown successful reconstitution of chemokine receptors in non-native systems. Due to their relative simplicity and successful use in the solubilization and purification of other native GPCRs, this study focused upon micelles. Micelles are also generally required for initial solubilization of membrane proteins before insertion into bicelles, vesicles or lipid cubic phases [29, 30]. Twelve different detergent/lipid combinations were tested side-by-side in order to identify the best micellar system for reconstitution of CCR1 (see Table 1 for summary of results). Of those tested, nine were able to solubilize CCR1 to varying extents, as assayed by Coomassie staining and western blotting of SDS-PAGE gels of partially purified protein (Figure 5a and b and Table 1, column 3). Not surprisingly, the detergents also varied in their ability to solubilize other contaminating proteins (Figure 5a). Further purification using an HA column or StrepII column of CCR1 constructs in DDM/cholesteryl hemisuccinate (CHS) micelles (lane 8 in Figure 5a/b) yielded pure protein on Coomassie-stained gels (Figure 5c). The yield of the N-terminally HA tagged variant was greater than that of the StrepII tagged variant, mainly because of tighter binding of the HA-tagged variant to the column. In the case of the StrepII tagged variant, ~40% of the protein was lost in the flow-through of the column, whilst only ~10% was lost for the HA-tagged variant. The average yield of purified CCR1 was 0.1–0.2mg per 109 cells, which equates to 1.4–2.8×106 purified receptors/cell. CCR1 contains a number of putative N-linked and O-linked glycosylation sites (Figure 1), but preliminary studies indicate that it is not glycosylated in this system (Hamel and Handel, unpublished observations). None of the three alkyl glucoside detergents tested, including octyl glucoside, were able to solubilize CCR1 (lane 1, Figure 5a/b). This is despite the fact that these detergents have been successfully used in the reconstitution of other G-protein coupled receptors, including rhodopsin [31]. In all three cases, western blots after incubation and ultracentrifugation in the presence of these detergents showed the presence of CCR1 in the pellet, confirming that the protein was not degraded, but was insoluble (data not shown).
Table 1.
Summary of the effects of different detergents upon the solubilization, oligomeric state and binding affinities of CCR1.
| Detergent/lipid system | Abbreviation | Properties of CCR1 in detergent system | ||
|---|---|---|---|---|
| Soluble? | Predominant oligomeric state* | KD for CCL14 binding | ||
| n-Octyl-β-D-glucopyranoside | OG | No | N.D. | |
| n-Decyl-β-D-glucopyranoside | DG | No | N.D. | |
| 5-Cyclohexyl-1-pentyl-β-D-maltoside | Cymal-5 | Yes | O | |
| 7-Cyclohexyl-1-heptyl-β-D-maltoside | Cymal-7 | Yes | O | |
| n-Dodecylphosphocholine | FC-12 | Yes | N.D. | >1uM |
| n-Tetradecylphosphocholine | FC-14 | Yes | M | >1uM |
| n-Hexadecylphosphocholine | FC-16 | Yes | N.D. | >1uM |
| 5-Cyclohexyl-1-2-pentylphosphocholine | Cyclofos-5 | Yes | O | |
| n-Decyl-β-D-maltopyranoside | DM | Yes | O | |
| n-Dodecyl-β-D-maltopyranoside | DDM | Yes | M | |
| n-Tetradecyl-β-D-maltopyranoside | TDM | Yes | O | |
| n-Dodecyl-β-D-maltopyranoside + Cholesteryl hemisuccinate | DDM/CHS | Yes | M | 21nM |
| n-Dodecyl-β-D-maltopyranoside + Cholesteryl hemisuccinate + 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propane sulfonate | DDM/CHS/CHAPS | Yes | M | 35nM |
| n-Octyl-β-D-glucopyranoside + Cholesteryl hemisuccinate | OG/CHS | No | N.D. | |
O: oligomer, M: monomer.
Multiple oligomeric states of CCR1 were observed to varying degrees in all of the detergent systems tested, but for the sake of clarity only the predominant state is shown in the table.
Figure 5. Purification of CCR1 in different lipid/detergent micelles.
a) Coomassie stained SDS-PAGE gel and b) western blot after solubilization and partial purification of CCR1 by Ni-affinity chromatography. The detergents/lipids used for solubilization were as follows: 1) 2% OG, 2) 0.2% DG, 3) 2% Cymal-5, 4) 1% Cymal-7, 5) 1% FC-14, 6) 2% Cyclofos-5, 7) 2% DM, 8) 1% DDM, 9) 1% TDM, 10) 1% DDM + 0.2% CHS, 11) 1% DDM + 0.02% CHS, 12) 2% OG + 0.02% CHS. Where possible, detergents were diluted 10-fold in column buffers after solubilization, but were always kept at concentrations >2-fold above the critical micelle concentration (CMC) values. C) Coomassie–stained gels of HA-His8 tagged (HAR1H) and StrepII-His8 tagged (SR1H) CCR1 after two-column purification.
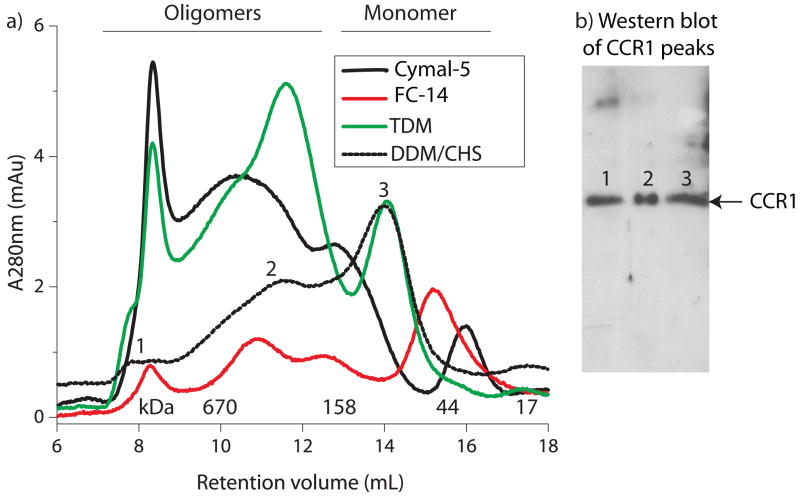
While it is becoming more apparent that oligomerization of many GPCRs, including chemokine receptors, is important in vivo [32], aggregation upon solubilization makes biophysical studies much more complex to analyze in vitro. As the oligomeric state of CCR1 in these micelle systems is unknown and cannot be determined easily using SDS-PAGE gels, CCR1 purified in nine different micelle systems was subjected to size exclusion chromatography. The chromatography traces from a representative set of five of these detergents are shown in Figure 6a and the western blot in Figure 6b illustrates that despite the large differences in apparent molecular weights in the size exclusion chromatograms, all of the peaks contain CCR1.
Figure 6. Representative size exclusion chromatography of CCR1 in different micelles.
a) CCR1 was purified by affinity chromatography, concentrated and run on a size-exclusion column as described in procedures. The different oligomeric states are indicated. b) SDS-PAGE and western blot analysis is unable to distinguish between the presence of monomers, dimers and higher order species of CCR1 solubilized in DDM/CHS.
Although the apparent molecular weight of monomeric CCR1 varied depending upon the micelle composition, it is clear that the oligomeric state of CCR1, and its tendency to aggregate, is highly dependent upon the detergents/lipids present in the environment (Table 1, column 4). For example, Cymal-5-solubilized CCR1 contained very little monomer and was predominantly oligomeric, with some very high molecular weight species (>700kDa) (presumably aggregates). In contrast, CCR1 solubilized in phosphocholine or DDM micelles contained a much larger proportion of monomeric CCR1, along with lower molecular weight oligomers, such as dimers and tetramers. For this reason, these detergents were used for further studies of CCR1 in solution.
High-affinity binding of chemokine ligands to CCR1 solubilized in detergent/lipid micelles
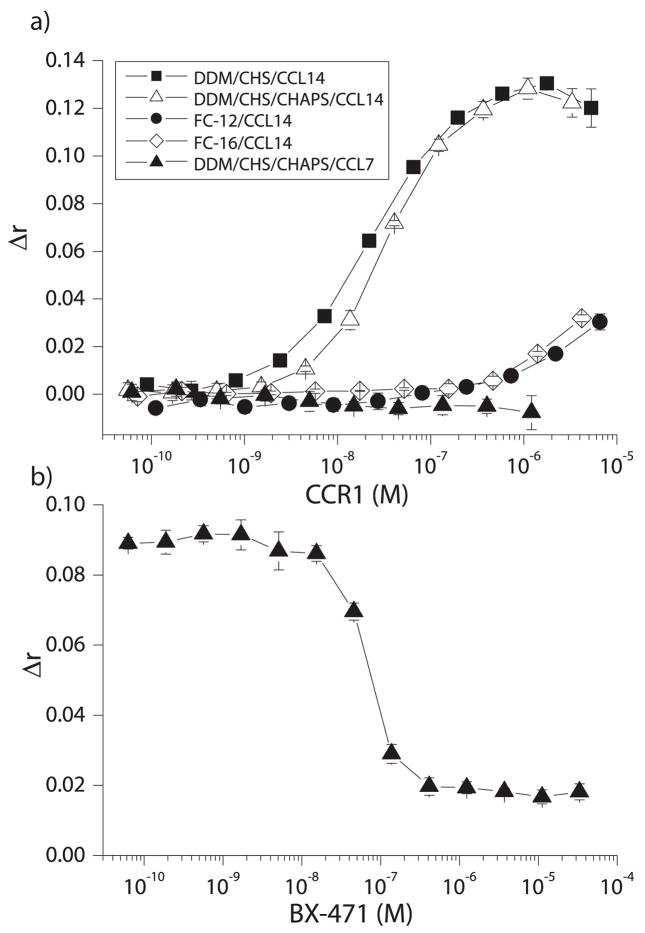
Binding affinities between CCR1 and its chemokine ligands were determined in solution using a fluorescence anisotropy assay. CCR1 solubilized in DDM/CHS or DDM/CHS/CHAPS bound CCL14 with high affinity (KD = 21nM and 35nM for DDM/CHS and DDM/CHS/CHAPS respectively) (Figure 7a and Table 1, column 5). This high-affinity binding was chemokine-specific, as no binding between CCL7 and CCR1 could be observed, even at CCR1 concentrations of >1μM. Similarly, the high affinity binding was detergent-specific, as little binding between CCL14 and CCR1 solubilized in phosphocholine (FC-12, FC-14 or FC-16) micelles was detected. Specificity between CCR1 and CCL14 was demonstrated by pre-incubation of a fixed concentration of CCR1 with increasing concentrations of the CCR1-specific small-molecule antagonist BX-471 prior to the addition of CCL14. In this case, BX-471 inhibited CCL14 binding to CCR1 in a dose-dependent manner, with an IC50 of 68nM (Figure 7b). The inability of BX-471 to fully compete off CCL14 indicates a low level of non-specific binding. Comparisons between binding curves using protein from CCR1-positive and CCR1-negative HEK293 cells indicated that at CCR1 concentrations where full binding was observed for CCR1-positive samples, only 11% of the total signal was due to non-specific effects (Figure S4).
Figure 7. Binding and competition studies of CCR1 solubilized in micelles.
a) Various concentrations of CCR1 solubilized in a number of different detergent/lipid micelles were incubated with a fixed concentration (5nM) of C-terminally fluorescein labeled chemokine (CCL7 or CCL14). Binding was monitored by increases in fluorescence anisotropy (Δr) values relative to those obtained in the absence of CCR1. b) 0.17μM CCR1 was incubated with increasing concentrations of the CCR1 inhibitor BX-471 for 15 minutes, before the addition of 5nM CCL14-fluorescein (n=3, shown as mean ± S.D.).
Stability studies of CCR1 in solution
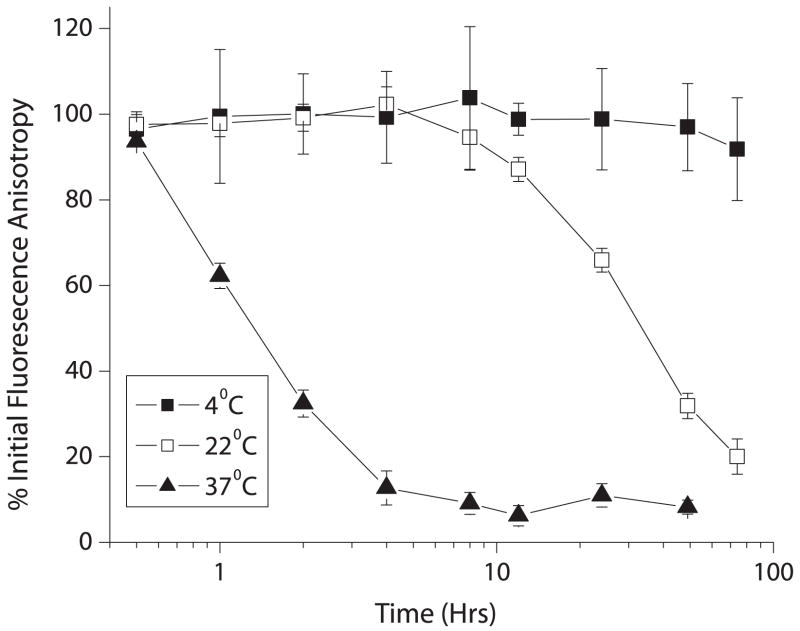
In order to study the stability of CCR1 over time in DDM/CHS/CHAPS micelles, samples were incubated for increasing amounts of time before measuring binding of CCL14 (Figure 8). CCR1 incubated at 37°C began to lose binding activity shortly after incubation. However, samples placed at 22°C retained full activity at 10 hours and CCR1 incubated at 4°C retained full binding, even after 72 hours of incubation.
Figure 8. Stability of CCR1 over time in micelles.
Samples of CCR1 in DDM/CHS micelles were placed at different temperatures for the indicated times, before mixing with CCL14-fluorescein and measuring binding by fluorescence anisotropy. Loss of function over time is indicated by decreasing values on the y-axis (n=3, shown as mean ± S.D.).
DISCUSSION
Chemokine receptors are associated with many diseases and as such represent attractive therapeutic targets. However, structural and functional studies have been hampered by many difficulties associated with expression and purification [6]. In order to address this problem, we used a systematic approach to identify cell systems capable of high-level expression of functional CCR1. We focused on HEK mammalian cells, due to their ability to faithfully reproduce post-translational modifications that may be necessary for correct function, expression, folding and stability of chemokine receptors in vivo. Also, HEK cells are desirable for cell-based studies, and avoid potential problems associated with differences in lipid membrane composition and/or G-proteins between different species. Using this approach, we identified the HEK293 cell expression system developed by Khorana et al [17] for high-level expression of rhodopsin, as an attractive system for the expression of CCR1. Interestingly, glycosylation is often reported to be necessary for stability, intracellular trafficking, expression at the cell surface and folding of GPCRs [33]. Preliminary studies indicate that CCR1 is not glycosylated in this system, yet this did not seem to hinder the ability of CCR1 to reach the cell surface or bind to chemokine ligands with high affinity.
This study highlights the importance of the lipid/detergent environment in the function, stability and oligomeric state of CCR1, and of membrane proteins in general. Of the micelle systems tested in this study, all but three were able to solubilize CCR1, yet despite this, CCR1 solubilized in these micelles behaved very differently with respect to both oligomeric state and the ability to bind CCL14 with high affinity. Interestingly, though FC-12 was not able to solubilize CCR1 in a functional form, previous studies have indicated that FC-12 solubilized-CCR5 binds the ligand CCL5, although with low (~1μM) affinity [34]. The differences observed between studies may well be due to the nature of the chemokine receptor and its individual requirements for function.
Of all of the micelle systems tested, DDM/CHS micelles provided the best environment for the function and stability of CCR1. This mixed micelle system has also been used to successfully reconstitute other GPCRs, including the non-signaling receptor D6 and β2-adrenergic receptor (β2-AR) [24, 35]. In the case of β2-AR the presence of CHS increased the ligand binding affinity and stability. Although these properties of CHS were not directly tested for CCR1, CHS helped to increase yields of soluble protein and maintain CCR1 in a non-aggregating state. The effect of CHAPS upon the binding affinity between CCR1 and CCL14 was also investigated in this study, as CHS/CHAPS played an important role in maintaining the functional state of the human adenosine A2a receptor solubilized in DDM micelles [36]. However, CHAPS made little difference to the ability of CCR1 in DDM/CHS micelles to bind CCL14.
Clearly much work needs to be done to thoroughly understand and identify the optimal membrane environment for CCR1, as many alternatives exist. Micelles, though useful because of their ease of use and amenability for spectroscopic studies, are not the best mimics of the membrane environment and alternatives such as bicelles, lipid vesicles and nanodiscs that are likely to maintain the receptor in a more-native like state are desirable. Though the success of these systems is also likely to be affected by the identity of the receptor under study, we hope to test them as alternate reconstitution systems for CCR1 and other chemokine receptors in the near future.
Previous studies have indicated that low expression levels of chemokine receptors may be at least in part, due to the presence of rare codons [37]. However, codon-optimization of CCR1 or CCR5 in the inducible HEK293 system had little effect on the expression of either receptor. This indicates that surface receptor levels in this system are likely to be limited by the ability of the receptors to travel to the cell surface and/or rates of degradation, and not by the presence of rare codons. Studies are underway to examine this further.
One of the most interesting findings of these studies is the stark contrast between the ability of solubilized CCR1 to bind CCL14(9-74) with nanomolar affinity, and little/no binding to CCL7 at concentrations above 1μM. The reason for this is not currently known, but could be due to a >40-fold difference in binding affinity between these ligands in vivo (Ki for inhibition of CCL5 binding to CCR1 by CCL14(9-74) =23pM, Kd for CCL7 binding to CCR1 ~1nM ) [26, 28]. It is possible chemokine receptor/ligand pairs that exhibit very high affinity in vivo (such as CCR1/CCL14) are less susceptible to the lipid/detergent environment, and so exhibit higher affinity binding in vitro, than those with weaker in vivo interactions. Environmental effects upon binding affinities are likely to include non-optimal detergent packing, incorrect hydrophobic thickness, and the absence of the correct lateral membrane pressure (micelles are more dynamic and less rigid than membranes), thereby leading to slight changes in helix packing, and consequentially binding. Many chemokine/receptor interactions are characterized by a high proportion of basic binding epitopes on the ligand side that presumably interact with acidic residues on the receptors, and it may be that modulations in the electrostatic environment caused by detergent adversely affects the affinity of specific chemokine receptor/ligand pairs. G-protein coupling is also necessary for high affinity interactions between CCR1 and CCL3 in insect cells [38], and therefore the loss of G-proteins during purification is also likely to affect binding affinities between CCR1 and its ligands. In this case, CCR1/CCL14 nanomolar binding may be retained whilst CCR1/CCL7 binding is lost if the CCR1/CCL14 interaction is less dependent upon the presence of G-proteins than the CCR1/CCL7 interaction. However, in the case of CCR1/CCL7, it is also possible that DDM and/or CHS directly bind to an important part of the CCR1/CCL7 interface that is not necessary for high affinity CCR1/CCL14 binding, thereby specifically reducing binding affinity for CCL7.
The absence of G-proteins, along with differences between properties of micelles and the native membrane are likely to be major reasons why CCR1/CCL14 binding in DDM/CHS micelles in vitro is 20nM compared with 23pM reported for in vivo binding. However, despite the decreased affinity between CCR1/CCL14 in vitro, observations that CCR1-negative cells show little non-specific binding and that BX-471 is able to displace CCL14 with an IC50 of 68nM (the reported IC50 for displacement of CCL3 by BX-471 is 10nM [21]) confirm that the binding between CCL14 and CCR1 in this system is indeed specific.
In conclusion, the HEK293 system used in this study may represent a general expression system for GPCRs, as it facilitates expression of rhodopsin [17], β2-AR [16], and all chemokine receptors we have tested to date. CCR1 solubilized from these cells and purified in detergent/lipid micelles binds the chemokine CCL14 with high-affinity in a novel fluorescence anisotropy assay, and binding is inhibited by the specific CCR1 antagonist BX-471. To our knowledge this is the first report of high affinity binding and specific inhibition between a chemokine ligand and its receptor in solution, and opens the way for biophysical experiments involving chemokines, receptors and small-molecule antagonists.
Supplementary Material
Figure S1. DNA sequences of CCR1, CCR2, CCR3 and CCR5 constructs. Tags were added to these constructs to facilitate purification, as described in Materials and Methods.Figure S2. Cell surface expression of CCR1/2/3 and 5 in HEK293 cells.Receptor variants (indicated by the numbers, e.g. CCR5=5) were transfected into HEK293 cells and expression levels were measured following induction by antibody staining and flow cytometry.Figure S3. Codon-optimization of chemokine receptors.Codon-optimized (solid lines) or non codon-optimized (dashed lines) variants of CCR1 (black) or CCR5 (red) were transfected into HEK293 cells. Cell surface expression following induction was detected using antibody staining and flow cytometry. Un-induced cells were used as controls.Figure S4. Binding between CCL14 and CCR1 is specific.HEK293 cells expressing CCR1 (CCR1-positive), or non-CCR1 expressing cells (CCR1-negative) were solubilized in DDM/CHS micelles and subjected to Ni-affinity chromatography. Binding to CCL14 was tested by fluorescence anisotropy.
Acknowledgments
This work was funded by a University of California AIDS Research Program fellowship to SJA (TF06-SD-501) and TMH (1D06-SD-206), and from NIH (RO1-AI37113 and R21 AI076961) to TMH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol. 2008;3:530–541. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- 2.Dalrymple MB, Pfleger KD, Eidne KA. G protein-coupled receptor dimers: functional consequences, disease states and drug targets. Pharmacol Ther. 2008;118:359–371. doi: 10.1016/j.pharmthera.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 4.Schlyer S, Horuk R. I want a new drug: G-protein-coupled receptors in drug development. Drug Discov Today. 2006;11:481–493. doi: 10.1016/j.drudis.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Sarramegn V, Muller I, Milon A, Talmont F. Recombinant G protein-coupled receptors from expression to renaturation: a challenge towards structure. Cell Mol Life Sci. 2006;63:1149–1164. doi: 10.1007/s00018-005-5557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCusker EC, Bane SE, O’Malley MA, Robinson AS. Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol Prog. 2007;23:540–547. doi: 10.1021/bp060349b. [DOI] [PubMed] [Google Scholar]
- 7.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 9.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 12.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 13.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 14.Allen SJ, Crown SE, Handel TM. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 15.Ribeiro S, Horuk R. The clinical potential of chemokine receptor antagonists. Pharmacol Ther. 2005;107:44–58. doi: 10.1016/j.pharmthera.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Chelikani P, Reeves PJ, Rajbhandary UL, Khorana HG. The synthesis and high-level expression of a beta2-adrenergic receptor gene in a tetracycline-inducible stable mammalian cell line. Protein Sci. 2006;15:1433–1440. doi: 10.1110/ps.062080006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves PJ, Kim JM, Khorana HG. Structure and function in rhodopsin: a tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc Natl Acad Sci U S A. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parody TR, Stone MJ. High level expression, activation, and antagonism of CC chemokine receptors CCR2 and CCR3 in Chinese hamster ovary cells. Cytokine. 2004;27:38–46. doi: 10.1016/j.cyto.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horuk R, Clayberger C, Krensky AM, Wang Z, Grone HJ, Weber C, Weber KS, Nelson PJ, May K, Rosser M, Dunning L, Liang M, Buckman B, Ghannam A, Ng HP, Islam I, Bauman JG, Wei GP, Monahan S, Xu W, Snider RM, Morrissey MM, Hesselgesser J, Perez HD. A non-peptide functional antagonist of the CCR1 chemokine receptor is effective in rat heart transplant rejection. J Biol Chem. 2001;276:4199–4204. doi: 10.1074/jbc.M007457200. [DOI] [PubMed] [Google Scholar]
- 21.Vaidehi N, Schlyer S, Trabanino RJ, Floriano WB, Abrol R, Sharma S, Kochanny M, Koovakat S, Dunning L, Liang M, Fox JM, de Mendonca FL, Pease JE, Goddard WA, 3rd, Horuk R. Predictions of CCR1 chemokine receptor structure and BX 471 antagonist binding followed by experimental validation. J Biol Chem. 2006;281:27613–27620. doi: 10.1074/jbc.M601389200. [DOI] [PubMed] [Google Scholar]
- 22.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horuk R. BX471: a CCR1 antagonist with anti-inflammatory activity in man. Mini Rev Med Chem. 2005;5:791–804. doi: 10.2174/1389557054867057. [DOI] [PubMed] [Google Scholar]
- 24.Blackburn PE, Simpson CV, Nibbs RJ, O’Hara M, Booth R, Poulos J, Isaacs NW, Graham GJ. Purification and biochemical characterization of the D6 chemokine receptor. Biochem J. 2004;379:263–272. doi: 10.1042/BJ20031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber M, Blair E, Simpson CV, O’Hara M, Blackburn PE, Rot A, Graham GJ, Nibbs RJ. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–2508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Detheux M, Standker L, Vakili J, Munch J, Forssmann U, Adermann K, Pohlmann S, Vassart G, Kirchhoff F, Parmentier M, Forssmann WG. Natural proteolytic processing of hemofiltrate CC chemokine 1 generates a potent CC chemokine receptor (CCR)1 and CCR5 agonist with anti-HIV properties. J Exp Med. 2000;192:1501–1508. doi: 10.1084/jem.192.10.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pease JE, Wang J, Ponath PD, Murphy PM. The N-terminal extracellular segments of the chemokine receptors CCR1 and CCR3 are determinants for MIP-1alpha and eotaxin binding, respectively, but a second domain is essential for efficient receptor activation. J Biol Chem. 1998;273:19972–19976. doi: 10.1074/jbc.273.32.19972. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Baruch A, Xu L, Young PR, Bengali K, Oppenheim JJ, Wang JM. Monocyte chemotactic protein-3 (MCP3) interacts with multiple leukocyte receptors. C-C CKR1, a receptor for macrophage inflammatory protein-1 alpha/Rantes, is also a functional receptor for MCP3. J Biol Chem. 1995;270:22123–22128. doi: 10.1074/jbc.270.38.22123. [DOI] [PubMed] [Google Scholar]
- 29.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Nollert P, Navarro J, Landau EM. Crystallization of membrane proteins in cubo. Methods Enzymol. 2002;343:183–199. doi: 10.1016/s0076-6879(02)43135-7. [DOI] [PubMed] [Google Scholar]
- 31.Stubbs GW, Smith HG, Jr, Litman BJ. Alkyl glucosides as effective solubilizing agents for bovine rhodopsin. A comparison with several commonly used detergents. Biochim Biophys Acta. 1976;426:46–56. doi: 10.1016/0005-2736(76)90428-4. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Norcross M. Dimerization of chemokine receptors in living cells: key to receptor function and novel targets for therapy. Drug Discov Today. 2008;13:625–632. doi: 10.1016/j.drudis.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Duvernay MT, Filipeanu CM, Wu G. The regulatory mechanisms of export trafficking of G protein-coupled receptors. Cell Signal. 2005;17:1457–1465. doi: 10.1016/j.cellsig.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Nisius L, Rogowski M, Vangelista L, Grzesiek S. Large-scale expression and purification of the major HIV-1 coreceptor CCR5 and characterization of its interaction with RANTES. Protein Expr Purif. 2008 doi: 10.1016/j.pep.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Yao Z, Kobilka B. Using synthetic lipids to stabilize purified beta2 adrenoceptor in detergent micelles. Anal Biochem. 2005;343:344–346. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 36.O’Malley MA, Lazarova T, Britton ZT, Robinson AS. High-level expression in Saccharomyces cerevisiae enables isolation and spectroscopic characterization of functional human adenosine A2a receptor. J Struct Biol. 2007;159:166–178. doi: 10.1016/j.jsb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirzabekov T, Bannert N, Farzan M, Hofmann W, Kolchinsky P, Wu L, Wyatt R, Sodroski J. Enhanced expression, native purification, and characterization of CCR5, a principal HIV-1 coreceptor. J Biol Chem. 1999;274:28745–28750. doi: 10.1074/jbc.274.40.28745. [DOI] [PubMed] [Google Scholar]
- 38.Bouvier M, Menard L, Dennis M, Marullo S. Expression and recovery of functional G-protein-coupled receptors using baculovirus expression systems. Curr Opin Biotechnol. 1998;9:522–527. doi: 10.1016/s0958-1669(98)80040-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. DNA sequences of CCR1, CCR2, CCR3 and CCR5 constructs. Tags were added to these constructs to facilitate purification, as described in Materials and Methods.Figure S2. Cell surface expression of CCR1/2/3 and 5 in HEK293 cells.Receptor variants (indicated by the numbers, e.g. CCR5=5) were transfected into HEK293 cells and expression levels were measured following induction by antibody staining and flow cytometry.Figure S3. Codon-optimization of chemokine receptors.Codon-optimized (solid lines) or non codon-optimized (dashed lines) variants of CCR1 (black) or CCR5 (red) were transfected into HEK293 cells. Cell surface expression following induction was detected using antibody staining and flow cytometry. Un-induced cells were used as controls.Figure S4. Binding between CCL14 and CCR1 is specific.HEK293 cells expressing CCR1 (CCR1-positive), or non-CCR1 expressing cells (CCR1-negative) were solubilized in DDM/CHS micelles and subjected to Ni-affinity chromatography. Binding to CCL14 was tested by fluorescence anisotropy.