Abstract
There is an increasing amount of emphasis being placed on serological biomarkers as tools for early detection of various cancers. In addition to the tumor-related circulating antigens under current investigation, autoantibodies to tumor-associated antigens are emerging as alternative candidates due to their potential high sensitivity and specificity. Already a number of specific autoantibodies have been identified and several groups have reported on the ability of panels of autoantibodies to discriminate malignant from non-malignant conditions. In this investigation we evaluate tumor-associated antigen autoantibody profiles in a group of healthy individuals. We identify a subset of individuals that demonstrate high levels of autoantibody production across the spectrum of tumor-associated antigens tested. We conclude that this observation is a result of undefined non-malignant autoimmune stimulation. Our findings may be an indication of factors present in the general population that may confound multiplex autoantibody-based diagnostic tests by reducing assay specificity. Such factors will require further characterization and the development of adequate controls in order to improve the performance of diagnostic tests.
Keywords: Tumor-associated antigens, autoantibody profiles, autoimmunity, early detection
Introduction
Immune surveillance of the early events of tumorigenesis may provide the key to early detection. Molecular alterations emanating from malignant cells stimulate a localized humoral immune response in nearby lymph nodes, which in turn leads to a systemic immune response. This response may be detected clinically in the form of circulating autoantibodies specific for tumor-associated antigens (TAA). An increasing amount of attention is being paid to the potential role of circulating antigens as cancer biomarkers. Such antigens are held to represent factors shed from the growing tumor as well as components of the host response. TAA autoantibodies offer several distinct advantages over antigenic biomarkers due to their inherent stability and specificity [1]. The nature of B-cell stimulation results in an amplified antibody response to a relatively small amount of antigen. This stimulation may stem from subtle changes such as antigen upregulation or altered glycosylation patterns [2]. Autoantibodies specific for TAAs have been detected in high titer in early stage cancer patients [3] and may indicate potential targets for immune therapy given their demonstrated activation of immune pathways [4–6].
Autoantibodies specific for oncogenic proteins such as p53 [7], her2/neu [3], MUC1 [8] and c-myc [9] have been previously detected in human cancers and considered as potential biomarkers. Serological expression cloning utilizing phage expression libraries, or SEREX, was first developed over 10 years ago [10] and has led to the identification of over 2000 autoantigens. While no single autoantibody has demonstrated the sensitivity and specificity required of a diagnostic test, advances based on the SEREX principle such as combinatorial phage display have enabled the development of multiplexed approaches. Investigations utilizing phage display have reported autoantibody panels that discriminate prostate [11], stage I NSCLC [12], and breast cancers [13] from controls with sensitivity/specificity of 88/82%, 90/90%, and 77/83% respectively. High throughput methods such as protein microarrays and glycan arrays, which screen for immunogenic alterations in glycosylation [14], have also been utilized to identify autoantibodies in ovarian [15] and breast cancers [2].
The greatest challenge encountered in the development of any diagnostic test based on serological biomarkers is the identification of individual markers highly specific for the malignant condition. The use of autoantibodies as biomarkers presents a unique set of challenges in that non-malignant conditions such as environmental factors, pathogen invasion, and autoimmune disease can trigger the production of a high-level of IgG and IgM autoantibodies which recognize various TAAs and thus reduce biomarker specificity [16–18]. We conducted an analysis of TAA autoantibodies in a large set of healthy control subjects. We identified a subgroup of individuals within our study set that demonstrated highly elevated levels of TAA autoantibodies for most of the antigens tested. We present these results as evidence of a potentially significant obstacle that must be overcome in order to advance the clinical relevance of autoantibody-based diagnostic methods.
Experimental
Materials and Methods
Study population and serum collection
Serum samples from 205 healthy controls were obtained for use in this study. 150 samples were collected as part of the Pittsburgh Lung Screening Study (PLuSS) [19] according to study protocols. An additional 55 samples were collected by the Early Detection Research Network (EDRN) according to a defined protocol [20]. Written and informed consent was obtained for each patient and all protocols were approved by the University of Pittsburgh IRB. The study group included active and non-active smokers and non-smokers. The characteristics of the study group are outlined in Table 1.
Table 1.
Characteristics of Study Population
| Description | Age | N | % |
|---|---|---|---|
| Male | 34–83 | 101 | 49.3 |
| Female | 36–81 | 104 | 50.7 |
| Smoking Status | N | % | |
| Active | 84 | 41.0 | |
| Former | 96 | 46.8 | |
| Never/Unkown | 25 | 12.2 |
Multiplexed bead-based TAA autoantibody assay development
Serum samples were tested for autoantibodies to 36 distinct tumor associated antigens chosen on the basis of published evidence (Table 2). The Luminex (Austin, TX) xMAP™ platform allows the simultaneous detection of up to 100 analytes based on the covalent attachment of specific capture molecules to internally-dyed spectrally distinct microbeads. Recombinant or native peptides corresponding to each target antigen were employed as capture probes and coupled to Luminex microbeads as previously described [21]. The individual microbead-antigen combinations were combined into multiplex panels in a stepwise fashion as each assay completed development and validation. Assay specificity was first evaluated by incubation of antigen-coupled microbeads with unrelated human IgG or IgM. Specificity was further evaluated by incubation of antigen-coupled microbeads with serum preincubated with antigen coated polystyrene beads (Sigma, St. Louis, MO) to remove specific autoantibodies, or Protein A/G Sepharose (EMD, Gibbstown, NJ) to absorb all IgG.
Table 2.
Tumor Associated Antigens Antigen
| Antigen | Supplier | Form | Antigen |
Supplier | Form |
|---|---|---|---|---|---|
| AFP | Bio Processing | Native | HCG |
Fitzgerald | Native |
| Akt1 | Invitrogen | Rec | Her2/neu |
R&D | Rec |
| AMACR* | Gift | Rec | H-Ras |
Jena Bioscience | Rec |
| AMACR Variant* | Gift | Rec | IL-6 |
Peprotech | Rec |
| CA125 | Bio Processing | Native | IL-8 |
Peprotech | Rec |
| CA 15-3 | Bio Processing | Native | K-Ras |
Jena Bioscience | Rec |
| CA 19-9 | Bio Processing | Native | MART-1 |
Lab Vision | Rec |
| CA 72-4 | Bio Processing | Native | Mesothelin |
Genway | Rec |
| CEA | Bio Processing | Native | Muc-1 |
Gift | Rec |
| c-myc (1-262) | Santa Cruz | Rec | Osteopontin |
R&D | Rec |
| c-myc (408-439) | EMD | Rec | P53 |
Santa Cruz | Native |
| CRP | Biodesign | Native | PDGF-BB |
US Biological | Rec |
| Cydin-B1 (1-433) | Santa Cruz | Rec | PDGFR-α |
Santa Cruz | Rec |
| Cydin-D1 (1-295) | Santa Cruz | Rec | PSA |
Bio Processing | Native |
| EGF | Peprotech | Rec | Survivin |
Alpha Diagnostics | Rec |
| EGFR | R&D | Rec | Transglutaminase |
Sigma | Native** |
| FasL | Peprotech | Rec | Tyrosinase |
Lab Vision | Rec |
| GplOO | Spring Bioscience | Rec | VEGF | ID Labs | Rec |
α-methyiacyi-CoA Racemase, see ref[31];
guinea pig; Rec - recombinant
Data collection and analysis
Assays were performed and validated as described previously [22]. Briefly, antigen-coated microbeads were blocked with bovine serum albumin for 1 hour, washed, and then incubated with serum diluted 1:100 in blocking buffer for 30 min at 4°C. This dilution was deemed optimal for antibody recovery based on titration (data not shown). Following this incubation, microbeads were washed and bound antibodies detected by phycoerythrin-conjugated donkey anti-human IgG/IgM (Jackson Laboratories, West Grove, PA). Fluorescence was measured on a Luminex 100 analyzer. The data was analyzed using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA) Standard statistical methods were used to establish relative fluorescence intensity distributions for each analyte and divide the data into percentiles. Values observed to be greater than the ninety-fifth percentile were considered exceptional.
Results
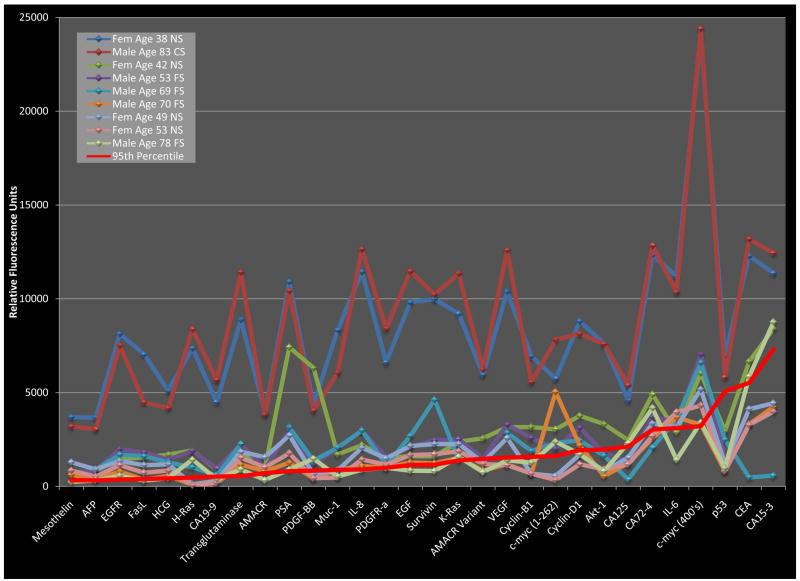
The analysis of our experimental results revealed a subset of individuals that demonstrated significantly elevated levels of autoantibodies to multiple TAAs tested (Figure1, Table 3). We established a statistical cutoff at the 95th percentile for each analyte tested and noted the distribution of results above that level. While outliers with respect to each analyte were observed intermittently throughout the study population, serum samples from nine individuals were found to contain autoantibody levels above the cutoff level for >40% of the antigens tested. Four of these samples were above cutoff levels for >75% of all tested antigens and many of the observed autoantibody intensities represented >10-fold increases over the cutoff levels. Each of these noteworthy subjects demonstrated autoantibody levels which fell below the cutoff value for at least one tested antigen, indicating that these results were not a product of sample evaporation. These observations in healthy control subjects suggest the influence of external stimuli of autoimmunity serving to confound our investigation.
Figure 1. Autoantibodies to tumor associated antigens.
Autoantibodies were measured by bead-based immunoassay in serum obtained from 205 healthy donors. Autoantibody levels from nine high-titer subjects are shown along with 95th percentile level for selected antigens. Solid lines connect measurements from individual subjects. Legend abbreviations: fem – female, NS – no smoking history, CS – current smoker, FS – former smoker.
Table 3.
Autoantibody levels of high-titer individuals
| Subject # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Description | 38 yof NS | 83 yom CS | 42 yof NS | 53 yom FS | 69 yom FS | 70 yom FS | 49 yof NS | 53 yof NS | 78 yom FS | ||
| Antigen | 95th Pctl | ||||||||||
| AFP | 322 | 3665 | 3039 | 835 | 829 | 882 | 369 | 905 | 523 | 319 | |
| Akt-1 | 1962 | 7624 | 7585 | 3320 | 1780 | 1630 | 542 | 805 | 910 | 814 | |
| AMACR | 699 | 3920 | 3823 | 1024 | 927 | NT | 805 | 1558 | 1014 | 332 | |
| AMACR variant | 1462 | 5950 | 6229 | 2561 | 1479 | NT | 838 | 1277 | 1190 | 756 | |
| CA125 | 2121 | 4587 | 5396 | 2465 | 1323 | 368 | 1235 | 1535 | 1171 | 2304 | |
| CA15-3 | 7302 | 11372 | 12445 | 8397 | 4099 | 575 | 4240 | 4429 | 3978 | 8756 | |
| CA19-9 | 524 | 4486 | 5667 | 892 | 948 | 583 | 177 | 289 | 194 | 382 | |
| CA72-4 | 3025 | 12336 | 12841 | 4890 | 3703 | 2070 | 2550 | 3346 | 2694 | 4174 | |
| CEA | 5518 | 12287 | 13178 | 6655 | 3361 | 471 | 3310 | 4113 | 3278 | 5862 | |
| c-myc (1-262) | 1614 | 5739 | 7811 | 3038 | 2001 | 2310 | 5028 | 552 | 335 | 2382 | |
| c-myc (408-439) | 3208 | 23978 | 24386 | 6031 | 7037 | 6594 | 3297 | 5171 | 4308 | 3398 | |
| CRP | 1202 | NT | NT | NT | NT | 1734 | NT | NT | NT | NT | |
| Cyclin-B1 | 1572 | 6938 | 5552 | 3162 | 2571 | 1884 | 678 | 673 | 679 | 1258 | |
| Cyclin-D1 | 1925 | 8794 | 8137 | 3762 | 3123 | 2445 | 2173 | 1697 | 1173 | 1714 | |
| EGF | 1130 | 9813 | 11461 | 1596 | 2095 | 2681 | 1282 | 2148 | 1663 | 836 | |
| EGFR | 362 | 8131 | 7515 | 1445 | 1973 | 1689 | 785 | 1349 | 1131 | 560 | |
| FasL | 390 | 7044 | 4480 | 1536 | 1808 | 1601 | 351 | 1125 | 724 | 305 | |
| gp100 | 4208 | NT | NT | 2790 | NT | NT | 8604 | 3650 | 1734 | 2244 | |
| HCG | 424 | 5100 | 4171 | 1692 | 1431 | 1281 | 560 | 1173 | 844 | 437 | |
| Her2/Neu | 996 | 657 | 900 | 996 | 222 | 242 | 324 | 439 | 322 | 2754 | |
| H-Ras | 467 | 7360 | 8378 | 1917 | 1892 | 1014 | 82 | 109 | 51 | 1426 | |
| IL-6 | 3108 | 11192 | 10372 | 2897 | 3395 | 3553 | 3667 | 3160 | 3955 | 1419 | |
| IL-8 | 897 | 11435 | 12641 | 2195 | 2957 | 2972 | 1089 | 2052 | 1427 | 882 | |
| K-Ras | 1376 | 9209 | 11345 | 2381 | 2508 | 1400 | 1528 | 2254 | 1866 | 1572 | |
| MART-1 | 7387 | NT | NT | 11075 | NT | NT | 7102 | 5480 | 1632 | 8656 | |
| Mesothelin | 317 | 3683 | 3190 | 728 | 723 | NT | 607 | 1283 | 841 | 230 | |
| Muc-1 | 876 | 8329 | 6051 | 1703 | 2027 | 2060 | 894 | 846 | 471 | 521 | |
| Osteopontin | 404 | 821 | 407 | 242 | 282 | 259 | 513 | 176 | 137 | 389 | |
| p53 | 5067 | 6973 | 5811 | 2943 | 1380 | 2485 | 806 | 1202 | 805 | 972 | |
| PDGF-BB | 831 | 4358 | 4039 | 6312 | 1175 | 1369 | 415 | 644 | 424 | 1473 | |
| PDGFR-a | 987 | 6613 | 8411 | 1543 | 1551 | 1128 | 989 | 1489 | 1154 | 980 | |
| PSA | 809 | 10909 | 10419 | 7417 | 2975 | 3150 | 1268 | 2712 | 1766 | 893 | |
| Survivin | 1150 | 9976 | 10230 | 1600 | 2454 | 4617 | 1225 | 2253 | 1712 | 817 | |
| Transglutaminase | 541 | 8890 | 11414 | 1801 | 2238 | 2257 | 1158 | 1842 | 1520 | 898 | |
| Tyrosinase | 1766 | NT | NT | 2406 | NT | NT | 2347 | 4550 | 3206 | 2238 | |
| VEGF | 1533 | 10377 | 12583 | 3152 | 3287 | 2856 | 1177 | 2576 | 1132 | 1386 | |
| # Markers Tested | 32 | 32 | 35 | 33 | 31 | 35 | 35 | 35 | 36 | ||
| # Outlying Markers | 31 | 31 | 31 | 26 | 23 | 20 | 18 | 16 | 16 | ||
| % Outlying | 97 | 97 | 89 | 79 | 74 | 57 | 51 | 46 | 44 |
All autoantibody values represent mean fluorescence intensity (MFI) determined by Luminex analysis. 95 percentile determined for each autoantibody by analysis of all subjects (N=205). Shaded values are above 95th percentile cutoff (outliers), shaded and bold values represent >10-fold increase over cutoff value. yom-year old male, yof-year old female. CS-current smoker, FS-former smoker, NS-no smoking history. NT-not tested.
Discussion
As was previously discussed, any effective serological biomarker-based screening test would require the use of combinations of biomarkers, as all individual biomarkers evaluated to date have lacked either sufficient sensitivity or specificity. The results we present here may represent a potential pitfall for multimarker approaches. We describe healthy normal individuals with elevated levels of numerous TAA autoantibodies which would serve to lower the specificity of any multimarker screening strategy based on those autoantibodies. Current clinical standards for cancer screening include stringent requirements for specificity. For example, given the low prevalence of ovarian cancer, it has been suggested that a screening strategy must achieve a minimum specificity of 99.6% and a sensitivity of >75% for early stage disease to avoid an unacceptable level of false-positive results [23, 24]. In our investigation the 9 individuals demonstrating elevated levels of >40% of tested autoantibodies represent 4.39% of our study population. If our study population is representative of the general population, this suggests a maximum diagnostic specificity of <96% for a test based on these autoantibodies, a level which no published study of this type has yet achieved. Clearly this is an observation that must be incorporated into continuing efforts to develop TAA autoantibody-based screening.
The 205 healthy subjects considered in this investigation represent the control arm of a larger study group that includes 815 patients diagnosed with a variety of benign and malignant conditions. The diseased group was comprised of conditions of the liver, esophagus, pancreas, lung, ovary, breast, and prostate and also melanoma. We found that sera from 24 (~2.9%) of these patients contained TAA autoantibody profiles similar to those of the 9 control subjects discussed above (data not shown). Considered together, these findings support the notion that these aberrant autoantibody profiles arise independent of our current set of clinical variables.
The source of the background level of autoimmunity observed in our investigation is unclear. One aspect or our study population that distinguishes it from the general population is the prevalence of smokers. In our study almost 88% of subjects were current or former smokers, while the CDC estimates the equivalent prevalence in the entire US to be almost 42% [25]. Epidemiologists have established causal links between cigarette smoking and autoimmune disorders such as system lupus erythematosus (SLE), rheumatoid arthritis (RA), multiple sclerosis (MS), Graves’ hyperthyroidism, and primary biliary cirrhosis (PBC) [26–29]. In attempts to characterize these links, researchers have shown that cigarette smoke has profound stimulatory effects on peripheral blood leukocytes, particularly neutrophils, macrophages and monocytes, and leads to increased cellular release of CRP, IL-6, fibrinogen, and matrix-metalloproteinases (reviewed in [30]). Although the role of smoking in autoimmunity is well established, its role in this investigation is not as straightforward, as four out of the nine exceptional subjects we identified are non-smokers. The nine-subject subset is equally nondescript with regards to gender and age, being comprised of five males and four females with ages ranging from 38–83. Thus, the observation we describe here is evidently multifactorial and warrants further investigation.
Conclusions
We report the findings outlined above not with the intent to discourage attempts to develop TAA autoantibody screening panels, but in the hope that our observation might lead to further refinement of those efforts. These refinements are certain to involve methods of controlling for background autoimmunity and the identification of tumor-associated antigens that interact with host immunity in a manner independent of any background humoral response. Given the inherent complexity and our limited understanding of the humoral response to tumorigenesis, it is not surprising that obstacles such as these will arise. However, the substantial promise and potential benefits of this type of diagnostic strategy remain clear.
Acknowledgments
The authors would like to thank Dr. Joel Weissfeld, Dr. Jeffrey Schragin, and Dr. Herbert Zeh for their assistance in obtaining serum samples. The authors would also like to thank Dr. James N. Mubiru for providing the AMACR peptides and Dr. John McKolanis for providing the muc-1 peptide.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4(4):1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS, Harris L, et al. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J Proteome Res. 2008;7(4):1490–1499. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Disis ML, Pupa SM, Gralow JR, Dittadi R, Menard S, Cheever MA. High-titer HER-2/neu protein-specific antibody can be detected in patients with early-stage breast cancer. J Clin Oncol. 1997;15(11):3363–3367. doi: 10.1200/JCO.1997.15.11.3363. [DOI] [PubMed] [Google Scholar]
- 4.Knutson KL, Disis ML. Tumor antigen-specific T helper cells in cancer immunity and immunotherapy. Cancer Immunol Immunother. 2005;54(8):721–728. doi: 10.1007/s00262-004-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA. Cancer vaccines based on the identification of genes encoding cancer regression antigens. Immunol Today. 1997;18(4):175–182. doi: 10.1016/s0167-5699(97)84664-6. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki H, Graziano DF, McKolanis J, Finn OJ. T cell-dependent antibody responses against aberrantly expressed cyclin B1 protein in patients with cancer and premalignant disease. Clin Cancer Res. 2005;11(4):1521–1526. doi: 10.1158/1078-0432.CCR-04-0538. [DOI] [PubMed] [Google Scholar]
- 7.Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res. 2000;60(7):1777–1788. [PubMed] [Google Scholar]
- 8.von Mensdorff-Pouilly S, Petrakou E, Kenemans P, van Uffelen K, Verstraeten AA, Snijdewint FG, van Kamp GJ, Schol DJ, Reis CA, Price MR, et al. Reactivity of natural and induced human antibodies to MUC1 mucin with MUC1 peptides and n-acetylgalactosamine (GalNAc) peptides. Int J Cancer. 2000;86(5):702–712. doi: 10.1002/(sici)1097-0215(20000601)86:5<702::aid-ijc16>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Koziol JA, Zhang JY, Casiano CA, Peng XX, Shi FD, Feng AC, Chan EK, Tan EM. Recursive partitioning as an approach to selection of immune markers for tumor diagnosis. Clin Cancer Res. 2003;9(14):5120–5126. [PubMed] [Google Scholar]
- 10.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A. 1995;92(25):11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353(12):1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 12.Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol. 2006;1(6):513–519. [PubMed] [Google Scholar]
- 13.Zhong L, Ge K, Zu JC, Zhao LH, Shen WK, Wang JF, Zhang XG, Gao X, Hu W, Yen Y, et al. Autoantibodies as potential biomarkers for breast cancer. Breast Cancer Res. 2008;10(3):R40. doi: 10.1186/bcr2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4(5):437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 15.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci U S A. 2007;104(44):17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Averbeck M, Gebhardt C, Emmrich F, Treudler R, Simon JC. Immunologic principles of allergic disease. J Dtsch Dermatol Ges. 2007;5(11):1015–1028. doi: 10.1111/j.1610-0387.2007.06538.x. [DOI] [PubMed] [Google Scholar]
- 17.Lu H, Goodell V, Disis ML. Humoral immunity directed against tumor-associated antigens as potential biomarkers for the early diagnosis of cancer. J Proteome Res. 2008;7(4):1388–1394. doi: 10.1021/pr700818f. [DOI] [PubMed] [Google Scholar]
- 18.Shoenfeld Y, Zandman-Goddard G, Stojanovich L, Cutolo M, Amital H, Levy Y, Abu-Shakra M, Barzilai O, Berkun Y, Blank M, et al. The mosaic of autoimmunity: hormonal and environmental factors involved in autoimmune diseases--2008. Isr Med Assoc J. 2008;10(1):8–12. [PubMed] [Google Scholar]
- 19.Wilson DO, Weissfeld JL, Fuhrman CR, Fisher SN, Balogh P, Landreneau RJ, Luketich JD, Siegfried JM. The Pittsburgh Lung Screening Study (PLuSS): outcomes within 3 years of a first computed tomography scan. Am J Respir Crit Care Med. 2008;178(9):956–961. doi: 10.1164/rccm.200802-336OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.EDRN Biological Specimens SOP. [ http://edrn.nci.nih.gov/resources/standard-operating-procedures/biological-specimens]
- 21.Gorelik E, Landsittel DP, Marrangoni AM, Modugno F, Velikokhatnaya L, Winans MT, Bigbee WL, Herberman RB, Lokshin AE. Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(4):981–987. doi: 10.1158/1055-9965.EPI-04-0404. [DOI] [PubMed] [Google Scholar]
- 22.Lokshin AE, Winans M, Landsittel D, Marrangoni AM, Velikokhatnaya L, Modugno F, Nolen BM, Gorelik E. Circulating IL-8 and anti-IL-8 autoantibody in patients with ovarian cancer. Gynecol Oncol. 2006;102(2):244–251. doi: 10.1016/j.ygyno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs IJ, Menon U. Progress and challenges in screening for early detection of ovarian cancer. Mol Cell Proteomics. 2004;3(4):355–366. doi: 10.1074/mcp.R400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Menon U, Jacobs IJ. Ovarian cancer screening in the general population. Curr Opin Obstet Gynecol. 2001;13(1):61–64. doi: 10.1097/00001703-200102000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention Smoking and Tobacco Use.
- 26.Bertelsen JB, Hegedus L. Cigarette smoking and the thyroid. Thyroid. 1994;4(3):327–331. doi: 10.1089/thy.1994.4.327. [DOI] [PubMed] [Google Scholar]
- 27.Karlson EW, Lee IM, Cook NR, Manson JE, Buring JE, Hennekens CH. A retrospective cohort study of cigarette smoking and risk of rheumatoid arthritis in female health professionals. Arthritis Rheum. 1999;42(5):910–917. doi: 10.1002/1529-0131(199905)42:5<910::AID-ANR9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 28.Parikh-Patel A, Gold EB, Worman H, Krivy KE, Gershwin ME. Risk factors for primary biliary cirrhosis in a cohort of patients from the united states. Hepatology. 2001;33(1):16–21. doi: 10.1053/jhep.2001.21165. [DOI] [PubMed] [Google Scholar]
- 29.Prummel MF, Wiersinga WM. Smoking and risk of Graves’ disease. JAMA. 1993;269(4):479–482. [PubMed] [Google Scholar]
- 30.Costenbader KH, Karlson EW. Cigarette smoking and autoimmune disease: what can we learn from epidemiology? Lupus. 2006;15(11):737–745. doi: 10.1177/0961203306069344. [DOI] [PubMed] [Google Scholar]
- 31.Mubiru JN, Valente AJ, Troyer DA. A variant of the alpha-methyl-acyl-CoA racemase gene created by a deletion in exon 5 and its expression in prostate cancer. Prostate. 2005;65(2):117–123. doi: 10.1002/pros.20277. [DOI] [PubMed] [Google Scholar]