Abstract
Background
Clostridium perfringens type A food poisoning (FP) is usually caused by C. perfringens type A strains that carry a chromosomal enterotoxin gene (cpe) and produce spores with exceptional resistance against heat and nitrites. Previous studies showed that the extreme resistance of spores made by most FP strains is mediated, in large part, by a variant of small acid soluble protein 4 (Ssp4) that has Asp at residue 36; in contrast, the sensitive spores made by other C. perfringens type A isolates contain an Ssp4 variant with Gly at residue 36.
Methodology/Principal Findings
The current study has further characterized Ssp4 properties and expression. Spores made by cpe-positive type C and D strains were found to contain the Ssp4 variant with Gly at residue 36 and were shown to be heat- and nitrite-sensitive; this finding may help to explain why cpe-positive type C and D isolates rarely cause food poisoning. Saturation mutagenesis indicated that both amino acid size and charge at Ssp4 residue 36 are important for DNA binding and for spore resistance. C. perfringens Ssp2 was shown to bind preferentially to GC-rich DNA on gel-shift assays, while Ssp4 preferred binding to AT-rich DNA sequences. Maximal spore heat and nitrite resistance required production of all four C. perfringens Ssps, indicating that these Ssps act cooperatively to protect the spore's DNA, perhaps by binding to different chromosomal sequences. The Ssp4 variant with Asp at residue 36 was also shown to facilitate exceptional spore survival at freezer and refrigerator temperatures. Finally, Ssp4 expression was shown to be dependent upon Spo0A, a master regulator.
Conclusions/Significance
Collectively, these results provide additional support for the importance of Ssps, particularly the Ssp4 variant with Asp at residue 36, for the extreme spore resistance phenotype that likely contributes to C. perfringens type A food poisoning transmission.
Introduction
Clostridium perfringens, a Gram-positive, anaerobic, sporeforming bacterium, can produce at least 17 different toxins. However, individual C. perfringens strains never produce this entire toxin repertoire. A commonly used system [1] exploits this variability in toxin production to classify individual C. perfringens isolates into types A–E, based upon their production of four typing toxins (alpha, beta, epsilon and iota toxins).
About 1–5% of type A isolates produce another toxin, named C. perfringens enterotoxin (CPE), which is responsible for causing the gastrointestinal (GI) symptoms of C. perfringens type A FP [2]. This FP currently ranks as the second most commonly reported bacterial foodborne disease in the USA and UK [3], [4], where (respectively) over 250,000 or 85,000 cases occur annually. Those cases usually resolve without long-term consequence, but C. perfringens type A FP can be fatal in the elderly or debilitated individuals. Consequently, C. perfringens ranks among the three or four most common bacterial causes of foodborne death [1], [3], [4]. In addition to FP, CPE-positive type A C. perfringens strains also cause about 5–15% of all cases of nonfoodborne human gastrointestinal diseases, including sporadic diarrhea and antibiotic-associated diarrhea [1], [2].
In type A isolates, the gene (cpe) encoding CPE can be either chromosomal or plasmid-borne [5], [6]. Most (∼75%) FP cases are caused by type A isolates carrying a chromosomal cpe gene (C-cpe) [5], [6], [7], [8], [9], [10]. Recent studies have provided at least three (possibly interrelated) explanations for this strong association between C-cpe isolates and FP. First, type A C-cpe isolates were found to be more prevalent than type A plasmid cpe (P-cpe) isolates in American retail meat products [11], which are important vehicles for C. perfringens type A FP [1]. Second, type A C-cpe isolates usually grow faster, and over a broader temperature range, than do type A P-cpe isolates [12], which should favor the multiplication of C-cpe isolates in foods so these bacteria can reach the food burden necessary for inducing disease. Finally, compared to the vegetative cells or (particularly) spores of type A P-cpe isolates, the cells/spores of type A C-cpe isolates were shown [12], [13], [14] to typically exhibit much more resistance against food safety-induced stresses such as heating, cold (refrigerator or freezer temperatures) storage, osmotic stress and food preservatives (e.g. nitrites). Since, i) spores of type A C-cpe isolates are present in retail foods [11] and ii) temperature abuse of foods during cooking or storage is the major underlying factor leading to C. perfringens type A FP outbreaks [1], the spore resistance phenotype of type A C-cpe isolates is likely to facilitate survival of these isolates in foods so they can later cause FP.
We recently identified [15] a major contributor to the exceptional spore resistance phenotype exhibited by the spores of most type A C-cpe isolates. Specifically, C-cpe isolates that produce resistant spores were found to express a variant of a novel small acid soluble protein named Ssp4. Whereas Gly is present at Ssp4 residue 36 in C. perfringens type A isolates producing sensitive spores, the Ssp4 residue 36 is an Asp in most, if not all, type A C-cpe isolates producing resistant spores. Inactivation of the gene (ssp4) encoding Ssp4 was shown to significantly increase the sensitivity of C. perfringens type A spores to both heat and nitrous acid (a fast-killing proxy assay for evaluating spore resistance against nitrite, an often used food preservative), directly demonstrating that Ssp4 plays an important role in spore resistance properties. Furthermore, when spores of those ssp4 null mutants were complemented to express Ssp4 with an Asp at residue 36 (i.e., a Ssp4 Asp variant), they exhibited greater heat and nitrous acid resistance than did spores of the same mutant complemented to express Ssp4 with Gly at residue 36 (i.e., a Ssp4 Gly variant). This result proved that the Ssp4 Asp variant is an important contributor to the exceptional resistance phenotype exhibited by spores made by most type A C- cpe isolates. It was also shown that the exceptional protection afforded spores by the Ssp4 Asp variant apparently involves, at least in part, tighter spore DNA binding by this Ssp4 Asp variant, compared to the Ssp4 Gly variant made by most C. perfringens isolates [15].
The goal of our current study was to characterize further the contributions of the Ssp4 Asp variant to C. perfringens spore resistance properties and to begin examining how C. perfringens regulates expression of the ssp4 gene during sporulation.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. All C. perfringens isolates were stored as stock cultures in Cooked Meat Medium (Sigma) at −20°C. The starter cultures were prepared from those stock cultures by overnight growth at 37°C in fluid thioglycolate broth (FTG) (Difco), as described previously [12], [15]. Sporulating cultures of C. perfringens were then prepared by inoculating 0.2 ml of the overnight FTG culture into 10 ml of Duncan-Strong (DS) sporulation medium [12]. After overnight incubation at 37°C, spores in the DS culture were purified as described previously [12]. Brain heart infusion (BHI) agar was used for plate count analyses [12].
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Sources or Refs |
| C. perfringens | ||
| SM101 | Food poisoning type A isolate, carries a chromosomal cpe gene | [18] |
| F4969 | GI disease type A isolate, carries a plasmid borne cpe gene | [6] |
| CN1793 | Type B isolate, toxigenic | UK, 1947 |
| NCTC8533 | Type B isolate, lamb dysentery | UK, 1950s |
| JGS1495 | Type C isolate, porcine | unknown |
| CN5388 | Type C isolate, human pigbel | unknown |
| CN4003 | Type D isolate, lamb | unknown, 1956 |
| JGS4138 | Type D isolate, goat | USA, 2002 |
| 853 | Type E isolate, calf with enteritis | North America |
| NCIB10748 | Type E isolate, calf with enteritis | Institut Pasteur, reference strain |
| IH101 | spo0A knock-out mutant derivative of SM101 | [17] |
| SM101::ssp4 | ssp4 knock-out mutant derivative of SM101 | [15] |
| F4969::ssp4 | ssp4 knock-out mutant derivative of F4969 | [15] |
| Plasmids | ||
| pDR81 | ssp2 fragment in the antisense direction to the ssp2 promoter | [16] |
| pMRS123 | spo0A ORF and ∼200 bp upstream sequence in pJIR751 | [17] |
| pJIR751 | C. perfringens/E. coli shuttle vector; Ermr | [17] |
| pCS | SM101 ssp4 ORF and ∼300 bp upstream sequence in pJIR751 | [15] |
| pCF | F4969 ssp4 ORF and ∼300 bp upstream sequence in pJIR751 | [15] |
| pD36E | pCS 36 amino acid site-directed mutagenesis D to E | This study |
| pD36N | pCS 36 amino acid site-directed mutagenesis D to N | This study |
| pD36K | pCS 36 amino acid site-directed mutagenesis D to K | This study |
Determination of the ssp4 sequence in non-type A C. perfringens isolates
DNA was isolated from C. perfringens strains CN1794 (type B), NCTC8533 (type B), JGS1495 (type C), CN5388 (type C), CN4003 (type D), JGS4138 (type D), 853(type E) and NCIB10748 (type E) using the MasterPure gram-positive DNA purification kit (Epicentre). The primers ssp4proF and ssp4proR [15] were added (at a 5 µM final concentration) to a PCR mixture containing 1 µl of purified DNA template and 25 µl 2×Taq mixture (NEB), with a total volume of 50 µl. Each sample was then placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 1 cycle of 95°C for 2 min, 35 cycles of 95°C for 30 s, 55°C for 40 s, and 68°C for 40 sec; and a single extension of 68°C for 5 min. The PCR products were cloned into a TOPO vector PCR2.1-TOPO (Invitrogen), which was sent for sequencing to the University of Pittsburgh Genomics Core Sequencing Facility. The ssp4 genes sequences were deposited in GenBank under accession numbers GQ222061 (CN1793); GQ222062 (NCTC8533); GQ222063 (JGS1495); GQ222064 (CN5388); GQ222065 (CN4003); GQ222066 (JGS4138); GQ222067 (strain 853) and GQ222068 (NBIC107481).
Measurement of spore resistance against heat and nitrous acid
The heat and nitrous acid resistance of spores were determined as described previously [13], [14]. Briefly, an aliquot of a DS spore culture was serially diluted in ddH2O, heated at 70°C for 20 min (to kill vegetative cells and promote spore germination), plated onto BHI agar, and incubated anaerobically overnight at 37°C to determine the initial Colony Forming Units (CFU)/ml of spores in the culture. Aliquots of the remaining DS culture were then heat-treated at 70°C for 20 min to kill vegetative cells, followed by a second heating at 100°C. At specified times, aliquots of those heated cultures were diluted and plated onto BHI agar. In other experiments, aliquots of DS cultures were heat-treated at 70°C for 20 min and then suspended in 100 mmol NaNO2, 100 mmol Na Acetate (pH 4.5) at room temperature for 1 h. Aliqouts of those nitrous acid-treated cultures were diluted and plated on BHI agar. After overnight anaerobic incubation (BD GasPack EZ Anaerobe Container systerm) at 37°C, the CFU/ml was counted and those results were used to calculate the decimal reduction times (D values), which is the treatment time needed to cause a 90% reduction in spore CFU/ml.
Site-directed mutagenesis of the ssp4 gene
Three site-directed mutations (D36K, D36E and D36N) were individually introduced into the ssp4 gene that we had separately cloned [15] into both pJIR751 (a C. perfringens-E. coli shuttle plasmid encoding erythromycin-resistance (Emr)) and pTrcHis A (Invitrogen). Each of those mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The reaction parameters were in accordance with the manufacturer's instructions. Each mutation was confirmed by DNA sequencing at the University of Pittsburgh Genomics Core Facility.
Mutated plasmid DNA resulting from each site-directed mutagenesis reaction was then transformed into XL1-Supercompetent Blue E. coli. The shuttle plasmids (named pJIR751-D36K, pJIR751-D36E and pJIR751-D36N) were separately electroporated into the SM101::ssp4 null mutant. Transformants were selected on BHI plates containing 40 mg/L of Em. Spore heat and nitrous acid resistance were then determined, as described above.
pTrcHisA-D36K, pTrcHis-D36E and pTrcHis-D36N were separately transformed into E. coli DH5α. Transformants were selected by growth on LB containing Amp (50 mg/L). The presence of the desired mutated ssp4 gene in each transformant was then confirmed by nucleotide sequencing. Overproduction and nickel affinity purification of each recombinant, His6-tagged rSsp4 mutant was performed as previously described [15]. The purified rSsp4 mutants were then used for Electomobility shift assay (EMSA) analyses as described later.
Comparison of low temperature survival for spores made by wild-type SM101 or F4969, their isogenic ssp4 null mutants, or complementing strains
The cold temperature (4°C or −20°C) resistance of spores produced by wild-type, ssp4 null mutants, or complementing strains of those mutants were determined as described previously [12]. Briefly, sporulating cultures were prepared for each isolate by overnight growth in DS medium. After determining the total number of spores present in an aliquot of each DS medium culture at the start (day 0) of the experiment, the remainder of the DS culture was divided into small tubes, half of which were incubated at 4°C and the other half at −20°C. Aliquots were removed from these small tubes after 6 months and surviving spore numbers (determined as described in a preceding section) were used to calculate the log reduction after each treatment.
Transformation of pDR81 into wild-type SM101 and an isogenic ssp4 mutant
Plasmid pDR81 [16], which encodes an ssp2 antisense gene that can inhibit ssp1, ssp2 and ssp3 transcription [16], was introduced by electroporation [2] into wild-type SM101 or SM101::ssp4. Emr (40 mg/L) transformants were then selected. The resultant SM101 and SM101::ssp4 transformants were designated as SM101 (pDR81) and SM101::ssp4 (pDR81), respectively.
Heat and nitrous acid resistance of spores made by SM101 (pDR81) and SM101::ssp4 (pDR81) were determined as described earlier.
Small acid soluble proteins (SASPs) extraction and Western blotting
To evaluate SASPs presence in spores, C. perfringens SASPs were extracted, as described previously [15], from 50 mg of dry washed spores produced by specified C. perfringens strains. The extracted proteins were subjected to SDS-PAGE and the separated proteins were then transferred onto a nitrocellulose membrane. The resultant blot was probed either with antiserum raised against recombinant C. perfringens Ssp4 [15] or with antiserum raised against a B. subtilis α/β-type SASP named SspC. This SspC antiserum has been shown previously to cross-react with C. perfringens Ssp1, Ssp2 and Ssp3 [16].
Spo0A production
To evaluate Spo0A production, C. perfringens strains were grown for 8 h at 37°C in DS medium and those cultures were then sonicated until >95% of the cells had lysed. Each culture lysate was then analyzed for the presence of Spo0A by Western blot using an antibody specific for B. subtilis Spo0A [17].
Electromobility shift assays (EMSA)
A 3′-biotin-labeled, AT-rich (72.8% AT) C. perfringens DNA sequence was prepared using a biotin 3′-end DNA labeling kit (Pierce) as described previously [15]. Similarly, a 3′-biotin-labeled, GC-rich probe was prepared consisting of a 55 bp sequence from a GC-rich (69.1% GC) C. perfringens genomic DNA sequence. For this purpose, the following two oliogonucleotides were synthesized (Integrated DNA Technologies, Coralville, IA): Label-D2 (5′CTGGCGACTCAGAAGGGGCTCGAACCCTCGACCTCCG-GCGTG-ACAGGCCGGCACT-3′) and Label-R2 (5′-AGTGCCGGCCTGTCACGCCGGAG-GTCGAGGGTTCGAGCCCCTTCTGAGTCGCCAG-3′) and 3′-biotin-end-labeled by the manufacturer's instructions using a biotin 3′-end DNA labeling kit (Pierce).
The AT-rich probe was used in a modified EMSA to compare the DNA binding of the three site-directed, His6-tagged rSsp4 mutants. AT-rich and GC-rich probes were used in a modified EMSA, as described previously [15], to compare rSsp4 and rSsp2 binding preferences. Briefly, 1 µl of probe was incubated with 50, 100 or 200 ng of purified His6-tagged rSsp at 37°C for 1 h. Bound rSsp was then fixed to the DNA probe by the addition of glutaraldehdye (final concentration of 0.01% (v/v)) by 15 min incubation at 37°C. Those mixtures were loaded into a 6% polyacrylamide gel and electrophoresed in 0.5×TBE (Tris-borate-EDTA) buffer at 4°C for 1 h. DNA-protein complexes were then transferred to a positive charge nylon membrane (Roche Applied Science), UV crosslinked and detected with a LightShift Chemiluminescent EMSA kit (Pierce).
Results
Comparison of the Ssp4 sequence and spore heat- and nitrite-resistance properties amongst non-type A C. perfringens strains
Our initial Ssp4 study [15] found that the Ssp4 protein produced by 13 different type A strains shares an identical sequence, except for variations at amino acids 36 and 72. Those 13 type A isolates produced an Ssp4 with either Gly or Asp at residue 36 and either Asn or Lys at residue 72. As described in the Introduction, the presence of Asp at Ssp4 residue 36 was shown to be important for helping to mediate the exceptional spore resistance properties exhibited by most type A C-cpe FP isolates.
To further evaluate Ssp4 sequence diversity amongst C. perfringens isolates, the current study sequenced the ssp4 ORF carried by eight strains belonging to C. perfringens type B, C, D or E (Tables 1 and 2). Those analyses revealed the presence of an identical ssp4 ORF in all eight surveyed non-type A isolates. Furthermore, the ssp4 ORF sequence present in these eight type B, C, D and E isolates identically matched the ssp4 sequence found in type A isolates (e.g. F4969) producing an Ssp4 with Gly present at residue 36 and Lys present at residue 72.
Table 2. Ssp4 sequence and spore resistance in various C. perfringens types.
| Strain | Types | Toxin gene | 3 6 res | 72 res | Heat resistance (min) | Chemical resistance (log reduction) |
| SM101 | A (FP) | cpe (chrom) | D | N | 59.1±1.3 | 1.1±0.4 |
| F4969 | A (NFP) | cpe (plasmid) | G | K | 0.5±0.0 | 4.0±0.5 |
| CN1794 | B | cpb, etx, cpb2 | G | K | ND | ND |
| NCTC8533 | B | cpb, etx, cpb2 | G | K | 1.4±0.5 | 4.2±0.7 |
| JGS1495 | C | cpb, cpb2 | G | K | NA | NA |
| CN5388 | C | cpb, cpe, cpb2 | G | K | 2.3±0.3 | 4.0±0.1 |
| CN4003 | D | etx, cpe, cpb2 | G | K | ND | ND |
| JGS4138 | D | etx, cpe, cpb2 | G | K | 2.7±0.6 | 5.3±0.1 |
| 853 | E | iota, cpe | G | K | ND | ND |
| NBIC10748 | E | iota, cpe, cpb2 | G | K | ND | ND |
The presence of the same ssp4 sequence in F4969 and the eight surveyed non-type A isolates suggested that the spores produced by non-type A isolates might resemble the spores made by F4969 in terms of their heat- and nitrous acid-sensitivity. This hypothesis was tested by phenotyping the spores produced by a type B, C and D isolate for their ability to withstand boiling and nitrous acid (no type E isolate in our collection produced suitable levels of spores to conduct phenotype analyses). Results of those experiments showed that, relative to the resistant spores made by most type A C-cpe FP isolates (e.g. SM101), the spores of the three tested non-type A isolates of C. perfringens exhibited significantly more sensitivity to heat and nitrous acid (Table 2). Furthermore, the resistance properties determined for spores made by non-type A isolates closely matched those of spores produced by P-cpe isolates (e.g. F4969) (Table 2).
Saturation mutagenesis of the SM101 ssp4 gene at the codon encoding Asp residue 36 and phenotyping the spore resistance properties of those Ssp4 mutants
The Table 2 results supported previous results [15] demonstrating that variations at Ssp4 residue 36 are important for the heat- and nitrous acid-resistance properties of C. perfringens spores. Specifically, those C-cpe isolates (e.g. SM101, a transformable derivative of FP isolate NCTC 8798 [18]) forming exceptionally resistant spores make an Ssp4 with Asp at residue 36, while this Ssp4 residue is Gly in heat- and nitrous acid-sensitive C. perfringnes spores, including F4969 and the non-type A isolates phenotyped in Table 2.
To evaluate which amino acid properties at Ssp4 residue 36 are important for helping to mediate the exceptional resistance phenotype of spores produced by most C-cpe FP isolates, site-directed mutagenesis was performed on the SM101 ssp4 gene cloned into either the pJIR751 shuttle plasmid (to allow testing of spore phenotypes) or the pTrcHis plasmid (to allow testing of DNA binding properties of rSsp4 mutants, which can be easily purified by nickel affinity chromatography due to their N-terminal, vector-encoded His6 sequence). These mutagenesis reactions created Ssp4 or rSsp4 variants where the natural Asp (D) present at residue 36 of the SM101 Ssp4 had been replaced by Glu (E), Lysine (K) or Asn (N).
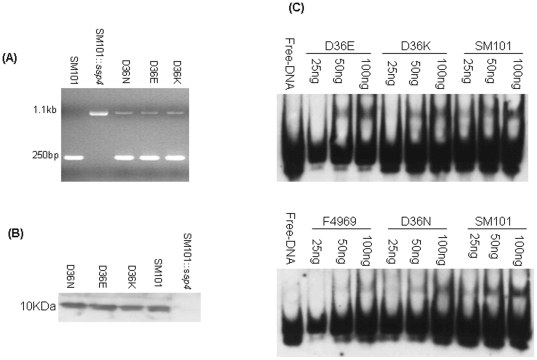
The pTrcHis plasmids carrying each rSsp4 mutant were separately transformed into E. coli, while the pJIR751 plasmids carrying each mutant ssp4 gene were separately transformed into a previously-created SM101 ssp4 null mutant (SM101::ssp4) [15]. The presence in each transformant of a plasmid carrying mutated ssp4 sequences was demonstrated by PCR (Fig. 1A) and the presence of the desired ssp4 ORF mutation in the transformant was confirmed by sequencing (not shown). Production of each Ssp4 mutant (Fig. 1B) or rSsp4 mutant (not shown) was demonstrated by Ssp4 Western blotting.
Figure 1. Site-directed mutagenesis of SM101 Ssp4 at residue 36 to change the natural codon encoding Asp (D) to encode Glu (E), Lysine (K) or Asn (N).
(A) Ssp4 ORF PCR results for: wild-type SM101; SM101::ssp4, SM101::ssp4 transformed with pJIR751 carrying a mutant ssp4 encoding one of the three site-directed Ssp4 mutants, i.e., D36N, D36E or D36K; (B) Western blot confirmation of expression of the Ssp4 D36N, D36E and D36K mutants. (C) DNA binding properties of purified recombinant His6-tagged rSsp4D36E, rSsp4D36K or rSsp4D36N and wild-type rSsp4 from SM101 or F4969. Top panel: Lane 1, free biotin-labeled C. perfringens DNA; lanes 8–10, indicated amounts of SM101 wild-type rSsp4 incubated with C. perfringens biotin-labeled DNA; lanes 2–4, indicated amounts of rSsp4D36E incubated with C. perfringens biotin-labeled DNA; lanes 5–7, indicated amounts of rSsp4D36K incubated with C. perfringens biotin-labeled DNA. Bottom panel: Lane 1, free biotin-labeled C. perfringens DNA lanes 2–4, indicated amounts of F4969 wild-type rSsp4 incubated with C. perfringens biotin-labeled DNA; lanes 5–7, indicated amounts of rSsp4D36N incubated with C. perfringens biotin-labeled DNA.
Experiments were then performed to evaluate the resistance properties of spores produced by SM101::ssp4 mutants after those null mutants had been complemented to express each Ssp4 variant with a residue 36 mutation. Control phenotypic comparisons first confirmed our previous observations [15] that inactivating the ssp4 gene in SM101 reduced spore heat and nitrous acid resistance properties (Table 3). As we had also reported previously [15], complementation of the SM101::ssp4 mutant to enable expression of Ssp4 with a wild-type Asp at residue 36 (SM101::ssp4-pCS) resulted in spores exhibiting exceptional resistant to both heat and nitrous acid. Similar complementation of this SM101::ssp4 mutant so it expressed wild-type F4969 Ssp4 with a Gly at residue 36 (SM101::ssp4-pCF) resulted in a much more limited increase in spore heat and nitrous acid resistance properties.
Table 3. Heat and chemical resistance of SM101 transformants producing site-directed mutants.
| SM101 | SM101::ssp4 | SM101::ssp4- pCS | SM101::ssp4- pCF | SM101::ssp4 -pD36E | SM101::ssp4- -pD36N | SM101::ssp4 -pD36K | |
| Heat resistance (D value) (min) | 59.1±1.3 | 8.7±1.9 | 44.7±1.8 | 16.4±0.6 | 41.9±2.6 | 24.0±1.7 | 40.8±1.3 |
| Chemical Resistance (log reduction) | 1.1±0.4 | 4.0±0.1 | 1.1±0.6 | 3.2±0.1 | 1.2±0.3 | 2.2±0.2 | 1.6±0.2 |
When the resistance properties of spores made by SM101::ssp4 transformants expressing the Ssp4D36E or Ssp4D36K mutants were tested (Table 3), those spores exhibited similar heat and nitrous acid resistance properties as SM101::ssp4-pCS spores containing the wild-type Ssp4 Asp variant. However, the spores produced by SM101::ssp4 transformants expressing the Ssp4D36N mutant had lower resistance against both heat and nitrous acid. Spores containing the Ssp4D36N mutant exhibited similar resistance properties as spores made by SM101::ssp4 transformed to produce the wild-type F4969 Ssp4 Gly variant.
To assess whether the resistance properties of spores containing Ssp4 with a site-directed mutation at residue 36 correlated with the DNA binding properties of their Ssp4 variant, wild-type SM101 rSsp4, the SM101 rSsp4 site-directed mutants, and wild-type F4969 rSsp4 were each purified and tested (Fig. 1C) for their DNA binding properties using an EMSA assay. Control EMSA analyses confirmed our previous report [15] that wild-type SM101 rSsp4 (with an Asp at residue 36) binds strongly to an AT-rich DNA probe, while F4969 rSsp4 (with a Gly at residue 36) binds less well to this DNA probe (Fig. 1C). Amongst the three SM101 rSsp4 variants with a mutated residue 36 amino acid, as created by site-directed mutagenesis for this study, Ssp4D36E and Ssp4D36K exhibited similar binding to the AT-rich DNA probe as did native SM101 rSsp4 with an Asp at residue 36, while Ssp4D36N exhibited weaker DNA binding.
Ssp4 preferentially binds to AT-Rich DNA
Results from Fig. 1 and our previous study [15] have demonstrated that Ssp4 can bind to an AT-rich probe mimicking C. perfringens DNA, which has a high (∼72%) overall AT content. Interestingly, the small acid soluble protein named SspC made by Bacillus spp. (another Gram-positive, sporeforming bacteria with low-GC% DNA) reportedly binds better to GC-rich DNA compared to AT-rich DNA [19], [20].
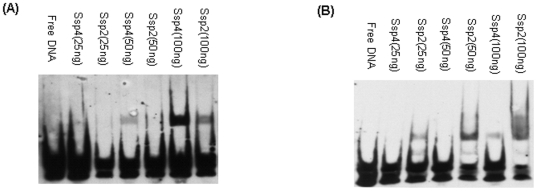
Therefore, the current study performed an EMSA analysis to compare the binding of C. perfringens SM101 rSsp4 to AT-rich vs. GC-rich DNA probes. The current study also examined the binding of C. perfringens SM101 rSsp2 (which has a very similar sequence to Bacillus Ssp, as well as C. perfringens Ssp1 and Ssp3) to the same AT-rich vs. GC-rich DNA probes. These EMSA analyses revealed (Fig. 2) that purified rSsp2 binds preferentially to a probe containing a GC-rich (69.1% GC) sequence of C. perfringens DNA vs. a probe containing an AT-rich (72.8% AT) sequence of C. perfringens DNA. In contrast, purified rSsp4 exhibited a binding preference for the AT-rich vs. the GC-rich C. perfringens DNA sequence (Fig. 2).
Figure 2. EMSA analysis of purified SM101 rSsp4 and rSsp2 binding to C. perfringens AT-rich biotin-labeled DNA (A) or CG-rich biotin-labeled DNA (B).
Free DNA, biotin-labeled C. perfringens DNA (no rSsp); Lane 2–6, 25–100 ng of rSsp4 or rSsp2, as indicated, incubated with a AT-rich (Panel A) or GC-rich (panel B) C. perfringens biotin-labeled DNA probe.
Ssp4 works in concert with other small acid soluble proteins (Ssp1, Ssp2, Ssp3) for maximal protection of C. perfringens spores against heat and nitrous acid
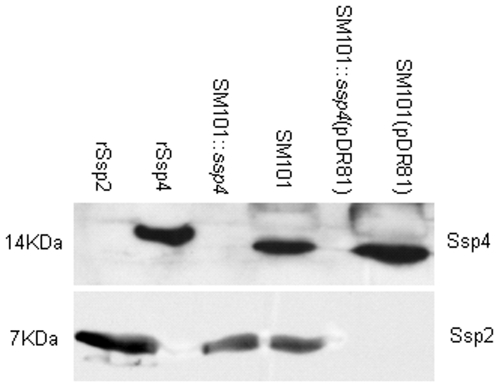
As mentioned above, C. perfringens produces at least four small acid soluble proteins. These include three proteins, named Ssp1–3, that differ from Ssp4 but share substantial sequence similarity with one another [16], [21], [22]. To explore why this bacterium produces so many different SASPs, an ssp2 antisense plasmid was transformed into our SM101::ssp4 null mutant. This antisense plasmid was shown previously to simultaneously block expression of Ssp1, Ssp2 and Ssp3 [16]. Consistent with those previous observations, Western blot analyses showed that neither SM101::ssp4 (pDR81) nor SM101(pDR81) produced Ssp1, Ssp2 or Ssp3 proteins (Fig. 3), although SM101(pDR81) still produced Ssp4.
Figure 3. Western blot analyses of purified rSsp2 or rSsp4 (10 ng) wild-type SM101, an SM101 ssp4 null mutant (SM101::ssp4), a wild-type SM101 transformant carrying the pDR81 plasmid encoding antisense RNA to inhibit production of Ssp1–3 (SM101(pDR81)) or the SM101 ssp4 null mutant transformed to carry the pDR81 plasmid (SM101::ssp4(pDR81)).
Top panel, Western blot probed with antiserum raised previously [15] against purified rSsp4; Bottom panel, Western blot probed with antiserum raised previously [16] against B. subtilis SspC, which is known to cross-react with C. perfringens Ssp1–3.
When heat and nitrous acid resistance were compared, wild-type SM101 spores exhibited about 7-fold higher resistance than spores of the ssp4 null mutant (Table 4). Although the SM101(pDR81) transformant still produced Ssp4, its spores showed a reduced heat resistance compared against wild type spores, indicating that (in addition to Ssp4) Ssp1, Ssp2 and Ssp3 are also important for the full development of SM101 spore heat resistance. Importantly, SM101::ssp4(pDR81), which does not produce any of the four known C. perfringens Ssps, produced spores with virtually no heat resistance at 100°C.
Table 4. Heat and chemical resistance of SM101 or SM101::ssp4 with or without the pDR81 antisense plasmid.
| SM101 | SM101(pDR81) | SM101::ssp4 | SM101::ssp4 (pDR81) | |
| Heat resistance D value (min) | 59.1±1.3 | 18.1±2.7 | 8.7±1.9 | 1.4±0.1 |
| Chemical Resistance (log reduction) | 1.1±0.4 | 2.5±0.3 | 4.0±0.1 | 4.8±0.7 |
Spore nitrous acid resistance properties for these C. perfringens strains showed a similar pattern of differences as described above for spore heat resistance differences (Table 4). Spores of wild-type SM101 exhibited a nitrous acid-induced log reduction in viability of only 1.1, while nitrous acid caused a 4 log reduction in spore viability for SM101::ssp4, confirming a role for Ssp4 in SM101 spore nitrous acid resistance. Decreased production of Ssp1, Ssp2 and Ssp3 also reduced SM101 spore resistance against nitrous acid, although this 2.5 log reduction in spore viability was less than observed after ssp4 gene inactivation. The strongest reduction in SM101 spore nitrous acid resistance (a 4.8 log reduction) was observed for SM101::ssp4 (pDR81), which does not produce any of the known C. perfringens Ssps. Collectively, these results (Table 4) indicate that Ssp4 works in combination with the three other Ssps to protect C. perfringens spores against heat and nitrous acid treatment.
Low temperature survival of spores produced by wild-type SM101 and F4969, their ssp4 null mutants and complemented strains
As confirmed in Table 2, inactivation of the ssp4 gene in SM101 or F4969 causes these isolates to produce spores with considerably less heat- and nitrous acid-resistance than their corresponding wild-type spores. In addition to heating and use of preservatives, storage of foods at low temperature (in refrigerators or freezers) is a very important food safety approach for controlling C. perfringens type A FP [12]. Therefore, the current study evaluated the involvement of Ssp4 for C. perfringens spore survival at 4°C and −20°C.
This study first confirmed previous conclusions [12] that wild-type SM101 spores exhibit exceptional survival at low temperatures, with only a 0.35 and 0.58 log reduction in spore viability measured after 6 months of storage at 4°C or −20°C, respectively (Table 5). Table 5 also shows, for the first time, that specific inactivation of the ssp4 gene in SM101 reduced spore viability upon low temperature storage, with 0.82 or 1.91 log reduction in spore viability measured after 6-month storage of ssp4 null mutant spores at 4°C or −20°C, respectively. These spore survival differences between wild type SM101 and its isogenic ssp4 null mutant were statistically significant (P<0.01) at both 4°C and −20°C. Demonstrating that the decreased low temperature resistance of spores made by the SM101 ssp4 null mutant was specifically due to inactivation of the ssp4 gene, complementation of this mutant with a shuttle plasmid carrying the wild-type SM101 ssp4 gene was able to substantially increase 6-month spore survival at both 4°C and −20°C. In contrast, transformation of the SM101::ssp4 mutant with a shuttle plasmid carrying the wild-type F4969 ssp4 gene more modestly increased 6-month spore survival of the SM101 ssp4 null mutant upon storage at low temperatures.
Table 5. Cold resistance of wild-type, ssp4 null mutants and complementing strains of SM101 and F4969.
| Stains | 4°C (log reduction after 6 month) | −20°C (log reduction after 6 month) |
| F4969 | 0.88±0.13 | 1.23±0.11 |
| F4969::ssp4 | 2.02±0.15 | 3.12±0.10 |
| F4969::ssp4(pCS) | 0.70±0.10 | 1.01±0.33 |
| F4969::ssp4(pCF) | 1.30±0.20 | 1.81±0.10 |
| F4969::ssp4(pJIR751) | 1.82±0.10 | 2.70±0.40 |
| SM101 | 0.35±0.10 | 0.58±0.17 |
| SM101::ssp4 | 0.82±0.18 | 1.91±0.35 |
| SM101::ssp4(pCS) | 0.40±0.13 | 0.65±0.20 |
| SM101::ssp4(pCF) | 0.50±0.14 | 1.22±0.56 |
| SM101::ssp4(pJIR751) | 1.03±0.42 | 1.86±0.67 |
Consistent with previous observations, wild-type F4969 spores exhibited poorer survival at 4°C or −20°C compared to wild-type SM101 spores (Table 5). Table 5 also shows that inactivation of the ssp4 gene in strain F4969 substantially decreased 6-month spore survival at both 4°C and −20°C. Spores of the F4969 ssp4 null mutant exhibited much better 6-month survival at these low temperatures when they were complemented with a shuttle plasmid carrying the SM101 ssp4 gene (Table 2). In contrast, complementation with the same shuttle plasmid carrying the wild-type F4969 ssp4 gene caused a lesser increase in 6-month spore survival of the F4969 null mutant upon storage at either 4°C or −20°C.
Collectively, these results (Table 5) demonstrate that Ssp4 is important for spore survival at low temperatures and that the SM101 Ssp4 variant is better at protecting spores against low temperature-induced lethality than F4969.
The role of Spo0A in regulating ssp4 expression
Our previous studies [21], [22] have shown that production of Ssp1–3 requires a C. perfringens isolate to possess a functional spo0A gene. Therefore, the current study investigated whether Ssp4 expression is also Spo0A-dependent.
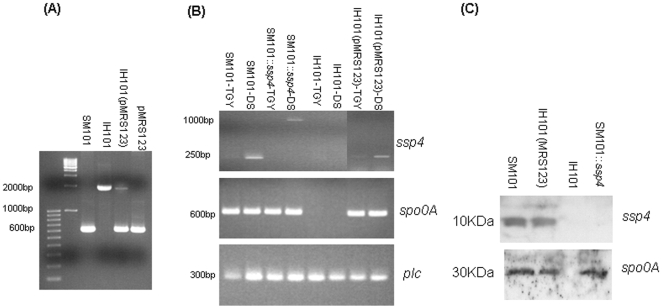
We first confirmed that C. perfringens IH101, a previously prepared spo0A null mutant of SM101 that cannot form spores [17], does not produce Spo0A. When Western blotting was used to compare by wild-type SM101 versus IH101 grown in Duncan-Strong (DS) sporulation medium (Fig. 4), the results obtained showed that DS cultures of SM101 and SM101::ssp4 both produce Spo0A, but DS cultures of IH101 do not produce Spo0A. Confirming that this phenotype was specifically due to inactivation of the spo0A gene in IH101, complementation of IH101 with a plasmid carrying the wild-type spo0A gene restored Spo0A expression.
Figure 4. The role of Spo0A in regulation of Ssp4 production.
(A) Total DNA isolated from SM101, spo0A null mutant IH101, and the complementing strain IH101(pMRS123), or plasmid pMRS123 alone, was subjected to PCR analysis using spo0A-specific internal primers CPP68 and CPP69 [17]. Note the larger size of the PCR product in IH101 due to insertional inactivation by chloramphenicol resistance gene catP. (B) Total RNA was isolated from SM101, SM101::ssp4, IH101 or IH101(pMRS123) grown as vegetative cultures in TGY or in DS sporulation-inducing medium, and then subjected to RT-PCR analysis using ssp4, spo0A or plc primers, as indicated. (C) Western blot analysis of Ssp4 and Spo0A production by SM101, IH101(MRS123), IH101 or SM101::ssp4 grown in DS medium.
As expected from our previous studies [15], Ssp4 was produced by DS cultures of wild-type SM101 but not by SM101::ssp4 [15]. As also reported previously [15], expression of Ssp4 by DS cultures of SM101::ssp4 was restored by complementation with a shuttle plasmid carrying the wild-type ssp4 gene. We now show that DS cultures of IH101 failed to produce Ssp4. However, DS cultures of the complementing strain IH101 (MRS123) did show Ssp4 expression, thereby demonstrating that Spo0A is specifically required for Ssp4 production.
Discussion
C. perfringens FP isolates generally possess two complimentary virulence traits, i.e, production of CPE and the ability to form spores that are highly resistant to food environment stresses, such as heat, low temperatures and preservatives such as nitrites [1]. Since our previous study [15] had identified a novel Ssp4 variant as a major contributor to the spore resistance properties of FP isolates, the current study sought to better characterize Ssp4 proteins and to begin exploring the regulation of Ssp4 expression.
Our previous study [15] had determined that the C. perfringens type A FP isolates forming resistant spores usually produce an Ssp4 with Asp at residue 36, while other type A C. perfringens isolates, including both P-cpe isolates and cpe-negative isolates, typically make an Ssp4 with Gly at residue 36. Cross-complementation approaches with ssp4 null mutants have also directly demonstrated that, 1) Ssp4 is an important mediator of spore resistance against heat and nitrous acid, and 2) the Ssp4 variant with Asp at residue 36 is better than the Ssp4 variant with Gly at residue 36 at protecting spores against heat and nitrites acid. In another previous study [12], we had shown that the spores of type A C-cpe isolates also exhibit exceptional survival at low temperatures, i.e., at 4°C and −20°C. The current study now reports that the strong low temperature resistance phenotype of spores made by FP isolates also involves Ssp4 and that the Ssp4 Asp variant is more against low temperature than the Ssp4 Gly variant. The exceptional low temperature resistance exhibited by spores containing the Ssp4 Asp variant is a likely contributor to C. perfringens FP transmission since meat and poultry products, common food vehicles for C. perfringens FP, are known to be contaminated with resistant spores of C-cpe isolates and are typically stored in refrigerators or freezers.
Determining that type A, C-cpe FP isolates typically produce spores whose resistance phenotype is mediated, in large part, by the Ssp4 Asp variant, while type A, P-cpe isolates produce spores whose sensitivity involves the Ssp4 Gly variant provided one explanation for the strong association between type A C-cpe isolates and FP. However, about 15% or 25% (respectively) of C. perfringens type C and D isolates also carry a cpe gene [23], [24] from which they produce similar amounts of an identical CPE protein as type A cpe-positive isolates. Interestingly, those cpe-positive type C or D isolates rarely, if ever, cause human FP. Results from the current study suggest that cpe-positive type C or D isolates may not commonly be involved in FP, at least in part, because they form spores that are sensitive to food environment stresses such as heating and preservatives. The cpe-positive type C and D isolates surveyed in this study also produce the same Ssp4 Gly variant as found in type A isolates producing sensitive spores. Since this same Ssp4 variant has been established as a major contributor to the sensitivity of type A isolates producing sensitive spores, the Ssp4 Gly variant is also likely to be an important factor behind the spore sensitivity of cpe-positive type C and D isolates, although this should be experimentally confirmed.
The current study also demonstrated that Ssp4 works in combination with Ssp1–3 to produce maximal spore resistance properties for C. perfringens type A C-cpe FP isolates. Furthermore, this work found that Ssp4 exhibits a preference for AT-rich DNA sequences, in contrast to Ssp2 (and most likely Ssp1 and 3 based upon their sequence similarities to Ssp2), which prefers binding to GC-rich DNA. This diversity in sequence binding preferences may help to explain why C. perfringens makes several Ssps. i.e., by producing multiple Ssps that bind to different chromosomal regions depending on their local AT% ratio, the entire chromosome can be maximally protected from damage induced by stresses such as heat or preservatives.
The properties of Ssp4 residue 36 that mediate spore resistance were also examined in the current study. These analyses revealed that spores retain exceptional heat and nitrous acid resistance if Lys or Glu were substituted for the Asp naturally found at Ssp4 residue 36 in C. perfringens type A FP isolates forming highly resistant spores. However, changing Ssp4 residue 36 from Asp to Asn produced sensitive spores resembling those made by isolates producing Ssp4 with Gly at residue 36. These results suggest that both the side chain length and presence of a charge at Ssp4 residue 36 may be important for mediating spore resistance properties. The strong resistance phenotype exhibited by spores carrying the mutant Ssp4 correlated with DNA binding properties of the corresponding purified rSsp4 mutant, supporting previous suggestions [15] that the DNA binding properties of Ssp4 variants are important determinants of spore resistance properties.
A bioinformatics search of Genbank revealed that other Clostridium spp. naturally carry ORFs encoding putative Ssp homologues with Glu (Cac4 of Clostridium acetobutylicum ATCC824, CnoI of Clostridium noyvi NT), Asp (Cac2 of Clostridium acetobutylicum ATCC824, Ssp2 of C. perfringens SM101, Cno2 of Clostridium noyvi NT, Cte2 of Clostridium tetani), Lys (Cac1 of Clostridium noyvi NT) or Gly (Ssp4 of C. perfringens ATCC3624) at the equivalent position as SM101 Ssp4 residue 36. The site-directed mutagenesis results of the current study might suggest that C. acetylobutylicum ATCC824, C. novyi NT and C. tetani E88 spores would exhibit substantial resistance against stresses such as heat, low temperature and food preservatives. However, this should be evaluated experimentally for two reasons. First, these isolates all carry ORFs encoding several different putative Ssps and it is clear from the current and previous studies [15] that no single Ssp fully determines spore resistance properties. Second, it is clear that Ssp4 variants are not the only contributor to variations in C. perfringens spore resistance properties. For example, the size of the spore core may also influence variations in C. perfringens spore resistance [25], [26]. It would also be of interest to evaluate whether other clostridial species exhibit intraspecies Ssp4 variants that influence spore resistance phenotypes, as occurs with C. perfringens.
Finally, the current results [22] revealed that, like Ssp1–3, expression of Ssp4 requires Spo0A. Spo0A is a master regulator of many genes expressed during sporulation [27] and late-stationary phase, so its involvement in Ssp4 expression is consistent with our previous finding that Ssp4 production is strongly sporulation-associated. Some Spo0A-regulated genes are regulated by Spo0A binding to sequences (0A boxes) located upstream of the ORF [27], [28]. A bioinformatics search detected potential 0A boxes upstream of all four C. perfringens ssp genes, including ssp4, in SM101. However, Ssp expression in C. perfringens may not only involve Spo0A regulation, as these bioinformatics searches also identified potential SigK binding sites upstream of the ssp1 and ssp4 genes of SM101. The presence of those SigK boxes could suggest that SigK, an alternative sigma factor, is also involved in the sporulation-associated regulation of some ssp genes. Further studies are underway to better understand how C. perfringens regulates Ssp expression.
Acknowledgments
The authors thank Dr. Masaya Fujita for supplying Spo0A antibody.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was generously supported by grant 2005-53201-15387 from the Ensuring Food Safety Program of the United States Department of Agriculture (to BAM) and by grant R37AI19844 from the National Institute of Allergy and Infectious Diseases (to BAM) and by grant from Army Research Office (to MRS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McClane BA. Clostridium perfringens in Food Microbiology Fundamentals and Frontiers (3rd Edition); In: Doyle MP, Beuchat LR, editors. Washington D.C.: ASM press; 2007. pp. 423–444. [Google Scholar]
- 2.Sarker MR, Carman RJ, McClane BA. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Molec Microbiol. 1999;33:946–958. doi: 10.1046/j.1365-2958.1999.01534.x. [DOI] [PubMed] [Google Scholar]
- 3.Adak GK, Long SM, O'Brien SJ. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut. 2002;51:832–841. doi: 10.1136/gut.51.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M, Painter J, Woodruff R, Braden C. Surveillance for foodborne-disease outbreaks - United States, 1998–2002. Morbidity and Mortality Weekly Report, CDC. 2006;55:1–42. [PubMed] [Google Scholar]
- 5.Cornillot E, Saint-Joanis B, Daube G, Katayama S, Granum PE, et al. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Molec Microbiol. 1995;15:639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 6.Collie RE, McClane BA. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. J Clin Microbiol. 1998;36:30–36. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparks SG, Carman RJ, Sarker MR, McClane BA. Genotyping of enterotoxigenic Clostridium perfringens isolates associated with gastrointestinal disease in North America. J Clin Microbiol. 2001;39:883–888. doi: 10.1128/JCM.39.3.883-888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyamoto K, Wen Q, McClane BA. Multiplex PCR genotyping assay that distinguishes between isolates of Clostridium perfringens type A carrying a chromosomal enterotoxin gene (cpe) locus, a plasmid cpe locus with an IS1470-like sequence or a plasmid cpe locus with an IS1151 sequence. J Clin Microbiol. 2004;41:1552–1558. doi: 10.1128/JCM.42.4.1552-1558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant K, Kenyon S, Nwafor I, Plowman J, Ohai C, et al. The identification and characterization of Clostridium perfringens by real-time PCR, location of enterotoxin gene, and heat resistance. Foodborne Pathog Dis. 2008;5:629–639. doi: 10.1089/fpd.2007.0066. [DOI] [PubMed] [Google Scholar]
- 10.Lahti P, Heikinheimo A, Johansson T, Korkeala H. Clostidium perfringens type A isolates carrying the plasmid-borne enterotocin gene (genotypes IS1151-cpe or IS1470-like-cpe) are a common cause of food poisonings. J Clin Microbiol. 2007;46:371–373. doi: 10.1128/JCM.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen Q, McClane BA. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl Environ Microbiol. 2004;70:2685–2691. doi: 10.1128/AEM.70.5.2685-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, McClane BA. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl Environ Microbiol. 2006;72:4561–4568. doi: 10.1128/AEM.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, McClane BA. Comparative effects of osmotic, sodium nitrite-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl Environ Microbiol. 2006;72:7620–7625. doi: 10.1128/AEM.01911-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarker MR, Shivers RP, Sparks SG, Juneja VK, McClane BA. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl Environ Microbiol. 2000;66:3234–3240. doi: 10.1128/aem.66.8.3234-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Li J, McClane BA. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathogens. 2008;4:e1000056. doi: 10.1371/journal.ppat.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raju D, Setlow P, Sarker MR. Antisense-RNA-mediated decreased synthesis of small, acid-soluble spore proteins leads to decreased resistance of Clostridium perfringens spores to moist heat and UV radiation. Appl Environ Microbiol. 2007;73:2048–2053. doi: 10.1128/AEM.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang IH, Waters M, Grau RR, Sarker MR. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin productioin in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol Lett. 2004;233:233–240. doi: 10.1016/j.femsle.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Melville SB. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J Bacteriol. 1998;180:136–142. doi: 10.1128/jb.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Setlow P. I will survive: DNA protection in bacterial spore. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Tennen R, Setlow B, Davis KL, Loshon CA, Setlow P. Mechanisms of killing of spores of Bacillus subtilis by iodine, glutaraldehyde and nitrous acid. J Appl Microbiol. 2000;89:330–338. doi: 10.1046/j.1365-2672.2000.01114.x. [DOI] [PubMed] [Google Scholar]
- 21.Raju D, Waters M, Setlow P, Sarker MR. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC Microbiol. 2006;8:50. doi: 10.1186/1471-2180-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raju D, Sarker MR. Production of small, acid-soluble spore proteins in Clostridium perfringens nonfoodborne gastrointestinal disease isolates. Can J Microbiol. 2007;53:514–518. doi: 10.1139/W07-016. [DOI] [PubMed] [Google Scholar]
- 23.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, et al. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun. 2006;74:5200–5210. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayeed S, Li J, McClane BA. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect Immun. 2007;75:2391–2398. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novak JS, Juneja V, McClane BA. An ultrastructural comparison of spores from various strains of Clostridium perfringens and correlations with heat resistance parameters. Int J Food Microbiol. 2003;86:239–247. doi: 10.1016/s0168-1605(02)00550-0. [DOI] [PubMed] [Google Scholar]
- 26.Orsburn B, Melville SB, Popham D. Factors contributing to heat resistance of Clostridium perfringens endospores. Appl Environ Microbiol. 2008;74:3328–3335. doi: 10.1128/AEM.02629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, et al. The Spo0A regulon of Bacillus subtilis. Molec Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 28.Greene AE, Spiegelman GB. The Spo0A protein of Bacillus subtilis inhibits transcription of the abrB gene without preventing binding of the polymerase to the promoter. J Biol Chemistry. 1996;271:11455–11461. doi: 10.1074/jbc.271.19.11455. [DOI] [PubMed] [Google Scholar]