Abstract
N-Acylethanolamines (NAEs) are lipids involved in several physiological processes in animal and plant cells. In brain, NAEs are ligands of endocannabinoid receptors, which modulate various signaling pathways. In plant, NAEs regulate seed germination and root development, and they are involved in plant defense against pathogen attack. This signaling activity is started by an enzyme called N-acylphosphatidylethanolamine (NAPE) synthase. This catalyzes the N-acylation of phosphatidylethanolamine to form NAPE, which is most likely hydrolyzed by phospholipase D β/γ isoforms to generate NAE. This compound is further catabolized by fatty amide hydrolase. The genes encoding the enzymes involved in NAE metabolism are well characterized except for the NAPE synthase gene(s). By heterologous expression in Escherichia coli and overexpression in plants, we characterized an acyltransferase from Arabidopsis thaliana (At1g78690p) catalyzing the synthesis of lipids identified as NAPEs (two-dimensional TLC, phospholipase D hydrolysis assay, and electrospray ionization-tandem mass spectrometry analyses). The ability of free fatty acid and acyl-CoA to be used as acyl donor was compared in vitro with E. coli membranes and purified enzyme (obtained by immobilized metal ion affinity chromatography). In both cases, NAPE was synthesized only in the presence of acyl-CoA. β-Glucuronidase promoter experiments revealed a strong expression in roots and young tissues of plants. Using yellow fluorescent protein fusion, we showed that the NAPE synthase is located in the plasmalemma of plant cells.
N-Acylethanolamines (NAEs)2 are bioactive lipids composed of an ethanolamine headgroup amide-linked to an acyl chain varying in length and degree of saturation. In animals, NAEs are involved in different physiological processes, such as neuroprotective action (1), embryo development (2), cell proliferation (3), apoptosis (4), nociception, anxiety, inflammation, appetite/anorexia, learning, and memory (for review, see Ref. 5). Most studies carried out with animal cells/tissues have focused on N-arachidonoylethanolamine (anandamide, NAE20:4), which is synthesized in brain neurons but also, under certain conditions, in macrophage cells (6). NAE20:4 binds CB1 cannabinoid receptors located in brain neurons (7) and also acts as ligand of vanilloid receptors for pain modulation (8). In addition, it has been shown that NAE20:4 also promotes food intake, whereas NAE18:0 and NAE18:1 exert anorexic effects by increasing satiety (9–11). NAE16:0 is accumulated during inflammation and has several anti-inflammatory effects (for a review, see Ref. 12).
In plants, NAEs are thought to be involved in various physiological functions. For example, because NAE levels observed in various dry seeds decline rapidly after imbibition, a possible role of these compounds in the regulation of seed germination has been proposed (13). It was further observed that the addition of 25 μm NAE12:0 to growth medium of Arabidopsis thaliana leads to a decrease in the size of the main and lateral roots and in root hair formation. This reduction in growth was associated with a modification of cytoskeletal organization (14). NAE12:0 is also able to delay cut Dianthus caryophyllus (carnation) senescence by decreasing oxidative damage and enhancing antioxidant defense (15), whereas NAE14:0 inhibits the elicitor-induced medium alkalinization and activates phenylalanine ammonia lyase gene expression involved in plant defense against pathogen attack (16).
Both in plant and animal cells (for a review, see Ref. 17), NAEs are formed by the hydrolysis (by PLDs) of N-acylphosphatidylethanolamine (NAPE). NAPE is an unusual derivative of phosphatidylethanolamine (PE) with a third fatty acid linked to the amine position of the ethanolamine headgroup. In animals, the formation of NAEs is catalyzed by a PLD with a high specificity toward NAPE (NAPE-PLD). In plants, PLDβ and PLDγ isoforms, but not PLDα, hydrolyzed NAPE into NAE in vitro, and this is thought to operate in response to several biotic and abiotic stresses. Both in animals and in plants, NAEs signaling is terminated by the action of fatty acid amide hydrolases, which hydrolyze NAEs to free fatty acid and ethanolamine. FAAH has been identified and characterized in mammals and plants (for a review, see Ref. 17). In Arabidopsis, FAAH has been shown to modulate NAE content. Moreover, lines overexpressing FAAH displayed enhanced seedling growth as well as increased cell size (18) and were also more susceptible to bacterial pathogens (19).
Although the role of NAEs and their catabolism have been extensively investigated, little is known about their precursors, the NAPEs. NAPEs represent a minor phospholipid class but are present in all tissues of plants and animals. The principal function of NAPEs is to serve as a precursor for the production of lipid mediator NAEs, but it has also been suggested that NAPEs could serve as a membrane stabilizer to maintain cellular compartmentalization during tissue damage (20). More recently, N-palmitoyl-PE was proposed to act as an inhibitor of macrophage phagocytosis through inhibition of the activation of Rac1 and Cdc42 (21).
In the animal and plant kingdoms, therefore, the signaling events mediated by NAEs appear to be involved in many physiological processes that have been extensively studied. The genes encoding the enzymes involved in the synthesis (from NAPEs) and the degradation of NAEs have been cloned and characterized. By contrast, little is known about the physiological roles of NAPEs or about the first step of this lipid signaling pathway, namely the N-acylation of PE to form NAPEs. In animals, the synthesis of NAPEs is catalyzed by an N-acyltransferase, where the O-linked acyl unit from a phospholipid donor is transferred to the ethanolamine headgroup of PE (22). Recently, a rat LRAT-like protein 1 or RLP1 was shown to display such an activity, but according to the authors, RLP-1 can function as a PE N-acyltransferase, catalytically distinguishable from the known Ca2+-dependent N-acyltransferase (23). However, a different situation is observed in plants. NAPE synthase activity was shown to directly acylate PE with free fatty acids (24, 25), but a gene encoding a NAPE synthase activity remained unidentified until now. The present work shows that the A. thaliana acyltransferase At1g78690p catalyzes the synthesis of NAPEs from PE and acyl-CoAs in vitro as well as in vivo when this enzyme is expressed in E. coli and overexpressed in plants.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
A tDNA insertion mutant of At1g78690 was identified in the SALK collection (SALK_029716). The location and orientation of the tDNA insert was confirmed by DNA sequencing of PCR products amplified with tDNA and insert-specific primers (LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′; LP1, 5′-TCTCCTGTGTATCCTCTCGTGTG-3′) and gene-specific primers (LP1 and RP1 (5′-TCAGATCTTGCTGCCCATTCC-3′)). Transgenic Arabidopsis was generated by the floral dip method with the Agrobacterium tumefaciens (strain C58C1) harboring the specific construct (26). Arabidopsis seeds were planted and grown on MS medium supplemented with 0.7% agar, 2.5 mm MES-KOH, pH 5.7.
Biological Molecular Constructs
The At1g78690 sequence was amplified using the PhusionTM High-Fidelity DNA polymerase by a set of sense and antisense primers containing the appropriate restriction sites for cloning the sequence of interest in the pET-15b vector (Novagen). The PCR product was purified with the UltraCleanTM GelSpinTM DNA extraction kit (MO BIO Laboratories, Inc.) according to the manufacturer's protocol. This product of purification and pET-15b vector were submitted to endonuclease enzymes (NcoI and XhoI; Biolabs), and the 5′ extremities were dephosphorylated (alkaline phosphatase; Biolabs). Then the vector was purified using the UltraCleanTM Standard Mini plasmid prep kit (MO BIO Laboratories) according to the manufacturer's protocol. The quantity and purity of this preparation were determined at 260 and 280 nm. All constructs were verified by sequencing. Finally, this insert was introduced into pET-15b vector by ligation (T4 DNA ligase of Biolabs). C41 (DE3) E. coli bacteria (Avidis, Saint-Beauzire, France) was further transformed with the pET-15b containing the At1g78690 sequence by the heat shock method. Independent clones were obtained by ampicillin selection.
To obtain At1g78690p tagged with His6 and the construction needed for tissue and cellular location, the cDNA of At1g78690 or the promoter of this gene (964 bp before the start codon) was amplified using the PhusionTM High-Fidelity DNA polymerase and a set of sense and antisense primers containing the attB region (respecting the reading frame for the different constructions). The PCR products were cloned in the PDONR221 vector using Gateway technology. After verification by sequencing, the pDONR221 containing the sequence of interest (cDNA or gene promoter) were purified as previously and used for the LR reaction with different destination vectors: pDESTTM17 (for His6-At1g78690p), pK7WG2D (for p35S:At1g78690), pK7YWG2 and pK7WGY2 (for p35S:YFP-At1g78690 in the C terminus and N terminus, respectively), or pKGWFS7 (for pAt1g78690:GUS).
Bacteria Growth Conditions
C41 (DE3) E. coli bacteria were grown overnight in LB medium with 0.1 mg/ml ampicillin at 37 °C at 200 rpm. The overnight culture was diluted (1:20), and the culture was continued in 250-ml flasks at 37 °C. When the culture reached an absorbance A600 of 0.6, expression of the At1g78690 was induced by the addition of 1 mm isopropyl β-d-thiogalactoside. The culture was then allowed to grow for 3 h at 30 °C. The cells were harvested by centrifugation, and the pellet was resuspended in 500 μl of water. The lipids were extracted according to Ref. 27.
NAPE Synthase Purification
A culture of E. coli C41-transformed At1g78690-His6 cDNA was obtained. After 3 h of isopropyl β-d-thiogalactoside induction, cells were harvested by centrifugation at 5000 rpm at 4 °C. The proteins were solubilized by incubation of the pellet with 10 mm phosphate buffer, pH 8, 0.5 m NaCl, 10 mm n-dodecyl-β-d-maltoside at 4 °C for 1 h. The solution was centrifuged for 20 min at 13,000 × g at 4 °C. The supernatant was used for protein purification.
The At1g78690-His6 was purified using the Ni SepharoseTM 6 Fast Flow kit (GE Healthcare) according to the manufacturer's protocol. The equilibration buffer was 20 mm phosphate buffer, 0.5 m NaCl, 25 mm imidazole, pH 8, and the NAPE synthase was eluted with 20 mm phosphate buffer, 0.5 m NaCl, 200 mm imidazole, pH 8. Protein concentration was determined using the Lowry method with bovine serum albumin as a standard.
Membrane Purification and Acyltransferase Assays
50 ml of E. coli culture were centrifuged, and the pellet was resuspended in 2.5 ml of 0.2 m Tris/HCl (pH 8) buffer. 2.5 ml of 0.2 m Tris/HCl, 1 m sucrose,1 mm EDTA (pH 8) buffer was added and gently mixed before the addition of 25 μl lysozyme (1 g/liter) and 10 ml of water. Digestion of cell walls was performed at 20 °C under gentle shaking for 30 min. Membranes were spun down by centrifugation (15 min at 150,000 × g, using a Hitachi centrifuge). The pellet was washed with 50 mm Tris/HCl (pH 8) buffer before centrifugation (15 min at 150,000 × g, using a Hitachi centrifuge) and resuspended in 50 mm Tris/HCl (pH 8) buffer. The protein concentration was determined using the Lowry method with BSA as a standard.
The NAPE synthase assays were conducted for 10 min at 30 °C in 100 μl of assay mixture (15 mm phosphate buffer, pH 8) containing 1 nmol of labeled acyl donor ([14C]palmitoyl-CoA (60 mCi/mmol), [14C]stearoyl-CoA (58 mCi/mmol), [14C]palmitic acid (56 mCi/mmol) or [14C]stearic acid (51 mCi/mmol)) and in the presence of 1 nmol of PE. The reaction was initiated by the addition of 50 μg of membrane proteins or 40 μg of purified enzyme. Reactions were stopped by the addition of 2 ml of chloroform/methanol (2:1, v/v) and 500 μl of water. The lipids were extracted as described below. The radioactivity incorporated into NAPE was quantified using a Storm 860 PhosphorImager (GE HealthCare).
Histochemical β-Glucuronidase (GUS) Assays
Complete seedlings or tissue cuttings were stained in multiwell plates. Histochemical assays for GUS activity were performed according to the protocol described in Ref. 28. The plants were observed with a Leica binocular loupe (Leica MZ16F) coupled with a camera (DFC 420), and data processing was done with Leica application suite version 2.5.0 R1.
Transient Expression System and Confocal Microscopy
Four-week-old tobacco (Nicotiana tabacum cv. Petit Havana) greenhouse plants grown at 22–24 °C were used for Agrobacterium tumefasciens (strain GV3101)-mediated transient expression as described in Ref. 29.
Lipid Analysis
Polar lipids were separated by two-dimensional TLC using chloroform/methanol/1-propanol/methyl acetate/0.25% KCl (solvent A; 10:4:10:10:3.6, by volume) for migration in the first dimension and using pyridine/chloroform/formic acid (25/15/3.5, by volume) for migration in the second dimension. The lipids were then visualized and quantified by GC, as described by Testet et al. (27).
Lipids were extracted from rosette leaf by the addition of 2 ml of hexane/2-propanol/water (20/55/25, by volume) and heated at 70 °C until total leaf discoloration. The solvent was evaporated to dryness under N2. The lipids were extracted according to Ref. 27. Plant lipids were separated on a Kieselgel G type 60 column. Chloroform, chloroform/methanol (6:1, v/v), and chloroform/methanol (2:1, v/v) were used to elute yellow pigments, chlorophyll + MGDG, and phospholipids, respectively. Each fraction was evaporated under N2 and further analyzed by TLC using solvent A.
Identification of NAPE
PLD Assays
The unidentified lipid synthesized by E. coli transformed with pET15b::At1g78690 was isolated as described previously. The lipid was evaporated to dryness and suspended in 1 ml of diethyl ether. The reaction mix was composed by 235 μl of MES-NaOH 40 mm, CaCl2 15 mm, with 62.5 units of Streptomyces chromofuscus PLD. The reaction was carried out at 32 °C with shaking (120 rpm) for 30 min. The reaction was stopped by the addition of 2 ml of chloroform/methanol (2:1, v/v). Then lipids were extracted and separated by TLC.
Electrospray Ionization Mass Spectrometry
Aliquots of lipids extracted from bacteria transformed by the cDNA of At1g78690 or E. coli (wild type) were dried gently under a stream of N2. The lipids were suspended in methanol solution. All experiments were performed using an ion trap mass spectrometer (LCQ Advantage; Thermo Fisher) fitted with an orthogonal electrospray ionization source, by analyzing negative ions. Methanolic lipid solutions were diluted at 1:10 and infused by means of a syringe pump at a flow rate of 300 μl/h (KD Scientific). The spray voltage was 4.5 kV, the capillary voltage was 10 V, and its temperature was 200 °C. Tandem mass spectrometry was performed in the ion trap by collision-induced dissociation with helium gas buffer at 0.1 pascals. Data were collected and analyzed with XcaliburTM software.
Real Time Reverse Transcription-PCR Conditions and Analysis
Total RNA from 3-week-old leaves of wild type (Col0), tDNA, and overexpressing At1g78690 lines was extracted using the RNeasy plant minikit (Qiagen). Reverse transcription-PCR was carried out with the SuperScript RT II kit (Invitrogen) using 1 μg of total RNA as template and oligo(dT) as primers. To determine the relative transcript levels of At1g78690, reverse transcription-PCR assays were performed using the following oligonucleotides: GGCCAAGTGCTAGACGAAAC as a 5′-primer and CAACCGCTTGGCTAAGAGTC as a 3′-primer. A standard curve was established using serially diluted cDNA. PCR efficiency ranged from 95 to 105%. All samples were assayed in triplicate wells. Real time PCR was performed on an iCyclerTM (Bio-Rad). Samples were amplified in a 25-μl reaction containing 1× SYBR Green Master Mix (Bio-Rad) and a 260 nm concentration of each primer. The thermal profile consisted of one cycle at 95 °C for 3 min followed by 42 cycles at 95 °C for 30 s and at 58 °C for 30 s. For each run, data acquisition and analysis were done using the iCyclerTMQ software (version 3.0a; Bio-Rad). The transcript abundance in tDNA and overexpressing samples relative to Col0 samples was determined using a comparative Ct method. The relative abundance of ACT2 (At1g49240) and EIF4 (At3g13920) mRNA in each sample was determined and used to normalize for differences in total RNA amounts.
RESULTS
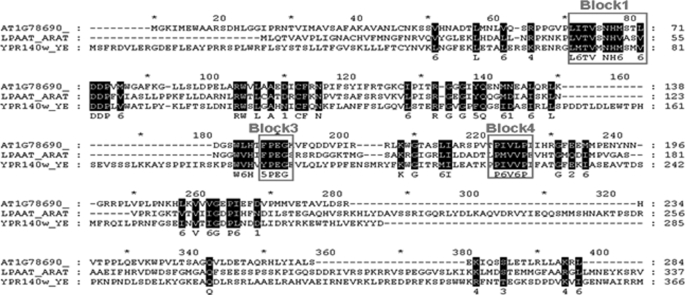
To identify and characterize lysolipid acyltransferases from A. thaliana other than the well characterized lysophosphatidic acid acyltransferase (30), we sought homologies between the lysophosphatidylcholine acyltransferase from S. cerevisiae, (YPR140wp) that we previously described (31) and open reading frames of A. thaliana. Two sequences were identified: At1g78690 and At3g05510. In the present paper, we describe results obtained for At1g78690p (the function of At3g05510p is completely different and will be described in a further article). The At1g78690p sequence presents three of the four conserved domains generally associated with glycerolipid acyltransferases (Fig. 1) and particularly the blocks I (NHX4D) and III (FPEGT) that might be the catalytic sites of these enzymes (32). The calculated molecular mass of At1g78690p is 31.74 kDa for 284 amino acids. The pHi is estimated at 8.84. The sequence analysis (TMpred program; available on the World Wide Web) of At1g78690p predicts the presence of two transmembrane helixes from amino acid 14 to 37 (score 671) and from amino acid 163 to 182 (score 526). However, other prediction sites do not mention these structures (HMMTOP, TMMOD, and PHOBIUS). The presence of myristoylation sites, prenylation sites, and glycosylphosphatidylinositol anchor addition sites was also sought using the NMT, PrePS, and Big-PI Plant Predictor programs (all available on the World Wide Web). The analysis of this sequence with these programs did not evidence post-translational modifications. In addition, no targeting signal was evidenced using WolfPSORT Prediction, Target P, and MITOPROT (all available on the World Wide Web).
FIGURE 1.
Amino acid sequence alignment of A. thaliana At1g78690p with various acyltransferase proteins. Proteins other than At1g78690p are lysophosphatidylcholine acyl-transferase from S. cerevisiae (Ypr140wp) and lysophosphatidic acid acyltransferase from A. thaliana (Slc1). Sequences were analyzed using the multiple alignment program ClustalW version 1.7. Blocks 1, 3, and 4 are characteristic of the acyltransferase family (32).
At1g78690 Synthesizes NAPEs When Expressed in E. coli
The complete cDNA of At1g78690p was transformed into the E. coli C41 strain. After induction with isopropyl β-d-thiogalactoside, the lipids were extracted and analyzed by TLC. As shown in Fig. 2A, the lipid extract of bacteria transformed with pET15b-At1g78690 contained an additional lipid class (RF = 0.68) compared with those of bacteria transformed with the empty plasmid. Following two-dimensional TLC, it appeared that this lipid co-migrated with NAPE standard (Fig. S1). This spot was further scraped from the TLC plate and submitted to hydrolysis catalyzed by S. chromofuscus PLD. After the reaction, lipids were extracted from the incubation mixture and separated on a TLC plate. As shown in Fig. 2B, the hydrolysis of the lipid of interest by PLD led to two products identified as NAE (RF = 0.93) (Fig. 2B) and phosphatidic acid (RF = 0.31). These results are in perfect agreement with the claim that At1g78690p catalyzes the synthesis of NAPE when expressed in E. coli. To check this assumption, the lipid extracts of bacteria transformed with the cDNA of At1g78690p or with the empty vector were further analyzed by mass spectrometry. In contrast with results obtained with the lipid extract from bacteria transformed with the empty vector, several pseudomolecular ions corresponding to NAPE species were detected in the lipid extract purified from bacteria expressing At1g78690p (Fig. S2; m/z 928–1010 mass range). These ions were further characterized by MS/MS, yielding characteristic fragment ions of NAPE species (33, 34). In agreement with the fatty acid composition of E. coli, only NAPEs containing C16:0, C16:1, C18:0, and C18:1 chains were detected. As an example, the MS/MS spectrum of the m/z 980.6 species (pseudomolecular ion [M − H]−) and its fragmentation scheme are shown in Fig. 2, C and D. Diagnostic fragment ions correspond to fatty acid carboxylate anions (palmitoleic acid at m/z 253.1 and oleic acid at m/z 281.1) and to the losses of these carboxylic acids. As already described for this type of compound (34), the loss of the sn-2-carboxylate was more abundant than the loss from the sn-1-position (m/z 727.3 > m/z 699.3, and m/z 253.1 > m/z 281.1; Fig. 2C). Furthermore, the characteristic neutral loss of the N-acylethanolamine moiety following the first loss of carboxylate was also observed (m/z 417.1 and 389.1), making it possible to assign the m/z 980.6 ion to N-stearic-1-oleic-2-palmitoleic-3-phosphatidylethanolamine. More generally, results obtained by MS and MS/MS analysis are consistent with the production of NAPEs in E. coli cells expressing At1g78690p. Therefore, we assumed that the At1g78690 gene codes for a NAPE synthase from A. thaliana, but this hypothesis remained to be confirmed both in vitro and in planta.
FIGURE 2.
Identification of the lipid synthesized following the expression of At1g78690p in E. coli. A, one-dimensional TLC of polar lipids from E. coli transformed by empty pET15b (control) or pET15b-At1g78690 plasmids. CL, cardiolipin. B, identification of NAPE by PLD hydrolysis assay. Lane 1, standard NAE; lane 2, standard NAPE; lane 3, lipid of interest incubated 1 h without PLD; lane 4, lipid of interest incubated 1 h with PLD. PA, phosphatidic acid; C, negative ion ESI tandem mass spectrum of N-stearic-1-oleic-2-palmitoleic-3-phosphatidylethanolamine (m/z 980.6). D, fragmentation pattern of NAPE, illustrated by N-stearic-1-oleic-2-palmitoleic-3-phosphatidylethanolamine. The m/z values refer to the expected m/z.
In Vitro Analysis of the At1g78690p Activity
The ability of At1g78690p to synthesize NAPE was analyzed by performing enzymatic assays with 14C-labeled free fatty acids (C16:0 or C18:0) or labeled acyl-CoAs (C16:0 or C18:0) and unlabeled PE as substrates. Initial experiments were carried out by using membranes purified from E. coli transformed with the empty vector pET15b (control) or with pET15b-At1g78690 plasmid (50 μg of proteins/assay; 10-min incubation). Results are shown in Table 1. Whatever the experimental conditions used, no significant amount (<1 pmol) of NAPE was synthesized by membranes purified from E. coli transformed with the empty vector. A similar result was obtained when membranes purified from bacteria expressing At1g78690p were incubated with PE and FFA. By contrast, the same membranes synthesized about 33 pmol of NAPE when palmitoyl-CoA or stearoyl-CoA was used as acyl donor.
TABLE 1.
Determination of the acyl donor
In vitro synthesis of NAPE was catalyzed with membranes from E. coli transformed by empty pET15b or pET-15b ::At1g78690 plasmids (50 μg of protein/assay), or IMAC-purified enzyme (40 μg of protein/assay). Assays were carried out with 1 nmol of [14C]palmitoyl-CoA, [14C]stearoyl-CoA, [14C]palmitic acid, or [14C]stearic acid and 1 nmol of PE (no exogenous PE was added in the case of E. coli membrane); qs 100 μl of phosphate buffer (10 mm, pH 8), 30 °C, 120 rpm. After a 10-min incubation, lipids were extracted and analyzed by TLC using chloroform/methanol/1-propanol/methyl acetate/0.25% aqueous KCl (10:4:10:10:3.6, v/v/v/v/v) as solvent followed by radioimaging. Values represent mean ± S.D. (n = 3).
| NAPE synthesized |
||||
|---|---|---|---|---|
| C16:0 FFA | C16:0-CoA | C18:0 FFA | C18:0-CoA | |
| pmol | ||||
| E. coli pET15b membranes | ||||
| 0 min | 0.09 ± 0.00 | 0.11 ± 0.09 | 0.16 ± 0.12 | 0.10 ± 0.09 |
| 10 min | 0.06 ± 0.04 | 0.26 ± 0.07 | 0.17 ± 0.02 | 0.12 ± 0.02 |
| E. coli pET15b ::At1g78690 membranes | ||||
| 0 min | 0.19 ± 0.04 | 0.08 ± 0.02 | 0.12 ± 0.11 | 0.01 ± 0.01 |
| 10 min | 1.23 ± 0.31 | 32.51 ± 1.19 | 0.07 ± 0.05 | 33.65 ± 0.97 |
| Purified protein | ||||
| 0 min | 0.11 ± 0.02 | 0.20 ± 0.06 | 0.02 ± 0.01 | 0.01 ± 0.02 |
| 10 min | 0.07 ± 0.02 | 1.82 ± 0.31 | 0.24 ± 0.04 | 3.72 ± 0.09 |
In a second set of experiments, we used the protein At1g78690p fused with a poly-His tag purified on an IMAC column. The NAPE synthase activity associated with the purified fraction was further determined in the presence of PE and FFA or acyl-CoA as above (40 μg proteins/assay, 10-min incubation). As in the case of in vitro assays catalyzed by E. coli membranes, when FFA was used as the acyl donor, no NAPE was detected in the reaction mixtures after a 10-min incubation (Table 1). By contrast, 1.82 and 3.72 pmol of NAPE were synthesized when the purified fraction was incubated with palmitoyl-CoA or stearoyl-CoA (Table 1). Moreover, since we previously showed that under some specific conditions, imidazole is able to catalyze a chemical (i.e. non-enzymatic) synthesis of NAPE (31), we checked that under the experimental conditions used with the purified fraction, the imidazole-containing buffer used to elute the protein did not induce a significant synthesis of NAPE (in fact, 0.09 ± 0.01 pmol of NAPE was observed without protein). The amount of NAPE synthesized by the purified enzyme could appear quite low, but it must be kept in mind that the presence of a poly-His tag can alter the enzyme activity. In addition, as often observed with membrane-bound enzymes using amphiphilic substrates (35), the activity of the solubilized enzyme was lower than the activity associated with biological membranes. Nevertheless, taken together, the present results clearly indicate that (i) At1g78690p is able to synthesize NAPE and (ii) at least in our experimental conditions, the acyl donors used to synthesize NAPEs are acyl-CoAs and not FFA like the NAPE synthase activity from cottonseeds (24, 25).
Study of At1g78690 in A. thaliana
To complete the functional characterization of the protein under study in planta, we purified homozygous lines of the At1g78690 mutant from the SALK tDNA mutant collection. The tDNA insertion in the SALK 029716 has been located in the gene promoter. In addition, we also generated overexpressing lines by placing the At1g78690 cDNA under the control of the cauliflower mosaic virus 35 S promoter (35S::At1g78690) in tDNA and Col0 plants. Four independent overexpressing lines were used (tDNA-At1NS oe-1, tDNA-AtNS oe2, Col0-AtNS oe1, and Col0-AtNS oe2) in experiments described below.
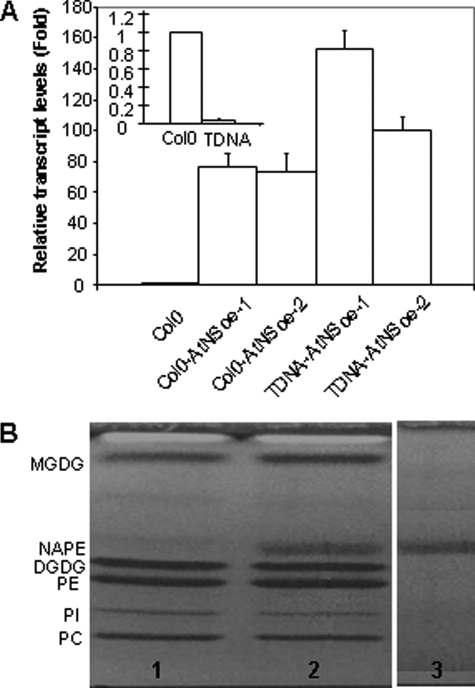
The transcript level of At1g78690 was measured in rosette leaves from 3-week-old plants of Col0, tDNA and overexpressing lines by semiquantitative reverse transcription-PCR. By comparison with that observed with Col0 plants, a weak (25-fold lower) and an elevated (from 70- to 150-fold higher) transcript level was measured in tDNA and in overexpressing lines respectively (Fig. 3A). Lipids were then extracted from rosette leaves from 3-week-old plants of tDNA, Col0, and overexpressing lines and further analyzed by TLC (Fig. 3B). As expected, the major glycerolipids in Col0 and tDNA plants were phosphatidylcholine, phosphatidylinositol, PE, digalactosyldiacylglycerol, and monogalactosyldiacylglycerol, and even in Col0 lines, no NAPE was detected. This result is in agreement with the low amounts of NAPE (∼1% of total phospholipids) detected in various plants by Chapman and Moore (33). However, large amounts of a compound co-migrating with standard NAPE were observed in the lipid extract from leaves of all of the overexpressing plants. To confirm the identity of this lipid, it was scraped from the TLC plates and analyzed by ESI/MS analysis. One NAPE species was unambiguously identified as a 16:0/16:0/16:0 NAPE by MS/MS (Fig. S3), and C16:0, C18:0, and C18:1 methyl esters were detected by GC-MS after transesterification of the fatty acids associated with this lipid. Interestingly, no shorter fatty acids, such as C12 or C14, were detected. Moreover, in the lipid extracts from all of the overexpressing lines studied, NAPE was the only additional lipid detected, and no overproduction of NAEs was evidenced.
FIGURE 3.
Expression of At1g78690 and lipid analysis in A. thaliana. A, determination of At1g78690 mRNA levels by real time PCR in leaves from tDNA, Col0, and overexpressing lines (3 weeks old). Values represent mean ± S.D. (n = 3). B, TLC analysis of lipid extract from A. thaliana. Lane 1, Col0; lane 2, tDNA-AtNS-oe1; lane 3, NAPE standard. PC, phosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; DGDG, Digalactosyldiacylglycerol; NAPE, N-acylphosphatidylethanolamine; MGDG, monogalactosyldiacylglycerol.
Subcellular Localization and Tissue Expression of At1g78690
To establish the subcellular location of the NAPE synthase, we generated fluorescent proteins by fusing YFP to the C terminus or the N terminus of the At1g78690 protein. Constructs were transiently expressed in tobacco leaf epidermal cells. The fusion proteins appeared to be associated with the plasma membrane. Whatever the construct analyzed, we never found any significant label associated with subcellular compartments other than plasmalemma. To confirm that NAPE synthase protein was located in the plasma membrane, we co-expressed At1g78690:YFP with the plasma membrane H+-ATPase (PMA4) from Nicotiana plumbaginifolia, fused with GFP. Fig. 4 clearly shows that both of the fusion proteins co-localized, thus strengthening the plasmalemmic location of At1g78690p.
FIGURE 4.
Localization of At1g78690-YFP in a tobacco leaf epidermal cell. A, plasmalemma marker PMA4-GFP (green); B, At1g78680-YFP (red); C, co-expression of At1g78690-YFP and PMA4-GFP in the plasmalemma of tobacco cell.
To investigate the cell type expression pattern of the NAPE synthase gene, we generated transgenic Arabidopsis lines that expressed the GUS reporter gene under the control of the NAPE synthase gene promoter. We further analyzed GUS expression in embryo of imbibed seed and 3-, 6- and 14-day-old seedlings, in roots, stems, and flowers. Results are shown in Fig. 5.
FIGURE 5.
Spatial expression patterns of NAPE synthase gene in transgenic Arabidopsis harboring the NAPE synthase promoter fused to the GUS gene. Promoter activity was visualized by histochemical GUS staining on embryo (A), 3-day-old seedlings (B), 6-day-old seedlings (C), leaves from 2-week-old plants (D), roots from 2-week-old plants (E), flower (F), cauline leaf and stem (G), and old leaf (H).
In the embryo of imbibed seed, expression was detected in the cotyledons and the hypocotyl. In 3- and 6-day-old germinating seedlings, expression was detected in the cotyledons, the young leaves, the shoot, and the root apical meristems and in the roots. No GUS activity was detected in the elongation zone of the primary root or in the hypocotyl. When expression in all of the rosettes from 2-week-old plants was compared, expression was preferentially associated with young leaves rather than with old leaves. GUS activity declined with leaf maturation but remained constant in roots during seedling development. In flowers, GUS activity was observed in the filament of stamens, in the style, and ovary with eggs of pistil. No activity was detected in the petals, sepals, pollen sacs, anther, or stigma.
DISCUSSION
We show in the present work that an acyltransferase from Arabidopsis (At1g78690p) catalyzes the synthesis of NAPE. The nature of the acyl donor was determined in vitro using membranes from E. coli transformed with At1g78690 and purified NAPE synthase obtained after IMAC chromatography. Palmitic acid, stearic acid, palmitoyl-CoA, and stearoyl-CoA were assayed as substrates. In both cases, NAPE was exclusively synthesized in the presence of acyl-CoAs. These results are not in agreement with those of Chapman and Moore (24), who suggested that the NAPE synthase associated with cottonseed (Gossypium hirsutum) microsomes uses free fatty acids rather than acyl-CoAs as substrate. The enzymologic properties of this enzyme were further determined on the purified enzyme (in DDM micelles) (36). It is therefore possible that plant cells contain an alternative enzyme like the one from cottonseed microsomes that is capable of the direct acylation of PE with FFA. In agreement, by centrifugation on discontinuous sucrose density gradient coupled to aqueous two-phase partitioning, Chapman and Sriparameswaran (25) found that NAPE synthase activity from cottonseed microsomes was associated with ER, plasma membrane, and Golgi membrane fractions. In contrast, following confocal microscopy observations, our study suggests that when expressed in tobacco leaves, At1g78690p is localized exclusively within the plasma membrane, a finding made with YFP fused both at the N-terminal and C-terminal regions of At1g78690p.
Regarding the spatial expression pattern, the use of a proAt1g78690::GUS construction clearly revealed a strong activity of the At1g78690 promoter in the embryo of imbibed seed. This observation is in agreement with results obtained by Chapman et al. (13), who showed that dry seeds contained high concentrations of NAE and that this content decreased sharply after 4 to 8 h of imbibition, while a strong synthesis of NAPE was observed. We also observed a strong expression of the NAPE synthase gene in roots. Since Blancaflor et al. (14) showed that the addition of a micromolar concentration of exogenous NAE12:0 disrupts normal root development in A. thaliana seedlings and since the overexpression of FAAH in A. thaliana enhanced root growth, we compared the root elongation of plants from the different lines for 15 days (tDNA, Col0, and overexpressing lines). No significant difference was observed (data not shown). Such a result is not necessarily in disagreement with previous data, because in the absence of exogenous NAE12:0, no phenotype was observed with plants overexpressing FAAH. Furthermore, their NAE content was reduced only in seeds and not in seedlings (18). Hence, the phenotype observed with these plants is probably due to a high(er) hydrolysis of the exogenous NAE12:0 rather than to a high(er) hydrolysis of NAE produced in roots. In addition, the 35S::At1g78690 lines studied in the present work did not contain NAE12:0, and it was previously shown that changes in root development were observed only when NAE12:0 but not NAE16:0 was added to the reaction medium (14). Moreover, even if high amounts of NAPE were produced in overexpressing lines, their NAE content could remain too low to induce any effect. This hypothesis is strengthened by the absence of increase in the amount of NAE in overexpressing lines, strongly suggesting that the NAE pathway is regulated by PLD (synthesis of NAE from NAPE) or by FAAH (hydrolysis of NAE) rather than by the NAPE synthase itself. In addition, not only did we not observe any difference in the root development, but we did not observe any clear phenotype in the different lines.
Supplementary Material
Acknowledgments
We thank Xavier Santarelli for assistance in purification of NAPE synthase.
This work was supported by the Conseil Regional d'Aquitaine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- NAE
- N-acylethanolamine
- PLD
- phospholipase D
- NAPE
- N-acylphosphatidylethanolamine
- PE
- phosphatidylethanolamine
- MES
- 4-morpholineethanesulfonic acid
- GC
- gas chromatography
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- FFA
- free fatty acid
- FAAH
- fatty amide hydrolase
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- IMAC
- immobilized metal ion affinity chromatography.
REFERENCES
- 1.Hansen H. S., Moesgaard B., Petersen G., Hansen H. H. (2002) Pharmacol. Ther. 95, 119–126 [DOI] [PubMed] [Google Scholar]
- 2.Paria B. C., Dey S. K. (2000) Chem. Phys. Lipids 108, 211–220 [DOI] [PubMed] [Google Scholar]
- 3.De Petrocellis L., Melck D., Bisogno T., Di Marzo V. (2000) Chem. Phys. Lipids 108, 191–209 [DOI] [PubMed] [Google Scholar]
- 4.Sarker K. P., Obara S., Nakata M., Kitajima I., Maruyama I. (2000) FEBS Lett. 472, 39–44 [DOI] [PubMed] [Google Scholar]
- 5.Simon G. M., Cravatt B. F. (2006) J. Biol. Chem. 281, 26465–26472 [DOI] [PubMed] [Google Scholar]
- 6.Liu J., Wang L., Harvey-White J., Osei-Hyiaman D., Razdan R., Gong Q., Chan A. C., Zhou Z., Huang B. X., Kim H. Y., Kunos G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felder C. C., Briley E. M., Axelrod J., Simpson J. T., Mackie K., Devane W. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7656–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar R. N., Chambers W. A., Pertwee R. G. (2001) Anaesthesia 56, 1059–1068 [DOI] [PubMed] [Google Scholar]
- 9.Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodríguez De Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., Piomelli D. (2003) Nature 425, 90–93 [DOI] [PubMed] [Google Scholar]
- 10.Nielsen M. J., Petersen G., Astrup A., Hansen H. S. (2004) J. Lipid Res. 45, 1027–1029 [DOI] [PubMed] [Google Scholar]
- 11.Terrazzino S., Berto F., Dalle Carbonare M., Fabris M., Guiotto A., Bernardini D., Leon A. (2004) FASEB J. 18, 1580–1582 [DOI] [PubMed] [Google Scholar]
- 12.Lambert D. M., Vandevoorde S., Jonsson K. O., Fowler C. J. (2002) Curr. Med. Chem. 9, 663–674 [DOI] [PubMed] [Google Scholar]
- 13.Chapman K. D., Venables B., Markovic R., Blair R. W., Jr., Bettinger C. (1999) Plant Physiol. 120, 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blancaflor E. B., Hou G., Chapman K. D. (2003) Planta 217, 206–217 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Guo W. M., Chen S. M., Han L., Li Z. M. (2007) J. Plant Physiol. 164, 993–1001 [DOI] [PubMed] [Google Scholar]
- 16.Tripathy S., Venables B. J., Chapman K. D. (1999) Plant physiol. 121, 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilaru A., Blancaflor E. B., Venables B. J., Tripathy S., Mysore K. S., Chapman K. D. (2007) Chem. Biodivers. 4, 1933–1955 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. S., Shrestha R., Kilaru A., Wiant W., Venables B. J., Chapman K. D., Blancaflor E. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12197–12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang L., Wang Y. S., Uppalapati S. R., Wang K., Tang Y., Vadapalli V., Venables B. J., Chapman K. D., Blancaflor E. B., Mysore K. S. (2008) Plant J. 56, 336–349 [DOI] [PubMed] [Google Scholar]
- 20.Sandoval J. A., Huang Z. H., Garrett D. C., Gage D. A., Chapman K. D. (1995) Plant Physiol. 109, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiratsuchi A., Ichiki M., Okamoto Y., Ueda N., Sugimoto N., Takuwa Y., Nakanishi Y. (2009) J. Biochem. 145, 43–50 [DOI] [PubMed] [Google Scholar]
- 22.Schmid H. H., Schmid P. C., Natarajan V. (1990) Prog. Lipid Res. 29, 1–43 [DOI] [PubMed] [Google Scholar]
- 23.Jin X. H., Okamoto Y., Morishita J., Tsuboi K., Tonai T., Ueda N. (2007) J. Biol. Chem. 282, 3614–3623 [DOI] [PubMed] [Google Scholar]
- 24.Chapman K. D., Moore T. S., Jr. (1993) Plant Physiol. 102, 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman K. D., Sriparameswaran A. (1997) Plant Cell Physiol. 38, 1359–1367 [Google Scholar]
- 26.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 27.Testet E., Laroche-Traineau J., Noubhani A., Coulon D., Bunoust O., Camougrand N., Manon S., Lessire R., Bessoule J. J. (2005) Biochem. J. 387, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beeckman T., Engler G. (1994) Plant Mol. Biol. Rep. 12, 37–42 [Google Scholar]
- 29.Chatre L., Brandizzi F., Hocquellet A., Hawes C., Moreau P. (2005) Plant Physiol. 13, 1244–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H. U., Li Y., Huang A. H. C. (2005) Plant Cell 17, 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Testet E., Akermoun M., Shimoji M., Cassagne C., Bessoule J. J. (2002) J. Lipid Res. 43, 1150–1154 [DOI] [PubMed] [Google Scholar]
- 32.Lewin T. M., Wang P., Coleman R. A. (1999) Biochemistry 38, 5764–5771 [DOI] [PubMed] [Google Scholar]
- 33.Chapman K. D., Moore T. S., Jr. (1993) Arch. Biochem. Biophys. 301, 21–33 [DOI] [PubMed] [Google Scholar]
- 34.Hansen H. H., Hansen S. H., Bjørnsdottir I., Hansen H. S. (1999) J. Mass Spectrom. 34, 761–767 [DOI] [PubMed] [Google Scholar]
- 35.Bessoule J. J., Lessire R., Cassagne C. (1989) Biochim. Biophys. Acta 983, 35–41 [DOI] [PubMed] [Google Scholar]
- 36.McAndrew R. S., Chapman K. D. (1998) Biochim. Biophys. Acta 1390, 21–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.