Abstract
To elucidate the mechanisms underlying peripheral neuropathic pain in the context of HIV infection and antiretroviral therapy, we measured gene expression in dorsal root ganglia (DRG) of rats subjected to systemic treatment with the anti-retroviral agent, ddC (Zalcitabine) and concomitant delivery of HIV-gp120 to the rat sciatic nerve. L4 and L5 DRGs were collected at day 14 (time of peak behavioural change) and changes in gene expression were measured using Affymetrix whole genome rat arrays. Conventional analysis of this data set and Gene Set Enrichment Analysis (GSEA) was performed to discover biological processes altered in this model. Transcripts associated with G protein coupled receptor signalling and cell adhesion were enriched in the treated animals, while ribosomal proteins and proteasome pathways were associated with gene down-regulation. To identify genes that are directly relevant to neuropathic mechanical hypersensitivity, as opposed to epiphenomena associated with other aspects of the response to a sciatic nerve lesion, we compared the gp120 + ddC-evoked gene expression with that observed in a model of traumatic neuropathic pain (L5 spinal nerve transection), where hypersensitivity to a static mechanical stimulus is also observed. We identified 39 genes/expressed sequence tags that are differentially expressed in the same direction in both models. Most of these have not previously been implicated in mechanical hypersensitivity and may represent novel targets for therapeutic intervention. As an external control, the RNA expression of three genes was examined by RT-PCR, while the protein levels of two were studied using western blot analysis.
Keywords: Neuropathic pain, HIV, Mechanical hypersensitivity, Microarray
1. Introduction
Peripheral nerve disorders are frequent complications of HIV disease. Distal symmetrical polyneuropathy (DSP) is the most common peripheral nerve disorder associated with HIV (Verma et al., 2005). About 40% of ambulatory HIV patients in the developed world population have peripheral neuropathy, a prevalence which has not altered with the advent of antiretroviral therapy (Smyth et al., 2007). Of these 74–93% have pain, which is severe in 37–43% of patients. In addition, 52% of people living with advanced HIV have peripheral neuropathy associated with symptoms such as pain (Simpson et al., 2006). The most common clinical features of DSP are measurable sensory abnormalities, most noticeably loss of sensitivity to thermal stimuli (Martin et al., 2003) and on-going, paroxysmal or stimulus evoked pain (Dalakas and Pezeshkpour, 1988, Martin et al., 1999). There are two predominant (and clinically similar) settings in which painful DSP may occur in HIV disease. First, a disease-related DSP associated with HIV disease, likely involving virally mediated mechanisms such as the interaction of the coat protein, gp120, with sensory neurones precipitating chemokine mediated neuronal damage (Oh et al., 2001, Keswani et al., 2003b, Melli et al., 2006, Wallace et al., 2007a) and; second a drug-induced DSP, particularly associated with the use of nucleoside reverse transcriptase inhibitors as part of an anti-retroviral therapy (Cherry et al., 2003, Cherry et al., 2006, Wallace et al., 2007b).
We have characterized a model of HIV-related neuropathy, termed gp120 + ddC (Wallace et al., 2007b), which combines perineural administration of the HIV-1 coat protein, glycoprotein 120 (gp120), with systemic treatment with the nucleoside reverse transcriptase inhibitor, 2′, 3′-dideoxycytidine (ddC/Zalcitabine). Gp120 + ddC treatment is associated with development of hypersensitivity to static punctate mechanical stimuli, without hypersensitivity to heat or cold, and thigmotaxis (anxiety-like behaviour) (Wallace et al., 2007b). This model represents one clinical scenario of DSP in the HIV patient treated with nucleoside reverse transcriptase inhibitors, whereby the sensory neurones are damaged by a combination of gp120 and ddC effects (Cherry et al., 2003). In this study, the gp120 + ddC model is used to elucidate the pathophysiology of DSP using gene microarrays of dorsal root ganglions.
To distinguish between genes that are directly related to the sensory dysfunction observed in the gp120 + ddC model and those that are related to other aspects of the pathophysiological response to a nerve lesion, we compared the gp120 + ddC-evoked gene expression with that measured in a model of traumatic neuropathic pain, the L5 spinal nerve transection model (L5 SNT), where hypersensitivity to a static mechanical stimulus is also observed. This is a well established and commonly employed model of peripheral nerve lesion in rats and results in hypersensitivity to mechanical, cold and heat stimuli. (Kim and Chung, 1992, Choi et al., 1994, Ringkamp et al., 1999, Bridges et al., 2001). Variants on this model have been utilized in previous microarray investigations (Wang et al., 2002, Valder et al., 2003, Lacroix-Fralish et al., 2006, Levin et al., 2007) making this an appropriate comparator model for this study. By comparing gene expression profiles in these two distinct models, we aim to highlight genes associated with HIV-related neuropathy as well as mechanical hypersensitivity in neuropathic pain conditions in general.
2. Methods
2.1. Animals
All experiments conformed to the British Home Office Regulations and IASP guidelines (Zimmermann, 1983). Male Wistar rats weighing 200-250 g at baseline were used for all experiments (B&K, Hull, UK) and were housed in a temperature-controlled environment, maintained on a 14:10 h light–dark cycle (experiments were performed during the light phase) and provided with feed and water ad libitum.
2.2. Experimental animals
2.2.1. Spinal nerve transection model (SNT)
Surgery was performed under general anaesthesia (1–2% isoflurane in 1% O2 and 1% N20), using standard aseptic techniques. SNT (n = 12) surgery was performed as previously described (Bridges et al., 2001). Using the transverse processes of L6 as a guide, the left paraspinal muscles were exposed and separated from the spinous processes. The L6 transverse process was then removed by hemi-laminectomy and the L5 spinal nerve exposed, ligated tightly with a 3–0 silk suture, and cut 1 ± 2 mm distal to the suture. Sham surgery (n = 12) was performed by exposing the L5 spinal nerve as described above, but not damaging it.
2.2.2. Gp120 + ddC model
Rats in this model were treated as described by Wallace and colleagues (2007b). In eight anaesthetized (1–2% isoflurane in 1% O2 and 1% N20) rats the left sciatic nerve was exposed to gp120 by placing a 5 mm × 2 mm piece of gel foam soaked in saline containing 200 ng HIV-1 gp120-MN (>95% pure; Immunodiagnostics, Bedford, MA, USA) in direct contact with the nerve, to form a pool of protein solution around the nerve that is left in place for 30 min. Following this, oxidized regenerated cellulose (Surgicell, Ethicon); previously soaked until saturation in the same saline-gp120 solution, was wrapped loosely around the sciatic nerve 2-3 mm proximal to the trifurcation so as not to cause any nerve constriction and left in situ. The nerve was gently manipulated back into place and the muscle and skin incisions closed with 4/0 silk sutures. Animals were injected on the day of surgery with ddC (Roche, Basel, Switzerland; 50 mg/kg in saline, i.p.) and injected thereafter three times per week. Fourteen days after the day of surgery animals were sacrificed and DRG tissue was collected. Sham surgery (n = 8) was performed by exposing the left sciatic nerve as described above, but exposing it to 0.1% rat serum albumin. The sham group also received saline as a control for ddC.
2.2.3. Varicella Zoster Virus (VZV) infection model
We used an additional model of neuropathic pain, the VZV infection model, for validation of the differential expression of three of our candidate genes by RT-PCR and immunoblot analysis. Rats in this model were treated as previously described by Hasnie and colleagues (2007). In brief, VZV (strain Dumas) was propagated on fibroblast (primary human embryonic lung) cells and harvested when cells exhibited 80% cytopathic effect (cpe) (equivalent to 104–105 plaque forming units). Animals (n = 16) were subcutaneously injected with 50 μl viral inoculum into the mid-plantar glabrous footpad of the left hind limb. Control animals (n = 16) received similar injection of uninfected fibroblast cells. Fourteen days after the injection, animals were sacrificed and L4 and L5 DRG tissue was harvested.
2.3. Behavioural reflex testing
Punctate mechanical hypersensitivity was assessed using graded “von Frey” nylon filaments (Alan Ainsworth, UCL, London), which were used to deliver a calibrated indentation pressure against the hairless skin of the hind paws. The withdrawal threshold was determined by sequentially increasing and decreasing the stimulus strength and threshold response was defined by the nominal bending force of the filament that evoked foot withdrawal at least three times in every five applications when delivered at a rate of 1 Hz (Chaplan et al., 1994). Baseline measurements were obtained for all animals on two separate occasions within the week prior to surgery. Following surgery, behavioural reflex tests were carried out in a blinded manner. Animals were re-tested on day 14 post-surgery/infection and only animals demonstrating a significant (paired t-test, p < 0.05) reduction in ipsilateral PWT compared to baseline values were retained for microarray analysis.
2.4. Tissue collection and microarray
Animals were sacrificed by administration of an overdose of pentobarbitone (100 mg/kg Animalcare Ltd., York, UK). Ipsilateral DRG from lumbar segments L4 and L5 were dissected on ice, and snap frozen in liquid nitrogen. Transection of the L5 spinal nerve in the SNT model was confirmed on post-mortem and only these animals were included in the microarray analysis. Prior to RNA preparation, for the gp120 + ddC model, ipsilateral L4 and L5 DRGs from two animals were pooled, to provide enough tissue for RNA preparation. For the SNT model, only ipsilateral L5 DRGs from three animals were collected and pooled. Four replicates per condition were prepared. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsband, CA, USA), and further purified using an RNeasy Mini kit (Qiagen, Crawley, UK), according to the manufacturer protocols. Affymetrix GeneChip® Rat Genome 230 2.0 arrays (Santa Clara, CA, USA) that study over 30,000 transcripts and variants from over 28,000 well-substantiated rat genes, were used for these experiments. Biotin labeled cRNA was generated using the Affymetrix Small Sample Labeling Protocol vII (http://www.affymetrix.com). Approximately 200 ng total starting RNA was used for each sample. Labeled cRNA fragmentation, as well as array hybridization, washing, and staining were performed as described in the Affymetrix GeneChip® Expression Analysis Technical Manual (http://www.affymetrix.com).
2.5. Data analyses
CEL files were obtained with Affymetrix Microarray Suite software. Data was analysed using R v2.3.1 and Bioconductor v1.8 packages (Gentleman et al., 2004) as follows: Quality control tests and RMA data normalization were performed using Simpleaffy (Wilson and Miller, 2005) and Affy (Gautier et al., 2004). The normalized data was filtered, using Genefilter (http://www.bioconductor.org/packages/1.8/bioc/html/genefilter.html), to remove probe sets with minimal expression levels (i.e., probe sets failing to have a signal higher than log2(100) in three or more arrays). Statistical analysis was performed using linear models and empirical Bayes methods implemented in the Limma package (Smyth, 2004, Smyth, 2005). Benjamini and Hochberg’s step-up method (Reiner et al., 2003) with false discovery rate (FDR) less than 0.1 was used to control for multiple testing. The threshold p value consistent with an FDR near 10% was identified as 0.03 for the SNT model (10.4% FDR) and 0.004 for the gp120 + ddC model (9.6% FDR). The lists of statistically significant genes were loaded into GeneSpring GX (v7.3.1) software (Agilent Technologies, Cheshire, UK), where a second filter (fold difference less than 1.2-fold) was applied to further reduce false positive results (Bakay et al., 2002). We chose 1.2-fold change, which is a moderate cut-off, to signify differential expression, because the two cycle amplification protocol used in this study is thought to suppress fold differences (see discussion). Finally, Venn diagrams were used to cross-compare data between models. The microarray data is available in MIAME-compliant (minimum information about a microarray experiment) format at the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) (Parkinson et al., 2007) under accession codes E-MEXP-974, E-MEXP-976.
2.5.1. Functional association analysis
Associations with the annotations of the Gene Ontology (GO) Consortium (Ashburner et al., 2000) were obtained, for the lists of significant probe sets (10% FDR and over 1.2-fold difference) that correspond to each model, using MAPPFinder 2.0, a part of the GenMAPP 2.1 application package (Dahlquist et al., 2002, Doniger et al., 2003). To ease the interpretation of results, output data were manually filtered, using criteria used by Doniger and colleagues (2003), to remove terms that represented the same genes (typically parent–child processes). For a process to be included in the results, it was required that the z score from the MAPPFinder statistics was higher than 2, with a permute p value less than 0.01, and that at least one gene changed significantly for this node (local results). Also, terms that (a) comprised of 5 or less genes; or (b) had more than 200 genes changed (nested results) were removed, because they were either too specific or too general for the data interpretation.
Pathway analysis was also performed using Gene Set Enrichment Analysis (GSEA) version 2.0 (Subramanian et al., 2005, Subramanian et al., 2007). A total of 253 gene sets were applied. These were obtained from the C2/Canonical Pathways collection of MSigDB version 2.1 (Subramanian et al., 2005), which contains gene sets collected from various sources such as online pathway databases, publications in PubMed, and knowledge of domain experts. Fourteen additional gene sets were generated by querying the Affymetrix NetAffx tool (https://www.affymetrix.com/analysis/netaffx/index.affx) with pain related key words. GSEA was run with default settings by using the gene_set permutation option and performing 1000 gene permutations for the determination of statistical significance. Significant FDR and p values were less than 25% and 0.01, respectively, in accordance with GSEA recommendations.
2.6. RT PCR
RT-PCR was performed as previously described (Boucher et al., 2000). The sequence of primers used is listed in Table 1. New pools of DRG RNA from SNT-, gp120 + ddC- and VZV-treated animals were used for these experiments. DRG RNA was extracted by using guanidine isothiocyanate. Total RNA (2 μg) from L4 and/or L5 DRGs of sham or treated animals (n = 3 per group) was treated with DNase I, and cDNA was synthesized with Superscript reverse transcriptase (Invitrogen, Paisley, UK) using random hexamers in a total volume of 20 μl. In order to identify the optimal conditions for a linear range of amplification in the quantitative RT-PCR analysis, the amount of each amplified product was first checked at 24, 27, 30, 33, 36, and 39 cycles. Portions of the reverse-transcribed solution (0.5 μl) of total RNA from treated or sham ganglia were used for PCR. Each PCR (94 °C, 30 s; 58 °C, 30 s; 72 °C, 1 min; 33 cycles, 30 μl) was started with the primer pair specific for genes of interest (see Table 1). The primer pairs for cyclophilin A (PpiA) were added to the PCR reaction 5 cycles after the start. PCR products (10 μl) were visualized on 1.5% agarose gels, using ethidium bromide. The intensity of each band was analysed using Image J software (http://rsb.info.nih.gov/ij/) and normalized to cyclophilin A. To test for differential expression, a Student’s t test with a significance level of p < 0.05 was used.
Table 1.
Primer sequences used for RT-PCR
| Gene name | 5′ primer | 3′ primer | Size (bp) |
|---|---|---|---|
| TrkB_FL | TCTATGAAGACTGGACCACGC | TTCTCCAAGCTCCCTCTTCAG | 449 |
| TrkB_T1 | TCTATGAAGACTGGACCACGC | CCAGTCACAGCTCACAACAAG | 422 |
| TrkB_T2 | TCTATGAAGACTGGACCACGC | AGTGGGCAAGGCTGAGTAATC | 399 |
| Npy | ACTGACCCTCGCTCTATCCC | AACGACAACAAGGGAAATGG | 400 |
| Pap/Reg2 | TGGCCTTCCCAGTCATGTC | AGATCTTGACAAGCTGCCACAG | 445 |
| Vgf | TACCCAGAACGAGGATTGCG | CAACAGTACCGCGGCCAG | 381 |
| PpiA | ACCCCACCGTGTTCTTCGAC | CATTTGCCATGGACAAGATG | 300 |
2.7. Western Blots
Protein extraction, SDS–PAGE, and Western blotting were performed as previously described (Okuse et al., 1997). DRG were isolated and snap frozen, and protein was extracted by homogenization in RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 μg/ml each aprotinin, leupeptin, pepstatin) followed by centrifugation at 10,000 g for 10 min. The protein concentration of the supernatant was determined by Bradford assay, and 50 μg protein was applied to each lane of 12% SDS–PAGE. Wet transfer was performed using Hybond ECL (Amersham), and the membrane was blocked for 60 min in 5% non-fat dried milk in TBS-T (50 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.1% Tween 20). The blot was probed with goat anti-Reg2/Pap (1:1000 dilution, R&D systems), rabbit anti-Atf3 (1:500 dilution, Santa Cruz Biotechnology) and anti-Actin (1:5000, Sigma) antibodies. Results were visualized with horseradish peroxidase-coupled anti-goat or anti-rabbit immunoglobulins (Dako) using ECL Western blotting detection system (Amersham).
3. Results
3.1. Mechanical hypersensitivity data
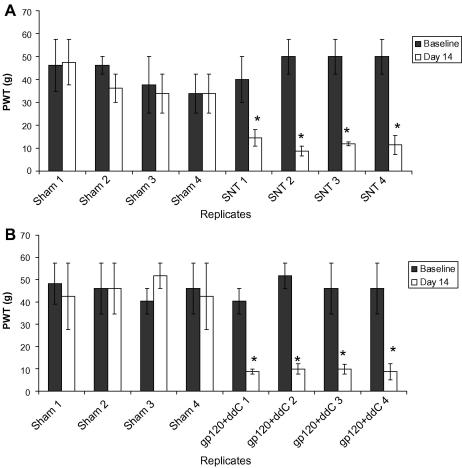
At day 14 post-intervention, all animals displayed hypersensitivity to a punctate mechanical stimulus, in the limb ipsilateral to treatment (Fig. 1). This has been previously established to be the time of maximal mechanical hypersensitivity in both models (Dowdall et al., 2005, Wallace et al., 2007b). On average there was an 80% reduction from baseline in gp120 + ddC-treated animals and a 75% reduction in SNT-treatment. Mechanical hypersensitivity was not observed in the contralateral paws of the treated animals (data not shown), nor in the ipsilateral paws of sham animals.
Fig. 1.
Paw withdrawal thresholds (PWT) to graded von Frey filaments at day 14 vs. baseline for (A) SNT and (B) gp120 + ddC. Sample size is three rats per replicate for A and two rats per replicate for B. ∗, Significant paired t test with p < 0.05 for comparing day 14 with the baseline value. Data are means ± SEM.
3.2. Model-specific differential expression of genes
The microarray experiment was conducted at one time point post-injury (day 14) and consisted of two conditions per model (treated animal versus sham) with four replicates per condition. To test for outlying chips, a number of quality control tests were carried out on the raw and RMA normalized data files (data not shown), which resulted in the exclusion of one of the replicates for the treated SNT animals.
We defined differentially expressed genes using criteria of 10% FDR and overall fold difference from sham >1.2 (see Section 2). Of the approximately, 30,000 transcripts queried on the Affymetrix GeneChip® Rat Genome 230 2.0 array, 656 probe sets were significantly disregulated in the gp120 + ddC DRGs. Of those, 243 were upregulated versus sham, whereas 413 were downregulated (supplementary tables 1 and 2). In SNT DRG, 2696 probe sets were upregulated versus sham and 2444 downregulated (supplementary tables 3 and 4). Moreover, considering fold changes alone, while only 40 probe sets showed over 2-fold expression differences between sham and treated animals in the gp120 + ddC model, 1593 probe sets showed over 2-fold difference in the SNT model. We interpret this as a reflection of the moderate amount of nerve damage induced in the sciatic nerve in the gp120 + ddC models, which is in contrast to the extensive trauma inflicted in the SNT model.
3.3. Comparison of SNT data with the literature
To verify the accuracy of the microarray results, we compared our SNT model results with two published microarray datasets describing differential gene expression (13 or 14 days after injury) in the DRG of rats subjected to spinal nerve ligation (SNL) (Wang et al., 2002, Valder et al., 2003). Our SNT model is a variation of the SNL model and thus our data are comparable to the SNL datasets. Both previous studies used rat RG-U34A arrays that contain 8799 probe sets and published lists of differentially expressed genes that show greater than 2-fold difference with p < 0.05. They compared ipsilateral to contralateral pools of L4, L5, and L6 DRG. Despite these analytical and methodological differences to our study, the data are expected to show strong similarities. Wang and colleagues (2002) identified 102 upregulated and 46 downregulated genes, while Valder and colleagues (2003) identified 98 upregulated and 113 downregulated genes. We merged these lists and identified probe sets for a total of 195 genes in the Rat Genome 230 2.0 array that we employed in this study. Hundred and fifty-nine of these probe sets (81.5%) are significant in our SNT dataset, with p < 0.04 (FDR < 0.1) and over 1.2-fold difference in the same direction as the previously reported studies. For a more detailed breakdown of these results, see supplementary Table 5. The correlation between the SNT results and published observations substantiates the accuracy of our data.
3.4. Gene Ontology results
We used MAPPFinder/GenMAPP and GSEA to perform a general search for pathways associated with gp120 + ddC treatment. With MAPPFinder/GenMAPP we found that 50% of the 243 significantly upregulated and 48% of the 413 downregulated genes in this model are linked to a GO term. Eleven GO functional annotation terms are over-represented in the upregulated gene list, neuropeptide signalling being the most significant (Table 2). Several other terms are related to organ development (e.g., tube morphogenesis, axonogenesis). Three GO terms are over-represented in the downregulated gene list (Table 2). They are all related to chloride channel activity (GABA-A receptor activity, chloride ion binding, chloride channel activity).
Table 2.
Functional categories of genes highlighted by MAPPFinder for the gp120 + ddC model
| GO name | GO type | No. of genes changing/no. of genes measured in the array | Z score | Permute p |
|---|---|---|---|---|
| Upregulated | ||||
| Neuropeptide signalling | P | 4/29 | 4.852 | 0 |
| Hyaluronic acid binding | F | 2/8 | 4.924 | 0.003 |
| Homophilic cell adhesion | P | 4/35 | 4.281 | 0.003 |
| Organ morphogenesis | P | 9/154 | 3.801 | 0.004 |
| Sex differentiation | P | 3/25 | 3.83 | 0.005 |
| Blood vessel morphogenesis | P | 6/83 | 3.727 | 0.005 |
| Regulation of epithelial cell proliferation | P | 2/7 | 5.315 | 0.006 |
| Electron carrier activity | F | 5/71 | 3.326 | 0.006 |
| Tube morphogenesis | P | 4/36 | 4.198 | 0.008 |
| Branching morphogenesis of a tube | P | 3/22 | 4.169 | 0.009 |
| Axonogenesis | P | 5/68 | 3.447 | 0.009 |
| Downregulated | ||||
| GABA-A receptor activity | F | 3/8 | 5.767 | 0 |
| Chloride ion binding | F | 3/17 | 3.577 | 0.005 |
| Chloride channel activity | F | 3/18 | 3.435 | 0.006 |
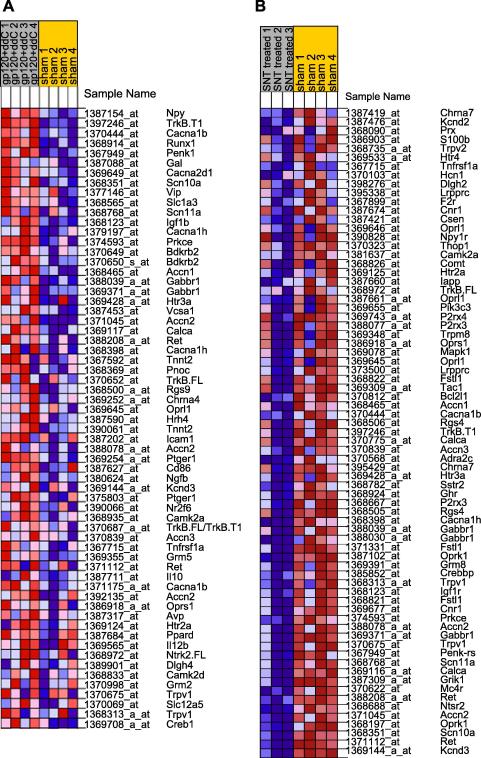
The GSEA software was used to obtain a more detailed classification of the data into various affected pathways. GSEA considers all of the genes in an experiment, not only those above an arbitrary cut-off in terms of fold-change or significance. When GSEA was applied to the gp120 + ddC dataset, 20 gene sets were found to be up-regulated (FDR < 2.5 and p < 0.01) (Table 3). These include five gene sets that involve calcium dependent signalling (G alpha q pathway, Wnt/Ca2+/cyclic GMP pathway, calcium regulation in cardiac cells pathway, Pyk2 pathway and calcium homeostasis) and three cell adhesion gene sets (cell adhesion molecule activity, Brentani cell adhesion and GO cell adhesion). This is in agreement with the MAPPFinder results. The upregulation enriched gene sets also include a list of pain related genes. The genes that contribute to the high enrichment score for this gene set are illustrated in Fig. 2. Twelve gene sets are downregulated in the gp120 + ddC model (Table 4). They include a gene set of ribosomal proteins, three gene sets related to proteasomes and two for RNA/DNA transcriptome reactome genes.
Table 3.
Gene sets associated with gene upregulation in gp120 + ddC using gene set enrichment analysis
| Name | Top 5 genes | Source | Size | NOM p-val | FDR q-val |
|---|---|---|---|---|---|
| CELL_ADHESION_MOLECULE_ACTIVITY | Ncam1, Mpz, Gp9, L1cam, Cdh1 | G0 | 103 | 0 | 0.064 |
| PELP1PATHWAY | Mapk3, Pelp1, Crebbp, Ep300, Src | BioCarta | 15 | 0 | 0.066 |
| BRENTANI_CELL_ADHESION | Itga7, Ncam1, Ank1, Jup, Timp2 | Brentani et al., Proc Natl Acad Sci USA, 2003, 100:13418–23 | 123 | 0 | 0.068 |
| ST_GAQ_PATHWAY | Itpr3, Dag1, Gnaq, Itpr2, Nfkb2 | STKE | 43 | 0 | 0.079 |
| ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Itpr3, Dag1, Itpr2, Slc6A13 | STKE | 31 | 0 | 0.094 |
| CELL_ADHESION | Ptprf, Ncam1, Gpr56, Atp2A2, Jup | G0 | 178 | 0 | 0.148 |
| PENG_RAPAMYCIN_UP | Kcnh2, Tp53, Aes, Ucp2, Cd37 | Peng et al., Mol. Cell Biol, 2002, 22:5575–84 | 153 | 0 | 0.157 |
| PAIN_RELATED | NpY, TrkB.T1, Cacna1b, Runx1, Penk-rs | NetAffx | 257 | 0 | 0.159 |
| CALCIUM_REGULATION_IN_CARDIAC_ CELLS | Itpr3, Cacna1b, Cacba1c, Atp2a2, Atp1b2 | GenMAPP | 243 | 0 | 0.172 |
| BRENTANI_SIGNALING | Stat3, Itga7, Wnt4, Arhgef12, Il6st | Brentani et al., Proc Natl Acad Sci USA, 2003, 100:13418–23 | 214 | 0 | 0.217 |
| AMINOSUGARS_METABOLISM | Hk1, Gck, Syb5r3, Hexa, Hk2 | KEGG | 15 | 0.002 | 0.158 |
| INFLAMMATORY_RESPONSE_PATHWAY | Lamc1, Lama5, Cd40lg, Cd86, Lamc2 | Broad Institute | 29 | 0.004 | 0.068 |
| ST_T_CELL_SIGNAL_TRANSDUCTION | Dag1, Plcg1, Grb2, Cd3d, Nfkb2 | STKE | 44 | 0.005 | 0.172 |
| PYK2PATHWAY | Mapk3, Plcg1, Gnaq, Jun, Grb2 | BioCarta | 63 | 0.005 | 0.206 |
| INSULIN_SIGNALING | Npy, Ptprf, Srebf1, Jun, Slc27a4 | Broad Institute | 138 | 0.006 | 0.214 |
| HIFPATHWAY | Jun, Ep300, Edn1, Hif1a, Creb1 | BioCarta | 19 | 0.006 | 0.074 |
| STARCH_AND_SUCROSE_METABOLISM | Hk1, Gusb, Gaa, Gck, Gp1 | KEGG | 30 | 0.007 | 0.164 |
| G2PATHWAY | Tp53, Gadd45a, Cdkn2d, Ep300, Cdc25b | BioCarta | 28 | 0.007 | 0.164 |
| CALCIUM_HOMEOSTASIS | Stat3, Casr, Cacna1c, Dnm1, Atp2a2 | NetAffx | 247 | 0.007 | 0.127 |
| ARAPPATHWAY | Cltb, Arfgef2, Arfgap1, Pscd1, Pscd2 | BioCarta | 22 | 0.009 | 0.119 |
Top 5 genes – top 5 ranking genes belonging to a gene set; source – source for the gene set; size – number of genes in a gene set; NOM p-val – nominal p value; FDR q-val – false discovery rate.
Fig. 2.
Heat map of the genes contributing most to the high enrichment score of the pain gene set for (A) gp120 + ddC and (B) SNT. Genes are ranked according to their expression under each experimental condition. For gp120 + ddC there is an association with upregulation, and thus the most differentially expressed genes are shown at the top of the heat map. For SNT there is an association with downregulation and thus the most differentially expressed genes are shown at the bottom of the heat map. Expression values are shown for each replicate within a condition and are represented as colours, where the range of colours (red, pink, light blue, dark blue) shows the range of expression values (high, moderate, low, and lowest).
Table 4.
Gene sets associated with gene downregulation in gp120 + ddC using gene set enrichment analysis
| Gene set name | Top 5 genes | Source | Size | NOM p-val | FDR q-val |
|---|---|---|---|---|---|
| PROTEASOME_DEGRADATION | Psmc6, Rps27a, Psma4, Psma1, H2afz | Broad Institute | 39 | 0 | 2.40e−04 |
| PROTEASOME | Psma2, Psma4, Psma1, Psma5, Psmb1 | GenMAPP | 18 | 0 | 3.60e−04 |
| RIBOSOMAL_PROTEINS | Rps24, Rps27a, Rps25, Rpl21, Rpl7 | GenMAPP | 86 | 0 | 7.21e−04 |
| STRIATED_MUSCLE_CONTRACTION | Tpm1, Ttn, Tnnt1, Tnni1, Myl2 | GenMAPP | 39 | 0 | 0.003 |
| PROTEASOMEPATHWAY | Psma2, Psma4, Psma1, Psma5, Psmb1 | BioCarta | 22 | 0 | 0.02 |
| ELECTRON_TRANSPORT_CHAIN | Cox6a2, Cox17, Nduf5, Cox7b, Ndyfs4 | Broad Institute | 80 | 0 | 0.021 |
| ROME_INSULIN_2F_UP | Dpysl3, Ppp1r12a, Sh3glb1, Canx, Tpm1 | Rome et al., J. Biol. Chem., 2003 | 204 | 0 | 0.161 |
| DNA_REPLICATION_REACTOME | Orc3l, Prim2a, Rfc1, Rps27a, Orc4l | GenMAPP | 34 | 0.002 | 0.075 |
| OXIDATIVE_PHOSPHORYLATION | Cox6a2, Ndufa5, Cox7b, Atp6v1d, Atp5o | KEGG | 48 | 0.003 | 0.066 |
| RNA_TRANSCRIPTION_REACTOME | Ccnh, Gtf2f2, Cdk7, Mnat1, Polr2g | GenMAPP | 28 | 0.009 | 0.115 |
| PORPHYRIN_AND_CHLOROPHYLL_ METABOLISM | Eprs, Blvra, Cp, Urod, Hmox1 | GenMAPP | 18 | 0.020 | 0.153 |
| UBIQUITIN_MEDIATED_PROTEOLYSIS | Ube2b, Ube2n, Ube2g1, Ube2d3 | GenMAPP | 20 | 0.022 | 0.1813 |
Top 5 genes – top 5 ranking genes belonging to a gene set; source – source for the gene set; size – number of genes in a gene set; NOM p-val – nominal p value; FDR q-val – false discovery rate.
In the SNT model data, 44% of the significantly upregulated and 43% of the downregulated genes are linked to a GO term. As shown in supplementary Table 6, the majority of biological process GO terms identified as significant in the list of upregulated genes are linked to neuroinflammation and immune system activation, highlighting the presence of dramatic neuroinflammation in SNT DRG. In contrast, multiple GO terms linked with ion transport (especially potassium ion transport), cell–cell signalling and cell communication are identified as significant in the list of downregulated genes. The obtained results agree with Wang et al., 2002, Valder et al., 2003 and provide another level of validation of our experimental results. The same pathways were also identified using GSEA analysis (supplementary tables 7 and 8). Interestingly, the gene set containing pain related genes is amongst those linked with down regulation in the SNT model. A detailed breakdown of the genes that contribute to the high enrichment score for this gene set are illustrated in Fig. 2.
3.5. Model cross comparisons
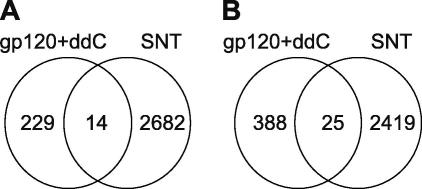
To identify genes that are relevant to mechanical hypersensitivity, we compared the lists of differentially expressed genes in both models, created using criteria of 10% FDR and overall >1.2-fold difference from sham, with Venn diagrams (see Fig. 3). We found 14 probe sets that are commonly upregulated in the DRGs of SNT and gp120 + ddC treated animals (Table 5). Only one of these is an EST of unknown function. The remaining probe sets correspond to annotated genes which can be subdivided into four groups on the basis of their function: signal peptides (Pap/Reg2, NpY, Scube1, Vgf), ion/protein transporters (Atp2B4, Dlg5, Slc4a8), transcription factors (Arnt2, Bptf, Irf7, Midn, Per2) and a cell differentiation molecule (Lama5) (Table 5).
Fig. 3.
Venn diagrams to cross-compare the lists of significant probe sets (>1.2-fold difference and 10% FDR) between the SNT and gp120 + ddC models. (A) comparison of upregulated gene lists; (B) comparison of downregulated gene lists.
Table 5.
Probe sets upregulated for the SNT and gp120 + ddC models
| Probe set ID | Gene title | Gene symbol | Fold increase SNT | Fold increase gp120 + ddc |
|---|---|---|---|---|
| 1373315_at | Aryl hydrocarbon receptor nuclear translocator 2 | Arnt2 | 1.4 | 1.3 |
| 1376911_at | ATPase, Ca2+ transporting, plasma membrane 4 (homo sapiens) (predicted) | Atp2B4_predicted | 2.9 | 1.3 |
| 1376182_at | Bromodomain PHD finger transcription factor (Mus) (predicted) | Bptf_predicted | 1.8 | 1.2 |
| 1377121_at | Discs, large homolog 5 (Drosophila) (predicted) | Dlg5_ predicted | 1.5 | 1.2 |
| 1383564_at | Interferon regulatory factor 7 | Irf7 | 3.4 | 1.2 |
| 1388932_at | Laminin, alpha 5 | Lama5 | 1.8 | 1.3 |
| 1377103_at | Midnolin (predicted) | Midn_ predicted | 1.6 | 1.3 |
| 1387154_at | Neuropeptide Y | Npy | 8.6 | 1.9 |
| 1368238_at | Pancreatitis-associated protein | Pap/Reg2 | 22.3 | 1.2 |
| 1368303_at | Period homolog 2 (Drosophila) | Per2 | 2.6 | 1.3 |
| 1371910_at | Signal peptide, CUB domain, EGF-like 1 (Mus) (predicted) | Scube1_ predicted | 1.6 | 1.4 |
| 1392238_at | Solute carrier family 4 (anion exchanger), member 8 (Mus) (predicted) | Slc4a8_predicted | 2.2 | 1.3 |
| 1368359_a_at | VGF nerve growth factor inducible | Vgf | 3.5 | 1.3 |
| 1393477_at | – | – | 5.0 | 1.5 |
–, Unknown.
Twenty-five probe sets corresponding to 24 genes or ESTs were downregulated in the DRGs of SNT and gp120 + ddC treated animals (Table 6). Thirteen of these have unknown function while the remaining eleven are implicated in a variety of processes varying from metabolism (Gla, Fpgt, St8sia3), to ion/protein transport (Efcbp1, Kif5a, Rab27b, Sv2b), transcriptional activation (Esrrg), mitochondrial function (Senp5) axon guidance (Robo2) and signal transduction (Nkiras1).
Table 6.
Probe sets downregulated for the SNT and gp120 + ddC models
| Probe set ID | Gene title | Gene symbol | Fold decrease SNT | Fold decrease gp120 + ddc |
|---|---|---|---|---|
| 1390892_at | DEP domain containing 1B (predicted) | Depdc1b_predicted | 2.0 | 1.3 |
| 1386120_at | Mus musculus EF hand calcium binding protein 1 (Mus musculus) (predicted) | Efcbp1_predicted | 1.7 | 1.4 |
| 1381445_at | Estrogen-related receptor gamma | Esrrg | 2.5 | 1.4 |
| 1376753_at | Fucose-1-phosphate guanylyltransferase | Fpgt | 1.7 | 1.3 |
| 1382063_at | Galactosidase, alpha | Gla | 1.7 | 1.3 |
| 1382787_at | Kinesin family member 5A | Kif5a | 2.0 | 1.7 |
| 1396215_at | Similar to RIKEN cDNA 2610022G08 | LOC502782 | 1.7 | 1.3 |
| 1396117_at | Leucine zipper protein 2 (predicted) | Luzp2_predicted | 2.0 | 1.7 |
| 1397555_at | Leucine zipper protein 2 (predicted) | Luzp2_ predicted | 1.4 | 2.0 |
| 1393847_at | NFKB inhibitor interacting Ras-like protein 1 (predicted) | Nkiras1_predicted | 1.7 | 1.3 |
| 1370122_at | RAB27B, member RAS oncogene family | Rab27b | 2.0 | 1.7 |
| 1393909_at | Similar to KIAA1841 protein (predicted) | RGD1305110_predicted | 1.4 | 1.7 |
| 1382812_at | Similar to Protein Njmu-R1 (predicted) | RGD1310429_predicted | 2.0 | 1.4 |
| 1378245_at | Similar to 6430514L14Rik protein (predicted) | RGD1311958_predicted | 1.7 | 1.3 |
| 1382632_at | Roundabout homolog 2 (Drosophila) | Robo2 | 2.5 | 1.7 |
| 1382477_at | SUMO/sentrin specific protease 5 (predicted) | Senp5_predicted | 1.4 | 1.4 |
| 1387435_at | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | St8sia3 | 1.4 | 1.3 |
| 1369627_at | Synaptic vesicle glycoprotein 2b | Sv2b | 1.7 | 2.5 |
| 1394412_at | Transmembrane protein 16C (predicted) | Tmem16c_ predicted | 3.3 | 1.4 |
| 1392045_at | Transmembrane protein 22 | Tmem22 | 2.0 | 1.3 |
| 1377917_at | Transcribed locus | – | 1.7 | 1.4 |
| 1383162_at | – | – | 1.3 | 1.4 |
| 1385972_at | – | – | 1.4 | 1.4 |
| 1392663_at | Transcribed locus | – | 1.7 | 1.3 |
| 1396676_at | Transcribed locus | – | 2.7 | 1.4 |
–, Unknown.
3.6. RT-PCR and immunoblotting results
We sought to confirm the gene expression changes of a selected subset of genes using semi-quantitative RT-PCR analysis. Cyclophilin A was used as internal control.
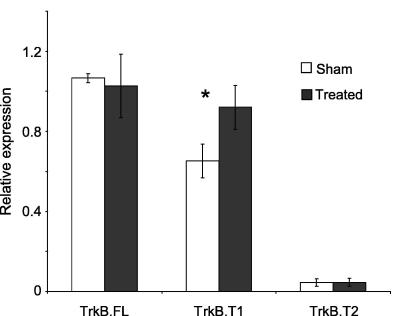
First, we measured the truncated isoform of TrkB (TrkB.T1), a receptor for BDNF (Huang and Reichardt, 2003) as this was the most upregulated gene in the gp120 + ddC DRG (supplementary information table 3). The Affymetrix GeneChip® Rat Genome 230 2.0 array also contains a probe set (1368972_at) that corresponds to the full length isoform of this gene (TrkB.FL). It showed no significant change in expression between treated and sham animals. We designed primers specific for TrkB.FL, for TrkB.T1 as well as for a second truncated isoform of TrkB (TrkB.T2) which is absent from the microarray. Only TrkB.T1 was found to be up-regulated in gp120 + ddC treated animals supporting the microarray findings (Fig. 4).
Fig. 4.
RT-PCR results for TrkB isoforms in gp120 + ddC treated animals. Normalized expression levels for each isoform are expressed as ratios to cyclophilin. Data are expressed as means ± SD. ∗, (p < 0.05) indicates statistical significance between treated and sham groups of animals (Student’s t test).
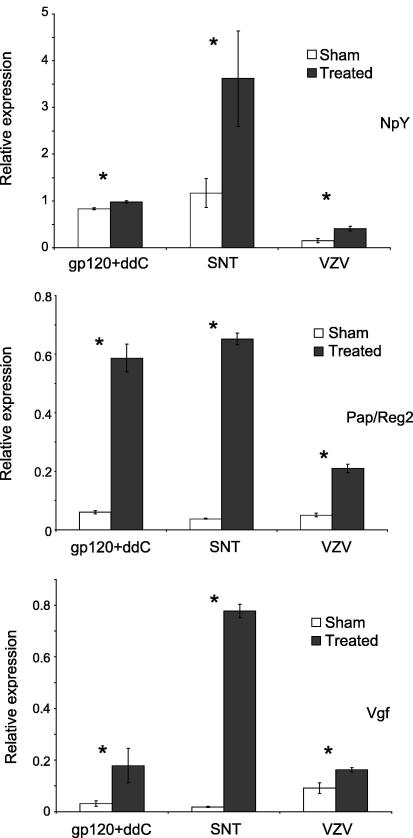
We also measured levels of NpY, Pap/Reg2 and Vgf, the three neuropeptides that are commonly upregulated in SNT and gp120 + ddC models, by RT-PCR. All three genes were found to be markedly upregulated in agreement with the microarray results (Fig. 5). As a further test for the link between these genes and mechanical hypersensitivity, we also measured the differential expression of these three genes in L4 and L5 DRG of rats infected with Varicella Zoster virus. This is a disparate animal model of neuropathic pain, used to model the mechanisms underlying post-herpetic neuralgia (Sadzot-Delvaux et al., 1990, Fleetwood-Walker et al., 1999, Dalziel et al., 2004, Garry et al., 2005), which is also associated with similar mechanical hypersensitivity and thigmotaxis behaviour as is observed in SNT and GP120 + ddC (Hasnie et al., 2007, Wallace et al., 2007b). All three genes were also upregulated in the VZV treated animals (Fig. 5).
Fig. 5.
RT-PCR results for Npy, Pap/Reg2 and Vgf in SNT, gp120 + ddC and VZV infected animals. Normalized expression levels for each gene are expressed as ratios to cyclophilin. Data are expressed as means ± SD. ∗, (p < 0.05) indicates statistical significance between treated and sham groups of animals (Student’s t test).
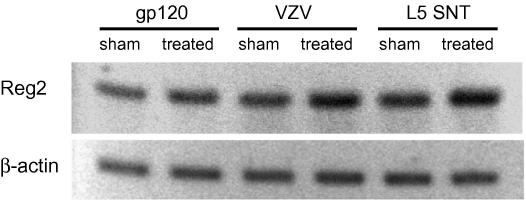
We further confirmed the upregulation of PAP/Reg2 using immunoblot analysis to test if the observed mRNA level changes are maintained at the protein level. Pap/Reg2 was found to be upregulated by 1.4-fold for gp120 + ddC, 1.4-fold for SNT and by 1.1-fold for VZV (Fig. 6) further validating our microarray results.
Fig. 6.
Immunoblots showing the induction of REG2/PAP protein expression in DRG from sham and SNT-, gp120 + ddC- and VZV-treated animals. The same membrane was re-probed with an anti β-ACTIN antibody.
4. Discussion
To elucidate the mechanisms underlying HIV DSP pain, we performed gene expression profiling in the L4 and L5 DRG of a recently developed rat model (gp120 + ddC) (Wallace et al., 2007b). We then focused our analysis towards the identification of genes that are directly relevant to presentation of mechanical hypersensitivity. To do so, we compared the gene profile of the gp120 + ddC model, with that of a model of direct sciatic nerve trauma (SNT) that although based on a distinct aetiology, shares the common outcome of mechanical hypersensitivity. As predicted, the models are associated with distinct sets of gene expression changes. However, we identified 39 genes/ESTs that are differentially regulated in the same direction in both models which may therefore underlie the common link of mechanical hypersensitivity.
Gp120 and nucleoside analogues are thought to affect cells by a variety of mechanisms including via cytokine and chemokine signalling pathways (Oh et al., 2001, Bhangoo et al., 2007). Accordingly, many biological process pathways overrepresented in the gp120 + ddC upregulated gene list involve signal transduction, such as neuropeptide signalling and the G alpha q pathway. Upregulated genes associated with these terms include TrkB.T1, Penk1, Celsr3, Nfkb2 and Itpr3. Interestingly, of these genes, the truncated isoform of TrkB (TrkB.T1), a receptor for BDNF (Huang and Reichardt, 2003), is the most upregulated gene in the gp120 + ddC model, which was further validated with RT-PCR. TrkB full length isoform (TrkB.FL)-BDNF signalling has been shown to protect neurons from gp120 induced cell death (Mocchetti and Bachis, 2004), but also thought to contribute to pain facilitation (Guo et al., 2006). TrkB.T1 is thought to modify the action of full length TrkB (TrkB.FL) signalling (Biffo et al., 1995, Eide et al., 1996, Haapasalo et al., 2001) and thus it is an interesting candidate for the development of hypersensitivity in the gp120 + ddC model.
DdC is associated with mitochondrial toxicity (Dalakas et al., 2001, Keswani et al., 2003a), while gp120 may cause axonal degeneration directly through activation of a mitochondrial caspase pathway and indirectly through neuronal apoptosis mediated by the activation of Schwann cells (Melli et al., 2006). GSEA analysis identified a gene set of ribosomal proteins as significantly associated with downregulation in the gp120 + ddC model. This set contains genes like Rps24, Rps27a, Rps25, and Rpl7.
The mitochondrial dysfunction induced by ddC alters calcium homeostasis in cultured DRG neurons (Werth et al., 1994) and in a model of ddC-associated painful peripheral neuropathy (Joseph et al., 2004). Gp120 is also associated with an increase in [Ca2+]i in subsets of cultured DRG cells through the activation of chemokines (Oh et al., 2001). Therefore, alterations in calcium activity could underlie a synergistic effect of a combination of gp120 and ddC exposure. Such synergism of the treatments is suggested by the fact that behavioural indices of pain are enhanced in the gp120 + ddC model as compared to either treatment alone (Wallace et al., 2007b). We find that several genes involved in calcium ion transport (Casr, Plcg1 and Atp2a2) and calcium binding (e.g., Bcan, Egfl4, Dag1, Stat3, Capn1) are upregulated in the gp120 + ddC model suggesting that changes in calcium regulation may contribute to the development of neuropathy and merit further investigation.
Comparison of the gp120 + ddC and SNT model DRG gene expression profiles indicates that the pathological processes inherent to each model influence multiple gene expression pathways. For example, pathways involved in signalling or cell–cell adhesion are mainly downregulated in the SNT model and upregulated in the gp120 + ddC model. Interestingly, certain genes implicated in neuropathy formation and/or maintenance showed opposing patterns of differential expression between models. For example; protein kinase C, epsilon (PKCε) and its signalling pathways have been implicated in primary afferent nociceptor sensitization (Khasar et al., 1999) and it is considered an important contributor to the development of painful diabetic neuropathy (Ahlgren and Levine, 1994), taxol-induced painful peripheral neuropathy (Dina et al., 2001b), painful alcoholic neuropathy (Dina et al., 2000, Dina et al., 2006), and inflammatory hyperalgesia (Khasar et al., 1999, Aley et al., 2000, Dina et al., 2001a, Parada et al., 2005). PKCε was down regulated in SNT DRG and up-regulated in gp120 + ddC DRG. The calcitonin-related polypeptide alpha (Calca) gene, that codes for CGRP, which has been implicated in the development of thermal hypersensitivity in mice (Mogil et al., 2005) and mechanical hypersensitivity in a traumatic model of rat (Jang et al., 2004). Calca was found to be downregulated in the SNT model, but showed no significant changes in expression for the gp120 + ddC model, which is in line with previous investigations (Wallace et al., 2007b). This suggests that although these genes have been deemed important in traumatic neuropathy, they may not be associated with neuropathic pain of multiple pathophysiological origins or the manifestation of mechanical hypersensitivity.
There are some limitations of our experimental approach which require discussion. First, this study focuses on a single time point, when mechanical hypersensitivity is well established. Thus the genes identified are potentially important for this phase of mechanical hypersensitivity. To fully identify candidate genes for hypersensitivity generation and maintenance, this study would have to be repeated at a number of time points, which is prohibitively expensive given the current cost of this technology. Secondly, these microarray data reflects changes in gene expression in the entire DRG, which is a heterogenous tissue, composed of many cell types. Thus, this study cannot link changes to specific cell types in the DRG. In situ hybridization and/or immunohistochemical experiments are required to further clarify this point. Thirdly, in an effort to minimize animal usage and reduce biological variability, we employed a 2 cycle amplification protocol to generate the microarray targets. The 2 cycle amplification protocol produces reproducible results with high correlation between amplified and non-amplified RNAs (Saghizadeh et al., 2003, Klur et al., 2004, Li et al., 2005). However, RNA amplification may cause slight distortion (mainly decrease) of the expression ratios (Diboun et al., 2006) and failure to detect transcripts from the low intensity range (van Haaften et al., 2006). Therefore, our data might suffer from a slight increase in false negative results. This offers an explanation as to why differential expression for ATF3, Galanin, CCL2 and GFAP were detected by immunohistochemistry (Wallace et al., 2007b) but not by microarrays for the gp120 + ddC model (which exhibits subtle gene changes). Finally, we used a model of HIV-related neuropathy that includes two insults to the nervous system, gp120 and ddC, instead of the insults been applied individually. This model is clinically relevant since the decision to commence antiretroviral therapy is taken at a time when there is a high viral load and therefore circulating gp120 levels are high, with patients being simultaneously exposed to the double neurotoxic insults of gp120 and antiretroviral drugs (Gazzard et al., 2006, Wallace et al., 2007b). However, whereas our model reflects the clinical situation, we cannot be certain whether it is gp120, ddC or the combination which drives the gene changes seen.
Despite the potential limitations of this microarray study, we identified several potential targets for mechanical hypersensitivity maintenance. Thirty-nine genes/ESTs are differentially regulated in the same direction in both of our models. Of these genes, NPY has previously been suggested to contribute to nerve injury-induced mechanical hyperalgesia (White, 1997, Ossipov et al., 2002, Intondi et al., 2007). However, the majority of genes highlighted by our experiment represent potential novel targets in pain processing, two of which are discussed in more detail below.
Vgf is upregulated in the DRG of both gp120 + ddC and SNT models. VGF expression is induced by neurotrophins such as nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) which are themselves important in the development, maintenance and normal functioning of neurons in the nervous system (Levi et al., 1985, Cho et al., 1989, Salton et al., 1991, Alder et al., 2003). The majority of experimental work on VGF peptide has focused upon its role as an energy homeostasis regulator (Hahm et al., 1999, Hahm et al., 2002, Watson et al., 2005, Bartolomucci et al., 2006) whilst the precise mechanism of VGF action and its role in pain requires further study. However, it is known that in the PNS, VGF is expressed mainly in small to medium diameter sensory neurons that project to the superficial laminae of the spinal cord (Ferri et al., 1992). VGF is a secreted polypeptide (Possenti et al., 1989), and thus may potentially mediate neuronal communication. Furthermore, another function of VGF is synaptic strengthening associated with learning ,memory and anti-depressant like behavioural effects (Alder et al., 2003, Thakker-Varia et al., 2007, Hunsberger et al., 2007). VGF has also been shown to be up-regulated in rat DRG at 2–28 days following axotomy (Xiao et al., 2002, Costigan et al., 2002). Although, axotomized rats do not develop mechanical hypersensitivity of the hind-paw due to deafferentation, VGF is also implicated as an important target in this model and clearly merits further investigation with regard to its role in pain.
Pap/Reg2 was also significantly up-regulated in both models. Reg-2 is a macrophage chemoattractant that is dynamically expressed in rat sensory neurons after peripheral nerve injury (Averill et al., 2002, Namikawa et al., 2005, Namikawa et al., 2006) and has been implicated in regeneration of motor and sensory neurons (Livesey et al., 1997, Namikawa et al., 2006). This regenerative role may account for the increase in SNT DRG as compared to the gp120 + ddC model. However, although largely expressed by injured neurons, Reg-2 has also been shown to be upregulated in uninjured axons also (Averill et al., 2002), which may implicate a non-regeneration associated role that may relate to mechanisms of hypersensitivity. The role of Reg-2 in pain is yet to be determined and our results suggest that this may be a good candidate to investigate for such properties.
This study has identified 39 potential mechanical hypersensitivity related genes/ESTs associated with the development of mechanical hypersensitivity and thigmotaxis following damage to the peripheral nervous system. Functional studies are now required to verify that the products of these genes are indeed involved in the generation and/or maintenance of neuropathic pain. Some of these genes may become targets for developing novel therapeutics to treat neuropathic pain at the level of the primary sensory neurone; where there is probably the most favourable prospect of developing analgesic drugs with a favourable therapeutic index in respect of CNS mediated adverse effects.
Acknowledgements
This work was funded by the Wellcome Trust (London Pain Consortium). We thank Caroline Johnston and Ilhem Diboun (Department of Biochemistry and Molecular Biology, University College London and London Pain Consortium) for advice and technical assistance. Thorsten Forster (Scottish Centre for Genomic Technology and Informatics, University of Edinburgh), Manuela Zucknick and Dr. Alexandra Lewin (Department of Biostatistics, St. Mary’s Hospital, Imperial College London) for helpful discussions. We also thank Professor Judy Breuer (Centre for Infectious Disease, Barts and The London, Queen Mary’s School of Medicine and Dentistry), for helping with the virological aspects of the VZV infection model.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ejpain.2008.05.011.
Supplementary data
References
- Ahlgren S.C., Levine J.D. Protein kinase C inhibitors decrease hyperalgesia and C-fiber hyperexcitability in the streptozotocin-diabetic rat. J Neurophysiol. 1994;72:684–692. doi: 10.1152/jn.1994.72.2.684. [DOI] [PubMed] [Google Scholar]
- Alder J., Thakker-Varia S., Bangasser D.A., Kuroiwa M., Plummer M.R., Shors T.J. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. 2003;23:10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aley K.O., Messing R.O., Mochly-Rosen D., Levine J.D. Chronic hypersensitivity for inflammatory nociceptor sensitization mediated by the epsilon isozyme of protein kinase C. J Neurosci. 2000;20:4680–4685. doi: 10.1523/JNEUROSCI.20-12-04680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averill S., Davis D.R., Shortland P.J., Priestley J.V., Hunt S.P. Dynamic pattern of reg-2 expression in rat sensory neurons after peripheral nerve injury. J Neurosci. 2002;22:7493–7501. doi: 10.1523/JNEUROSCI.22-17-07493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakay M., Chen Y.W., Borup R., Zhao P., Nagaraju K., Hoffman E.P. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics. 2002;3:4. doi: 10.1186/1471-2105-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomucci A., La C.G., Possenti R., Locatelli V., Rigamonti A.E., Torsello A. TLQP-21, a VGF-derived peptide, increases energy expenditure and prevents the early phase of diet-induced obesity. Proc Natl Acad Sci USA. 2006;103:14584–14589. doi: 10.1073/pnas.0606102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo S.K., Ren D., Miller R.J., Chan D.M., Ripsch M.S., Weiss C. CXCR4 chemokine receptor signaling mediates pain hypersensitivity in association with antiretroviral toxic neuropathy. Brain Behav Immun. 2007;21:581–591. doi: 10.1016/j.bbi.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffo S., Offenhauser N., Carter B.D., Barde Y.A. Selective binding and internalisation by truncated receptors restrict the availability of BDNF during development. Development. 1995;121:2461–2470. doi: 10.1242/dev.121.8.2461. [DOI] [PubMed] [Google Scholar]
- Boucher T.J., Okuse K., Bennett D.L., Munson J.B., Wood J.N., McMahon S.B. Potent analgesic effects of GDNF in neuropathic pain states. Science. 2000;290:124–127. doi: 10.1126/science.290.5489.124. [DOI] [PubMed] [Google Scholar]
- Bridges D., Ahmad K., Rice A.S. The synthetic cannabinoid WIN55,212-2 attenuates hyperalgesia and allodynia in a rat model of neuropathic pain. Br J Pharmacol. 2001;133:586–594. doi: 10.1038/sj.bjp.0704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplan S.R., Bach F.W., Pogrel J.W., Chung J.M., Yaksh T.L. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Cherry C.L., McArthur J.C., Hoy J.F., Wesselingh S.L. Nucleoside analogues and neuropathy in the era of HAART. J Clin Virol. 2003;26:195–207. doi: 10.1016/s1386-6532(02)00118-x. [DOI] [PubMed] [Google Scholar]
- Cherry C.L., Skolasky R.L., Lal L., Creighton J., Hauer P., Raman S.P. Antiretroviral use and other risks for HIV-associated neuropathies in an international cohort. Neurology. 2006;66:867–873. doi: 10.1212/01.wnl.0000203336.12114.09. [DOI] [PubMed] [Google Scholar]
- Cho K.O., Skarnes W.C., Minsk B., Palmieri S., Jackson-Grusby L., Wagner J.A. Nerve growth factor regulates gene expression by several distinct mechanisms. Mol Cell Biol. 1989;9:135–143. doi: 10.1128/mcb.9.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Yoon Y.W., Na H.S., Kim S.H., Chung J.M. Behavioral signs of ongoing pain and cold allodynia in a rat model of neuropathic pain. Pain. 1994;59:369–376. doi: 10.1016/0304-3959(94)90023-X. [DOI] [PubMed] [Google Scholar]
- Costigan M., Befort K., Karchewski L., Griffin R.S., D’Urso D., Allchorne A. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Dalakas M.C., Pezeshkpour G.H. Neuromuscular diseases associated with human immunodeficiency virus infection. Ann Neurol. 1988;23 Suppl.:S38–S48. doi: 10.1002/ana.410230713. [DOI] [PubMed] [Google Scholar]
- Dalakas M.C., Semino-Mora C., Leon-Monzon M. Mitochondrial alterations with mitochondrial DNA depletion in the nerves of AIDS patients with peripheral neuropathy induced by 2′, 3′-dideoxycytidine (ddC) Lab Invest. 2001;81:1537–1544. doi: 10.1038/labinvest.3780367. [DOI] [PubMed] [Google Scholar]
- Dalziel R.G., Bingham S., Sutton D., Grant D., Champion J.M., Dennis S.A. Allodynia in rats infected with varicella zoster virus – a small animal model for post-herpetic neuralgia. Brain Res Brain Res Rev. 2004;46:234–242. doi: 10.1016/j.brainresrev.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Diboun I., Wernisch L., Orengo C.A., Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina O.A., Aley K.O., Isenberg W., Messing R.O., Levine J.D. Sex hormones regulate the contribution of PKCepsilon and PKA signalling in inflammatory pain in the rat. Eur J Neurosci. 2001;13:2227–2233. doi: 10.1046/j.0953-816x.2001.01614.x. [DOI] [PubMed] [Google Scholar]
- Dina O.A., Chen X., Reichling D., Levine J.D. Role of protein kinase Cepsilon and protein kinase A in a model of paclitaxel-induced painful peripheral neuropathy in the rat. Neuroscience. 2001;108:507–515. doi: 10.1016/s0306-4522(01)00425-0. [DOI] [PubMed] [Google Scholar]
- Dina O.A., Barletta J., Chen X., Mutero A., Martin A., Messing R.O. Key role for the epsilon isoform of protein kinase C in painful alcoholic neuropathy in the rat. J Neurosci. 2000;20:8614–8619. doi: 10.1523/JNEUROSCI.20-22-08614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina O.A., Messing R.O., Levine J.D. Ethanol withdrawal induces hyperalgesia mediated by PKCepsilon. Eur J Neurosci. 2006;24:197–204. doi: 10.1111/j.1460-9568.2006.04886.x. [DOI] [PubMed] [Google Scholar]
- Doniger S.W., Salomonis N., Dahlquist K.D., Vranizan K., Lawlor S.C., Conklin B.R. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdall T., Robinson I., Meert T.F. Comparison of five different rat models of peripheral nerve injury. Pharmacol Biochem Behav. 2005;80:93–108. doi: 10.1016/j.pbb.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Eide F.F., Vining E.R., Eide B.L., Zang K., Wang X.Y., Reichardt L.F. Naturally occurring truncated trkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J Neurosci. 1996;16:3123–3129. doi: 10.1523/JNEUROSCI.16-10-03123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri G.L., Levi A., Possenti R. A novel neuroendocrine gene product: selective VGF8a gene expression and immuno-localisation of the VGF protein in endocrine and neuronal populations. Brain Res Mol Brain Res. 1992;13:139–143. doi: 10.1016/0169-328x(92)90053-e. [DOI] [PubMed] [Google Scholar]
- Fleetwood-Walker S.M., Quinn J.P., Wallace C., Blackburn-Munro G., Kelly B.G., Fiskerstrand C.E. Behavioural changes in the rat following infection with varicella-zoster virus. J Gen Virol. 1999;80(Pt. 9):2433–2436. doi: 10.1099/0022-1317-80-9-2433. [DOI] [PubMed] [Google Scholar]
- Garry E.M., Delaney A., Anderson H.A., Sirinathsinghji E.C., Clapp R.H., Martin W.J. Varicella zoster virus induces neuropathic changes in rat dorsal root ganglia and behavioral reflex sensitisation that is attenuated by gabapentin or sodium channel blocking drugs. Pain. 2005;118:97–111. doi: 10.1016/j.pain.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy – analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gazzard B., Bernard A.J., Boffito M., Churchill D., Edwards S., Fisher N. British HIV Association (BHIVA) guidelines for the treatment of HIV-infected adults with antiretroviral therapy (2006) HIV Med. 2006;7:487–503. doi: 10.1111/j.1468-1293.2006.00424.x. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Robbins M.T., Wei F., Zou S., Dubner R., Ren K. Supraspinal brain-derived neurotrophic factor signaling: a novel mechanism for descending pain facilitation. J Neurosci. 2006;26:126–137. doi: 10.1523/JNEUROSCI.3686-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapasalo A., Koponen E., Hoppe E., Wong G., Castren E. Truncated trkB. T1 is dominant negative inhibitor of trkB.TK+-mediated cell survival. Biochem Biophys Res Commun. 2001;280:1352–1358. doi: 10.1006/bbrc.2001.4296. [DOI] [PubMed] [Google Scholar]
- Hahm S., Fekete C., Mizuno T.M., Windsor J., Yan H., Boozer C.N. VGF is required for obesity induced by diet, gold thioglucose treatment, and agouti and is differentially regulated in pro-opiomelanocortin- and neuropeptide Y-containing arcuate neurons in response to fasting. J Neurosci. 2002;22:6929–6938. doi: 10.1523/JNEUROSCI.22-16-06929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm S., Mizuno T.M., Wu T.J., Wisor J.P., Priest C.A., Kozak C.A. Targeted deletion of the Vgf gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron. 1999;23:537–548. doi: 10.1016/s0896-6273(00)80806-5. [DOI] [PubMed] [Google Scholar]
- Hasnie F.S., Breuer J., Parker S., Wallace V., Blackbeard J., Lever I. Further characterization of a rat model of varicella zoster virus-associated pain: Relationship between mechanical hypersensitivity and anxiety-related behavior, and the influence of analgesic drugs. Neuroscience. 2007;144:1495–1508. doi: 10.1016/j.neuroscience.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E.J., Reichardt L.F. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Hunsberger J.G., Newton S.S., Bennett A.H., Duman C.H., Russell D.S., Salton S.R. Antidepressant actions of the exercise-regulated gene VGF. Nat Med. 2007;13:1476–1482. doi: 10.1038/nm1669. [DOI] [PubMed] [Google Scholar]
- Intondi A.B., Dahlgren M.N., Eilers M.A., Taylor B.K. Intrathecal neuropeptide Y reduces behavioral and molecular markers of inflammatory or neuropathic pain. Pain. 2007 doi: 10.1016/j.pain.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.H., Nam T.S., Paik K.S., Leem J.W. Involvement of peripherally released substance P and calcitonin gene-related peptide in mediating mechanical hyperalgesia in a traumatic neuropathy model of the rat. Neurosci Lett. 2004;360:129–132. doi: 10.1016/j.neulet.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Joseph E.K., Chen X., Khasar S.G., Levine J.D. Novel mechanism of enhanced nociception in a model of AIDS therapy-induced painful peripheral neuropathy in the rat. Pain. 2004;107:147–158. doi: 10.1016/j.pain.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Keswani S.C., Chander B., Hasan C., Griffin J.W., McArthur J.C., Hoke A. FK506 is neuroprotective in a model of antiretroviral toxic neuropathy. Ann Neurol. 2003;53:57–64. doi: 10.1002/ana.10401. [DOI] [PubMed] [Google Scholar]
- Keswani S.C., Polley M., Pardo C.A., Griffin J.W., McArthur J.C., Hoke A. Schwann cell chemokine receptors mediate HIV-1 gp120 toxicity to sensory neurons. Ann Neurol. 2003;54:287–296. doi: 10.1002/ana.10645. [DOI] [PubMed] [Google Scholar]
- Khasar S.G., Lin Y.H., Martin A., Dadgar J., McMahon T., Wang D. A novel nociceptor signaling pathway revealed in protein kinase C epsilon mutant mice. Neuron. 1999;24:253–260. doi: 10.1016/s0896-6273(00)80837-5. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Chung J.M. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. 1992;50:355–363. doi: 10.1016/0304-3959(92)90041-9. [DOI] [PubMed] [Google Scholar]
- Klur S., Toy K., Williams M.P., Certa U. Evaluation of procedures for amplification of small-size samples for hybridization on microarrays. Genomics. 2004;83:508–517. doi: 10.1016/j.ygeno.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Lacroix-Fralish M.L., Tawfik V.L., Tanga F.Y., Spratt K.F., Deleo J.A. Differential spinal cord gene expression in rodent models of radicular and neuropathic pain. Anesthesiology. 2006;104:1283–1292. doi: 10.1097/00000542-200606000-00025. [DOI] [PubMed] [Google Scholar]
- Levi A., Eldridge J.D., Paterson B.M. Molecular cloning of a gene sequence regulated by nerve growth factor. Science. 1985;229:393–395. doi: 10.1126/science.3839317. [DOI] [PubMed] [Google Scholar]
- Levin M.E., Jin J.G., Ji R.R., Tong J., Pomonis J.D., Lavery D.J. Complement activation in the peripheral nervous system following the spinal nerve ligation model of neuropathic pain. Pain. 2007 doi: 10.1016/j.pain.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Li L., Roden J., Shapiro B.E., Wold B.J., Bhatia S., Forman S.J. Reproducibility, fidelity, and discriminant validity of mRNA amplification for microarray analysis from primary hematopoietic cells. J Mol Diagn. 2005;7:48–56. doi: 10.1016/S1525-1578(10)60008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey F.J., O’Brien J.A., Li M., Smith A.G., Murphy L.J., Hunt S.P. A Schwann cell mitogen accompanying regeneration of motor neurons. Nature. 1997;390:614–618. doi: 10.1038/37615. [DOI] [PubMed] [Google Scholar]
- Martin C., Pehrsson P., Osterberg A., Sonnerborg A., Hansson P. Pain in ambulatory HIV-infected patients with and without intravenous drug use. Eur J Pain. 1999;3:157–164. doi: 10.1053/eujp.1999.0111. [DOI] [PubMed] [Google Scholar]
- Martin C., Solders G., Sonnerborg A., Hansson P. Painful and non-painful neuropathy in HIV-infected patients: an analysis of somatosensory nerve function. Eur J Pain. 2003;7:23–31. doi: 10.1016/s1090-3801(02)00053-8. [DOI] [PubMed] [Google Scholar]
- Melli G., Keswani S.C., Fischer A., Chen W., Hoke A. Spatially distinct and functionally independent mechanisms of axonal degeneration in a model of HIV-associated sensory neuropathy. Brain. 2006;129:1330–1338. doi: 10.1093/brain/awl058. [DOI] [PubMed] [Google Scholar]
- Mocchetti I., Bachis A. Brain-derived neurotrophic factor activation of TrkB protects neurons from HIV-1/gp120-induced cell death. Crit Rev Neurobiol. 2004;16:51–57. doi: 10.1615/critrevneurobiol.v16.i12.50. [DOI] [PubMed] [Google Scholar]
- Mogil J.S., Miermeister F., Seifert F., Strasburg K., Zimmermann K., Reinold H. Variable sensitivity to noxious heat is mediated by differential expression of the CGRP gene. Proc Natl Acad Sci USA. 2005;102:12938–12943. doi: 10.1073/pnas.0503264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namikawa K., Fukushima M., Murakami K., Suzuki A., Takasawa S., Okamoto H. Expression of Reg/PAP family members during motor nerve regeneration in rat. Biochem Biophys Res Commun. 2005;332:126–134. doi: 10.1016/j.bbrc.2005.04.105. [DOI] [PubMed] [Google Scholar]
- Namikawa K., Okamoto T., Suzuki A., Konishi H., Kiyama H. Pancreatitis-associated protein-III is a novel macrophage chemoattractant implicated in nerve regeneration. J Neurosci. 2006;26:7460–7467. doi: 10.1523/JNEUROSCI.0023-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.B., Tran P.B., Gillard S.E., Hurley R.W., Hammond D.L., Miller R.J. Chemokines and glycoprotein120 produce pain hypersensitivity by directly exciting primary nociceptive neurons. J Neurosci. 2001;21:5027–5035. doi: 10.1523/JNEUROSCI.21-14-05027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuse K., Chaplan S.R., McMahon S.B., Luo Z.D., Calcutt N.A., Scott B.P. Regulation of expression of the sensory neuron-specific sodium channel SNS in inflammatory and neuropathic pain. Mol Cell Neurosci. 1997;10:196–207. doi: 10.1006/mcne.1997.0657. [DOI] [PubMed] [Google Scholar]
- Ossipov M.H., Zhang E.T., Carvajal C., Gardell L., Quirion R., Dumont Y. Selective mediation of nerve injury-induced tactile hypersensitivity by neuropeptide Y. J Neurosci. 2002;22:9858–9867. doi: 10.1523/JNEUROSCI.22-22-09858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C.A., Reichling D.B., Levine J.D. Chronic hyperalgesic priming in the rat involves a novel interaction between cAMP and PKCepsilon second messenger pathways. Pain. 2005;113:185–190. doi: 10.1016/j.pain.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Parkinson H., Kapushesky M., Shojatalab M., Abeygunawardena N., Coulson R., Farne A. ArrayExpress – a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possenti R., Eldridge J.D., Paterson B.M., Grasso A., Levi A. A protein induced by NGF in PC12 cells is stored in secretory vesicles and released through the regulated pathway. EMBO J. 1989;8:2217–2223. doi: 10.1002/j.1460-2075.1989.tb08345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Ringkamp M., Eschenfelder S., Grethel E.J., Habler H.J., Meyer R.A., Janig W. Lumbar sympathectomy failed to reverse mechanical allodynia- and hyperalgesia-like behavior in rats with L5 spinal nerve injury. Pain. 1999;79:143–153. doi: 10.1016/s0304-3959(98)00186-9. [DOI] [PubMed] [Google Scholar]
- Sadzot-Delvaux C., Merville-Louis M.P., Delree P., Marc P., Piette J., Moonen G. An in vivo model of varicella-zoster virus latent infection of dorsal root ganglia. J Neurosci Res. 1990;26:83–89. doi: 10.1002/jnr.490260110. [DOI] [PubMed] [Google Scholar]
- Saghizadeh M., Brown D.J., Tajbakhsh J., Chen Z., Kenney M.C., Farber D.B. Evaluation of techniques using amplified nucleic acid probes for gene expression profiling. Biomol Eng. 2003;20:97–106. doi: 10.1016/s1389-0344(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Salton S.R., Fischberg D.J., Dong K.W. Structure of the gene encoding VGF, a nervous system-specific mRNA that is rapidly and selectively induced by nerve growth factor in PC12 cells. Mol Cell Biol. 1991;11:2335–2349. doi: 10.1128/mcb.11.5.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson D.M., Kitch D., Evans S.R., McArthur J.C., Asmuth D.M., Cohen B. HIV neuropathy natural history cohort study: assessment measures and risk factors. Neurology. 2006;66:1679–1687. doi: 10.1212/01.wnl.0000218303.48113.5d. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. In: Bioinformatics and computational biology solutions using R and bioconductor. Gentleman Vcsdriwh R., editor. Springer; New York: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- Smyth K., Affandi J.S., McArthur J.C., Bowtell-Harris C., Mijch A.M., Watson K. Prevalence of and risk factors for HIV-associated neuropathy in Melbourne, Australia 1993–2006. HIV Med. 2007;8:367–373. doi: 10.1111/j.1468-1293.2007.00478.x. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker-Varia S., Krol J.J., Nettleton J., Bilimoria P.M., Bangasser D.A., Shors T.J. The neuropeptide VGF produces antidepressant-like behavioral effects and enhances proliferation in the hippocampus. J Neurosci. 2007;27:12156–12167. doi: 10.1523/JNEUROSCI.1898-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valder C.R., Liu J.J., Song Y.H., Luo Z.D. Coupling gene chip analyses and rat genetic variances in identifying potential target genes that may contribute to neuropathic allodynia development. J Neurochem. 2003;87:560–573. doi: 10.1046/j.1471-4159.2003.02016.x. [DOI] [PubMed] [Google Scholar]
- van Haaften R.I., Schroen B., Janssen B.J., van E.A., Debets J.J., Smeets H.J. Biologically relevant effects of mRNA amplification on gene expression profiles. BMC Bioinformatics. 2006;7:200. doi: 10.1186/1471-2105-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Estanislao L., Simpson D. HIV-associated neuropathic pain: epidemiology, pathophysiology and management. CNS Drugs. 2005;19:325–334. doi: 10.2165/00023210-200519040-00005. [DOI] [PubMed] [Google Scholar]
- Wallace V.C., Blackbeard J., Pheby T., Segerdahl A.R., Davies M., Hasnie F. Pharmacological, behavioural and mechanistic analysis of HIV-1 gp120 induced painful neuropathy. Pain. 2007 doi: 10.1016/j.pain.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace V.C., Blackbeard J., Segerdahl A.R., Hasnie F., Pheby T., McMahon S.B. Characterization of rodent models of HIV-gp120 and anti-retroviral-associated neuropathic pain. Brain. 2007;130:2688–2702. doi: 10.1093/brain/awm195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sun H., Della P.K., Benz R.J., Xu J., Gerhold D.L. Chronic neuropathic pain is accompanied by global changes in gene expression and shares pathobiology with neurodegenerative diseases. Neuroscience. 2002;114:529–546. doi: 10.1016/s0306-4522(02)00341-x. [DOI] [PubMed] [Google Scholar]
- Watson E., Hahm S., Mizuno T.M., Windsor J., Montgomery C., Scherer P.E. VGF ablation blocks the development of hyperinsulinemia and hyperglycemia in several mouse models of obesity. Endocrinology. 2005;146:5151–5163. doi: 10.1210/en.2005-0588. [DOI] [PubMed] [Google Scholar]
- Werth J.L., Zhou B., Nutter L.M., Thayer S.A. 2′,3′-Dideoxycytidine alters calcium buffering in cultured dorsal root ganglion neurons. Mol Pharmacol. 1994;45:1119–1124. [PubMed] [Google Scholar]
- White D.M. Intrathecal neuropeptide Y exacerbates nerve injury-induced mechanical hyperalgesia. Brain Res. 1997;750:141–146. doi: 10.1016/s0006-8993(96)01340-6. [DOI] [PubMed] [Google Scholar]
- Wilson C.L., Miller C.J. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- Xiao H.S., Huang Q.H., Zhang F.X., Bao L., Lu Y.J., Guo C. Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc Natl Acad Sci USA. 2002;99:8360–8365. doi: 10.1073/pnas.122231899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M. Ethical guidelines for investigations of experimental pain in conscious animals. Pain. 1983;16:109–110. doi: 10.1016/0304-3959(83)90201-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.