Abstract
Microcephalin (MCPH1) is a BRCT-domain containing protein involved in the cellular response to DNA damage that has been implicated in autosomal recessive primary microcephaly. MCPH1 is recruited to sites of DNA double strand breaks by phosphorylated histone H2AX (γH2AX) but the mechanism by which MCPH1 contributes to the repair process remains to be determined. Here we show that MCPH1 binds to BRCA2 and regulates the localization of BRCA2 and Rad51 at sites of DNA damage. The interaction occurs through the N-terminus of BRCA2 and the C-terminal BRCT domains of MCPH1. Disruption of the interaction between MCPH1 and BRCA2 has no effect on the ability of BRCA2 to form a complex with Rad51 but is associated with substantially reduced levels of both BRCA2 and Rad51 at sites of DNA double strand breaks. Uncoupling of MCPH1 from BRCA2 also interferes with Rad51 and BRCA2 dependent homologous recombination repair activity. These results suggest that the role of MCPH1 in the DNA damage response is in part associated with the ability to localize BRCA2 to sites of DNA double stand breaks.
Keywords: Microcephalin, BRCA2, DNA repair, homologous recombination
Introduction
Microcephalin (MCPH1) is a member of the BRCA1 C-terminal (BRCT) domain family of proteins that are involved in the cellular response to DNA damage (1, 2). MCPH1 contains a single N-terminal and two C-terminal BRCT domains. Biallelic mutations in the MCPH1 gene are associated with primary microcephaly (3) (OMIM 251200) and premature chromosome condensation syndrome (4, 5) (OMIM 606858). Mutations in other DNA damage response genes including Nijmegen breakage syndrome (NBS1) (6), ataxia-telangiectasia (A-T) (7) and ataxiatelangiectasia and Rad3-related protein (ATR) (8) are also associated with microcephaly, suggesting that MCPH1 participates in the NBS1, ATM and ATR-associated DNA damage response signaling pathways. Indeed, MCPH1 is rapidly recruited to nuclear foci following DNA damage (9). The localization of MCPH1 along with MDC1 (9), 53BP1 and γH2AX (10-12) to sites of DNA double strand breaks is dependent on an interaction between the C-terminal BRCT domains of MCPH1 and γH2AX (11, 12), but does not appear to involve MDC1 or 53BP1 (11). MCPH1 deficient cells display defects in intra-S and G2/M phase cell cycle checkpoint activation in response to irradiation resulting in nuclear fragmentation, chromosomal instability and centrosome amplification (9, 10, 13, 14).
Other domains of MCPH1also appear to have roles in mediating chromosome instability. Specifically, the single N-terminal BRCT domain in MCPH1 is required for centrosome localization of the protein (12) and also has been implicated in rescue of the premature chromosome condensation (PCC) phenotype observed in MCPH1 deficient cells from individuals with primary microcephaly (15). In addition, a central domain of MCPH1 that interacts with Condensin II through the CAPG2 subunit is required for homologous recombination activity in response to DNA double strand breaks in mouse embryonic fibroblasts reconstituted with MCPH1 (15).
Together these findings suggest that MCPH1 is an important mediator of the response to DNA damage and maintenance of chromosomal stability. The recent observation that MCPH1 exhibits reduced expression in breast tumors and is associated with genomic instability and metastases (14) is in keeping with these results and suggests that MCPH1 may have a role in tumor suppression. Further studies aimed at fully understanding the mechanisms by which MCPH1 influences DNA damage signaling and repair may clarify the role of this protein in cancer.
Here we investigate the role of MCPH1 in the DNA repair process at sites of DNA damage. We find that BRCA2 interacts with the C-terminal BRCT domains of MCPH1 and that this interaction controls the enrichment of BRCA2 and Rad51 at sites of DNA double strand breaks after irradiation. We show that MCPH1 does not influence formation of BRCA2-Rad51 complexes, suggesting that the interaction of MCPH1 with BRCA2 is required for recruitment and/or retention of these BRCA2-Rad51 complexes at sites of damage. We also verify that the interaction between MCPH1 and BRCA2-Rad51 complexes mediates BRCA2 and Rad51-dependent homologous recombination repair of DNA damage.
Materials and Methods
Plasmids and antibodies
FLAG-tagged BRCA2 fragments were cloned in pEV3S-FLAG3 vector. MCPH1 wildtype and deletion mutants were cloned in pCS3−6xMyc vector. GST-tagged BRCA2 fragments (B2F1-B2F7) in pEGB and FLAG-tagged fragments (B2F1-B2F7) in pcDNA3.1 were provided by Dr. J. Chen (Yale University). The anti-MCPH1 rabbit polyclonal antibody was raised against residues 92−214 of MCPH1. The anti-BRCA2 rabbit polyclonal antibody was raised against the C-terminal of BRCA2 (residues 2418−3201). The antisera were affinity purified with AminoLink Plus immobilization and purification kit (Pierce). Anti-γH2AX, 53BP1, MDC1 and Rad51 polyclonal antibodies were gifts from Dr. J. Chen (Yale University). Mouse anti-Flag (M2) antibody was purchased from Sigma. Mouse anti-HA antibody was purchased from Roche. Mouse anti-Myc antibody was purchased from Santa Cruz.
Short interfering RNA
siRNAs against MCPH1 were synthesized by Dharmacon Inc. The siRNA duplexes were as follows: MCPH1 siRNA1 sense strand, 5’C TCT CTG TGT GAA GCA CCA; MCPH1 siRNA2 sense strand, 5’AA GCT CAG AAG AGA GGC GT. BRCA2 siRNAs (SMARTpool) were purchased from Dharmacon. Transfections were performed with 200nM siRNA using FuGene-6 (Roche) according to the manufacturer's instructions.
Co-immunoprecipitation
293T cells were purchased from American Tissue Type Culture (Manassas, VA) and maintained in DMEM supplemented with 10% bovine serum and 1% penicillin/streptomycin at 37°C in 5% CO2 (v/v). VU423 FANCD1 (FA-D1) and A913 423/2−33 BRCA2-reconstituted FANCD1 cells were grown under similar conditions (16). Cells were lysed with NETN buffer (20mM Tris-HCl, pH 8.0, 100mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) containing protease inhibitors (CompleteTM, Boehringer Mannheim Biochemicals Inc.) 2hr after exposure to 10Gy of irradiation, or in the absence of irradiation (data not shown). Whole cell lysates obtained after centrifugation were incubated with 2μg antibody and Protein A or G sepharose beads (Amersham Biosciences) for 2h at 4°C. For GST-pull downs of GST-tagged proteins, cleared cell lysates were incubated with Glutathione agarose beads (Sigma).
Immunofluorescence
293T cells cultured on glass coverslips were irradiated (10Gy), incubated for 3hr and analyzed for formation of DNA damage repair foci. Cells were fixed with cold methanol for 20 min, permeabilized in 0.05% Triton-X 100 for 15 min, incubated in blocking buffer (16) and incubated with primary antibodies diluted in blocking buffer for 1hr at room temperature. After washing in PBS, cells were incubated with Alexa Fluor 488 and 568 conjugated Goat anti-mouse and Goat anti-rabbit IgG secondary antibodies (Molecular Probes). Cells were co-stained with 4’6-diamidino-2-phenylindole (DAPI). After a further wash in PBS coverslips were mounted in Prolong (Molecular Probes). Cells were visualized and imaged using a Zeiss LSM510 confocal microscope. For each experiment 100 nuclei transfected with expression constructs or siRNAs were evaluated. Effects on foci were consistently observed in greater than 80% of transfected cells in each experiment.
Homologous Recombination Repair Assay
The homologous recombination repair assay was carried out as described previously (17). Briefly, the efficiency of homology directed repair was assessed using an I-SceI expression plasmid (pcBASce) and an I-SceI repair reporter plasmid (DR-GFP) composed of two differentially mutated GFP genes, one of which contains a unique I-SceI restriction site. Here, Hela cells or V-C8 BRCA2 null hamster lung fibroblast cells that were stably transfected with a single copy of DR-GFP (16) were co-transfected with I-SceI and BRCA2 plasmids or MCPH1 siRNAs and the number of GFP-expressing cells was assessed by flow cytometry after 48hr. Transfection efficiency was defined by immunofluorescence analysis of FLAG-tagged BRCA2 constructs and was used to normalize homologous recombination repair levels. Statistical significance relative to wildtype BRCA2 or vector control was determined using paired t-tests and Fisher's Protected Least Significant Difference pair-wise comparison procedure.
Results
MCPH1 interacts with BRCA2
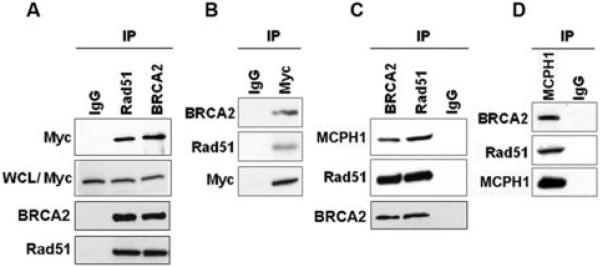
While it has been noted that MCPH1 is one of the first signaling proteins recruited to nuclear foci following DNA damage, there is also evidence that MCPH1 functions downstream of Chk1 and BRCA1 (13) and that MCPH1 is retained at sites of damage beyond the initial recognition stage (12). This suggests that MCPH1 contributes to DNA repair at multiple levels. To elucidate the involvement of MCPH1 in the response to DNA double strand breaks we explored potential interactions between MCPH1 and components of DNA damage signaling and repair pathways. Extracts from 293T cells ectopically expressing Myc-tagged MCPH1 were exposed to 10Gy of irradiation and were screened for interactions between MCPH1 and a series of proteins involved in the DNA repair process. We reproducibly found that myc-tagged MCPH1 co-immunoprecipitated with BRCA2 and Rad51 (Fig. 1A and B). In separate experiments, a significant amount of endogenous MCPH1 associated with endogenous BRCA2 and Rad51 (Fig. 1C and D). These interactions were also evident in the absence of irradiation. Together these results suggest that MCPH1 interacts with BRCA2-Rad51 complexes in the presence and absence of DNA damage.
Figure 1. MCPH1 Interacts with BRCA2 and Rad51.
(A) Whole cell lysates (WCL) from 293T cells transfected with Myc-MCPH1 and treated with 10Gy of irradiation were immunoprecipitated (IP) with anti-BRCA2 and anti-Rad51 antibodies and immunoblotted with anti-Myc (MCPH1) antibody. (B) Lysates immunoprecipitated with Myc were immunoblotted for BRCA2, Rad51, and Myc-tagged MCPH1. (C and D) Interactions between endogenous BRCA2, Rad51 and MCPH1 in 293T cells was assessed by co-immunoprecipitation and immunoblotting.
The MCPH1 BRCT domains are required for interaction with the N-terminus of BRCA2
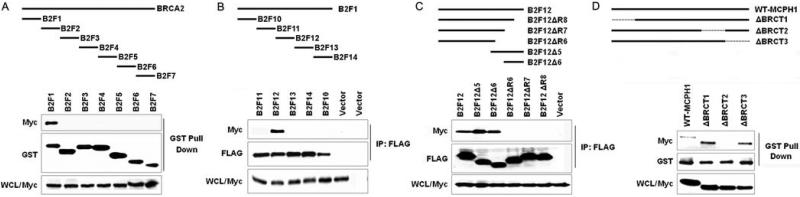
To identify regions of BRCA2 that are responsible for the MCPH1 interaction, we generated a series of constructs encoding fragments of BRCA2 (Fig. 2A and supporting information (SI) Table S1). The fragment B2F1, encompassing BRCA2 residues 1−472, accounted for the interaction between BRCA2 and MCPH1 (Fig. 2A). Deletion mutants of B2F1 mapped the MCPH1 interaction region to residues 234−335 (B2F12) (Fig. 2B). Further deletion analysis within B2F12 showed that residues 294−335 in B2F12Δ6 were sufficient for the interaction (Fig. 2C). However, deletion of residues 293−323 in B2F12.3 disrupted the interaction (Fig. S1), as did deletion of residues 325−335 in B2F12ΔR6, B2F12ΔR7 and B2F12ΔR8 (Table S1 and Fig. 2C). The combined results suggest a complex MCPH1 interaction site located between residues 294−335 of BRCA2. We also generated deletion mutants of MCPH1 that lacked each of the three BRCT domains (Fig. 2D and Table S1) and assessed their ability to interact with BRCA2. GST-tagged B2F1 failed to interact with MCPH1 in the absence of the MCPH1 BRCT2 domain (residues 642−720) (Fig. 2D). In contrast, deletion of the BRCT1 (residues 1−96) and BRCT3 domains (residues 753−823) had no effect on the interaction (Fig. 2D). These findings indicate that a previously undefined BRCA2 N-terminal domain and the MCPH1 BRCT2 domain are necessary for the interaction of BRCA2 with MCPH1.
Figure 2. An N-terminal region of BRCA2 and MCPH1 C-terminal BRCT domain are required for the MCPH1-BRCA2 interaction.
(A) Plasmids encoding seven GST-tagged fragments of BRCA2 were expressed in 293T cells with Myc-tagged MCPH1. GST pull-downs from 293T lysates were immunoblotted with anti-Myc and anti-GST antibodies. (B and C) Plasmids encoding FLAG-tagged fragments of B2F1 (residues 1−472) and FLAG-tagged deletion mutants of B2F12 (residues 234−335) were co-transfected with Myc-tagged MCPH1 into 293T cells. Lysates (WCL) were inmmunoprecipitated with FLAG-agarose beads and immunoblotted with anti-Myc and anti-FLAG antibodies. (D) Plasmids encoding Myc-tagged full-length and deletion mutants of MCPH1 were co-transfected into 293T cells with GST-tagged B2F1. GST pull-downs were immunoblotted with anti-GST and anti-Myc antibodies. Dotted lines represent deleted regions.
Localization of BRCA2 at sites of DNA damage is dependent on MCPH1
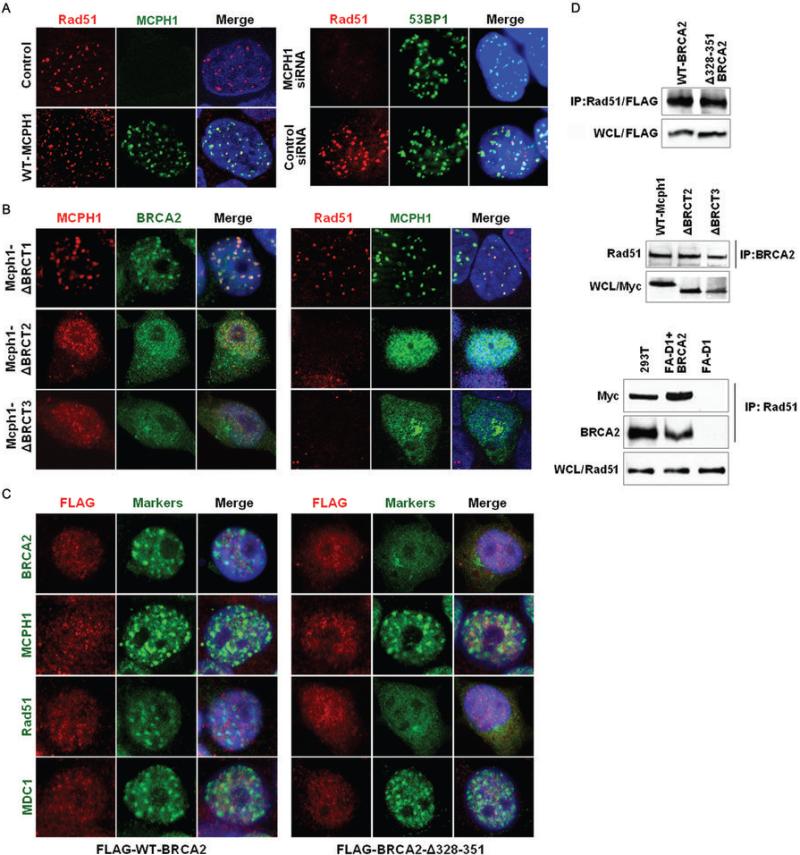
While MCPH1 is one of the first proteins recruited to sites of radiation induced DNA damage, the interaction of MCPH1 with BRCA2 and Rad51 suggests that MCPH1 may function at multiple levels within the DNA damage signaling pathway. Here, we focused on determining the influence of MCPH1 on distal DNA damage signaling pathways. Specifically, we evaluated whether MCPH1 influenced the presence of BRCA2 and Rad51 at sites of DNA damage. As expected, MCPH1 was rapidly recruited to DNA damage foci after irradiation, where it co-localized with 53BP1, γH2AX (Fig. S2a,c), and Rad51 (Fig. 3A) in all cells examined. However, when MCPH1 was depleted by siRNAs (Fig. S2b,c), a substantial reduction in the number and intensity of Rad51 foci (Fig. 3A) and BRCA2 foci (Fig. S2d) was consistently observed. In contrast, the MCPH1 siRNAs had little influence on recruitment of 53BP1 (Fig 3A and Fig. S2c) and MDC1 (Fig. S2d) to foci, suggesting that the proximal DNA damage response pathway remained intact. This is consistent with recent findings that MCPH1 functions in an γH2AX-dependent but MDC1-independent DNA damage response pathway (11). Together, these results indicate that MCPH1 is an important mediator of the BRCA2 and Rad51 associated double strand DNA break repair process.
Figure 3. The MCPH1-BRCA2 interaction is required for localization of Rad51 at sites of DNA damage.
(A) MCPH1 colocalizes with Rad51 after DNA damage. 293T cells either not transfected (Control) or transfected with a Myc-tagged MCPH1 expression construct (WTMCPH1) were stained with anti-Rad51 (Red) and anti-Myc (Green) antibodies 3hr after exposure to 10Gy of radiation to visualize ionizing radiation induced nuclear foci. Likewise, 293T cells transfected with MCPH1 siRNA for 48hr were exposed to 10Gy of radiation and stained with anti-Rad51 (Red) and anti-53BP1 (Green) antibodies after 3hr to visualize depletion of Rad51 foci. (B) Cells transfected with plasmids encoding Myc-tagged MCPH1 deletion constructs were treated and evaluated for MCPH1 and BRCA2 foci by staining for Myc (Red) and BRCA2 (green). Separately, the same cells were evaluated for Rad51 and Myc-tagged MCPH1 foci with Myc (green) and Rad51 (Red). (C) 293T cells transfected with FLAG-tagged wildtype BRCA2 (FLAG-WT-BRCA2) or FLAG-tagged BRCA2-Δ328−351 deletion mutant were incubated for 48h, exposed to 10 Gy of radiation and stained after 3hr for FLAG (Red) and BRCA2, MCPH1, Rad51 or MDC1 (Green). (D) Lysates (WCL) from 293T cells transfected with Flag-tagged wildtype BRCA2 (WT-BRCA2) and the Flag-tagged BRCA2-Δ328−351 deletion construct were co-immunoprecipitated with anti-Rad51 antibody and immunoblotted with anti-FLAG antibody. Lysates from cells expressing wildtype (WT-MCPH1) and deletion mutants of MCPH1 were co-immunoprecipitated with anti-BRCA2 antibody and immunoblotted with Rad51 antibody and with anti-Myc antibody to verify MCPH1 expression. VU423 FANCD1 cells (FA-D1) and A913 423/2−33 FANCD1 cells reconstituted with BRCA2 (FAD1+BRCA2) were transfected with Myc-MCPH1, immunoprecipitated with anti-Rad51 antibody and immunoblotted with anti-Myc and anti-BRCA2 antibodies. Lysates were immunoblotted with anti-Rad51 antibody to verify Rad51 expression.
To determine whether the influence of MCPH1 on DNA damage repair foci is dependent on the interaction of MCPH1 with BRCA2 and Rad51 we assessed the impact of MCPH1 mutants with deletions of the N-terminal BRCT1 and C-terminal BRCT2 and BRCT3 domains on radiation foci. Absence of the N-terminal BRCT domain had no effect on localization of MCPH1, BRCA2 or Rad51 to DNA damage foci (Fig. 3B). This is consistent with the proposed involvement of this MCPH1 domain in centrosome regulation and PCC but not DNA repair (12, 15). In contrast, the BRCT2 and BRCT3 deletion mutants did not localize to foci, but instead displayed a dispersed nuclear and cytoplasmic expression pattern (Fig. 3B and Fig. S3). The absence of the BRCT2 and BRCT3 deletion mutants from the foci had no effect on recruitment of 53BP1 and γH2AX (Fig. S3) but substantially reduced the number and intensity of BRCA2 and Rad51 foci (Fig. 3B). These results indicate that MCPH1 can influence the DNA damage response signaling pathway distal to γH2AX and are consistent with a direct effect of MCPH1 on the recruitment and/or retention of BRCA2 and Rad51 at the DNA repair foci.
MCPH1 does not influence formation of BRCA2-Rad51 complexes
To determine the influence of MCPH1 on the established interaction between BRCA2 and Rad51, we generated a deletion mutant (BRCA2-Δ328−351) of full-length BRCA2 (Table S1) that disrupts part of the MCPH1 interaction site defined by the deletion mapping studies (Fig. 2C and 2D). Similarly to BRCA2 siRNA, this mutant had no influence on the localization of MCPH1, MDC1 and 53BP1 to sites of DNA damage (Fig. 3C and Fig. S4a,b), but substantially reduced the number and intensity of BRCA2 and Rad51 foci (Fig. 3C and Fig. S4b). Likewise, the B2F12 fragment that contains the MCPH1 interaction site and binds to MCPH1, inhibited the localization of MCPH1, BRCA2 and Rad51 to DNA damage foci (Fig. S5a) in a dominant negative manner. In contrast, the B2F12ΔR6 construct, in which the MCPH1 interaction domain is disrupted, had no influence on MCPH1, BRCA2 or Rad51 focus formation (Fig. S5B). These results suggest that the presence of Rad51 and BRCA2 at DNA damage foci is dependent on the interaction between BRCA2 and MCPH1.
Importantly, we also found that the BRCA2-Δ328−351 deletion mutant retained the ability to co-immunoprecipitate with Rad51 following DNA damage (Fig. 3D). Similarly, deletion of the MCPH1 BRCT2 and BRCT3 domains that mediate the interaction with BRCA2 and the presence of BRCA2 and Rad51 at foci had no effect on the BRCA2-Rad51 complex (Fig. 3D). We further explored this effect using BRCA2 deficient FANCD1 cells and FANCD1 cells reconstituted with wildtype BRCA2. Following DNA damage, MCPH1 co-immunoprecipitated with Rad51 from the BRCA2 reconstituted cells but not from the BRCA2 deficient cells (Fig. 3D), suggesting that the Rad51 interaction with MCPH is dependent on BRCA2. Together, these results indicate that MCPH1 interacts with and controls the presence of the BRCA2-Rad51 complex at sites of DNA damage.
MCPH1 influences BRCA2-dependent homologous recombination repair
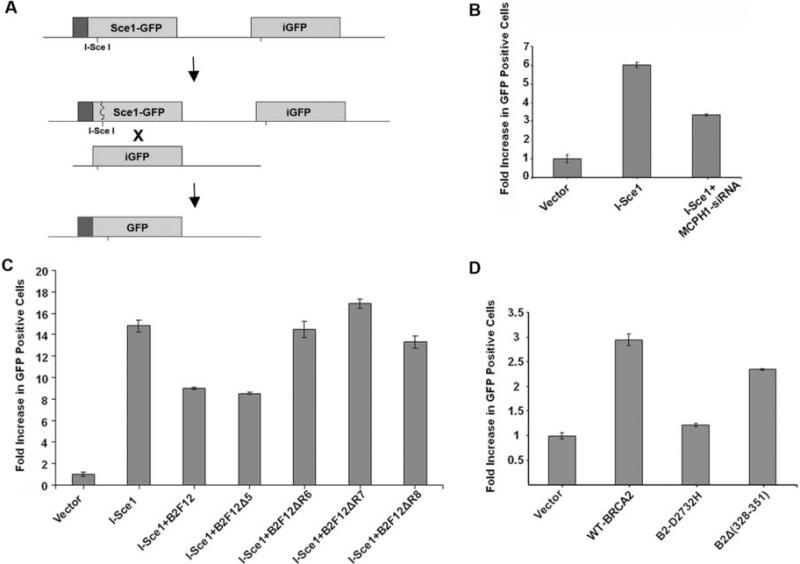
Given the direct involvement of the BRCA2-Rad51 complex in homologous recombination repair of DNA double strand breaks, these results suggest that MCPH1 may influence homologous recombination repair. We evaluated the influence of MCPH1 on homologous recombination using an in-vivo assay (18-22) that depends on repair of a double strand break at a unique I-Sce1 restriction endonuclease site (Fig. 4A). Depletion of MCPH1 from HeLa cells by siRNA significantly reduced homologous recombination repair activity relative to controls (p=0.002) suggesting that MCPH1 levels influence this form of DNA damage repair (Fig. 4B). To determine whether the interaction of MCPH1 with BRCA2 accounts for the influence of MCPH1 on homologous recombination we evaluated a series of BRCA2 deletion constructs (Fig. 2 and Table S1) using the in-vivo assay. Expression of B2F12 and B2F12Δ5, that contain the MCPH1 interaction domain, dominant negatively inhibited homologous recombination activity (p=0.002) (Fig. 4C). In contrast, three BRCA2 fragments (B2F12ΔR6, B2F12ΔR7 and B2F12ΔR8) that do not interact with MCPH1 did not significantly disrupt homologous recombination activity (Fig. 4C). To confirm these effects in the context of full length BRCA2, the influence of the BRCA2-Δ328−351 deletion mutant on homologous recombination activity relative to wildtype BRCA2 and a known deleterious missense mutant (D2723H) was assessed in BRCA2-null V-C8 cells containing the DR-GFP construct. Consistent with the results described above, the BRCA2-Δ328−351 deletion mutant caused a partial but statistically significant reduction in activity relative to wildtype BRCA2 (p=0.009) similarly to D2723H (p=0.001) (Fig 4D). These data indicate that the interaction between MCPH1 and BRCA2 mediates the homologous recombination repair activity of the BRCA2-Rad51 complex.
Figure 4. The MCPH1-BRCA2 interaction influences homologous recombination repair.
(A) Diagram of the homologous recombination repair reporter assay. (B) Homologous recombination repair activity in a HeLa DR-GFP cell line is reduced relative to vector controls when MCPH1 is depleted by MCPH1 siRNA. (C) Homologous recombination repair activity in a HeLa DR-GFP cell line following co-transfection of I-Sce1 plasmid and BRCA2 deletion constructs with the MCPH1 interaction domain (B2F12, B2F12Δ5) and without the interaction domain (B2F12ΔR6−8). (D) Homologous recombination repair activity in a V-C8 DR-GFP cell line, using BRCA2-Δ328−351 and BRCA2-D2723H full-length mutant constructs.
Discussion
Although several studies suggest that MCPH1 is one of the first proteins recruited to sites of irradiation-induced foci where it is thought to mediate the response to DNA damage (10-14), little is known about how MCPH1 influences DNA repair. Recent studies have suggested that MCPH1 controls the cell cycle response to DNA damage by binding and mediating Chk1 and Cdc25a activity (13) and by regulating both the S and G2/M checkpoints. In addition, the interaction between MCPH1 and Condensin II is apparently not associated with premature chromosome condensation but is required for homologous recombination repair of DNA double strand breaks (15), suggesting that MCPH1 may have a role in the process of chromosome condensation and perhaps chromatin remodeling in response to DNA damage.
Here we report that the presence of BRCA2-Rad51 complexes at sites of DNA double strand breaks is dependent on an interaction between the N-terminus of BRCA2 and the C-terminal tandem-BRCT domains of MCPH1. Importantly, BRCA2 and Rad51 form complexes in the absence of MCPH1, albeit not at sites of DNA damage, suggesting that MCPH1 is required for the recruitment and/or retention of BRCA2-Rad51 complexes to repair foci. We also show that MCPH1 can regulate homologous recombination repair of DNA double strand breaks in a BRCA2-dependent manner. These findings are consistent with the observation that MCPH1−/− MEFs exhibit defects in homologous recombination repair of DNA damage (15). Thus, MCPH1 appears to contribute to homologous recombination repair through interactions with BRCA2 and with the Condensin II complex (15).
Our findings extend our knowledge of MCPH1 and suggest that it forms an important link between the initial sensors and effectors of the DNA damage response, as defined by its rapid recruitment to DNA repair foci through an interaction with γH2AX, and the process of DNA repair, as established by its ability to regulate the localization of BRCA2-Rad51 complexes to sites of damage. Further efforts are needed to better understand how MCPH1 integrates these components of the DNA damage response signaling pathway.
Interestingly, it has recently been shown that Akt1 can repress homologous recombination through cytoplasmic retention of BRCA1 and Rad51 (23). It remains to be seen whether Akt1 signaling influences MCPH1-dependent recruitment and/or retention of BRCA2-Rad51 complexes at sites of damage. Furthermore, MEFs deficient in the SIRT1 histone deacetylase have been shown to exhibit significantly reduced recruitment of BRCA1, Rad51 and 53BP1 to DNA double strand breaks and reduced homologous recombination activity (24). Given the role of SIRT1 in chromatin remodeling and the recent finding that SIRT1 localizes to DNA breaks to promote repair (25), further studies aimed at understanding the relationship between SIRT1 and components of the DNA signaling pathway such as MCPH1 and BRCA2 are needed.
The ability of MCPH1 to regulate BRCA2 activity suggests that down-regulation or mutation of MCPH1 could lead to defects in DNA repair similar to those associated with BRCA2 inactivation. Importantly, down-regulation of MCPH1 has been observed in a number of tumors. Thus, MCPH1 may function as a tumor suppressor. Whereas mutation of both MCPH1 alleles is associated with premature chromosome condensation (PCC) and primary microcephaly (5), mutation of a single allele may predispose or contribute to cancer development. Future studies aimed at detecting mutations in this gene and studies using knockout-mice will test whether MCPH1 functions as a tumor suppressor in vivo. In addition, since MCPH1 appears to mediate BRCA2 function, it may be possible to use PARP inhibitors or DNA crosslinking agents that appear to be particularly effective against BRCA2 deficient cells to treat tumors with reduced levels of MCPH1.
Acknowledgements
Financial support: This work was supported in part by National Institutes of Health grants CA102701 and CA116167 to F.J.C.
Footnotes
Conflicts of Interest: None
References
- 1.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Altschul SF, Bork P. BRCA1 protein products ... Functional motifs. Nat Genet. 1996;13:266–8. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 3.Jackson AP, Eastwood H, Bell SM, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–42. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neitzel H, Neumann LM, Schindler D, et al. Premature chromosome condensation in humans associated with microcephaly and mental retardation: a novel autosomal recessive condition. Am J Hum Genet. 2002;70:1015–22. doi: 10.1086/339518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trimborn M, Bell SM, Felix C, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–6. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varon R, Vissinga C, Platzer M, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–76. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 7.Gilad S, Chessa L, Khosravi R, et al. Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am J Hum Genet. 1998;62:551–61. doi: 10.1086/301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem. 2004;279:34091–4. doi: 10.1074/jbc.C400139200. [DOI] [PubMed] [Google Scholar]
- 10.Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci U S A. 2005;102:15105–9. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282:35416–23. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffers LJ, Coull BJ, Stack SJ, Morrison CG. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 2008;27:139–44. doi: 10.1038/sj.onc.1210595. [DOI] [PubMed] [Google Scholar]
- 13.Alderton GK, Galbiati L, Griffith E, et al. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol. 2006;8:725–33. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 14.Rai R, Dai H, Multani AS, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–57. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood JL, Liang Y, Li K, Chen J. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283:29586–92. doi: 10.1074/jbc.M804080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lingle WL, Barrett SL, Negron VC, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–83. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohashi A, Zdzienicka MZ, Chen J, Couch FJ. Fanconi anemia complementation group D2 (FANCD2) functions independently of BRCA2- and RAD51-associated homologous recombination in response to DNA damage. J Biol Chem. 2005;280:14877–83. doi: 10.1074/jbc.M414669200. [DOI] [PubMed] [Google Scholar]
- 18.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu K, Hinson SR, Ohashi A, et al. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65:417–26. [PubMed] [Google Scholar]
- 20.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 21.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plo I, Laulier C, Gauthier L, Lebrun F, Calvo F, Lopez BS. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res. 2008;68:9404–12. doi: 10.1158/0008-5472.CAN-08-0861. [DOI] [PubMed] [Google Scholar]
- 24.Wang RH, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberdoerffer P, Michan S, McVay M, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–18. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]