SUMMARY
Background
In childhood acute lymphoblastic leukemia (ALL) genetic subtypes are recognized that determine the risk-group for further treatment. However, 25% of precursor BALL are currently genetically unclassified and have an intermediate prognosis. The present study used genome-wide strategies to reveal new biological insights and advance the prognostic classification of childhood ALL.
Methods
A classifier based on gene expression in ALL cells from 190 newly diagnosed pediatric cases was constructed using a double-loop cross-validation method and next, was validated on an independent cohort of 107 newly diagnosed pediatric ALL cases. Hierarchical cluster analysis using classifying gene probe sets then revealed a novel ALL subtype for which underlying genetic abnormalities were characterized by comparative genomic hybridization-arrays and molecular cytogenetics.
Findings
The prediction accuracy of the classifier was median 90% in the discovery cohort and 87.9% in the independent validation cohort. A significant part of the currently genetically unclassified cases clustered with BCR-ABL-positive cases in both the discovery and validation cohort. These BCR-ABL-like cases represent 15–20% of ALL cases and have a highly unfavorable outcome (5-year disease-free survival 59.5%, 95%CI: 37.1%–81.9%) compared to other precursor B-ALL cases (84.4%, 95%CI: 76.8%–92.1%; P=0.012), similar to the poor prognosis of BCR-ABL-positive ALL (51.9%, 95%CI: 23.1%–80.6%), as was confirmed in the validation cohort. Further genetic studies revealed that the BCR-ABL-like subtype is characterized by a high frequency of deletions in genes involved in B-cell development (82%), including IKAROS, E2A, EBF1, PAX5 and VPREB1, compared to other ALL cases (36%, p=0.0002). BCR-ABL-like leukemic cells were median >70-times resistant to L-asparaginase (p=0.001) and 1.6-times more resistant to daunorubicin (p=0.017) compared to other precursor B-ALL cases whereas the toxicity of prednisolone and vincristine did not significantly differ.
Interpretation
Classification by gene expression profiling identified a novel subtype of ALL not detected by current diagnostic procedures but which comprises the largest group of patients with a high-risk of treatment failure. New treatment strategies are needed to improve outcome for this novel high-risk subtype of ALL.
Funding
Dutch Cancer Society, Sophia Foundation for Medical Research, Pediatric Oncology Foundation Rotterdam, Center of Medical Systems Biology of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research, American National Institute of Health, American National Cancer Institute and American Lebanese Syrian Associated Charities.
Keywords: microarray, gene expression profiling, classification, genotype, novel subtype, class discovery, ALL
INTRODUCTION
In childhood acute lymphoblastic leukemia (ALL) several prognostically unfavorable subgroups are known: T-lineage ALL (T-ALL; ~15% of all cases) and the precursor B-lineage subtypes with chromosomal translocations creating MLL-rearrangements or the BCR-ABL gene fusion, each found in <5% of cases.1–3 Prognostically favorable precursor B-subtypes are TEL-AML1-positive ALL (20–25% of cases), hyperdiploid ALL (>50 chromosomes; ~25% of cases) and E2A-rearranged ALL (often E2A-PBX1-positive; ~5% of cases). About 25% of cases are currently genetically unclassified ALL (B-other).2–4 A large absolute number of relapses currently occurs in the group of patients with genetically unclassified ALL, indicating the need for new biological insights and treatment options for these patients.5
Genome-wide analyses that quantify gene expression (mRNA) levels in ALL cells has provided new insights into the genetic subtypes of ALL and the biological basis of drug resistance.6,7 Two leading studies showed that newly diagnosed pediatric ALL cases can be assigned to lineage and genetic subtypes using gene expression classifiers with an accuracy for predicting the subtype of >95%.8,9 Once these findings are validated in independent cohorts of patients, they will offer new approaches for the classification of ALL and guide treatment decisions.
However, standard procedures to select gene probe sets for classification can result in over-fitting and can lead to over-interpretation of the clinical value of the gene expression classifier as diagnostic tool.10 The present study aimed to critically analyze whether gene expression signatures improve genetic classification of ALL using a double-loop cross-validation method, with further validation in an independent cohort of patients. This revealed that classification of known subtypes of ALL is feasible, with highest prediction accuracy for TALL, TEL-AML1 positive, hyperdiploid and E2A-rearranged ALL subtypes. Importantly, we identified a new high-risk subtype of ALL that comprises 15–20% of all precursor B-ALL cases whose gene expression pattern is most similar to that of BCR-ABL-positive ALL. Leukemia cells with this BCR-ABL-like gene expression pattern exhibited a high percentage of abnormalities in genes involved in B-cell development, were in vitro more resistant to L-asparaginase and daunorubicin and patients had an unfavorable treatment outcome.
MATERIALS and METHODS
Patients and leukemic cell samples
Bone marrow and peripheral blood samples were collected from children with newly diagnosed ALL (prior to initial therapy) enrolled in the German Cooperative ALL (COALL)-92/97 and Dutch Childhood Oncology Group (DCOG)-ALL8/9 studies as approved by institutional review boards and after written informed consent was obtained. Leukemic samples were processed as previously reported.11 Patients characteristics (gender, age, WBC, immunophenotype, BCR-ABL translocation, MLL-rearrangement) were collected by COALL and DCOG study centers in Hamburg and The Hague, respectively. Cases were analyzed for the presence of a TEL-AML1 fusion (FISH and RT-PCR), an E2A-rearrangement (split-signal FISH), E2A-PBX1 fusion (RT-PCR) and ploidy status (DNA-index and/or number of chromosomes) in our laboratory.
Affymetrix GeneChip: data acquisition and processing
Total RNA was extracted out of samples containing >90% leukemic cells using Trizol reagents (Gibco BRL, Breda, NL) and RNA integrity was checked with Agilent’s Bio-analyzer. Next, cDNA and biotinylated cRNA was synthesized according to manufacturer’s guidelines. The COALL cohort (n=190; 180 COALL-92/97 and 10 DCOG-ALL-9 cases of the Erasmus MC-Sophia Children’s Hospital) was hybridized to Affymetrix U133A GeneChips at St. Jude Children’s Research Hospital, Memphis TN. Later in time, the DCOG cohort (107 DCOG-ALL-8 cases) was processed and hybridized to Affymetrix U133 plus 2.0 GeneChips at Erasmus MC, Rotterdam. Data-acquisition was performed using Affymetrix Microarray Analysis Suite 5.0 (COALL cohort) and GCOS 1.0 (DCOG cohort) software. Using the probe sets common to both array types, the two datasets were jointly normalized using the variance-stabilizing normalization procedure which in essence leaves the independence of both datasets intact.12 All included samples had a ratio between 3′probe/5′probe for β-actin or glyceraldehyde-3-phosphate dehydrogenase smaller than three, suggesting a minimal breakdown of RNA/cRNA during the experimental procedure. The data discussed in this paper have been deposited in NCBI’s Gene Expression Omnibus13 and are accessible through GEO Series accession number GSE13351 and GSE13425 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13351 or =GSE13425).
Prediction model for classification of pediatric ALL by gene expression signatures
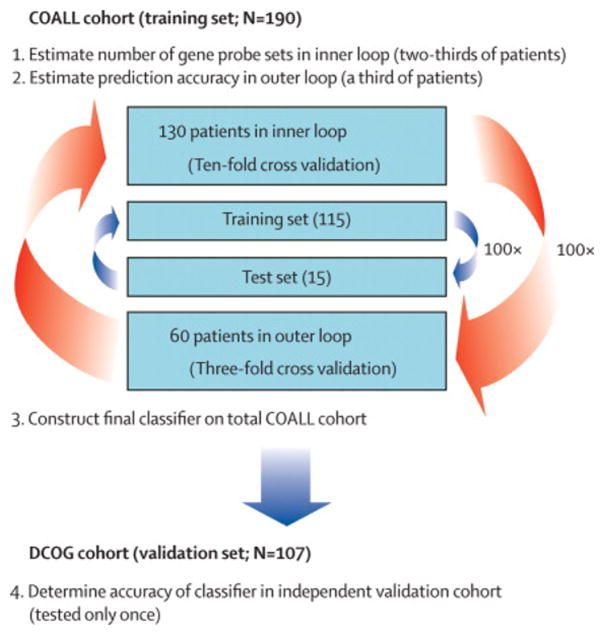
A combined double-loop cross-validation approach in a training cohort with validation of obtained gene expression classifier in an independent validation cohort was chosen to avoid over-fitting of the data.14 The double-loop cross validation method used an inner loop to establish the minimal number of probe sets needed to classify ALL subtypes and an outer loop to determine the predictive value of the constructed prediction model (classifier). A schematic description of the classifier-building approach is given in Figure 1A. In short, the inner loop was applied to 2/3 (n=130) of the COALL cohort to determine the number of probe sets needed for the prediction of 6 known subtypes, i.e. T-ALL, TEL-AML1-positive, hyperdiploid, E2A-rearranged, BCR-ABL-positive and MLL-rearranged ALL. The proportions of cases in each subtype was kept similar as in the entire set of patients (Table S-I, supplement). In each of 100 runs of this inner loop, patients were randomly assigned to the inner-training (9/10) or the inner-test (1/10) group (10-fold cross-validation). To start, top-50 probe sets most discriminative for each subtype were selected by rank of Wilcoxon’s test P-values. T-ALL-associated probe sets were selected by comparing T-ALL to precursor B-ALL cases whereas precursor B-ALL subtype-associated probe sets were selected by comparing the subtype to all other precursor B-ALL cases (e.g. TEL-AML1-positive versus non-TEL-AML1 precursor B-ALL). Since the number of BCR-ABL-positive and MLL-rearranged cases was too limited to yield statistically significant probe sets (supplementary Table S-II, P>0.05), we used the top-50 probe sets of the decision tree list of Ross et al. as starting set for both subtypes.9 Next, the minimal number of probe sets that optimally classified the patients was obtained by backwards selection starting with 300 probe sets (50 probe sets × 6 subtypes) using a global test which facilitates analyzing groups of probe sets thereby lowering multiple testing errors.15 The optimal number of probe sets determined in the inner loop served as input for a learning algorithm, i.e. radial-kernal support vector machine, that enabled the construction of a prediction model (classifier) which can be used to classify single patients into subtypes. The median sensitivity of this model was estimated via 3-fold cross-validation by applying the trained classifier to the remaining 1/3 (60 cases) of the COALL cohort (100 iterations; Table I-A). The final gene expression classifier, trained on all 190 COALL cases, was used to determine the prediction accuracy in the independent group of 107 DCOG cases (Table I-B, Figure 1A).
Figure 1. Schematic view of the approach to determine a gene expression signature that enabled classification of pediatric ALL in known immunophenotypic and genetic subtypes and which resulted in the discovery of the poor-prognostic BCR-ABL-like ALL subtype.
(A), Outline for construction and validation of a gene expression signature that enabled classification of pediatric ALL. A double-loop cross validation method was used to determine the number of gene probe sets that most optimally predicted cases assigned to the inner loop-test set (blue arrows) of the COALL cohort. Next, this number of probe sets was used to construct a classifier that becomes tested in the outer loop to estimate the prediction accuracy (red arrows). Upon construction of the final classifier in the total COALL cohort, the true accuracy of this classifier is determined using the independent DCOG validation cohort which is tested only once.
(B), Flow diagram of the discovery of the BCR-ABL-like subtype in pediatric ALL. The different steps taken that lead to the identification of the novel BCR-ABL-like subtype have been summarized.
Table I.
Diagnostic test values for the classification of pediatric ALL by a gene expression signature consisting of 110 gene probe sets
| Table I-A: Diagnostic test values for the classification of 190 COALL cases by 3-fold cross-validation approacha | |||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Accuracy | |
| Per subtype: | % | % | % | % | % |
| T-lineage ALL | 100 | 100 | 100 | 100 | 100 |
| (100-100) | (100-100) | (100-100) | (100-100) | (100-100) | |
| Precursor B-ALL: | |||||
| TEL-AML1-positive | 100 | 97.8 | 93.3 | 100 | 98.3 |
| (100-100) | (95.7–97.8) | (87.5–93.3) | (100-100) | (96.7–98.3) | |
| Hyperdiploid | 100 | 97.8 | 92.6 | 100 | 96.7 |
| (92.9–100) | (95.7–97.8) | (86.7–93.3) | (97.8–100) | (95.0–98.3) | |
| E2A-rearranged | 100 | 100 | 100 | 100 | 98.3 |
| (75.0–100) | (98.2–100) | (80.0–100) | (98.2–100) | (98.3–100) | |
| BCR-ABL-positive | 0 | 100 | 0 | 98.3 | 98.3 |
| (0-0) | (100-100) | (0-0) | (98.3-98.3) | (98.3-98.3) | |
| MLL-rearranged | 0 | 100 | 0 | 98.3 | 98.3 |
| (0-0) | (100-100) | (0-0) | (98.3-98.3) | (98.3-98.3) | |
| All patientsd: | |||||
| Overall values | 93.5 | 78.6 | 93.6 | 80.0 | 90.0 |
| (93.5–95.7) | (78.6–85.7) | (93.2–95.6) | (76.4–84.6) | (88.3–91.7) | |
| Table I-B. Diagnostic test values for independent validation group of 107 DCOG casesb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Accuracy | ||||||
| n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | |
| Per subtype: | ||||||||||
| T-lineage ALL | 15/15 | 100 | 92/92 | 100 | 15/15 | 100 | 92/92 | 100 | 107/107 | 100 |
| Precursor B-ALL: | ||||||||||
| TEL-AML1-positive | 24/24 | 100 | 81/83 | 97.6 | 24/26 | 92.3 | 81/81 | 100 | 105/107 | 98.1 |
| Hyperdiploid | 28/28 | 100 | 74/79 | 93.7 | 28/33 | 84.8 | 74/74 | 100 | 102/107 | 95.3 |
| E2A-rearranged | 2/2 | 100 | 104/104–105c | 99.0 or 100c | 2–3/3c | 66.7 or 100c | 104/104 | 100 | 106–107/107c | 99.1 or 100c |
| BCR-ABL-positive | 0/1 | 0 | 106/106 | 100 | 0/0 | 0 | 106/107 | 99.1 | 106/107 | 99.1 |
| MLL-rearranged | 0/4 | 0 | 103/103 | 100 | 0/0 | 0 | 103/107 | 96.3 | 103/107 | 96.3 |
| All patientsd: | ||||||||||
| Overall values | 69/74 | 93.2 | 25/33 | 75.8 | 69/77 | 89.6 | 25/30 | 83.3 | 94/107 | 87.9 |
The median value and 25th–75th percentile (in parenthesis) for sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and accurary are given of 100 iterations that include 130 cases to build the classifier and 60 other patients to determine the diagnostic test values in each interation (3-fold cross validation).
DCOG cohort was used to independently validate the predictive value of classification by gene expression signature (tested only once). The number of predicted cases (n) out of total number per subtype (N) as well as percentage for each diagnostic test value is given.
The specificity, PPV and accuracy are 100% for E2A-rearranged cases if the B-other case with an E2A-rearranged subclone (21% positive cells) is included as true positive case, see Table S-VI, supplement.
The overall values are based on the classification of all cases, including the remaining B-other group.
R (version 2.3.1) and the R packages vsn, e1071, globaltest, limma, multtest and marray were used to run the above-mentioned analyses.16 Probe sets that were most discriminative for the 6 ALL subtypes were used to perform a hierarchical clustering of patients using GeneMaths 2.0 software (Applied Maths, Sint-Martens-Latem, Belgium).
Definitions of diagnostic test values
Sensitivity: the percentage of cases correctly classified out of true positive cases.
Specificity: the percentage of cases correctly predicted out of true negative cases.
Prediction accuracy: the percentage of correctly classified negative and positive cases out of the total number of cases.
Positive predictive value: the percentage of positive test cases that is true positive for the predicted subtype.
Negative predictive value: the percentage of negative test cases that is true negative for the predicted subtype (supplementary Table S-III)
Genetic characterization of novel BCR-ABL-like subtype
DNA was extracted using Trizol reagent (Gibco BRL, Life Technologies, Breda, The Netherlands) out of 44 BCR-ABL-like, 15 BCR-ABL-positive and 25 control precursor B-ALL (without BCR-ABL translocation and non-hyperdiploid) samples containing >90% leukemic cells, and stored at 4°C. Five μg patient and 5 μg reference DNA (pooled DNA of 10 males, Promega, Madison WT) was digested with AluI (10 U) and RsaI (10 U) (Invitrogen) for 2 hours at 37°C and subsequently labeled o/n at 37°C with Cy-3 or Cy-5 using the Agilent labeling kit (Palo Alto, CA). Patient and reference DNA were pooled and mixed with 25 μg human Cot-1 DNA (Invitrogen) and blocking agent in a final volume of 250 μL hybridization buffer (Agilent Technologies). The hybridization mixtures were denatured at 95°C for 3 minutes, incubated at 37°C for 30 minutes, and hybridized to Agilent human genome comparative genomic hybridization (CGH)-microarrays (44A CGH-arrays containing 105.000 60-mer probes) for 42 hours at 65°C. The array slides were washed in 0.5xSSC/0.005% Triton X-102 at room temperature for 5 minutes, followed by 5 minutes at 37°C in 0.1xSSC/0.005% Triton X-102. Slides were dried and scanned using a 2565AA DNA microarray scanner (Agilent Technologies). Microarray images were analyzed using Feature Extraction software (v9.5; Agilent Technologies) and data were imported into array-CGH analytics software v3.5 (Agilent Technologies). Cy5/Cy3 and dye-swapped Cy3/Cy5 ratios for each probe were subsequently plotted into chromosome-specific profiles. Genomic loss and gain was identified as a minimum of three adjacent probes deviating beyond the threshold of 0.8 for single copy loss and 1.8 for bi-allelic loss. Known large-scale copy number polymorphisms were not considered disease-related. Array-CGH analysis was performed in duplicate for each patient to minimize false positive results.
Bacterial artificial chromomose (BAC) clones for the detection of dic(9;20) were obtained from BAC/PAC Resource Center (Children’s Hospital, Oakland, USA). BAC DNAs were isolated using DNA MiniPrep plasmid kit (Promega) and labeled with biotin-16-dUTP/digoxigenin-11-dUTP (Roche) by nick translation. A combination of chromosome 9p13.2 BAC clones (RP11-397D12 and RP11-405L18 were used in combination with 20q11.21 BAC clones (RP5-1184F4, RP5-836N17 and centromere probe). Cytospins slides were fixated in methanol, treated with RNAse and pepsin, and post-fixed with formaldehyde, before being denatured for 2 min 15 s in 70% formamide/2xSSC at 72°C. Fluorescence in situ hybridization probes were denatured for 8 min at 72°C and hybridized overnight at 37°C in a moist chamber. Slides were washed in 50% formamide/2xSSC and 2xSSC at 50°C, 4 min each. After dehydration through an ethanol series (70, 85 and 96%), they were mounted with antifade containing 4′6-diamino-2-phenyl indol (DAPI) as counterstain. For each sample a minimum of 100 interphase cells were scored. Images were captured using an epifluorescence microscope (Zeiss Axioplan 2, Sliedrecht, The Netherlands) using MacProbe software (version 4.3, Applied Imaging, Newcastle upon Tyne, UK).
In vitro drug toxicity-assay
Cytotoxicity of prednisolone (Bufa Pharmaceutical Products, Uitgeest, The Netherlands), vincristine (TEVA Pharma, Mijdrecht, The Netherlands), L-asparaginase (Paronal, Christiaens, Breda, The Netherlands), and daunorubicin (Cerubidine, Rhône-Poulenc Rorer, Amstelveen, The Netherlands) was determined by the 4-day in vitro MTT drug resistance assay as previously reported.6,11 The drug concentration lethal to 50% of the leukemic cells (LC50) was used as the measure of cellular drug resistance and these values have been previously shown to be associated with treatment outcome in children with ALL.11,17
Disease-free survival analysis
The probability of disease-free survival (pDFS) was calculated using the method of Kaplan and Meier, using relapse as event.18 The standard error (SE) was determined according to Peto et al.19 Cox proportional hazard analysis was used for univariate and multivariate analyses of potential prognostic factors.
Role of funding source
The funding resources had no role in study design, data collection, analysis, interpretation and writing or submission of the manuscript. All authors had full access to all the data in this study and had the final responsibility for the decision to submit for publication.
RESULTS
Table S-I (supplement) summarizes the number of patients used to generate the prediction model (COALL cohort, n=190) and to independently validate the accuracy of this model for subtype classification (DCOG cohort, n=107). The percentage of patients per subtype in both cohorts represents the general distribution of cases in pediatric ALL.2,3
The gene probe sets required for classification of cases into known immunophenotypic and genetic subtypes of pediatric ALL were selected using a double-loop cross-validation approach (see Figure 1A). The 10-fold cross-validation inner loop revealed that a minimum of 108 probe sets yielded the highest sensitivity to correctly classify patients into one of six known subtypes of ALL, i.e. T-ALL, TEL-AML1-positive ALL, hyperdiploid, E2A-rearranged, MLL-rearranged and BCR-ABL positive ALL. Each subtype was represented by 18 probe sets chosen based on ranking in P-values (Wilcoxon’s test). Three probe sets discriminative for the TEL-AML1-subtype had identical P-values to the top 18th probe set and were therefore included. One probe set was selected for both MLL-rearranged and T-ALL. Hence, in total 110 probe sets, representing 90 unique genes and 3 expressed sequence tags, were discriminative for the 6 ALL subtypes. Our list of probe sets only partially overlapped with the previously reported lists by Yeoh et al. and Ross et al., i.e. 24% (26 probe sets) and 28% (31 probe sets), respectively (see supplementary Table S-IV).8,9
Next, the diagnostic test values of the 110 probe set-based classifier were estimated in the 3-fold cross-validation outer loop (Figure 1A). The outer loop (100 iterations) revealed a median sensitivity of 93.5% and median classification accuracy of 90.0%, using 110 probe sets (Table I-A). All T-ALL, TEL-AML1-positive ALL, hyperdiploid and E2A-rearranged ALL cases were correctly identified (100% sensitivity; Table I-A). Next, the gene expression classifier was applied to the independent cohort of 107 DCOG patients (tested only once). The sensitivity of the gene expression classifier was 93.2% (69/74) whereas the accuracy to correctly classify both positive (6 subtypes) and negative (B-other) cases was 87.9% (94/107; Table I-B).
The negative predictive value was only affected by misclassified MLL-rearranged and BCR-ABL-positive cases in both the COALL (80%) and DCOG (83.3%) validation cohorts. Importantly, the negative predictive value for T-ALL, TEL-AML1-positive, hyperdiploid and E2A-rearranged cases was 100% (Table I). In addition, the specificity to classify non-MLL and non-BCRABL-positive cases as negative for these translocations was 100% (Table I and S-V, supplement). Interestingly, MLL-rearranged and BCR-ABL-positive leukemias clustered in distinct groups based on their gene expression pattern in both the patient cohorts (Figure 2). This indicates that MLL-rearranged and BCR-ABL-positive cases have a distinct gene expression pattern that enables discrimination by hierarchical clustering which visualizes similarity in gene expression patterns amongst multiple cases but that, on the other hand, the individual gene expression levels are insufficient for correct subtype prediction by the classifying model.
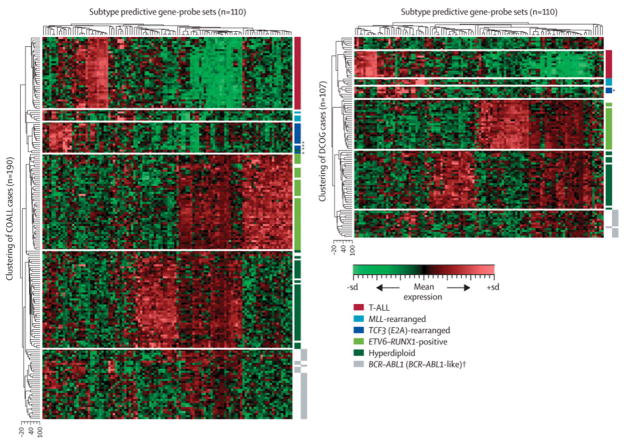
Figure 2. Clustering of ALL subtypes by gene expression profiles.
Hierarchical clustering of 190 COALL (A) and 107 DCOG (B) study patients using 110 gene probe sets that were selected to classify pediatric ALL. Heat map shows which gene probe sets are relatively over-expressed (in red) and which gene probe sets are relatively under-expressed (in green) compared to the mean expression of all gene probe sets, see scale bar. *, cases with E2A-rearranged subclone (15–26% positive cells). T-ALL, red; MLL-rearranged, light blue; E2A-rearranged, dark blue; TEL-AML1 positive, light green; hyperdiploid, dark green; BCR-ABL positive, yellow; novel BCR-ABL-like; yellow/dotted; white, cases with unknown or other genetic abnormalities.
The positive predictive value to correctly identify patients having one of the 6 major subtypes was 93.6% and 89.6% in the COALL and DCOG-validation groups respectively (Table I). The majority of B-other cases being falsely classified as TEL-AML1-positive, hyperdiploid or E2A-rearranged could partly be explained by the presence of genetic abnormalities linking them to these subtypes, as described in Table S-VI (supplement).
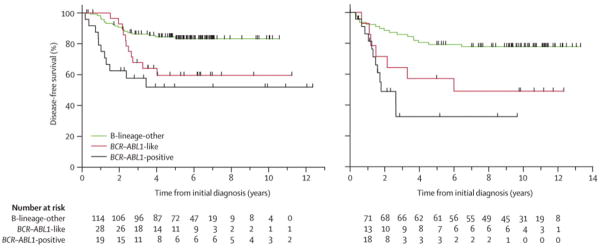
Hierarchical clustering using the 110 classifying gene probe sets showed that the major subtypes of pediatric ALL separated in distinct clusters by their gene expression signature (Figure 2). Remarkably, the gene expression pattern of 19.5% (30/154) of precursor B-ALL patients, representing 30/44 (68%) of B-other cases negative for known genetic subtypes (including BCR-ABL fusion), resembled that of the poor prognostic BCR-ABL-positive cases in the COALL cohort. Analysis of the dendrogram and heatmap obtained by hierarchical clustering revealed that the gene expression pattern of these 30 cases was most similar to that of BCR-ABL-positive cases and most dissimilar from the other subtypes of ALL (Figure 2A). Intriguingly, the relapse-rate of these 30 cases was significantly higher compared to other precursor B-ALL cases, i.e. 37% versus 16% (p=0.020; Table II). In addition, these patients had a highly unfavorable 5-year disease-free survival of 59.5% (95%CI: 37.1%–81.9%) compared to 84.4% (95%CI: 76.8%–92.1%) for patients with other precursor B-ALL in the COALL cohort (p=0.012), which was similar to the dismal prognosis of BCR-ABL-positive cases (51.9% (95%CI: 23.1%–80.6%; Figure 3A). This observation prompted us to further investigate the existence of this apparent new prognostic group, which we called BCR-ABL-like ALL, in the independent DCOG cohort (see Figure 1B). The clustering pattern of the BCR-ABL-like cases was confirmed in the independent DCOG cohort where this group comprised 15.2% (14/92) of precursor B-ALL cases, representing 14/33 (42%) of B-other cases (Figure 2B). Also in this DCOG validation group (being treated according to a different protocol than the COALL discovery cases), the relapse-rate of BCR-ABL-like cases was significantly higher compared to other precursor B-ALL cases, i.e. 50% versus 22% (p=0.046; Table II) and, similar to the COALL discovery cohort, the 5-year disease-free survival of 57.1% (95%CI: 31.2%–83.1%) for BCR-ABL-like cases was unfavorable compared to the 79.2% (95%CI: 70.2%–88.3%) observed for other precursor B-ALL cases (p=0.026; Figure 3B) and resembled that of BCR-ABL-positive DCOG cases (5-yr pDFS 32.5%, 95%CI: 2.3%–62.7%).
Table II.
Clinical characteristics of the novel BCR-ABL-like subtype of ALL
| Clinical feature | Group | Unit | COALL cohort | DCOG cohort | ||||
|---|---|---|---|---|---|---|---|---|
| BCR-ABL-like | B-other1 | P-value4 | BCR-ABL-like | B-other1 | P-value4 | |||
| N=30 | N=119 | N=14 | N=77 | |||||
| Age at diagnosis | all patients | median (years) | 6.0 | 4.5 | 0.14 | 7.4 | 4.0 | 0.15 |
| <10 years | %(N) | 67% (20) | 78% (93) | 71% (10) | 88% (68) | |||
| ≥10 years | %(N) | 33% (10) | 22% (36) | 0.23 | 29% (4) | 12% (9) | 0.11 | |
| WBC at diagnosis | all patients | median (/nl) | 46.3 | 21.0 | 0.004 | 49.0 | 15.6 | 0.010 |
| <25/nl | %(N) | 33% (10) | 55% (65) | 29% (4) | 61% (47) | |||
| ≥25/nl | %(N) | 67% (20) | 45% (54) | 0.043 | 71% (10) | 39% (30) | 0.039 | |
| Risk group | LR | %(N) | 17% (5) | 44% (52) | - | - | ||
| HR | %(N) | 83% (25) | 56% (67) | 0.006 | - | - | ||
| SR | %(N) | - | - | 7% (1) | 26% (20) | |||
| MR | %(N) | - | - | 50% (7) | 61% (47) | |||
| HR | %(N) | - | - | 36% (5) | 9% (7) | 0.0325 | ||
| NEL/NIT2 | %(N) | - | - | 7% (1) | 4% (3) | |||
| Follow-up | CCR3 | %(N) | 63% (19) | 84% (100) | 50% (7) | 78% (60) | ||
| relapse | %(N) | 37% (11) | 16% (19) | 0.020 | 50% (7) | 22% (17) | 0.046 | |
precursor B-ALL cases excluding BCR-ABL-like and BCR-ABL-positive cases
NEL/NIT; not eligible/not in treatment protocol
CCR, continuous complete remission
Mann-Whitney U test for continuous variables and Chi-square for categorical variables
NEL/NIT category excluded
Figure 3. Kaplan-Meier estimates for the probability of disease-free survival (pDFS) in children with precursor B-ALL.
(A), pDFS of COALL cohort precursor B-ALL cases; The COALL precursor B-ALL cohort consisted of 145 COALL-92/97 treated and 9 DCOG ALL-9 treated Sophia Children’s Hospital-patients. As a reference, data of 22 BCR-ABL positive cases enrolled in the COALL-92/97 protocol were included. Univariate analysis of pDFS comparing 30 BCR-ABL-like to 119 remaining precursor B-ALL cases (excluding 5 BCR-ABL positives): P=0.012. Kaplan-Meier curves were virtually the same when the 9 Sophia patients were excluded from this analysis.
(B), pDFS of DCOG cohort precursor B-ALL cases. The DCOG precursor B-ALL cohort consisted of 92 DCOG-ALL8 treated children. As a reference, data of 25 BCR-ABL positive cases enrolled in DCOG ALL 7, 8 and 9 were included. Univariate analysis of pDFS comparing 14 BCR-ABL-like and 77 remaining precursor B-ALL cases (excluding BCR-ABL positive case): P=0.026. Numbers at the bottom indicate patients at risk.
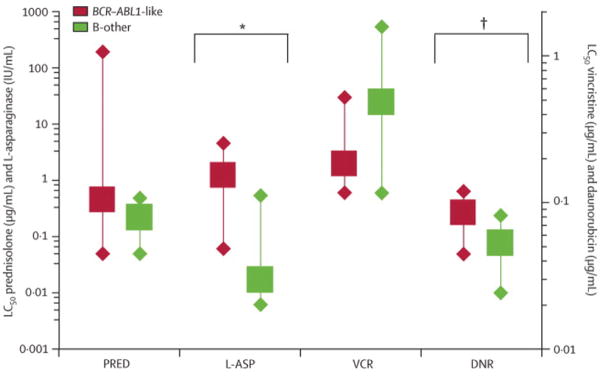
The clinical risk factor age of patients with BCR-ABL-like ALL did not differ from the age of other precursor B-ALL cases, but BCR-ABL-like cases had a 2- to 3-times higher white blood cell count at diagnosis (p=0.004 and p=0.010 for COALL- and DCOG-patients, respectively). Despite the fact that the proportion of BCR-ABL-like cases with high-risk criteria is higher than found for other precursor B-ALL subtypes (p=0.006 and p=0.032 for COALL and DCOG cohorts, respectively), still 17% and 57% of BCR-ABL-like cases was defined as low-risk and medium-risk based on current risk criteria of the COALL and DCOG protocols, respectively (Table II). The BCR-ABL-like leukemic cells were median 73-times (p=0.001) and 1.6-times (p=0.017) more resistant to L-asparaginase and daunorubicin compared to other precursor B-ALL cases, respectively, whereas no significant difference was seen in cytotoxic effect of prednisolone and vincristine (Figure 4).
Figure 4. In vitro cytotoxicity of 4 major drugs used in the treatment of pediatric ALL compared between BCR-ABL-like and other precursor B-ALL cases.
LC50 left Y-axis: PRED, prednisolone; L-ASP, L-asparaginase. Right Y-axis: VCR, vincristine; DNR, daunorubicin. Box represents median LC50 value, whiskers indicate 25th and 75th percentiles. BCR-ABL-like data are depicted in red, those of other precursor B-ALL cases are depicted in green. Comparison between BCR-ABL-like and B-other group: p=0.001 for L-asparaginase (*) and p=0.017 for daunorubicin (**), other drugs p>0.05 (2-sided).
Multivariate analysis including initial white blood cell count, age at diagnosis and genetic subtypes revealed that the BCR-ABL-like subtype was an independent risk factor associated with poor clinical outcome in both COALL-treated (P=0.012, Hazard ratio 5.3, 95% CI 1.4–19.4) and DCOG-treated cases (P=0.038, Hazard ratio 3.6, 95% CI 1.1–12.1). All together these data suggest that the BCR-ABL-like group represents a relative large poor prognostic subtype which is not being recognized by current diagnostic markers.
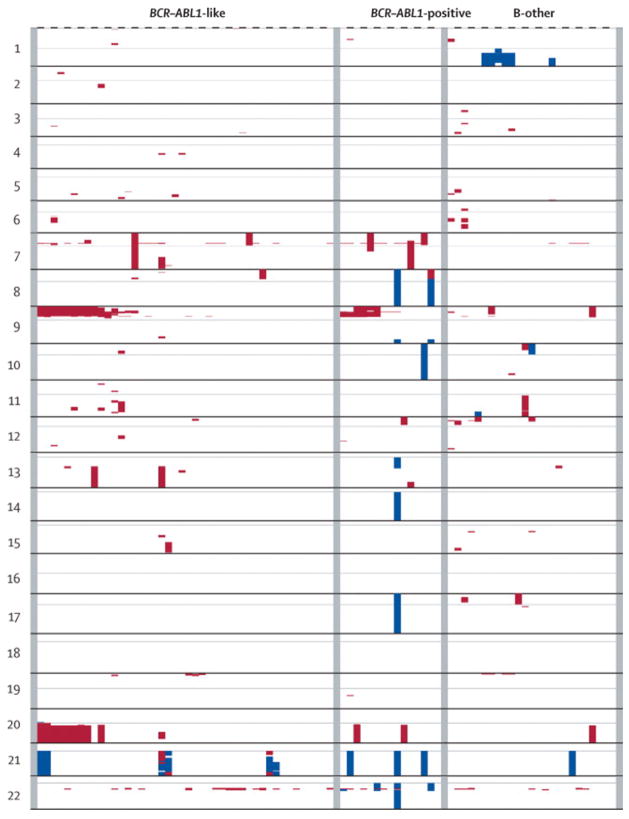
Analysis of available karyotypic data of BCR-ABL-like cases did not reveal a common genetic denominator (Supplementary Table S-VII). All cases were negative for the TEL-AML1 translocation, MLL-rearrangement, E2A-rearrangement, hyperdiploidy and BCR-ABL translocation. Further genetic characterization by array-CGH revealed seven recurrent deletions and one recurrent amplification in BCR-ABL-like cases (affected region >0.5 Mb; Table III, Figure 5). The percentage of patients with recurrent abnormalities was higher in both BCR-ABL-like (66%, p=0.001) and BCR-ABL-positive cases (80%, p=0.0009) compared to BCR-ABL-negative precursor B-ALL cases (24%). Deletions in chromosome 9p and 20q were most frequent, whereas recurrent amplifications were restricted to a duplication of chromosome 21q21-q22. In 8/9 cases with combined chromosome 9p and 20q deletion, the affected loci were conserved (Table S-VIII, supplement) which may be indicative of a dicentric chromosome dic(9;20). The presence of a dic(9;20) was indeed confirmed in 5/6 cases for whom material was available (Figure S-1 and Table S-VIII supplement). The BCR-ABL-like cases with and without dic(9;20) did not differ in their (unfavorable) outcome. The most common abnormality in chromosome 21 was an intrachromosomal amplification (iAMP21) which was found in 3/44 BCR-ABL-like cases compared to 0/25 control cases. The presence of an iAMP21 is known to be associated with a poor prognosis and in the present study all 3 cases relapsed.20 However, the low incidence of both dic(9;20) and iAMP21 indicates that other genetic abnormalities are contributing to the BCR-ABL-like subtype.
Table III.
Summary of recurrent genetic abnormalities found in BCR-ABL-like, BCR-ABL-positive and other precursor B-ALL cases
| Recurrent abnormality1 | BCR-ABL-like | BCR-ABL-positive | B-other2 | |||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| del(7)(p12) | 6/44 | 14 | 3/15 | 20 | 0/25 | 0 |
| del(9)(p)3 | 15/44 | 34 | 6/15 | 40 | 3/25 | 12 |
| del(11)(q22-q23) | 3/44 | 7 | 0/15 | 0 | 1/25 | 4 |
| del(13)(q14) | 3/44 | 7 | 0/15 | 0 | 1/25 | 4 |
| del(19)(p13) | 4/44 | 9 | 0/15 | 0 | 0/25 | 0 |
| del(20)(q13) | 10/44 | 23 | 2/15 | 13 | 1/25 | 4 |
| del(22)(q11) | 9/44 | 20 | 4/15 | 27 | 2/25 | 8 |
| all recurrent deletions | 25/44 | 57 | 12/15 | 80 | 5/25 | 20 |
| dup(21)(q21-q22) | 6/44 | 14 | 3/15 | 20 | 1/25 | 4 |
| all recurrent amplifications | 6/44 | 14 | 3/15 | 20 | 1/25 | 4 |
| Total4 | 29/44 | 66 | 12/15 | 80 | 6/25 | 24 |
| P=0.001* | P=0.0009* | |||||
Definition of recurrent genetic abnormality: deletion or amplification covering at least 0.5 Mb and found in at least 3 BCR-ABL-like cases
Precursor B-ALL cases excluding BCR-ABL-like, BCR-ABL-positive and hyperdiploid cases
Most common affected area in chromosome 9p covers 9p13.2–9p21.3 region but also complete loss of 9p was observed
Patients can have more than one genetic abnormality, hence, the total sum of cases with genetic abnormalities does not equal the sum of individual lesions
P-value compared to B-other group
Figure 5. Genome-wide copy number alterations in pediatric precursor B-ALL.
Genome-wide copy number data are visualized for BCR-ABL-like patients (n=44), BCR-ABL-positive (n=15) and other precursor B-ALL cases (B-other, n=25). Deletions are visualized in red, whereas amplifications are shown in blue. Centromere is indicated by grey line per chromosome.
Detailed analysis of chromosome 9p deleted areas revealed that 5 cases had a break in the PAX5 gene (9p13.2). Because this gene encodes a transcription factor essential to normal B-cell development, we investigated whether other genes involved in B-cell development were affected by focal deletions (<0.5 Mb). As shown in Table IV, 82% of BCR-ABL-like cases had one or more deletions in B-cell development genes compared to only 36% in other precursor B-ALL cases (p=0.0002). Affected genes include the transcription factors IKAROS (IKZF1), E2A (TCF3), EBF1 and PAX5 and the pre-B-cell receptor (pre-BCR) surrogate light chain VPREB1 (see supplemental Table S-VII), whereas no deletions were found in other B-cell transcription factors (PU.1, BCL11A, E2-2, FOXP1, LEF1). Also in true BCR-ABL-positive cases a high frequency of abnormalities in IKAROS, PAX5 and VPREB1 was found (80%, p=0.0098), which may explain the similarity in gene expression signature between BCR-ABL-like and BCR-ABL-positive leukemias.
Table IV.
High frequency of deletions in B-cell development genes characteristic for BCR-ABL-like and BCR-ABL-positive ALL
| Gene | Chromosomal location | BCR-ABL-like | BCR-ABL-positive | B-other1 | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| IKAROS/IKZF1 | 7p12.2 | 17/44 | 39 | 11/15 | 73 | 4/25 | 16 |
| E2A/TCF3 | 19p13.3 | 3/44 | 7 | 0/15 | 0 | 0/25 | 0 |
| EBF1 | 5q33.3 | 6/44 | 14 | 0/15 | 0 | 0/25 | 0 |
| PAX5 | 9p13.2 | 16/44 | 36 | 6/15 | 40 | 3/25 | 12 |
| VPREB1 | 22q11.22 | 15/44 | 34 | 6/15 | 40 | 6/25 | 24 |
| Total2 | 36/44 | 82 | 12/15 | 80 | 9/25 | 36 | |
| P=0.0002* | P=0.0098* | ||||||
precursor B-ALL cases excluding BCR-ABL-like, BCR-ABL-positive and hyperdiploid cases
Patients can have more than one gene deleted, hence, the total sum of patients with deleted genes does not equal the sum of individual genes No aberrations were found in other transcription factors, including PU.1, BCL11A, E2-2, FOXP1 and LEF1.
P-values compared to B-other group
Discussion
In this study genome-wide gene expression arrays were used to select gene probe sets that enabled the classification of patients into the major subtypes of pediatric ALL. The constructed classifier had a predictive accuracy of 90% in the discovery cohort of 190 cases with newly diagnosed ALL, and validation of this classifier in an independent cohort of 107 patients yielded a true predictive accuracy of 87.9%. Hierarchical clustering revealed that a significant part of currently genetically unclassified precursor B-ALL cases clustered together with BCR-ABL-positive ALL cases in both the COALL discovery and DCOG validation cohort. These so-called BCR-ABL-like cases had a poor prognosis, similar as BCR-ABL-positive cases. Genetic characterization by comparative genomic hybridization-arrays and molecular cytogenetics revealed that >80% of these cases have deletions in genes involved in B-cell development.
Several studies have reported a high classification accuracy of 90–95% for gene expression signatures in children with newly diagnosed ALL.8,9,21,22 However, in most studies low abundant probe sets are eliminated by filtering procedures based on percentage of present calls or fold-change differences which affects the false-discovery rate of selected probe sets. Also important is that no independent sets of patients have been used for validation of these prediction models to date.14,23 To minimize over-fitting and hence over-interpretation of the value of classification by gene expression profiles, we used a double-loop cross-validation approach to construct a prediction classifier that was then validated in an independent group of patients. The high similarity in percentages of sensitivity, specificity, accuracy, positive and negative predictive values between the COALL discovery and DCOG validation cohorts re-assured us that indeed our applied strategy avoids over-fitting of the data. Moreover, the robustness of the present classifier is emphasized by the fact that the gene expression profiles of COALL and DCOG cases were generated using two different versions of the Affymetrix GeneChip and samples and arrays were processed in two different locations. The overall accuracy of our gene expression classifier was ~88%, and all T-ALL, TEL-AML1-positive, hyperdiploid and E2A-rearranged cases were correctly classified (i.e., 100% sensitivity) which resembled the data previously reported using other strategies for probe set selection and classifier construction.8,9 In contrast, the sensitivity to classify BCR-ABL-positive and MLL-rearranged cases was 0%. These small but prognostically important subtypes are always classified as “B-other” ALL cases and never falsely classified as T-ALL, TEL-AML1-positive, hyperdiploid or E2A-rearranged ALL. The positive and negative predictive value of the gene expression classifier should be high if the predicted subtype is going to be used to determine treatment. These values were 100% for T-ALL and E2A-rearranged ALL (including the case with an E2A-rearranged subclone which was missed by routine diagnostics; Table I and supplementary Table S-VI) and hence each of these cases was correctly classified. The positive predictive value for TEL-AML1-positive and hyperdiploid cases was not 100%, indicating that some negative cases are falsely assigned as TEL-AML1-positive or hyperdiploid subtype. On the other hand, the negative predictive value for both subtypes was 100%, indicating that none of the true TEL-AML1 positive and hyperdiploid cases was falsely assigned to another subtype and, moreover, each predicted negative case was a true negative case. Therefore, if the gene expression classifier is used as diagnostic tool, additional genetic verification of TEL-AML1 fusion and hyperdiploidy can be restricted to only those cases that are predicted to belong to these subtypes. This reduces the number of patients to be screened for the TEL-AML1 fusion and/or ploidy by ~50%. Additional cytogenetic analysis to identify MLL-rearranged and BCR-ABL-positive cases is only mandatory for cases classified as Bother ALL, which comprises only one third of patients. Before this diagnostic approach becomes widespread, it will be important to prospectively compare classification by gene expression with conventional diagnostic techniques to determine the clinical and technical success rate as new diagnostic tool, which is currently being performed in a Dutch nationwide setting.
Detailed studies on discriminative genes between genetic subtypes of ALL may reveal more insight in the biology of each subtype. A gene that warrants further studies is the erythropoietin receptor that is 7.4-fold higher expressed in TEL-AML1-positive cases compared to other precursor B-ALL cases (see supplementary Table S-IV) confirming other gene expression classification studies.8,9,24 The erythropoietin receptor is thought to be restricted to myeloid-lineage committed progenitor cells. The increased expression in TEL-AML1-positive ALL emphasizes that this gene may have either other non-erythropoietin linked functions or that TEL-AML1-positive cells also have myeloid characteristics. In correspondence, TEL-AML1-positivity has indeed been associated with expression of myeloid markers in pediatric ALL.25 In case of hyperdiploidy, one would expect that most of the selected genes are located on chromosomes often amplified in this subtype, e.g. chromosome 4, 6, 10, 14, 17, 18, 21 and X.26 Twelve out of 18 selected probe sets indicative for hyperdiploidy are indeed located on these chromosomes. However, the expression level of these genes does not reflect a gene-dosage effect since these levels are >1.5-fold higher in the hyperdiploid cases. One of the most discriminative genes associated with hyperdiploidy is the gene encoding for SH3-binding protein 5 (SH3BP5). This gene, located on chromosome 3p24, is median 9.7-fold higher expressed in hyperdiploid cases compared to other precursor B-ALL cases (supplementary Table S-IV). Based on the presence of specific functional domains, SH3BP5 has been suggested to be an adaptor protein transducing signals derived from the Bruton’s tyrosine kinase (BTK) receptor. If functionality becomes proven, this receptor and/or its downstream pathway may be targeted by specific compounds, such as LFM-A13.27
Besides its use to classify patients in known subtypes by the gene expression classifier (i.e., class prediction) the same gene probe sets can also be used to discover entities based on similarities in gene expression patterns using hierarchical clustering (i.e., class discovery). An important finding in the current study was the identification of a new ALL subtype with a gene expression pattern resembling that of BCR-ABL-positive ALL. These BCR-ABL-like cases comprised 15–20% of precursor B-ALL cases in both the German discovery (COALL) and Dutch validation (DCOG) cohorts. This newly recognized group had a highly unfavorable prognosis, with 5-year disease-free survival estimates of 60%, comparable to that of ALL cases with the BCR-ABL gene fusion in both the COALL- and DCOG-study cohorts (Figure 3). Moreover, the number of BCR-ABL-like cases was 5-fold higher than the number of BCR-ABL-positive cases and thus comprises by far the largest poor prognostic subgroup in childhood ALL. Using the current diagnostic criteria, a considerable percentage of BCR-ABL-like cases is assigned to low-risk and medium-risk treatment schemes (Table II). The fact that BCR-ABL-like cases are in vitro not increased resistant to prednisolone compared to other precursor B-ALL cases suggests that these patients will not be recognized by a poor prednisone window response, e.g. as given in I-BFM-based protocols. Previous studies indicated that both in vitro resistance as well as a poor clinical window response to prednisone and L-asparaginase are linked to an unfavorable outcome in pediatric ALL.17,28 The present finding that BCR-ABL-like cases have a poor clinical outcome and are in vitro resistant to L-asparaginase (and to a lesser extend to daunorubicin) suggest that these patients should receive more intensified therapy using currently applied drugs or new, more targeted drugs for which the biology of the BCR-ABL-like subtype needs further study. The use of the (BCR)ABL-directed tyrosine kinase inhibitor Imatinib may not be indicated because BCR-ABL-like leukemia cells do not have elevated ABL mRNA expression. In contrast, BCR-ABL-positive cases have a 2–4-fold higher ABL mRNA expression than other precursor B-ALL cases (data not shown).
Genetic characterization of this novel high-risk subtype of ALL revealed that >80% of BCR-ABL-like cases have one or more abnormalities in genes involved in B-cell development, including IKAROS (IKZF1), E2A (TCF3), EBF1, PAX5 and VPREB1. A recent study by Mullighan et al. suggests that PAX5 abnormalities (mutations, deletions and translocations) located on chromosome 9p13 occur in ~30% of all precursor B-ALL cases.29 The observed abnormalities in PAX5 were shown to reduce the transcriptional activity of PAX5 protein and reduced the formation of surface immunoglobulin M (sIgM) characteristic for more differentiated precursor B-cells.29 In contrast to the BCR-ABL-like subtype we have identified, these PAX5 abnormalities were not associated with an unfavorable prognosis. Moreover, the deletions observed in the present BCR-ABL-like group not only affect the PAX5 gene locus but often include larger deleted regions on chromosome 9p and also deletions in other genes involved in B-cell development. This implies that the BCR-ABL-like group comprises a different subset of ALL patients than those identified by PAX5 abnormalities.
In conclusion, our study has developed and validated a new gene expression classifier that identifies the major subtypes of childhood ALL with a high level of accuracy and sensitivity. Importantly, we identified a new high-risk subtype which gene expression pattern is most similar to that of ALL cases containing the BCR-ABL gene fusion. This BCR-ABL-like subtype is characterized by abnormalities in B-cell development genes, indicating a defective (pre)B-cell receptor signaling pathway. The use of affected genes as diagnostic markers for the BCR-ABL-like subtype is yet to be investigated. Because the new BCR-ABL-like subtype represents the largest poor-prognostic subtype of childhood ALL, improving the treatment of this high-risk leukemia will have a major impact on the overall cure rate of childhood ALL.
Supplementary Material
A) The genomic position of BAC clones used for FISH analysis is depicted. (B) FISH analysis using RP11-397D12 (green) and RP5-836N17 (red) confirms a dic(9;20) in patient #2 (see also Table S-VIII, supplement).
Acknowledgments
We highly appreciate the contribution of members and hospitals participating in the COALL and DCOG study groups for pediatric ALL for providing patient samples. This study has been funded by the Dutch Cancer Society (grants EUR 2005-3662, EMCR 2005-3313 and EMCR 2007-3718; MLDB, RP), Sophia Foundation for Medical Research (grant SSWO-456, MLDB, RP), the Pediatric Oncology Foundation Rotterdam (MLDB, RP) and the Center of Medical Systems Biology established by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (RXDM). This work was also supported in part by NIH grant R37 CA36401 (WEE), NIH Pharmacogenomics Network grant U01 GM61393 (WEE) and Cancer Center Support Grant CA 21765 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities (ALSAC). The funding resources had no role in study design, data collection, analysis, interpretation, writing and submission of the manuscript.
Footnotes
Contribution of authors
MLDB, RXDM, WEE and RP designed the experimental set-up, analysed and interpretated data, and wrote the manuscript. MLDB and MHC performed microarray experiments, MLDB and MvS performed and analysed array-CGH experiments. LJCMVZ and HBB reviewed cytogenetic data. MLDB and RXDM performed the statistical analysis of data, PJVDS provided bio-informatical resources. MLDB, STCJMP, JGCAMBG, GE, MAH, GEJS and WAK contributed by collecting and processing of patient samples. All authors read, revised and approved this manuscript.
Conflict of interest statement
The authors do not report conflict of interests related to this paper except the submission of a patent application (PCT/NL08/050373) for classification of leukemia by gene expression signatures by MLDB and RP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pieters R, Schrappe M, De Lorenzo P, et al. A treatment protocol for infants younger than one year of age with acute lymphoblastic leukemia (Interfant-99): an observational study and multicentre randomised trial. Lancet. 2007;370:240–50. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–48. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 3.Pieters R, Carroll WL. Biology and treatment of acute lymphoblastic leukemia. Pediatr Clin North Am. 2008;55:1–20. doi: 10.1016/j.pcl.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Raimondi SC, Hancock ML, et al. Immunologic, cytogenetic and clinical characterization of childhood acute lymphoblastic leukemia with the t(1;19)(q23;p13) or its derivative. J Clin Oncol. 1994;12:2601–6. doi: 10.1200/JCO.1994.12.12.2601. [DOI] [PubMed] [Google Scholar]
- 5.Möricke A, Reiter A, Zimmermann M, et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111:4477–89. doi: 10.1182/blood-2007-09-112920. [DOI] [PubMed] [Google Scholar]
- 6.Holleman A, Cheok MH, Den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 7.Hulleman E, Kazemier KM, Holleman A, et al. Inhibition of glycolysis modulates prednisolone resistance in acute lymphoblastic leukemia cells. Blood. 2009:113. doi: 10.1182/blood-2008-05-157842. in press. (Oct 31st 2008 pre-published on-line) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 9.Ross ME, Zhou X, Song G, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–9. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 10.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–92. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 11.Den Boer ML, Harms DO, Pieters R, et al. Patient stratification based on prednisolone-vincristine-asparaginase resistance profiles in children with acute lymphoblastic leukemia. J Clin Oncol. 2003;21:3262–8. doi: 10.1200/JCO.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 12.Huber W, Von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 13.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci U S A. 2002;99:6562–6. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goeman JJ, Van der Geer SA, De Kort F, Van Houwelingen JC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 16.R Development Core Team. R: a language and enviroment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. URL http://www.R-project.org. [Google Scholar]
- 17.Kaspers GJL, Pieters R, Van Zantwijk CH, Van Wering ER, Van der Does-van den Berg A, Veerman AJP. Prednisolone resistance in childhood acute lymphoblastic leukemia: vitro-vivo correlations and cross-resistance to other drugs. Blood. 1998;92:259–66. [PubMed] [Google Scholar]
- 18.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–81. [Google Scholar]
- 19.Peto R, Pike MC, Armitage P, et al. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moorman AV, Richards SM, Robinson HM, et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21) Blood. 2007;109:2327–30. doi: 10.1182/blood-2006-08-040436. [DOI] [PubMed] [Google Scholar]
- 21.Willenbrock H, Juncker AS, Schmiegelow K, Knudsen S, Ryder LP. Prediction of immunophenotype, treatment response, and relapse in childhood acute lymphoblastic leukemia using DNA microarrays. Leukemia. 2004;18:1270–7. doi: 10.1038/sj.leu.2403392. [DOI] [PubMed] [Google Scholar]
- 22.Van Delft FW, Bellotti T, Luo Z, et al. Prospective gene expression analysis accurately subtypes acute leukaemia in children and establishes a commonality between hyperdiploidy and t(12;21) in acute lymphoblastic leukemia. Br J Haematol. 2005;130:26–35. doi: 10.1111/j.1365-2141.2005.05545.x. [DOI] [PubMed] [Google Scholar]
- 23.Pawitan Y, Krishna Murthy KR, Michiels S, Ploner A. Bias in the estimation of false discovery rate in microarray studies. Bioinformatics. 2005;21:3865–72. doi: 10.1093/bioinformatics/bti626. [DOI] [PubMed] [Google Scholar]
- 24.Fine BM, Stanulla M, Schrappe M, et al. Gene expression patters associated with recurrent chromosomal translocations in acute lymphoblastic leukemia. Blood. 2004;103:1043–9. doi: 10.1182/blood-2003-05-1518. [DOI] [PubMed] [Google Scholar]
- 25.Baruchel A, Cayuela JM, Ballerini P, et al. The majority of myeloid-antigen-positive (My+) childhood B-cell precursor acute lymphoblastic leukemias express TEL-AML1 fusion transcripts. Br J Haematol. 1997;99:101–6. doi: 10.1046/j.1365-2141.1997.3603174.x. [DOI] [PubMed] [Google Scholar]
- 26.Raimondi SC, Pui CH, Hancock ML, Behm FG, Filatov L, Rivera GK. Heterogeneity of hyperdiploid (51–67) childhood acute lymphoblastic leukemia. Leukemia. 1996;10:213–24. [PubMed] [Google Scholar]
- 27.Uckun FM, Zheng Y, Cetkovic-Cvrlje M, et al. In vivo pharmacokinetic features, toxicity profile, and chemosensitizing activity of alpha-cyano-beta-hydroxy-beta- methyl-N-(2,5-dibromophenyl)propenamide (LFM-A13), a novel antileukemic agent targeting Bruton’s tyrosine kinase. Clin Cancer Res. 2002;8:1224–33. [PubMed] [Google Scholar]
- 28.Appel IM, Kazemier KM, Boos J, et al. Pharmacokinetic, pharmacodynamic and intracellular effects of PEG-asparaginase in newly diagnosed childhood acute lymphoblastic leukemia: results from a single agent window study. Leukemia. 2008;22:1665–79. doi: 10.1038/leu.2008.165. [DOI] [PubMed] [Google Scholar]
- 29.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) The genomic position of BAC clones used for FISH analysis is depicted. (B) FISH analysis using RP11-397D12 (green) and RP5-836N17 (red) confirms a dic(9;20) in patient #2 (see also Table S-VIII, supplement).