Abstract
Most pathogenic Proteus species are primarily associated with urinary tract infections, especially in persons with indwelling catheters or functional/anatomic abnormalities of the urinary tract. Urinary tract infections caused by Proteus vulgaris typically form biofilms and are resistant to commonly used antibiotics. The Rts1 conjugative plasmid from a clinical isolate of P. vulgaris carries over 300 predicted open reading frames, including antibiotic resistance genes. The maintenance of the Rts1 plasmid is ensured in part by the HigBA toxin-antitoxin system. We determined the precise mechanism of action of the HigB toxin in vivo, which is distinct from other known toxins. We demonstrate that HigB is an endoribonuclease whose enzymatic activity is dependent on association with ribosomes through the 50 S subunit. Using primer extension analysis of several test mRNAs, we showed that HigB cleaved extensively across the entire length of coding regions only at specific recognition sequences. HigB mediated cleavage of 100% of both in-frame and out-of-frame AAA sequences. In addition, HigB cleaved ∼20% of AA sequences in coding regions and occasionally cut single As. Remarkably, the cleavage specificity of HigB coincided with one of the most frequently used codons in the AT-rich Proteus spp., AAA (lysine). Therefore, the HigB-mediated plasmid maintenance system for the Rts1 plasmid highlights the intimate relationship between host cells and extrachromosomal DNA that enables the dynamic acquisition of genes that impart a spectrum of survival advantages, including those encoding multidrug resistance and virulence factors.
Toxin-antitoxin (TA)3/addiction/suicide modules typically include an autoregulated operon encoding a labile antitoxin and a more stable toxic protein (1). TA toxins facilitate stress survival (chromosomal) or plasmid maintenance and post-segregational killing (extrachromosomal; reviewed in Refs. 1, 2). Most chromosomal TA toxins inhibit cell growth by reversibly targeting either protein translation or DNA replication; their cognate antitoxins prevent toxin activity during periods of optimal growth but enable finely tuned control of TA module toxicity during relatively short periods of environmental stress. However, prolonged stress leads to a point of no return and cell death (3–5).
There are six confirmed chromosomal TA loci in Escherichia coli K12 cells: dinJ-yafQ, relBE, yefM-yoeB, mazEF, chpBI-BK, and hipBA. The toxins MazF and ChpBK are sequence-specific endoribonucleases that cleave free mRNA (6–10). The RelE toxin interacts with the ribosome and induces mRNA cleavage with a preference for the UAG stop codon (11–13). The YafQ toxin is a ribosome-associated endoribonuclease that cleaves in-frame AAA codons that are followed by either an A or G in the subsequent codon (14). The YoeB toxin inhibits translation at the initiation step, apparently by destabilization of the initiation complex (15). HipA toxin is a kinase whose mechanism of action is not known (16, 17).
Although the mechanism of action of many E. coli chromosomal and plasmid-derived toxins has been determined, the precise function of the HigB toxin has not been characterized. The higBA TA module is not present in E. coli K12; it resides on the Rts1 plasmid that typically replicates in Proteus spp. and imparts kanamycin resistance as well as temperature-sensitive post-segregational killing at 42 °C (18, 19). Interestingly, one or more chromosomal counterparts of higBA have been reported for several pathogens, including Vibrio cholerae, Streptococcus pneumoniae, E. coli CFT073, and E. coli O157:H7 (20). Some characterization of the two V. cholerae HigBA modules has been performed. First, one of the two higBA modules was shown to possess the general characteristics of TA systems by demonstration of toxin-antitoxin interaction, module organization/regulation, HigB toxicity, and rescue of toxicity with the cognate HigA antitoxin (21). Overexpression of HigB derived from two individual higBA modules encoded in V. cholerae or from Rts1 leads to inhibition of protein synthesis through translation-dependent mRNA cleavage in a manner similar to, but distinct from, RelE (22).
HigB is a member of the RelE family of toxins, including RelE, YafQ, and YoeB (20). In this study, we have identified the precise mode of action of HigB from Rts1. HigB associated with the 50 S ribosomal subunit, and this HigB-ribosome complex cleaved within mRNA coding regions at all AAA triplet sequences, both in-frame and out-of-frame. HigB appeared to be responsible for the mRNA cleavage activity of the HigB-ribosome complex because a HigB H92Q mutant lacked mRNA cleavage activity but remained associated with the ribosome. Finally, the cleavage specificity of HigB on plasmid Rts1 coincided with the sequence (AAA, lysine) of either the most abundant or the second most abundant codon in its Proteus host.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Reagents
The E. coli strains BL21(DE3) (F− ompT hsdSβ(rβ-mβ) dcm gal (DE3) tonA) (Novagen) and BW25113 (lacIq rrnBT14 Δlac-ZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) were used for all protein expression and toxicity studies. E. coli K12 Mach1 T1 cells (ΔrecA1398 endA1 tonA Φ80ΔlacM15 ΔlacX74 hsdR(rk+mk+); Invitrogen) were used for all cloning experiments. The higBA module was cloned from the Rts1 plasmid (a generous gift from Dr. Roger Woodgate, National Institutes of Health). The E. coli BW25113 strain with deleted chpBI-BK, hipBA, mazEF, relBE, yefM-yoeB, and dinJ-yafQ modules (BW25113 Δ6) was a generous gift from Qian Tan and Yoshi Yamaguchi of the M. Inouye laboratory. Plasmids used in this study include pBAD24 (23), pET28a (Novagen), and pBAD/Myc-HisA (Invitrogen). The higBA operon was PCR-amplified from the Rts1 vector with 5′ BamHI/HindIII 3′ ends and cloned into the corresponding sites of pET28a to create pET28a-higBA(His)6 with the His6 tag fused onto higB. The higB ORF was PCR-amplified from the Rts1 vector with 5′ NdeI/Xho 3′ ends and cloned into the corresponding sites of pBAD24 to create pBAD24-higB. The relE ORF was PCR-amplified from BW25113 cells with 5′ NdeI/Xho 3′ ends and cloned into the corresponding sites of pBAD24 to create pBAD24-relE. The higA ORF was PCR-amplified from the Rts1 vector with 5′ BamHI/HindIII 3′ ends and cloned into the corresponding sites of pET28a to create pET28a-higA(His)6. The higB ORF was PCR-amplified from the Rts1 vector with 5′ Nco/HindIII 3′ ends, and an additional GC on the 5′ end to maintain frame was cloned into the corresponding sites of pBAD/Myc-HisA to create pBAD/Myc-HisA-higB(His)6. The higB ORF was PCR-amplified from the Rts1 vector with 5′ NdeI/Xho 3′ ends and a mutated H92Q amino acid and cloned into the corresponding sites of pBAD24 to create pBAD24-higB(H92Q). All bacterial liquid cultures were grown in M9 minimal media (M9) supplemented with either 0.2% glucose or 0.21% glycerol at 37 °C, unless otherwise noted. The working concentrations of ampicillin, kanamycin, and chloramphenicol were 100, 40, and 34 μg/ml, respectively. The accuracy of the DNA sequences of PCR products used for cloning was confirmed by automated DNA sequence analysis. Polyclonal antibodies used for Western analysis were produced in rabbits using the HigBA complex as antigen (Pocono Rabbit Farm and Laboratory, Canadensis, PA).
DNA, RNA, and Protein Synthesis in Vivo
[35S]Methionine (24), [methyl-3H]thymidine, and [methyl-3H]uracil (22) incorporation studies were carried out as described previously.
Analysis of mRNA Levels in E. coli
Total RNA was extracted using the hot phenol method as described previously (25). The radiolabeled DNA fragments used for Northern analysis were derived from PCR products, including open reading frames of the E. coli genes lpp (major outer membrane lipoprotein), ompA (outer membrane porin protein A), ompF (outer membrane porin protein F), rpsA (ribosomal protein subunit A), or tufA (EF-Tu). RNA levels were quantified with a phosphorimager, and all sample values were normalized to their respective control (i.e. the wild type uninduced values).
Ribosome Profile Analysis
Ribosomal extracts were prepared as described previously (14); equivalent amounts of total RNA from S30 extracts were layered onto each continuous sucrose gradient.
Primer Extension
Primer extension analysis was carried out as described previously (14). Primers used were as follows: lpp, 5′-TTACTTGCGGTATTTAGTAGCC-3′; ompA3, 5′-CCGCCAGCGAAGACCGGAGAAACGCCGG-3′; ompA5, 5′-TCAGAACCGATGCGGTCGGTGTAACCC-3′; ompF, 5′-AAACCAAGACGGGCATAGGTC-3′; rpsA, 5′-CGTCAACTTCGTCACCTACC-3′; and tufA, 5′-GAGAAGTGTTGATGGTGATACC-3′.
Recombinant HigA and HigB
The higA(His)6-pET28a and higB(His)6-pBAD/Myc-HisA constructs were transformed into BL21(DE3) and BW25113 cells, respectively, and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside (higA(His)6-pET28a) or 0.2% arabinose (higB(His)6-pBAD/Myc-HisA) for 6 h. Cells were disrupted using a French press; the protein extracts were applied to nickel-nitrilotriacetic acid resin (Qiagen), and the His6-tagged proteins were purified as recommended by Qiagen. The protein content of each eluted fraction was visualized by 17.5% SDS-PAGE followed by Coomassie staining.
Determination of HigB(His)6 Activity in Vitro
Prokaryotic cell-free protein synthesis was carried out with the E. coli T7 S30 extract system for circular DNA (Promega); the HigA only, HigB only, and HigB/HigA samples contained 2 μg of HigA(His)6, 0.5 μg of HigB(His)6, or 2 μg of HigA(His)6 plus 0.5 μg of HigB(His)6, respectively. The HigB/HigA samples were preincubated for 30 min at 4 °C prior to the addition of total RNA and the samples incubated for an additional 30 min at 37 °C.
Total cellular RNA was isolated from BW25113 cells as described above. 10 μg of total RNA was added to a reaction containing increasing amounts of pure recombinant HigB in 10 mm Tris, pH 7.8.
RESULTS
Expression of HigB Inhibits Translation
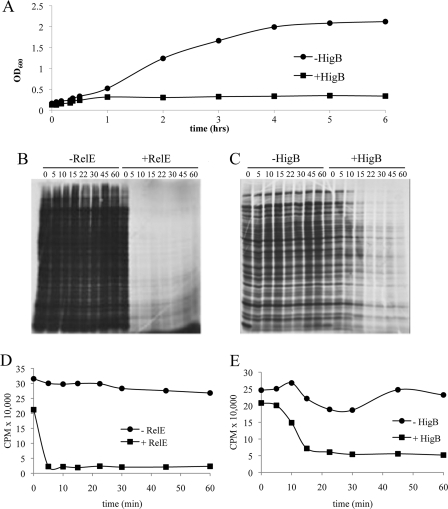
Although it has been reported that HigB inhibits protein synthesis through translation-dependent mRNA cleavage (22), we characterized the enzymatic activity of HigB (92 amino acids, 10.7 kDa) from the Rts1 plasmid in more detail and compared its properties to family member RelE. We first expressed relE or higB in the arabinose-inducible pBAD24 plasmid. We found that the high toxicity of either RelE or HigB precluded the isolation of a stable strain containing either toxin gene in pBAD24 under noninducing conditions with glycerol as the carbon source. Therefore, in this study we used glucose under noninducing conditions and arabinose/glycerol for induction to minimize leaky toxin expression. As an additional precaution, all experiments were performed with strains that were recently transformed with the relevant toxin-expressing plasmid to preclude the accumulation of genomic suppressors of toxicity. To test whether induction using this unconventional method (glucose to arabinose/glycerol) was effective, we monitored growth after induction of higB or relE at mid-logarithmic phase. Expression of either higB (Fig. 1A) or relE (data not shown) resulted in growth arrest by 30 min. Interestingly, although both toxins arrested growth at approximately the same time, striking differences in [35S]methionine incorporation were observed upon toxin induction (Fig. 1, B–E). RelE expression led to an extremely rapid shut down of translation (to ∼5% of wild type within 5 min), whereas a more gradual decrease in protein synthesis was noted upon HigB expression; only at the 22-min time point did the effect of HigB remain constant (where translation levels dropped to 20% of wild type levels). This and previously published data (22) are consistent with a model whereby RelE and HigB cause growth arrest by blocking translation. In fact, in vivo incorporation of radiolabeled precursors of DNA or RNA revealed that expression of HigB did not lead to significant defects in DNA replication or RNA transcription (supplemental Fig. 1).
FIGURE 1.
Expression of HigB leads to translation arrest. A, growth profile of HigB-induced (arabinose +) or uninduced (arabinose −) BW25113 cells containing pBAD24-higB grown at 37 °C in M9 medium containing either glucose (uninduced) or glycerol (induced). B and C, [35S]methionine incorporation of RelE- or HigB-induced versus uninduced cells in vivo. Equivalent amounts of cell lysate, derived from equal culture volumes, were subjected to SDS-PAGE, followed by autoradiography. D and E, quantification of [35S]methionine incorporation into RelE- or HigB-induced (arabinose +) or uninduced (arabinose −) cells. Counts/min were the result of trichloroacetic acid-precipitated counts taken from a single aliquot per time point. Time points correspond to those in B; cell samples for experiments in D and E were derived from the growth-profile experiment shown in A.
Expression of HigB Leads to Rapid and Extensive Degradation of Several mRNAs in Vivo
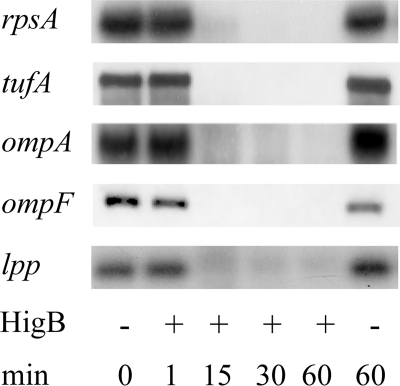
HigB or RelE expression results in degradation of lpp mRNA only when the start codon is intact (11, 22). To determine whether HigB affects the stability of several transcripts, we performed Northern analysis for five E. coli mRNAs: lpp, ompA, ompF, rpsA, and tufA (Fig. 2). Although no degradation was detectable immediately after (1 min) induction of HigB expression, degradation was extensive after 15 min; after 60 min, degradation of all five transcripts were complete or nearly complete. In comparison, the steady state levels of the respective control mRNAs (cells harboring the plasmid but not expressing HigB) were stable. No visible degradation products were detected below the full-length mRNA in any of the +HigB lanes.
FIGURE 2.
Expression of HigB decreases steady state levels of several mRNAs in vivo. RNA was extracted from HigB-expressing cells followed by Northern analysis. No visible degradation products were detected below the full-length mRNA in any of the +HigB lanes.
To ensure that the mRNA degradation we observed upon HigB induction was not due to activation of other TA toxins resulting from the inherent stress of HigB expression, we expressed HigB in an E. coli strain (“Δ6”) in which each of the six known chromosomal TA modules has been deleted (mazEF, relBE, chpBI-BK, hipBA, yefM-yoeB, and dinJ-yafQ). The levels of full-length lpp, ompA, and rpsA transcripts were compared in Δ6 and BW25113 cells before and after induction of the toxin. We found no difference in the steady state levels of each of the three mRNAs in the two strains after HigB induction (supplemental Fig. 2). The same experiments were also performed with RelE, and no difference was observed (supplemental Fig. 2). Therefore, HigB or RelE expression appears to lead to global degradation of cellular mRNA in vivo, and this effect does not enlist the action of other toxins.
HigB Associates with the Ribosome through the 50 S Ribosomal Subunit
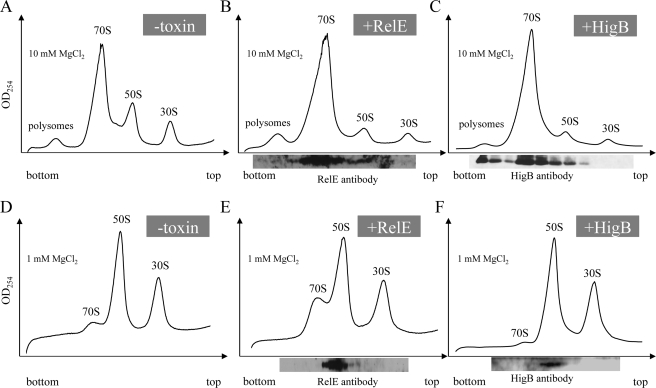
Mutagenesis of the lpp AUG start codon to AAG prevents lpp mRNA degradation in vivo, revealing that the action of HigB is translation-dependent (as is family member RelE) (22). These data suggest that HigB acts through interaction with the ribosome. Therefore, we analyzed ribosome profiles from cells overexpressing HigB for 60 min (when cells were completely growth-arrested and cellular mRNA degradation in Fig. 2 was maximal) to assess whether toxin expression altered the profile (Fig. 3, compare A and C). We also used Western analysis of fractions spanning the sucrose gradient to determine whether we were able to identify an association of HigB with the ribosome (Fig. 3C). We did not observe additional peaks or substantial changes in polysome, 70 S, 50 S, or 30 S sedimentation. However, HigB led to the increase in the relative abundance the 70 S monosome because we reproducibly noted a higher relative ratio of 70 S monosome compared with both 50 S/30 S ribosomal subunit peaks upon HigB induction (compare Fig. 3, A and C). In extracts/gradients containing the standard 10 mm Mg2+, HigB associated primarily with 70 S monosomes, but it was also detected with the 50 S ribosomal subunit and polysomes. However, HigB was not associated with the 30 S ribosomal subunit (Fig. 3C). In addition, buffer conditions favoring dissociation of the 70 S ribosome to its component subunits (1 mm Mg2+) confirmed that HigB associates with only the 50 S ribosomal subunit and not the 30 S. Thus, HigB associates with the ribosome through the 50 S ribosomal subunit in vivo (Fig. 3F).
FIGURE 3.
HigB and RelE associate with ribosomes in vivo. A–C, ribosomes prepared and fractionated in 10 mm Mg2+; D–F, 1 mm Mg2+. Ribosome profiles of wild type BW25113 cells (A and D), RelE (B and E), and HigB (C and F) are shown. Western blots (shown below fraction numbers tested) in B, C, E, and F were performed using antibody directed against either RelE or HigB. All 10 mm Mg2+ ribosome profiles were repeated three times.
Other members of the RelE family, YafQ (14) and YoeB (15), associate with ribosomes through the 50 S subunit. Therefore, we analyzed the ribosome profile for RelE association with these fractions just as we had for HigB to determine whether this is a common property of RelE family members (Fig. 3, compare A and B). The ribosome profile for RelE was similar to that for HigB, with a higher relative ratio of 70 S to 50/30 S peaks, and as with the other three RelE family members tested, it interacted with the ribosome through the 50 S subunit (Fig. 3, B and E).
HigB Facilitates Ribosome-dependent Cleavage of mRNA at A-rich Sequences
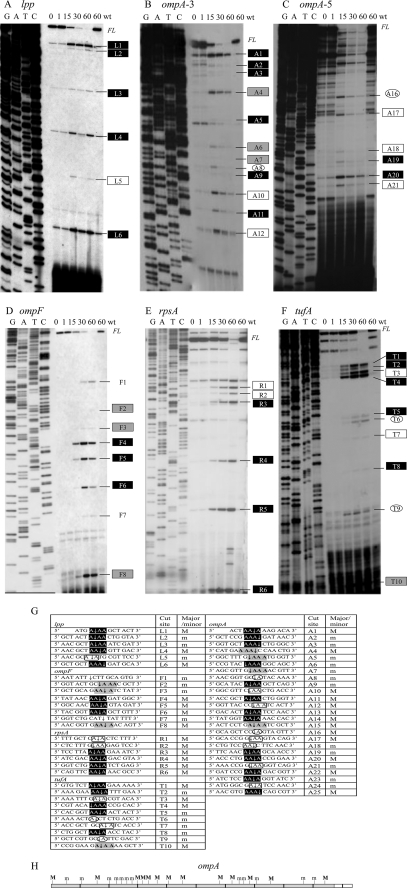
Because HigB associated with the ribosome and facilitated extensive mRNA cleavage, we determined the sequence specificity and frame dependence of its cleavage activity and recognition sequence. We isolated total RNA from HigB induced and uninduced cells and performed primer extension analysis on the five mRNAs (lpp, ompA, ompF, rpsA, and tufA) that were degraded upon HigB expression in Fig. 2. We initially used a primer ∼150 nucleotides downstream of the 5′ end of the translation start site for each mRNA and then performed additional primer extension experiments to enable nearly complete coverage of lpp (237 bp) and ompA (1041 bp; Fig. 4, A–F). In total, we detected 55 HigB-specific cleavage products among all mRNAs (Fig. 4G). Of these, 30 were major sites representing relatively abundant cleavage products, and 25 were minor sites whose intensities were approximately ≤25% that of the major sites. Note that HigB is a very potent toxin; even though we used glucose prior to induction to minimize leaky expression of the toxin gene, some HigB-mediated cleavage was detected even in the uninduced time points. Therefore, we were careful to choose only those primer extension products that increased with induction time relative to the uninduced control.
FIGURE 4.
HigB-mediated sequence-specific, frame-independent mRNA cleavage. Primer extension analysis of the following: A, lpp; B, ompA-3; C, ompA-5; D, ompF; E, rpsA; and F, tufA mRNAs extracted from cells expressing HigB. Exact cleavage positions of mRNAs identified near the top of the gel were determined with closer primers (data not shown); FL, full-length products; wt, wild type. Labeled cut sites correspond to those reported in G. Note that some HigB-mediated cleavage was detected even in the uninduced time points; only those products that increased with induction time relative to the uninduced control were highlighted. G, summary of HigB cleavage sites (spacing denotes translational frame) as follows: in-frame AAAs (black rectangle); out-of frame AAAs (gray rectangle); AA (open rectangle); and single A (open oval). The two unmarked sites for ompF correspond to putative RNase E sites. H, location of major and minor cleavage products for the ompA mRNA. Cleavage sites in the region corresponding to the white box at 3′ end could not be detected; this region was where the terminal primer annealed. The drawing is to scale, with each black line demarcating 100 nucleotides.
Analysis of the sequence and location of all cleavage sites revealed several features of HigB-mediated cleavage as follows. 1) 5′-AAA-3′ is the primary recognition consensus sequence (highlighted in black and in gray in Fig. 4G). We found that HigB cleaved 100% of the AAA sequences covered in our primer extension analyses. The regions we tested contained 38 AAA sites, and we documented cut sites for all of them (23 major and 15 minor).
2) HigB targets both in-frame and out-of-frame AAA or AAA-containing sequences for cleavage (compare black versus gray highlighted sequences in Fig. 4G).
3) HigB also cleaved stretches of four or more A-rich sequences; moreover, HigB cleaved within the first three AAAs in A4 or A5 stretches in six of the seven examples we uncovered.
4) The location of the cleavage site in the AAA site (or the first three As in A4 or A5 sequences) was not fixed but instead exhibited a preference for cleavage after the second A, i.e. 5′-AA↓A-3′. More specifically, 45% were 5′-AA↓A-3′, 29% 5′-A↓AA-3′, 13% 5′↓AAA-3′, and 13% 5′-AAA↓-3′.
5) HigB cleaves throughout the length of the mRNA (see illustration for ompA, Fig. 4H) with no apparent pattern for major and minor site distribution.
6) HigB also recognized AA sequences with much lower efficiency (20% of the 55 cut sites; open rectangles in Fig. 4G) and occasionally cut at single A sequences (4/55, 7%; open ovals, Fig. 4G).
7) Two of the 55 HigB-specific cleavage sites (both in ompF) did not cut at an A residue. Instead, the sequences 5′-AAUAUU↓CUUGC-3′ (F1) and 5′-GCAU↓UAUUUUU-3′ (F7) have similarity to RNase E consensus sites (26, 27). RNase E is a single strand-specific endoribonuclease required for rapid mRNA decay and accurate RNA processing in E. coli. Therefore, HigB cleavage of ompF and other mRNAs may be assisted by RNase E and the degradasome (28).
8) We did not detect cleavage sites in the 5′-untranslated regions upstream of the five mRNAs, corroborating our data demonstrating HigB ribosome association and earlier studies demonstrating that lpp mRNA must have an intact start Met codon for HigB-mediated degradation (11).
HigB Does Not Cleave RNA in Vitro
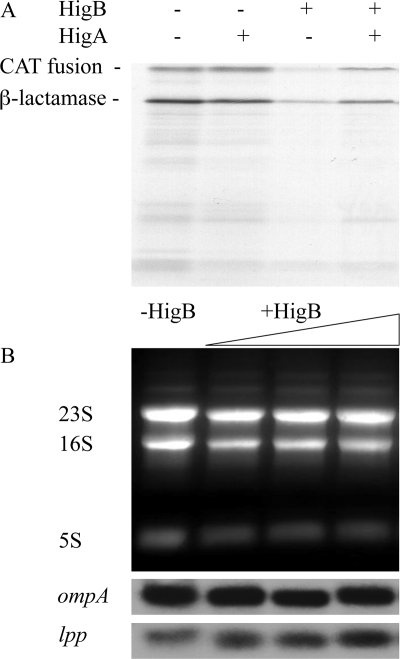
To better characterize the mechanism of HigB-mediated mRNA cleavage, we assessed whether HigB possessed endoribonuclease activity in vitro. As a first step, we purified recombinant HigB toxin and confirmed that it was active using a coupled in vitro transcription/translation assay with or without the addition of HigB toxin or with both HigB toxin/HigA antitoxin. Recombinant HigB inhibited in vitro translation, and the effects of HigB were reversed by addition of the antitoxin HigA (Fig. 5A). Therefore, these recombinant proteins were active after purification.
FIGURE 5.
HigB inhibits translation but does not degrade mRNA in vitro. A, recombinant HigB inhibits coupled in vitro transcription/translation (3rd lane); HigA can rescue the inhibition and reconstitute translation (4th lane) to normal levels (as shown in 1st lane); the product of the chloramphenicol acetyltransferase (CAT) fusion template (39 kDa) includes one of the two major reaction products, and the other β-lactamase (28 kDa) product is present at high levels in T7 S30 extracts because of transcription from the T7 promoter upstream of the chloramphenicol acetyltransferase fusion that reads through into the ampicillin resistance gene. B, HigB does not cleave rRNA or mRNA in vitro. 1st lane, 10 μg of total RNA; 2nd, 3rd, and 4th lanes, increasing amounts of purified HigB(His)6 were added to 10 μg of total RNA.
We next tested if the same HigB preparation was able to cleave total RNA purified from E. coli K12. No cleavage was detected in either mRNA or rRNA after a 30-min incubation of total RNA with increasing concentrations of HigB. There was also no effect on the RNA when the HigA antitoxin was added to the HigB (Fig. 5B and data not shown). When the incubation time was increased to 6 h, we also did not detect RNA cleavage. These results suggest that the HigB-mediated in vivo cleavage activity is dependent upon interaction with the ribosome.
HigB(H92Q) Abolishes Toxicity and mRNA Cleavage but Not Ribosome Binding
X-ray crystal structures of RelE, YoeB, and accompanying mutagenesis studies (29, 30) implicated a highly conserved histidine near their carboxyl termini as important for their toxicity. However, subsequent studies on YoeB demonstrated that mutagenesis of this conserved histidine (H83Q) did not lead to a loss in toxicity or a reduction in its primary function as an inhibitor of translation initiation (15). In contrast, mutagenesis of the corresponding residue in YafQ (31) (H87Q) abolished YafQ toxicity and endoribonuclease activity in vivo (14).
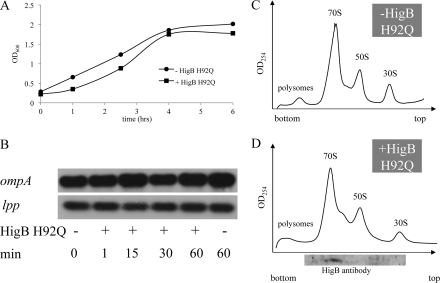
Because HigB is in the RelE family, we aligned all family members and noted that HigB from the Rts1 plasmid and E. coli CFT073 also contained a highly conserved histidine (His-92 for Rts1 and His-90 for E. coli CFT073) at the carboxyl-terminal end of their primary sequences. Because active HigB did not exhibit detectable ribonuclease activity in vitro, it may mediate mRNA cleavage only upon association with the ribosome in vivo. Alternatively, as suggested for RelE (13), there was a formal possibility that the HigB mRNA cleavage activity we documented in vivo was because of an enhancement of the intrinsic mRNA cleavage activity of the ribosome (32). Therefore, to differentiate between these possible modes of action, we created an H92Q mutant in Rts1 HigB and examined the growth rate of the mutant strain upon induction of HigB(H92Q). Unlike wild type HigB, induction of HigB(H92Q) no longer resulted in growth arrest (Fig. 6A), indicating that His-92 is essential for HigB-mediated toxicity. We then determined the effect of HigB(H92Q) expression on the steady state levels of ompA and lpp transcripts and found no difference compared with the wild type control (cells harboring the plasmid but not expressing HigB(H92Q)) (Fig. 6B), i.e. mRNA was no longer degraded in cells expressing the HigB(H92Q) mutant. Finally, ribosome profile analysis revealed that the mutant HigB(H92Q) toxin retained its ability to associate with the ribosome (Fig. 6, C and D). Therefore, the endoribonuclease activity of the HigB-ribosome complex appears to be conferred by HigB, and this activity requires His-92.
FIGURE 6.
HigB(H92Q) expression results in loss of toxicity (A) and mRNA cleavage (B) but not ribosome binding (C and D).
DISCUSSION
The HigBA TA system is essential for plasmid maintenance of the kanamycin-resistant Rts1 plasmid (19). Notably, although the E. coli K12 chromosome contains only a higA homolog, its pathogenic relatives E. coli CFT073 and E. coli O157:H7 appear to possess one or more copies of the entire module (20). In fact, the chromosomes of many pathogenic bacteria contain one or more higBA modules; this is also true for many bacteria with slow growth rates or characteristically long dormant periods (20). Toward a better understanding of how this toxin imparts mRNA degradation, a concomitant block in protein synthesis, and growth arrest (21, 22), we determined that the ribosome-HigB complex targets actively translated mRNA for cleavage at A-rich sequences. HigB cuts 100% of in-frame and out-of-frame AAA sequences in the coding regions of mRNAs (including the first three AAAs of stretches of 4 or more As). HigB also cuts AA sequences but with much lower efficiency (20%), and only occasionally cuts at single As.
The conjugative Rts1 plasmid was obtained from a clinical isolate of Proteus vulgaris (33). Analysis of the available codon usage data for P. vulgaris (323 coding regions, the majority comprising genes of the Rts1 plasmid) revealed that HigB cleaves at a target sequence that corresponds to the most common codon, AAA (lysine 41.0/1000 codons). The second most common codon, GAA (glutamic acid 38.3/1000 codons), also contains an AA HigB cleavage site.
However, Rts1 can also replicate in Proteus mirabilis (34), which causes the majority of Proteus urinary tract infections. Analysis of the codon usage data for the complete genome of P. mirabilis revealed that HigB cleaves at a target sequence that corresponds to the second most common codon, AAA, encoding lysine (46.5/1000 codons) for the P. mirabilis genome (leucine is the most used codon at 48.6/1000 codons). Although leucine is also the most common codon in E. coli K12 (52.82/1000 codons), AAA is represented at lower frequency (33.62/1000 codons) than in P. mirabilis.
Although the complete genome sequence of P. vulgaris has not been reported, it is AT-rich (61%) (35) as is the genome of P. mirabilis (also 61% AT) (36). For comparison, the E. coli K12 genome has a lower AT content (49%) (37). Curiously, the sequenced P. mirabilis strain contains a plasmid distinct from Rts1 that is designated pHI4320. Not only is AAA lysine the most common codon in this plasmid, it is almost twice as abundant compared with its host genome (76.3/1000 codons). Perhaps Rts1 has adapted to have a selective advantage when a Proteus strain contains both Rts1 and pHI4320 plasmids such that any free HigB would selectively degrade mRNA of its competitor plasmid. More specifically, if only Rts1 is cured, not only would the remaining stable HigB toxin degrade host mRNA, it should also degrade the competitor plasmid even more efficiently, ensuring an advantage for retention of Rts1 over pHI4320. In summary, these data suggest that the HigB cleavage site exploits the Achilles' heel of its host (i.e. A-rich sequences both in-frame or out-of-frame) to maximize its potency when conditions lead to plasmid loss and warrant post-segregational killing.
Studies in E. coli K12 have also linked the presence of an AAA codon at +2 to high translation initiation efficiency (38, 39). AAA is also the most common codon at +2, represented in ∼10% of all E. coli K12 mRNAs containing AUG or GUG start codons (39). Purine-rich sequences, especially adenine-rich ones, are correlated with single strand regions in 16 S rRNA (40). Thus the higher translational efficiency when A-rich sequences are found 3′ of the translation start site may be due to the absence of mRNA secondary structure. The high AT content of the P. vulgaris and P. mirabilis genomes suggests that these strains may also contain a higher than average number of AAA codons early in the mRNA. In fact, we surveyed 100 random open reading frames from the P. mirabilis genome and found an even higher percentage (17%) of AAAs at +2 than E. coli K12. Therefore, HigB acts on multiple levels of its characteristically AT-rich host cells. First, it targets one of the most abundant codons (AAA) of its host cell for cleavage. Second, it also cleaves all AAA-containing sequences independent of frame. Third, it cleaves specific AAs (∼20% of the total) and occasionally cleaves specific single As. Fourth, targeting highly expressed mRNAs containing AAA at +2 may enhance the potency of HigB. In concert, this multitiered mechanism ensures rapid and efficient post-segregational killing of hosts that lose the Rts1 plasmid.
The analysis of the molecular mechanism of HigB completes the characterization of all four representative members of the RelE family as follows: RelE, YafQ, and YoeB from E. coli K12 and HigB from plasmid Rts1. All four family members act through the association with the 50 S ribosomal subunit of the 70 S ribosome. However, each imparts toxicity through distinct mechanisms. YafQ possesses ribonuclease activity in vitro and in vivo but acquires its frame-dependent, sequence-specific activity only upon association with the ribosome in vivo (14). HigB only acquires ribonuclease activity upon association with the ribosome and cleaves its recognition sequences on translating mRNAs both in- and out-of-frame. In contrast, neither RelE nor YoeB alone exhibits endoribonuclease activity in vitro (13, 15), and mRNA cleavage is only secondary for YoeB-mediated inhibition of translation initiation in vivo (15). The in vivo mRNA cleavage activity associated with RelE has been linked to the endonucleolytic activity of the ribosome (32).
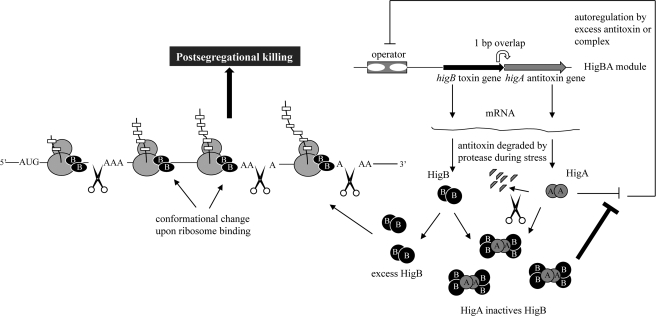
We propose the following model of HigB-mediated post-segregational killing (Fig. 7). Because we documented HigB-mediated cleavage throughout the length of mRNAs (see Fig. 4H), HigB appears to associate with actively translating 70 S ribosomes through the 50 S ribosomal subunit. HigB may also associate with the 50 S ribosomal subunit as it assembles into a 70 S monosome based on the following. 1) It can cleave mRNA very early, at the second codon of lpp (Fig. 4G); and 2) it was also detected in the 50 S fraction in 10 mm Mg2+ profiles (Fig. 3C). The interaction of HigB with ribosomes appears to cause an allosteric change in HigB that activates its site-specific endoribonuclease activity. HigB then travels with the translating ribosome and cleaves the mRNA when it reaches the first major or minor A-rich consensus sequence; however, our data do not enable us to distinguish if HigB cleaves at the first consensus site it reaches or subsequent ones. This extensive cleavage results in rapid mRNA degradation and inhibition of translation elongation. As a consequence, the tmRNA system for rescuing stalled ribosomes is overwhelmed, resulting in slower ribosome recycling evidenced by the reproducibly higher 70 S peak (that we propose represents stalled ribosomes) as well as the presence of HigB in the disome fraction in Fig. 3C (because ribosomes without bound HigB will not be able to continue translating past the stalled ribosome at the HigB cut site). The high AT content of the Proteus host genomes are remarkably complementary to the overall mechanism of HigB action and may portend the existence of a similar adaptation for chromosomal HigB counterparts.
FIGURE 7.
Features of HigB-HigA function in E. coli. Toxin and antitoxin mRNAs are synthesized from an autoregulated operon, and the open reading frame of the toxin overlaps the antitoxin gene by 1 bp (19). HigBA is distinct from other TA systems because the toxin gene precedes the antitoxin. Both HigB/HigA and HigA autoregulate the operon by binding to the operator consisting of two subsites (subsite one includes the palindrome 5′-TATTGCACATCGTGTAATA-3′, and subsite two includes the palindrome 5′-GTATTACACACCATGTAATAC-3′) (18). HigA/HigB-mediated repression is 4-fold higher than that by HigB alone (18). HigB associates with actively translating 70 S ribosomes through the 50 S ribosomal subunit. The interaction of HigB with ribosomes appears to cause an allosteric change in HigB that activates its site-specific endoribonuclease activity (represented by a change in HigB shape). HigB then travels with the translating ribosome and cleaves the mRNA when it reaches the first major or minor A-rich consensus sequence. The tmRNA system cannot accommodate the extensive mRNA degradation, resulting in slower ribosome recycling (shown by HigB disomes). This unchecked block in translation elongation leads to bacterial cell death (i.e. post-segregational killing for cells that have lost the Rts1 plasmid).
Supplementary Material
Acknowledgments
We thank Roger Woodgate for providing the Rts1 plasmid and Yoshi Yamaguchi for the construction and use of the BW25113 Δ6 strain. We also thank the members of the laboratory for manuscript proofreading; Yonglong Zhang for the RelBE polyclonal antibody; Koichi Inoue for guidance; and Masayori Inouye for extensive advice and expertise.
This work was supported, in whole or in part, by National Institutes of Health Grants GM74097 and U54 Grant GM74958 (NE Structural Genomics Consortium). This work was also supported by Takara Bio, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- TA
- toxin-antitoxin
- ORF
- open reading frame.
REFERENCES
- 1.Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 2.Engelberg-Kulka H., Glaser G. (1999) Annu. Rev. Microbiol. 53, 43–70 [DOI] [PubMed] [Google Scholar]
- 3.Amitai S., Yassin Y., Engelberg-Kulka H. (2004) J. Bacteriol. 186, 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelberg-Kulka H., Hazan R., Amitai S. (2005) J. Cell Sci. 118, 4327–4332 [DOI] [PubMed] [Google Scholar]
- 5.Pedersen K., Christensen S. K., Gerdes K. (2002) Mol. Microbiol. 45, 501–510 [DOI] [PubMed] [Google Scholar]
- 6.Christensen S. K., Pedersen K., Hansen F. G., Gerdes K. (2003) J. Mol. Biol. 332, 809–819 [DOI] [PubMed] [Google Scholar]
- 7.Muñoz-Gómez A. J., Santos-Sierra S., Berzal-Herranz A., Lemonnier M., Díaz-Orejas R. (2004) FEBS Lett. 567, 316–320 [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Zhang J., Hara H., Kato I., Inouye M. (2005) J. Biol. Chem. 280, 3143–3150 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., Inouye M. (2003) Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Zhu L., Zhang J., Inouye M. (2005) J. Biol. Chem. 280, 26080–26088 [DOI] [PubMed] [Google Scholar]
- 11.Christensen S. K., Gerdes K. (2003) Mol. Microbiol. 48, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 12.Christensen S. K., Mikkelsen M., Pedersen K., Gerdes K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003) Cell 112, 131–140 [DOI] [PubMed] [Google Scholar]
- 14.Prysak M. H., Mozdzierz C. J., Cook A. M., Zhu L., Zhang Y., Inouye M., Woychik N. A. (2009) Mol. Microbiol. 71, 1071–1087 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Inouye M. (2009) J. Biol. Chem. 284, 6627–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correia F. F., D'Onofrio A., Rejtar T., Li L., Karger B. L., Makarova K., Koonin E. V., Lewis K. (2006) J. Bacteriol. 188, 8360–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumacher M. A., Piro K. M., Xu W., Hansen S., Lewis K., Brennan R. G. (2009) Science 323, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Q. B., Hayashi T., Murata T., Terawaki Y. (1996) Biochem. Biophys. Res. Commun. 225, 679–684 [DOI] [PubMed] [Google Scholar]
- 19.Tian Q. B., Ohnishi M., Tabuchi A., Terawaki Y. (1996) Biochem. Biophys. Res. Commun. 220, 280–284 [DOI] [PubMed] [Google Scholar]
- 20.Pandey D. P., Gerdes K. (2005) Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budde P. P., Davis B. M., Yuan J., Waldor M. K. (2007) J. Bacteriol. 189, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen-Dalsgaard M., Gerdes K. (2006) Mol. Microbiol. 62, 397–411 [DOI] [PubMed] [Google Scholar]
- 23.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirashima A., Inouye M. (1973) Nature 242, 405–407 [DOI] [PubMed] [Google Scholar]
- 25.Sarmientos P., Sylvester J. E., Contente S., Cashel M. (1983) Cell 32, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 26.Ehretsmann C. P., Carpousis A. J., Krisch H. M. (1992) Genes Dev. 6, 149–159 [DOI] [PubMed] [Google Scholar]
- 27.Redko Y., Tock M. R., Adams C. J., Kaberdin V. R., Grasby J. A., McDowall K. J. (2003) J. Biol. Chem. 278, 44001–44008 [DOI] [PubMed] [Google Scholar]
- 28.Carpousis A. J. (2007) Annu. Rev. Microbiol. 61, 71–87 [DOI] [PubMed] [Google Scholar]
- 29.Kamada K., Hanaoka F. (2005) Mol. Cell 19, 497–509 [DOI] [PubMed] [Google Scholar]
- 30.Takagi H., Kakuta Y., Okada T., Yao M., Tanaka I., Kimura M. (2005) Nat. Struct. Mol. Biol. 12, 327–331 [DOI] [PubMed] [Google Scholar]
- 31.Motiejūnaite R., Armalyte J., Markuckas A., Suziedeliene E. (2007) FEMS Microbiol. Lett. 268, 112–119 [DOI] [PubMed] [Google Scholar]
- 32.Hayes C. S., Sauer R. T. (2003) Mol. Cell 12, 903–911 [DOI] [PubMed] [Google Scholar]
- 33.Terawaki Y., Takayasu H., Akiba T. (1967) J. Bacteriol. 94, 687–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terawaki Y., Rownd R. (1972) J. Bacteriol. 109, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falkow S., Ryman I. R., Washington O. (1962) J. Bacteriol. 83, 1318–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson M. M., Sebaihia M., Churcher C., Quail M. A., Seshasayee A. S., Luscombe N. M., Abdellah Z., Arrosmith C., Atkin B., Chillingworth T., Hauser H., Jagels K., Moule S., Mungall K., Norbertczak H., Rabbinowitsch E., Walker D., Whithead S., Thomson N. R., Rather P. N., Parkhill J., Mobley H. L. (2008) J. Bacteriol. 190, 4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blattner F. R., Plunkett G., 3rd, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., Mau B., Shao Y. (1997) Science 277, 1453–1474 [DOI] [PubMed] [Google Scholar]
- 38.Brock J. E., Paz R. L., Cottle P., Janssen G. R. (2007) J. Bacteriol. 189, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenström C. M., Jin H., Major L. L., Tate W. P., Isaksson L. A. (2001) Gene 263, 273–284 [DOI] [PubMed] [Google Scholar]
- 40.Wang H. C., Hickey D. A. (2002) Nucleic Acids Res. 30, 2501–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.