Abstract
Antibodies to rat theilovirus (RTV) have been detected in rats for many years because of their serologic crossreactivity with strains of Theiler murine encephalomyelitis virus (TMEV) of mice. Little information exists regarding this pathogen, yet it is among the most common viruses detected in serologic surveys of rats used in research. In the study reported here, a novel isolate of RTV, designated RTV1, was cultured from the feces of infected rats. The RTV1 genome contained 8094 nucleotides and had approximately 95% identity with another rat theilovirus, NSG910, and 73% identity with TMEV strains. In addition, the genome size of RTV1 was similar to those of TMEV strains but larger than that reported for NSG910. Oral inoculation of Sprague–Dawley (SD) and CD male rats (n = 10 each group) with RTV1 revealed that SD rats were more susceptible than CD rats to RTV1 infection. At 14 d postinoculation, 100% of SD rats shed virus in the feces, and 70% were positive for RTV serum antibodies. By 56 d postinoculation 30% of SD rats continued to have detectable virus in the feces, and 90% had seroconverted. In contrast, in inoculated CD rats RTV was detected only in the feces at 14 d postinoculation, at which time 40% of CD rats were fecal positive. By 56 d postinoculation only 20% of CD rats had detectable RTV serum antibodies. Our data provide additional sequence information regarding a rat-specific Cardiovirus and indicate that SD rats are more susceptible than CD rats to RTV1 infection.
Abbreviations: RACE, rapid amplification of cDNA ends; RTV, rat theilovirus; SD, Sprague Dawley; TMEV, Theiler murine encephalomyelitis virus
For decades it has been known that rats used in research can develop antibodies to a Cardiovirus that is antigenically similar to Theiler murine encephalomyelitis virus (TMEV) of mice.4,6,10,12,13,20 Recent reports on the prevalence of antibodies in rats to this Cardiovirus vary from approximately 0.6% of sera tested from research rats in North America10 to 54.4% in a survey of 18 Brazilian research facilities.3,6,20 Multiple designations have been used to identify the Cardiovirus that infects rats, including Theiler-like virus of rats,13 Theiler murine encephalomyelitis virus (TMEV),20 rat enterovirus,1 rat encephalomyelitis virus,7 rat cardiovirus,15 and recently rat theilovirus.2 We have elected to refer to the virus as rat theilovirus (RTV), consistent with 1 of the cited references,2 to indicate the relation of the rat virus to TMEV of mice and to identify it as a rat-specific agent.
The first report of natural infection of rats with a Cardiovirus was in 1964 with the discovery of MHG virus.12 The finding resulted from an isolated observation in which a few rats in a large research colony displayed clinical signs indicative of central nervous system deficits, including incoordination, torticollis, circling, and tremors. The MHG virus recovered from infected rats was antigenically crossreactive with TMEV strain GDVII and had physical properties consistent with viruses in the Picornaviridae family. The virus was propagated in cell culture, and neurologic disease was reproduced when virus was inoculated into suckling mice and suckling rats.12 Subsequent serologic studies using crossneutralization, complement fixation, and hemagglutination inhibition assays further substantiated the antigenic relatedness between MHG virus and multiple strains of TMEV.4,11 In addition, sera from ‘normal’ rats contained antibodies to the newly identified Theiler-like virus of rats, suggesting widespread infection of the virus in research rat colonies.12 More recently in Japan, a Theiler-like virus was isolated after intracranial inoculation of newborn Wistar rats with intestinal homogenates from TMEV GDVII-seropositive rats.13 Inoculated rats did not develop clinical signs of infection, but virus was cultivated in BHK21 cells from brain homogenates of the 10-d-old Wistar rats inoculated intracranially. Physiochemical properties of the virus, designated NSG910, were consistent with those of the Cardiovirus genus. Sequence analysis also showed that NSG910 was a Cardiovirus in the family Picornaviridae that was related to, but distinct from MHG virus, and strains of TMEV. This report served to further document the existence of a unique Cardiovirus of rats closely related to, but distinct from, TMEV strains.13 In a recent report from Brazil, neonatal mice and rats inoculated with intestinal homogenates from rats with antibodies to TMEV strain GDVII developed neurologic signs of flaccid hindlimb paralysis and tremors. In addition, brain homogenates from the affected animals were positive by RT-PCR for cardioviral RNA.20
Picornavirus virions are approximately 30 nm in diameter, nonenveloped, with icosahedral symmetry and a single-stranded, positive-sense RNA genome.19 Encephalomyocarditis virus and Theilovirus are 2 species of Cardiovirus in the Picornaviridae family. Encephalomyocarditis virus species includes mengovirus, Maus Elberfeld virus, and Columbia SK virus.7 Strains of Theilovirus species include TMEV, Vilyuisk virus, and RTV.13,18,22 Most often studied are the TMEV strains, which are classified according to their neurovirulence after intracerebral inoculation. Included are the highly neurovirulent GD VII and FA strains23 and the less virulent, more persistent DA, BeAn 8386, WW, and TO (Theiler original) strains.9,17,22 Studies have shown that the virus replicates in the alimentary tract and is shed in the feces of infected mice.15,19 Mice rarely show clinical disease when infected under natural conditions; however, neurologic manifestations have been reported.21,24
Sentinel animals typically are used to survey rodent colonies for the presence or absence of infectious agents. Outbred stocks are frequently used as sentinels because of their vigor, relatively low cost, and ability to mount a robust humoral immune response to infectious agents.8,14 Sprague–Dawley (SD) and CD rats are 2 stocks that are commonly used as sentinels for rat colonies. The origins of the SD rat (Rattus norvegicus) date back to the 1920s as a result of mating Wistar stock with a hybrid rat stock of unknown origin. In the 1950s, an SD breeding stock was cesarean derived in an effort to improve microbiologic status. This nucleus of cesarean-derived rats formed the foundation of the CD rat stock.25 Because SD and CD rat stocks have a common ancestry, they frequently are considered to be interchangeable for the purpose of sentinel animals.
In the studies reported here, we isolated and propagated a novel strain of Theilovirus, referred to as RTV1, from the feces of infected SD rats. The entire genome of RTV1 was sequenced and compared with those of isolates of TMEV and NSG910, the only other isolate of RTV to be sequenced in its entirety. In addition, we evaluated the susceptibility of SD and CD outbred rats to RTV1 after oral inoculation with the virus.
Materials and Methods
Viral isolation.
Neonatal SD rats (Harlan Sprague Dawley, Indianapolis, IN) were exposed biweekly to approximately 240 ml dirty bedding from a single cage of rats that had antibodies that reacted with TMEV strain GDVII and parvoviral antigens and were fecal positive for rat parvovirus and rat minute virus by PCR. Fecal pellets (30 cm3) from 3-wk-old SD rat pups were homogenized manually in Tris–EDTA buffer (50 mM Tris, 1 mM EDTA, pH 8.7; Sigma–Aldrich, St Louis, MO) at a total volume of 80 ml. Aliquots of the fecal homogenate were sonicated (Model 100, Fisher Scientific, Pittsburgh, PA) on ice at the maximum setting for 6 cycles (30 s on, 30 s off) followed by centrifugation at 10,000 × g for 10 min at 4 °C. The resultant supernatant was subjected to precipitation and concentration by addition of 1 M NaCl and 8% (w/v) polyethylene glycol 8000 (Fischer Scientific, Fair Lawn, NJ). After overnight incubation, the precipitated material was pelleted by centrifugation at 10,000 × g for 15 min at 4 °C, resuspended in 4 ml PBS, passed sequentially through 0.45- and 0.22-μm filters, and frozen at −20 °C. Thawed fecal filtrates (250 μl) were placed on monolayers of BHK21 cells (70% confluent; CCL10, American Type Culture Collection, Manassas, VA) for 1 h at 37 °C, after which the inocula were poured off, cells were washed in PBS, and maintained in DMEM (SH30243.02, HyClone, Logan, UT) supplemented with 10% FBS (S11150, Atlanta Biologicals, Atlanta, GA) and 10 μg/ml ciprofloxacin in an atmosphere of 5% CO2 and 37 °C. Cultures were split 1:20 when cells reached confluency, and cultures were monitored for cytopathic effect by using an inverted microscope.
Plaque purification and propagation.
RTV1 was plaque purified as previously described5 with a few modifications. BHK21 cells maintained in DMEM supplemented with 10% FBS and 10 μg/ml ciprofloxacin were used to cultivate virus. Culture plates (diameter, 6 cm) were seeded with 1 × 106 cells, inoculated with virus when the cells reached 60% to 70% confluency, and incubated at 33 °C for 1 h. Inocula were removed and agarose overlay media was added to plates, which were incubated at 33 °C until plaques were visible after 48 to 72 h of incubation. Single plaques were isolated and used to generate stocks of virus that were tittered according to standard methods.
RTV1 was propagated by using plaque-purified virus to inoculate BHK21 cells (at approximately 80% confluency) in 75-cm2 tissue culture flasks (Corning, Lowell, MA) at a multiplicity of infection of 0.1. Cultures were incubated at 33 °C and 5% CO2 for 1 h then the inoculum was replaced with fresh growth medium for additional incubation. When cytopathic effect was greater than or equal to 90%, 3 freeze–thaw cycles were performed on cells and supernatant combined, the cellular material was pelleted at 3000 × g for 15 min at 4 °C, and the virus-containing supernatant was harvested and stored at −80 °C until used for animal inoculations or until further concentrated and purified.
Electron microscopy.
RTV1 was concentrated and purified by polyethylene glycol and salt precipitation followed by cesium chloride gradient ultracentrifugation as previously described.5 To determine the ultrastructure of the virus, viral preparations were pelleted by centrifugation at 41,000 rpm for 1 h at 4 °C in a SW41Ti rotor (Beckman Coulter, Fullerton, CA). Chilled 1% ammonium acetate was used to resuspend the virus overnight at 4 °C. A 7-µl sample was placed on a 300-mesh carbon-coated copper grid and allowed to settle for 5 min. Excess solution was wicked off, and a drop of 1.3% phosphotungstic acid (pH 7.0) was added and the virus particles stained for approximately 10 min. The grids were evaluated at 80 kV in a transmission electron microscope (1200EX, JEOL, Tokyo, Japan).
Sequencing.
The genomic sequence of the RTV1 isolate was obtained by using RT-PCR and nucleotide sequencing. RNA was extracted from the supernatant of BHK21 cell cultures infected with plaque-purified RTV1 using a QIAmp Viral RNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Twelve oligonucleotide primer pairs were designed by using Discover Studio Gene software package (Accelrys, San Diego, CA) and the nucleotide sequence of NSG190 (GenBank accession no. AB090161) as a reference to generate overlapping amplicons that were 600 to 800 bp in length (Table 1). Primer pairs spanned from nucleotide position 112 to 7929 of the 8021-nucleotide genome of NSG910. RT-PCR was performed on extracted RNA with primer pairs at concentrations of 0.6 µM each by using a 1-step RT-PCR kit with Q Solution (Qiagen) according to the manufacturer's protocol. Thermocycler parameters consisted of an initial reverse transcription step of 50 °C for 30 min followed by 1 cycle at 95° for 15 min to activate the DNA polymerase and 40 cycles consisting of denaturation at 94 °C for 1 min, annealing at 49 °C for 1 min, and extension at 72 °C for 2 min with a final extension at 72 °C for 10 min. Amplified products were separated on agarose gels containing ethidium bromide and purified with Montage PCR Centrifugal Filter Devices (Millipore, Bedford, MA) or Zymogen gel DNA Recovery Kit (Zymo Research, Orange, CA). Sequence information for the termini of the viral genomic RNA were obtained through rapid amplification of cDNA ends (RACE) by using 5′ and 3′ RACE Systems kit (Invitrogen, Carlsbad, CA) and associated gene specific primers according to the manufacturer's protocol (Table 2). Sequencing of purified products was done in forward and reverse directions for the 12 internal primer sets and in duplicate for the termini by the University of Missouri DNA Core Facility (Columbia, MO). Sequence data were assembled by using Sequencher Software (Gene Codes, Ann Arbor, MI). Discrepancies in base identification were resolved through manual editing by using associated chromatograms. Sequence homologies were evaluated by using ClustalW alignment and phylogenetic analysis with the Discover Studio Gene software package (Accelrys, San Diego, CA).
Table 1.
Primer pairs used for sequencing the RTV1 genome
| Primer sequence (5′ to 3′) |
|||
| Sense | Antisense | Amplicon size (bp) | Location (nt) |
| CGTCCTCCCTATTCCAACTAC | GGGCTTGAGAAAGACCATTC | 781 | 112–892 |
| TGTAGCGACCTCACAGTAG | GGTCAAGAAGTAAAGGAGCC | 765 | 733–1497 |
| GGAACATACTTGGTAATGCTGC | TACTGGAGGGGCGTAAGAAC | 760 | 1438–2197 |
| TGGATGGTTTACCCACACC | GCTAGTGTAACTAGTCTGGCG | 789 | 2049–2837 |
| AACAGGCAATGCAGGCAAC | ACTGTTGGATGTGGCACTG | 692 | 2725–3416 |
| TGCTTTCTTGCTTTTCTCACC | TTCAAACCCATAGTCATGTAGC | 796 | 3341–4136 |
| AACCATGCCCCCTTTTCCAC | TTTGAACCACGACATGAGCC | 753 | 3990–4742 |
| TCCTCCAACCTCTCCTCTTC | CGAACGTAATCCGCCTATC | 746 | 4607–5352 |
| TTTTGCCCAACATGGCTC | AGCTACACAGACTTCATAGTCC | 785 | 5215–5999 |
| CGCAAAAAGAGGCGCTAAG | GGCAGGAAAATTTCATGGGC | 789 | 5891–6679 |
| ATTACACAGGAACTTGTGGAAG | CAGTGGAATGTGAGGCATC | 793 | 6531–7323 |
| CGCTTTCAGACAAACCCTG | CTGGAACAACTACTCCAATCC | 754 | 7176–7929 |
nt, nucleotides
Table 2.
Primers used for sequencing of the 3′ and 5′ genomic termini
| RACE protocol | Primer | Primer sequence (5′ to 3′) |
| 3′ RACE | GSP1 | TTGGTTAAAGAAAGATTGGCCC |
| 5′ RACE | GSP1 | GCTGCATCCACGTTTTTCG |
| GSP2 | AGGGGCAGCACATACCCTTTTC | |
| GSP3 | AACCCTGTCATATTCCAAGTGG |
GSP, gene-specific primer
Rats and sample collection.
Four-week-old male CD rats were acquired from Charles River Laboratories (Charlotte, NC) and 4-wk-old male SD rats were acquired from Harlan Sprague Dawley (Indianapolis, IN). Rats were documented to be free of endoparasites, ectoparasites, Mycoplasma pulmonis, Helicobacter spp., known enteric and respiratory bacterial pathogens, antibodies to RTV, Sendai virus, pneumonia virus of mice, rat corona viruses, reovirus, rat parvoviruses, lymphocytic choriomeningitis virus, Hantaan virus, mouse adenovirus, Encephalitozoon cuniculi, and Clostridium piliforme. The animal care and procedures were approved and in accordance with the University of Missouri's Animal Care and Use Committee. Rats were monitored daily, and neither analgesics nor supportive care was needed throughout the course of the study.
Rats were separated into a control group (n = 6) and an experimental group (n = 10) for each rat stock. All rats were pair-housed for the entire study. Oral gavage with a 20-gauge ball-point needle was used to administer inoculum in all experiments. Experimental rats each were inoculated with 2.5 × 106 PFU of RTV1. Control groups were inoculated with uninfected BHK21 cell lysates, processed in the same manner as infected BHK21 cell cultures. Prior to inoculation on day 0 and on days 14, 28, and 56 thereafter, blood was collected from the lateral saphenous veins and feces were collected directly from the rats. Serum was separated and stored at −20 °C until antibody evaluation, and fecal samples were stored at −80 °C until processed for RT-PCR evaluation. Rats were euthanized by an inhaled overdose of carbon dioxide 56 d after inoculation. Terminal blood samples were obtained by cardiocentesis.
RT-PCR.
RNA was extracted from fecal and tissue samples by using magnetic beads (MagAttract RNA tissue Mini M48 kit, Qiagen) and a robotic workstation (BioRobot M48 Workstation, Qiagen) according to the manufacturer's protocols. Briefly, 1 fecal pellet or approximately 20 mg of mesenteric lymph node was placed in a sterile 2-ml tube with a 5 mm steel ball and 400 μl buffer RLT added (Qiagen). All mixtures were disrupted and homogenized in a tissue lyser (Qiagen). Fecal samples were agitated at 30 Hz for 10 s and tissue samples at 20 Hz for 2 min. Lysates from feces were centrifuged for 10 min at 3000 × g and those from and lymph node were spun for 10 min at 13,000 × g; RNA was extracted from the resulting supernatants.
Two oligonucleotide primers (212f, 5′ ATT TTC CGG CCC AGG CTA AGA G 3′; 392r, 5′ CTT AGA TCT CCA ACC ACG TCG C 3′) were designed to conserved sequences in the 5′ untranslated region of TMEV strains (GenBank accession nos. X56019, M20301, M20562, and U33045) and RTV-NSG910 (AB090161) by using the Prime program (GCG Wisconsin package, Genetics Computer Group, Madison, WI). Primer specificity was verified by using the Basic Local Alignment Search Tool (BLAST, National Center for Biotechnology Information, Bethesda, MD). RTV1 RT-PCR was performed by using a 1-step RT-PCR kit with Q Solution (Qiagen) according to the manufacturer's directions (see Sequencing) and by using primers 212f and 392r and an annealing temperature of 63 °C. RT-PCR products were subjected to electrophoresis in agarose gels containing ethidium bromide and were visualized under UV light. Positive RT-PCR reactions produced a 185-nucleotide product. All samples were tested in duplicate.
Multiplex fluorescent immunoassay.
TMEV strain GDVII viral particles were purified by cesium chloride gradient centrifugation and covalently coupled to carboxylated polystyrene microspheres (Luminex, Austin, TX) at a concentration of 2 µg protein per 106 microspheres and then stored at 4 °C until use. TMEV strain GDVII mouse antigen is cross-reactive with and routinely used by the Research Animal Diagnostic Laboratory for detection of antiRTV antibodies. A LiquiChip workstation (Qiagen) was used to process the rat serum samples for detection of the anti-RTV antibodies per manufacturers’ protocol. Baseline values were set according to that used by the Research Animal Diagnostic Laboratory at the University of Missouri. The following serologic parameters were used to delineate antiRTV antibody status: sera with values greater than 250 in multiplexed fluorescent immunoassays were classified as positive for antiRTV antibodies; sera with values of 125 to 250 were classified as intermediate and potentially containing antiRTV antibodies; and sera with values of less than 125 were classified as negative. Sera in the intermediate range were further tested by the Research Animal Diagnostic Laboratory at the University of Missouri by an indirect fluorescent antibody assay that used GDVII as an antigen and were classified as positive or negative based on this assay.
Histologic preparations.
Histologic sections were prepared from brain, duodenum, jejunum, ileum, cecum, kidney, liver, spleen, and lung samples collected at termination of the study. Tissues from all experimental and control rats were fixed in 10% neutral buffered formalin for 24 h, trimmed, embedded in paraffin, cut into 5-µm sections, and stained with hematoxylin and eosin. Stained sections were surveyed microscopically for lesions consistent with pathologic processes.
Statistical analysis.
Seroconversion and fecal shedding data for experimentally inoculated rat stocks were compared by using the Fisher Exact test (SigmaStat 3.5, Systat Software, San Jose, CA). A P value of less than 0.05 was considered significant.
Results
Isolation and ultrastructure of RTV1.
RTV1 was isolated from the feces of 3-wk-old SD rats that had been exposed since birth to dirty bedding from rats with serum antibodies against TMEV antigens. BHK21 cells inoculated with filtered fecal preparations from these rats displayed 100% cytopathic effect (rounding and detachment of cells) 24 h after the third and fourth passages. RTV1 was plaque purified and cultivated in subconfluent monolayers of BHK21 cells. Inoculating BHK21 cells with plaque-purified RTV1 at a multiplicity of infection of 0.1 and incubating cultures in an atmosphere of 5% CO2 at 33 °C produced visible cytopathic effect of cultured cells starting at 24 h postinoculation, with 90% to 100% cytopathic effect reached by 48 h postinoculation. These cell culture conditions produced RTV1 viral titers of 2.5 × 106 PFU/ml.
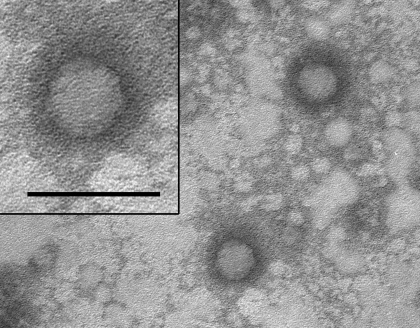
Cesium chloride ultracentrifuged preparations of RTV1 were negatively stained and subjected to transmission electron microscopy. RTV1 viral particles were approximately 30 nm in diameter with icosahedral symmetry (Figure 1), consistent with morphology previously reported for picornaviruses.19
Figure 1.
Transmission electron micrograph of negatively stained RTV1 particles cultivated in BHK21 cells and purified by cesium chloride gradient centrifugation. Viral particles are approximately 30 nm in diameter with icosahedral symmetry. Bar, 50 nm.
Analysis of RTV1 genome.
The full-length nucleotide sequence of RTV1 was obtained by aligning overlapping RT-PCR fragments that spanned the RTV genome and 5′ and 3′ RACE products of the viral termini. Overlapping RT-PCR products were sequenced in forward and reverse directions, and RACE products were sequenced in duplicate. The complete RTV1 genome (GenBank accession no. EU542581) contained 8094 nucleotides, which had approximately 95% nucleotide identity with NSG910 and 73% identity with TMEV strains GD7, BeAn, and DA. Consistent with NSG190 and other Picornaviridae, RTV1 contains a single, large 6924-nucleotide open reading frame from positions 1050 to 7973, which codes for the viral polyprotein. In addition, a 5′ untranslated region was present, as was a 3′ untranslated region that terminated with a polyA tail. Interestingly, the RTV1 genome was 73 nucleotides greater than that reported for NSG910, with 71 of these nucleotides being present in a single stretch at the 5′ untranslated region of the genome. Sequence alignments of the first 125 nucleotides of the 5′ untranslated region of RTV with those of the TMEV strains GDVII, BeAn, DA (NC001366, M16020, and M20301) and the first 54 nucleotides of NSG910 (AB090161) are presented in Figure 2. The 71 terminal 5′ nucleotides of RTV1 share 69% to 71% sequence identity with the 3 TMEV strains, indicating that, although not reported for NSG910, homologous regions to these additional 5′ bases of RTV are present in other closely related Picornaviridae. Similar to other strains in the Theilovirus species, a poly C tract was not present in the 5′ terminus.17
Figure 2.
Alignment of the first 125 nucleotides of the 5′ untranslated regions of TMEV strains GDVII, BeAn (8386), and DA and RTV strains NSG910 and RTV1. Dashes indicate gaps in sequence.
RTV1 RT-PCR sensitivity.
Sensitivity of the RT-PCR assay was evaluated by using RNA extracted from cesium-chloride–purified cell culture preparations of RTV1. RNA concentration was determined by spectrophotometry. Log-fold serial dilutions, from 100 pg to 100 ag RTV1 RNA, were used to evaluate the detection limits of the RTV RT-PCR. RT-PCR products were detected at 1 fg RTV1 RNA (data not shown). Based on an approximate molecular weight of 4.55 ag per virion (calculated based on a single-stranded RTV genome size of 8094 and an average molecular mass of 339 Da/nucleotide) and the assumption that samples were pure, this primer set detected as few as 220 viral particles per amplification reaction.
Susceptibility of SD and CD rats to RTV.
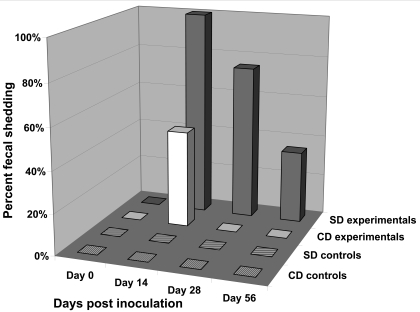
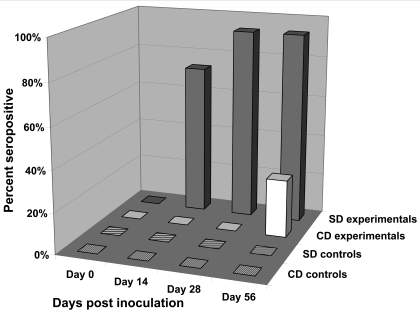
To evaluate the susceptibility of 2 outbred stocks of rats to RTV1, groups of 4-wk-old male SD and CD rats were inoculated by oral gavage with BHK21 cell lysates containing 2.5 × 106 PFU of RTV1 (n = 10 rats per stock) or uninfected BHK21 cell lysates (n = 6 rats per stock). The feces of rats were monitored for viral shedding by a RTV RT-PCR assay and serum samples were monitored for antibodies to RTV by using a TMEV antigen-based multiplexed fluorescent immunoassay (Figures 3 and 4). In SD rats at 14 d postinoculation, 100% of rats were shedding RTV1 in the feces, and 70% of the rats were positive for RTV serum antibodies. RTV fecal shedding steadily declined to 30% at the last time point evaluated (day 56 postinoculation), at which time 90% of inoculated SD rats were seropositive for RTV. In contrast, RTV was detected in the feces of only 40% of the inoculated CD rats at 14 d postinoculation (Figure 3) and not at any other time point. Moreover, serum antibodies to RTV in inoculated CD rats were not detected until 56 d postinoculation and then in only 20% of rats (Figure 4). Statistically significant (P < 0.05) differences in RTV fecal shedding between inoculated SD and CD rats were detected on postinoculation days 14 and 28, and although the difference on postinoculation day 56 was not statistically significant, a trend in viral shedding was evident. Statistically significant (P < 0.05) differences in seroconversion to RTV between inoculated SD and CD rats were detected at all postinoculation time points. All sham-inoculated rats tested negative for RTV by fecal RT-PCR testing and serologic analysis throughout the study.
Figure 3.
Percentages of SD and CD rats shedding RTV1 in the feces after oral inoculation with RTV1 (experimental groups) or uninfected BHK21 cell lysates (control groups). The presence of RTV1 in feces was detected by an RTV RT-PCR assay. SD experimental rats, n = 10; CD experimental rats, n = 10; SD control rats, n = 6; and CD control rats, n = 6.
Figure 4.
Percentages of SD and CD rats developing antibodies to RTV1 after oral inoculation with RTV1 (experimental groups) or uninfected BHK21 cell lysates (control groups). Serologic responses were detected by multiplexed fluorescent immunoassays using TMEV strain GDVII virus as antigen. SD experimental rats, n = 10; CD experimental rats, n = 10; SD control rats, n = 6; and CD control rats, n = 6.
At the end of the study (56 d postinoculation), the mesenteric lymph nodes from rats were evaluated for RTV by RT-PCR, and the brain, duodenum, jejunum, ileum, cecum, kidney, liver, spleen and lung were evaluated for histopathologic lesions. The mesenteric lymph nodes from all experimental and control SD and CD rats tested negative for RTV, and no lesions attributable to RTV infection were noted in any of the organs examined. In addition, no clinical signs of disease occurred in any of the rats inoculated with RTV.
Discussion
Previous studies involving rat theilovirus strains MHG and NGS910 revealed that rats infected with these viruses developed antibodies that crossreact with antigens prepared from TMEV isolates that infect mice.4,13 These findings, in conjunction with sequence data from the NGS910 isolate, validated the existence of a distinct rat Cardiovirus that is related to, but different from mouse TMEV isolates. A paucity of information exists regarding RTV, yet according to serologic data, it is among the most prevalent viral pathogens infecting rats used in biomedical research.6,10 In the current study, a novel isolate of RTV was cultured directly from the feces of rats and sequenced in its entirety. The genome size of RTV1 is comparable to those reported for TMEV isolates but is larger than that reported for NSG910, the only other RTV isolate to be sequenced. The RTV1 genome contains an additional 71-nucleotide fragment in the noncoding region at the 5′ end of the genome that is absent from NSG910. As such, this fragment maintains an approximate 70% sequence identity with the homologous fragment of strains of TMEV, further documenting the relationship of RTV1 to TMEV.
The SD and CD rat stocks are 2 of the most commonly used groups of outbred rats in research and are often viewed as biologically comparable with one another. The relationship between SD and CD rats stems from the derivation of the CD stock from the original SD stock in the 1950s. Both the SD and CD rats are used for research applications that range from nutrition to aging studies. However, differences between the 2 stocks in lifespan and morbidity have been documented.16 In our study, we found differences between SD and CD rats in susceptibility to RTV1 infection, as evidenced by differences in RTV1 fecal shedding and the development of antibodies to RTV1 after oral inoculation with the virus. At 14 d postinoculation, all inoculated SD rats shed RTV1 in their feces. In addition, the percentage of SD rats shedding virus steadily declined over time until the end of the study (day 56 postinoculation), when 30% of the SD rats remained positive for virus in the feces. In contrast, RTV1 was detected in the feces of inoculated CD rats only at 14 d postinoculation, at which point only 40% of inoculated rats shed virus in the feces. Similarly, SD rats developed antibodies to RTV1 more quickly than did CD rats: 70% of inoculated SD rats were seropositive at 14 d postinoculation. In contrast, the first seropositive CD rats were not detected until day 56 postinoculation, at which point only 20% of CD rats were seropositive compared with 90% of the inoculated SD rats. The reason for this difference is unknown but is likely due to genetic differences that exist between these 2 rat stocks. Because of the greater susceptibility of SD rats compared with CD rats to oral infection with RTV, the SD rat likely would be the better choice of stock for sentinel detection of RTV infections in research rat colonies. In circumstances that require use of CD rats as sentinels, a longer postexposure period should be used and a larger number of rats should be tested to allow serologic detection of RTV. Our data suggest the potential value of assessing possible differential susceptibilities to RTV infection in additional stocks and strains of rats.
Two of the CD rats and 1 of the SD rats orally inoculated with RTV shed virus at only 1 time point examined (postinoculation day 14) and failed to develop detectable antibodies to RTV by the end of the study (postinoculation day 56). However, each of these rats was pair-housed with a rat that shed virus and seroconverted. Perhaps the virus present in the feces of the rats that failed to mount a humoral response to the virus was due to coprophagy of feces from infected cagemates but the virus did not cause active infections in these rats. Alternatively, these rats may have been actively infected but failed to develop a detectable humoral immune response by the last time point examined. The serologic values from multiplexed fluorescent immunoassays for these 3 experimental rats were similar to the values obtained from the sham-inoculated control rats (data not shown).
In rare circumstances, overt disease has occurred in murine hosts infected with Theilovirus spp.12,21,24 We did not observe any clinical or histologic signs of disease in any of the rats inoculated with RTV1. However, our study does not preclude the possibility of RTV1 causing disease in other rat strains or of additional virulent strains of RTV. Regardless, like numerous other murine pathogens, RTV may confound, alter, or invalidate research findings even in the absence of overt disease. Investigation into the existence of additional strains of RTV and the potential effects of RTV in infected rats warrant further study.
In conclusion, we present the entire genomic sequence of a novel isolate of RTV (RTV1). The genome size is larger than the NSG910 isolate and comparable in size to genomes reported for strains of TMEV. In addition, we found a distinct differential susceptibility to infection between SD and CD rats experimentally inoculated with RTV1. SD rats exhibited prolonged fecal shedding compared with that of CD rats, and a greater number of SD rats shed virus in the feces and developed antibodies to RTV than did CD rats over the 8 wk study. These data suggest that SD rats may be more effective than CD rats for use as sentinels to detect this pathogen in rat colonies.
Acknowledgments
We would like to thank Beth Bauer for work with isolation of RTV1, Greg Purdy for cell culture and viral purification, Cheryl Jensen and the Electron Microscopy Core Facility at the University of Missouri–Columbia for electron microscopy, and Don Connor and Howard Wilson for graphics.
This work was supported by funds from a National Institutes of Health Postdoctoral Training in Comparative Medicine grant (T32-RR07004) and the Research Animal Diagnostic Laboratory (RADIL).
References
- 1.Charles River Laboratories 2005. Research models and services catalog. Wilmington (MA): Charles River Laboratories [Google Scholar]
- 2.Charles River Laboratories 2008. Research models and services catalog. Wilmington (MA): Charles River Laboratories [Google Scholar]
- 3.Gilioli R, Sakurada JK, Andrade LA, Kraft V, Meyer B, Rangel HA. 1996. Virus infection in rat and mouse colonies reared in Brazilian animal facilities. Lab Anim Sci 46:582–584 [PubMed] [Google Scholar]
- 4.Hemelt IE, Huxsoll DL, Warner AR., Jr 1974. Comparison of MHG virus with mouse encephalomyelitis viruses. Lab Anim Sci 24:523–529 [PubMed] [Google Scholar]
- 5.Hsu CC, Wobus CE, Steffen EK, Riley LK, Livingston RS. 2005. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol 12:1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby RO, Lindsey JR. 1997. Health care for research animals is essential and affordable. FASEB J 11:609–614 [DOI] [PubMed] [Google Scholar]
- 7.King AMQ, Brown F, Christian P, Hovi T, Hyypiä T, Knowles NJ, Lemon SM, Minor PD, Palmenberg AC, Skern T, Stanway G. 2000. Picornaviridae. : Van Regenmortel MHV, Fauquet CM, Bishop DHL, Calisher CH, Carsten EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, Virus taxonomy. Seventh report of the international committee for the taxonomy of viruses. New York: Academic Press; p 657–673 [Google Scholar]
- 8.Lipman NS, Homberger FR. 2003. Rodent quality assurance testing: use of sentinel animal systems. Lab Anim 32:36–43 [DOI] [PubMed] [Google Scholar]
- 9.Lipton HL. 1978. Characterization of the TO strains of Theiler's mouse encephalomyelitis viruses. Infect Immun 20:869–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livingston RS, Riley LK. 2003. Diagnostic testing of mouse and rat colonies for infectious agents. Lab Anim 32:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lussier G, Descoteaux JP. 1986. Prevalence of natural virus infections in laboratory mice and rats used in Canada. Lab Anim Sci 36:145–148 [PubMed] [Google Scholar]
- 12.McConnell SJ, Huxsoll DL, Garner FM, Spertzel RO, Warner AR, Jr, Yager RH. 1964. Isolation and characterization of a neurotropic agent (MHG virus) from adult rats. Proc Soc Exp Biol Med 115:362–367 [DOI] [PubMed] [Google Scholar]
- 13.Ohsawa K, Watanabe Y, Miyata H, Sato H. 2003. Genetic analysis of a Theiler-like virus isolated from rats. Comp Med 53:191–196 [PubMed] [Google Scholar]
- 14.Otto G, Franklin CL. 2006. Medical management and diagnostic approaches. : Suckow MA, Weisbroth SH, Franklin CL, The laboratory rat. Burlington (MA): Elsevier Academic Press; p 547–564 [Google Scholar]
- 15.Percy DH, Barthold SW. 2001. Pathology of laboratory rodents and rabbits. Ames (IA): Iowa State University Press [Google Scholar]
- 16.Pettersen JC, Morrissey RL, Saunders DR, Pavkov KL, Luempert LG, 3rd, Turnier JC, Matheson DW, Schwartz DR. 1996. A 2-year comparison study of Crl:CD BR and Hsd:Sprague-Dawley SD rats. Fundam Appl Toxicol 33:196–211 [DOI] [PubMed] [Google Scholar]
- 17.Pevear DC, Calenoff M, Rozhon E, Lipton HL. 1987. Analysis of the complete nucleotide sequence of the picornavirus Theiler's murine encephalomyelitis virus indicates that it is closely related to cardioviruses. J Virol 61:1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard AE, Strom T, Lipton HL. 1992. Nucleotide sequence identifies Vilyuisk virus as a divergent Theiler's virus. Virology 191:469–472 [DOI] [PubMed] [Google Scholar]
- 19.Racaniello VR. 2001. Picornaviridae: The viruses and their replication. : Knipe DM, Howley PM, Fields Virology. Philadelphia: Lippincott Williams and Wilkins; p 685–722 [Google Scholar]
- 20.Rodrigues DM, Martins SS, Gilioli R, Guaraldo AM, Gatti MS. 2005. Theiler's murine encephalomyelitis virus in nonbarrier rat colonies. Comp Med 55:459–464 [PubMed] [Google Scholar]
- 21.Rozengurt N, Sanchez S. 1993. A spontaneous outbreak of Theiler's encephalomyelitis in a colony of severe combined immunodeficient mice in the UK. Lab Anim 27:229–234 [DOI] [PubMed] [Google Scholar]
- 22.Theiler M. 1937. Spontaneous encephalomyelitis of mice, a new virus disease. J Exp Med 65:705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theiler M, Gard S. 1940. Encephalomyelitis of mice. I. Characteristics and pathogenesis of the virus. J Exp Med 72:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson R, Harrison VM, Myers FP. 1951. A spontaneous epizootic of mouse encephalomyelitis. Proc Soc Exp Biol Med 77:262–266 [DOI] [PubMed] [Google Scholar]
- 25.White WJ, Lee CS. Development and maintenance of the Crl CD(SD) IGS BR rat breeding system [Internet]. [cited 2008 March 28]. Available from: http://www.criver.com/flex_content_area/documents/rm_rm_a_igs_rat_breeding_system.pdf