Abstract
Antiviral immune responses are initiated through Toll-like receptors (TLRs) and RIG-I (retinoic acid-inducible gene-I)-like RNA helicases that recognize nucleic acids from distinct viruses. In this study, we show that the tyrosine kinase c-Src participates in antiviral responses induced by the cytoplasmic RNA helicase RIG-I. Sendai virus (SV), which is recognized by RIG-I, induced c-Src phosphorylation. Functional impairment of c-Src through chemical inhibition or transient expression of a c-Src kinase-inactive mutant attenuated production of endogenous antiviral proteins after SV infection or after expression of RIG-I or its adapter protein MAVS. Importantly, SV-stimulated synthesis of antiviral proteins was significantly impaired in cells treated with c-Src small interfering RNA and in cells from c-Src-deficient mice. In addition, we found that c-Src interacted with components of the RIG-I pathway: RIG-I, MAVS, and TRAF3 (tumor necrosis factor receptor-associated factor-3). The interaction between c-Src and TRAF3 was found to occur within the RING domain of TRAF3. Taken together, our results suggest that c-Src enhances RIG-I-mediated signaling, acting at the level of TRAF3.
Sensing of viral infections by the innate immune system depends on pattern recognition receptors (PRRs),2 such as Toll-like receptors (TLRs) and the DEX(D/H) box RNA helicases RIG-I (retinoic acid-inducible gene I) and MDA-5 (melanoma differentiation-associated protein-5). Transmembrane TLRs, localized to endosomes, and cytoplasmic RNA helicases bind viral nucleic acids and initiate distinct intracellular signaling pathways to produce type I interferons (IFNs), thereby mounting antiviral responses (1, 2). These PRRs differ with respect to their cellular expression, viral recognition, and subcellular localization. TLR3, TLR7/8, and TLR9 respond to nucleic acids in endosomes, whereas RIG-I and MDA-5 bind cytoplasmic nucleic acids from RNA viruses. Thus, TLRs and RIG-I-like helicases provide immune protection against distinct viruses (3, 4). Regarding the subcellular localization of nucleic acid-binding TLRs, it has been shown that the large parts of TLR3 and TLR9 are localized in the endoplasmic reticulum with stimulus-dependent translocation to endosomes (5, 6). Due to their homology with TLR9, TLR7 (in mice) and TLR8 (in humans) are also believed to bind viral nucleic acids present in endosomes. The relative importance and involvement of TLRs and RIG-I like helicases are cell type-specific (7). Plasmacytoid dendritic cells appear to utilize the TLR system, whereas, for example, fibroblasts, macrophages, and monocyte-derived dendritic cells signal through the RNA helicase RIG-I.
RIG-I and MDA-5 contain an amino-terminal caspase recruitment domain (CARD) that associates with the CARD-containing mitochondrial protein MAVS (mitochondrial antiviral signaling protein) (also called VISA, Cardif, or IPS-1; see Ref. 8). Both the CARD domain and mitochondrial anchoring domain of MAVS have been shown to be essential for antiviral signaling events that activate nuclear factor-κB (NF-κB) and IFN-β production (9).
Binding of viral nucleic acids to TLRs or to DEX(D/H) box RNA helicases in endosomes or cytosol stimulates recruitment of specific adapter proteins and signaling components that induce intracellular signaling pathways converging on TBK1 (TRAF family member-associated NF-κB activator-binding kinase 1). TBK1 is a noncanonical IκB-kinase-related kinase that phosphorylates and activates the transcription factor IRF-3, thus integrating signaling from both endosomal and cytosolic receptors recognizing viral RNA or DNA (10, 11). IRF-3, along with NF-κB, is critical for transcription of type I interferons (12).
TLRs may associate with different cytoplasmic Toll-interleukin-1 receptor domain-containing adapters (13). TLR-dependent induction of type I IFNs in response to viral infections occurs through the adapter protein MyD88 (myeloid differentiation factor 88) (TLR7/8 and TLR9) or TRIF (Toll-interleukin-1 receptor domain-containing adapter inducing interferon-β) (TLR3). It was recently shown that IFN-β production and NF-κB activation through TLRs are mediated by TRAF3 (tumor necrosis factor receptor-associated factor-3) and TRAF6, respectively (14, 15). TRAF3-mediated activation of TBK1 and IRF-3 is necessary for RIG-I/MAVS-dependent IFN-β induction (14, 15). Hence, different TLR adapters and either TRAF3 or TRAF6 regulate signal specificity and gene transcription.
The Src family kinases are nonreceptor tyrosine kinases that regulate diverse arrays of cellular responses, including immune responses, integrin signaling, motility, and carcinogenesis. We have previously shown that the tyrosine kinase c-Src is implicated in double-stranded RNA-elicited TLR3-dependent activation of the transcription factors IRF-3 and STAT1. Moreover, we found that c-Src acted downstream of the TLR3 adapter protein TRIF. In this study, we have investigated if c-Src also contributes to TLR-independent antiviral responses mediated through RIG-I. Our results show that c-Src enhances RIG-I-mediated signaling to IRF-3 activation and production of antiviral proteins. Moreover, we show that c-Src associates with the RING domain of TRAF3, thus providing a molecular explanation for its involvement in both RIG-I and TLR-dependent signaling.
EXPERIMENTAL PROCEDURES
Reagents
The monoclonal antibody to FLAG (M2) was from Sigma. Antibodies against phosphorylated forms of kinases and against STAT1, IRF3, and Src were purchased from Cell Signaling Technology. Antibodies to TRAF3 and α-tubulin were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against MAVS were from Alexis biochemicals (immunoprecipitation) and Cell Signaling Technology (immunoblotting). PP2 and PP3 were purchased from Calbiochem. Sendai virus (Cantell strain) was bought from Charles River Laboratories.
Plasmids
The TRAF3-FLAG construct and the Gal4/IRF-3 system were gifts from Dr. Hans Hacker (School of Medicine, University of California, San Diego) and Dr. Tom Maniatis (Harvard University, Cambridge, MA), respectively. The plasmids for luciferase reporter constructs containing the full IFN-β p125 promoter, MAVS-FLAG, RIG-I-FLAG, RIG-I ΔC-FLAG, TBK1-FLAG, and IRF3-FLAG have been described previously. The TRAF3 Δ1.1, TRAF3 Δ2.2, TRAF3 Δ3, TRAF3 Δ6.4, TRAF3 Δ7, TRAF3 Δ9.4, TRAF3 Δ10.2, and pSG5-FLAG TRAF3 constructs were generous gifts from Dr. Carl F. Ware (16). SrcK297R and Src wild-type in pUSE were purchased from Upstate Biotechnology, Inc.
Transfections
For reporter gene studies, HEK293 cells were seeded in 96-well plates 24 h prior to transfection with Genejuice (Novagen) following the manufacturer's instructions. Forty ng of each plasmid (Gal4/IRF-3, Gal4-luc, IFN-β p125 promoter) were used. The Renilla luciferase pRL-TK vector (Promega) was in some experiments cotransfected for normalization. Cell lysates were prepared, and reporter gene activity was measured using the luciferase assay system or the dual luciferase assay system (Promega). All of the reporter gene experiments were repeated at least twice. When increasing amounts of inactive SrcK297R were used, the total amount of vector backbone DNA was kept constant by adding empty vector.
Immunoprecipitation
Cells were seeded into 6-well plates or 100-mm plates 24 h prior to transfection or treatment. After 24 h of transfection or the indicated stimulation, the cells were lysed in 500 μl or 1 ml of lysis buffer containing 50 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, 0,5% Triton X-100, 2 mm EDTA, 40 mm glycerophosphate, 100 mm NaF, 200 μm Na3VO4, 10 μg/ml leupeptin, 1 μm pepstatin, and 1 mm phenylmethylsulfonyl fluoride. Lysates were clarified by centrifugation and incubated with 1–4 μg of antibody overnight at 4 °C. The immunocomplexes were recovered by brief centrifugation after a 1-h incubation with protein G-Sepharose (Nycomed; Amersham Biosciences) at 4 °C, washed four times with lysis buffer, resuspended in NuPage sample buffer, and assayed on NuPage gels (Invitrogen). Representative results from 2–4 separate experiments are shown.
Immunoblotting
Following treatment, cells were washed once in PBS and lysed in lysis buffer, as described above. Cell lysates were clarified by centrifugation, separated by SDS-PAGE, and electrophoretically transferred to Hybond-C nitrocellulose membranes (GE Healthcare) using the NuPAGE gel system from Invitrogen. The membranes were blocked in Tris-buffered saline containing 5% nonfat dry milk and 0.25% Tween 20 for 1 h at 22 °C before incubation with the indicated antibodies overnight at 4 °C. After washing three times with Tris-buffered saline containing 0.25% Tween 20, immunoreactive proteins were detected using horseradish peroxidase-conjugated secondary antibody and ECL detection reagent (Pierce).
RNA Interference
Small interfering RNA (siRNA) oligonucleotides targeting c-Src and TRAF3 were synthesized by Dharmacon (c-Src Smart pool siRNA) and Santa Cruz Biotechnology, respectively. siRNA duplexes were transfected into cells in 6-well plates or confocal culture dishes using Lipofectamine RNAi max (Invitrogen) or X-tremeGENE (Roche Applied Science) siRNA transfection reagents according to the manufacturer's instructions. Chloramphenicol acetyltransferase (CAT) siRNA (Qiagen) was used as a control siRNA. 48 h after transfection, further experiments were performed.
Real Time PCR
Total RNA was isolated from cell lysates using the RNA isolation kit (Qiagen) and stored at −80 °C. cDNA was synthesized from RNA using cDNA synthesis kit (Bio-Rad). cDNA was subjected to quantitative real time PCR (qRT-PCR) analysis on a Chromo4 using the iQ SYBR Green Supermix (Bio-Rad). The specificity of amplification was assessed by melting curve analysis. Relative quantification was performed using the 2−ΔΔCt method (17), and the data were normalized against β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. The following primer pairs have been used: for humans, β-actin (5′-agcctcgcctttgccga (forward) and 5′-ctggtgcctggggcg (reverse)), IFN-β (5′-gccgcattgaccatctatgaga (forward) and 5′-gagatcttcagtttcggaggtaac (reverse)), IP-10 (5′-tcgaaggccatcaagaattt (forward) and 5′-gctcccctctggttttaag (reverse)), GAPDH (5′-gaaggtgaaggtcggagtc (forward) and 5′-gaagatggtgatgggatttc (reverse)); for mice, β-actin (5′-acccacactgtgcccatcta (forward) and 5′-gccacaggattccataccca (reverse)), IFN-β (5′-actgcctttgccatccaag (forward) and 5′-gcagttgaggacatctccca (reverse)), IP-10 (5′-tcatcctgctgggtctgagtgg (forward) and 5′-cgctttcattaaattcttgatggtc (reverse)), GAPDH (5′-ggaagggctcatgaccaca (forward) and 5′-ccgttcagctctgggatgac (reverse)). The primer pair used for amplification of the SV P-protein has been reported previously (18). The results presented are representative of 2–4 independent experiments.
Confocal Microscopy
Confocal microscopy studies were performed with a Zeiss Axiovert 100-M inverted microscope equipped with an LSM 510 laser-scanning unit and a 1.4 numerical aperture ×63 Plan-Apochromat oil immersion objective. Cells were seeded on glass bottom 35-mm tissue culture dishes (Matek). The cell chamber was heated to 37 °C using a Tempcontrol Digital 37-2 device (Warner Instruments). To minimize photobleaching, laser power was typically 20% under maximum, and the pinhole was set to 0.8–1.2. Multitracking was used for dual or triple color imaging. The Zeiss LSM Image browser version 3 was used for acquisition, and processing was completed using Adobe Illustrator CS2 and Microsoft Office Visio Professional 2003.
Immunofluorescence
Cells were fixed in phosphate-buffered saline containing 1% paraformaldehyde for 30 min or in 100% methanol for 1 min on ice. Nonspecific antibody sites were blocked with phosphate-buffered saline containing 2.5% bovine serum albumin, 20% A+ serum, and 0.2% saponin for 30 min at room temperature. For staining, cells were incubated with antibodies diluted in phosphate-buffered saline containing 2.5% bovine serum albumin, 20% A+ serum, and 0.2% saponin for 30 min at room temperature.
RESULTS
The Tyrosine Kinase c-Src Is Implicated in Type I Interferon Production in Response to SV Infection
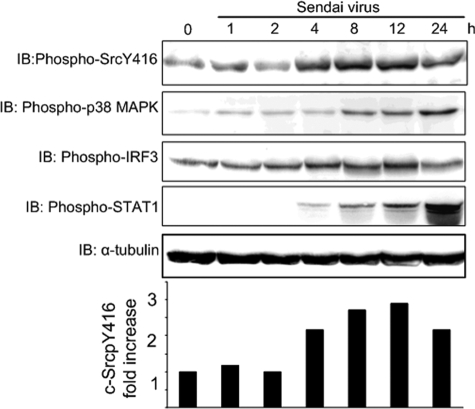
Viral nucleic acids may initiate immune responses through different cellular sensors, such as TLRs or RIG-like helicases (9). We have previously shown that c-Src is activated by double-stranded RNA and is implicated in double-stranded RNA-stimulated, TLR3-mediated IRF-3 activation (6). To elucidate if c-Src is additionally involved in IFN-β production through the cytosolic receptor RIG-I, we examined if c-Src is implicated in SV-induced responses in HEK293 cells. It has previously been shown that down-regulation of RIG-I mRNA using RIG-I siRNA reduces SV-elicited IFN-β production in HEK293 cells by ∼80% (19). Moreover, the same authors showed that transfection of full-length RIG-I synergistically enhanced IFN-β production after SV infection and that a dominant negative mutant of RIG-I abolished SV elicited IFN-β production. Hence, these results suggest that SV-elicited antiviral gene induction is mediated by RIG-I in HEK293 cells. To examine if c-Src is activated through RIG-I, HEK293 cells were infected with SV for various times, and c-Src activation was assessed by immunoblotting using an antibody specific for phosphorylated c-Src (SrcpY416), since c-Src activation depends on autophosphorylation of Tyr416 (20). Band intensities were quantified, and -fold induction of c-Src phosphorylation relative to medium-treated cells is shown in the lower panel of Fig. 1. SV stimulated c-Src phosphorylation after 4 h of viral infection, giving maximal induction after 8–12 h of infection. We also compared the kinetics of SV-induced c-Src activation with phosphorylation and activation of p38 mitogen-activated protein kinase, IRF-3, and STAT1, which displayed kinetics and amplitude of activation different from c-Src phosphorylation (Fig. 1). SV-elicited STAT1 phosphorylation was markedly delayed relative to c-Src activation, which is expected, since STAT1 is activated subsequently to type I IFN production (21). Epidermal growth factor has previously been shown to induce c-Src phosphorylation in HEK293 cells (22). We also compared SV- and epidermal growth factor-stimulated c-Src phosphorylation in HEK293 cells. SV stimulated c-Src phosphorylation to a slightly higher extent than epidermal growth factor (comparing 6 and 14 h of SV infection with 15 min of epidermal growth factor treatment; data not shown).
FIGURE 1.
The tyrosine kinase c-Src is activated during SV infection. HEK293 cells were exposed to SV (300 HAU/ml) for various times and analyzed by immunoblotting for activating phosphorylation of c-Src, p38, IRF-3, and STAT1. Immunoblotting (IB) for α-tubulin was performed to assess protein loading. Band intensities were quantified using the Kodak image analysis software, and -fold induction of c-Src phosphorylation relative to medium-treated cells is shown in the lower panel.
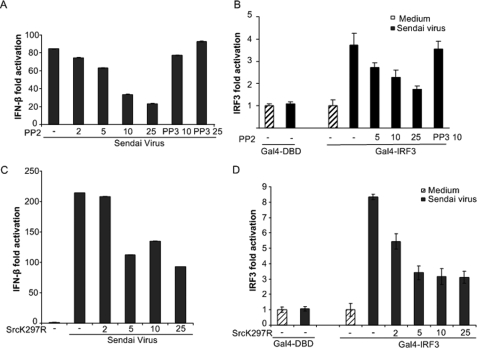
Next, we examined the effect of the Src kinase inhibitor PP2 on SV-induced IFN-β promoter activation. HEK293 cells were transfected with an IFN-β luciferase reporter gene and treated with PP2 or its inactive analogue PP3 prior to SV infection and analysis of IFN-β promoter-regulated luciferase activity. The Src kinase inhibitor PP2 dose-dependently attenuated IFN-β reporter gene transcription, whereas PP3 had no effect (Fig. 2A). The transcription factor IRF-3 is necessary for IFN-β production (12). To further explore the role of c-Src in SV-stimulated antiviral responses, we examined the effect of c-Src inhibition on SV-elicited IRF-3 activation using a Gal4-based IRF-3-dependent reporter assay, in which the DNA-binding domain of Gal4 has been cloned in frame with IRF-3 lacking its DNA binding domain (Gal4-IRF3). HEK293 cells were transfected with Gal4-IRF3 and a luciferase reporter under control of the Gal4 upstream activation sequence prior to treatments. As shown in Fig. 2B, the Src kinase inhibitor PP2 significantly reduced SV-stimulated IRF-3 activation. Next, we examined the effect of ectopic expression of a kinase-inactive variant of c-Src (K297R), acting as a dominant negative protein, on SV-induced activation of IFN-β and IRF-3 reporters. Kinase-inactive c-Src dose-dependently reduced induction of IFN-β and IRF-3 activation after SV infection (Fig. 2, C and D). Hence, these results indicate that RIG-I-mediated c-Src activation contributes to IRF-3 activation and IFN-β production.
FIGURE 2.
Inhibition of c-Src function attenuates SV-regulated IFN-β-production and IRF-3 activation. HEK293 cells were transiently transfected with luciferase reporter genes containing the IFN-β promoter (A and C) or Gal4-based IRF-3 reporter constructs (B and D). In A and B, at 24 h posttransfection, cells were pretreated with the indicated concentrations (μm) of PP2 or PP3 prior to SV infection (300 HAU/ml) for 6 h. In C and D, cells were cotransfected with various concentrations of a c-Src kinase-inactive variant (K297R) prior to infection with SV (300 HAU/ml) for 12 h. The indicated amounts (ng) of SrcK297R in pUSE were added to 96-well culture plates. Total amounts of DNA in the transfections were kept constant by adding the empty vector of kinase-inactive c-Src. Cells were lysed, and luciferase reporter gene activity was measured.
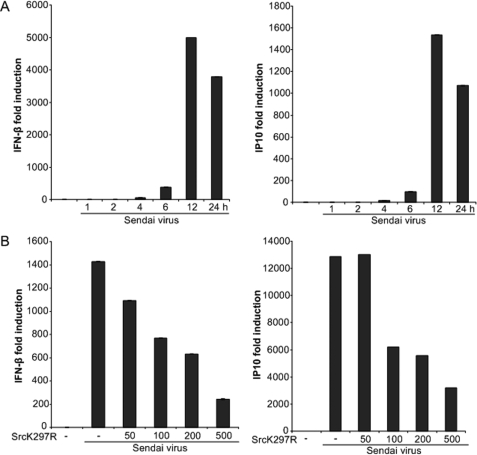
To test the biological relevance of the reporter-based findings, we analyzed the effect of kinase inactive c-Src on SV-stimulated transcription from endogenous IFN-β and IP10 promoters using qRT-PCR analysis. First, we examined the time-dependent induction of IFN-β and IP10 mRNA levels after SV infection. Considerable IFN-β and IP10 mRNA induction was observed after 6 h, with maximal induction at 12 h of SV infection (Fig. 3A). Subsequently, HEK293 cells were transfected with kinase-inactive c-Src prior to SV infection, isolation of total RNA, and qRT-PCR analysis of IFN-β and IP10 mRNAs. The SV-stimulated induction of IFN-β and IP10 were significantly reduced by transfection of increasing amounts of kinase-inactive Src (Fig. 3B), consistent with a role for c-Src in SV-mediated immune responses. As a control, we checked that transfection of the kinase-inactive c-Src did not affect expression of the housekeeping gene GAPDH when compared with β-actin (data not shown).
FIGURE 3.
SV-induced expression of endogenous IRF-3-dependent genes is regulated by c-Src. A, HEK293 cells were infected with SV (300 HAU/ml) for various times, and total RNA was analyzed for IFN-β and IP10 mRNA by qRT-PCR analysis. B, HEK293 cells were transfected with various concentrations of kinase-inactive c-Src (K297R). The indicated amounts (ng) of SrcK297R in pUSE were added to 6-well culture plates. Total amounts of DNA in the transfections were kept constant by adding the empty vector of kinase-inactive c-Src. 24 h after transfection, cells were exposed to SV (300 HAU/ml) for 12 h, and IFN-β and IP10 mRNA levels were determined by qRT-PCR. IFN-β and IP10 mRNA levels were compared with β-actin and GAPDH mRNA, respectively, in the sample, as described under “Experimental Procedures.”
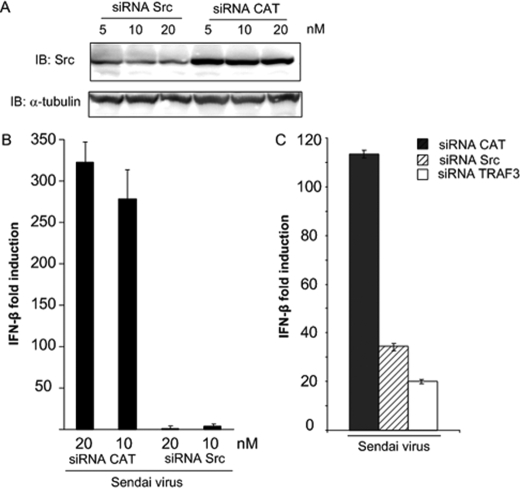
To further ascertain whether c-Src modulates SV-mediated responses, we used siRNA to down-regulate c-Src mRNA prior to assessment of IFN-β production. HEK293 cells were transfected with SMART pool siRNA against c-Src or CAT siRNA as a nonspecific control prior to SV infection. The efficacy of c-Src siRNA oligonucleotides was examined by immunoblotting of c-Src protein. Treatment with c-Src siRNA strongly, although not completely, reduced c-Src protein expression when compared with cells treated with the nonspecific control (Fig. 4A). Analysis by qRT-PCR showed that SV-stimulated IFN-β production was strongly attenuated in cells transfected with c-Src siRNA compared with cells transfected with CAT (Fig. 4B).
FIGURE 4.
Down-regulation of c-Src by c-Src siRNAs impairs IFN-β induction in response to SV. A, HEK293 cells were transfected with 5, 10, or 20 nm c-Src SMART pool siRNA or CAT siRNA using Lipofectamine RNAi Max. After 48 h, protein expression was analyzed by immunoblotting for c-Src or α-tubulin. B, HEK293 cells were transfected with 10 or 20 nm c-Src SMART pool siRNA or siRNA specific to CAT using Lipofectamine RNAi Max. 40 h posttransfection, cells were infected with SV (300 HAU/ml for 9 h), and IFN-β mRNA levels were assessed by qRT-PCR. C, HEK293 cells were transfected with 10 nm each of c-Src SMART pool siRNA, siRNA specific to CAT, or TRAF3 using X-tremeGENE. Subsequently, cells were infected with SV (300 HAU/ml), and IFN-β mRNA levels were assessed by qRT-PCR.
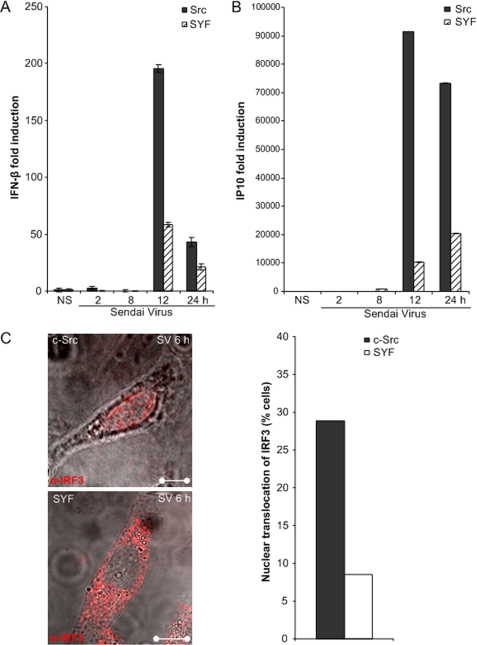
To further confirm the role of c-Src in TLR-independent signaling, we analyzed transcription of antiviral genes after SV infection of embryonic fibroblasts derived from Src/Yes/Fyn (SYF) triple-deficient mice. SYF cells in which the wild type c-src gene has been introduced with a retroviral vector (denoted “c-Src”; see Ref. 23) were used for comparison with triple kinase-deleted cells. The RNA helicase RIG-I, but not TLRs, has previously been shown to be critical for SV-elicited antiviral responses in mouse embryonic fibroblasts (7). As assessed by qRT-PCR, SV-elicited transcription of IFN-β and IP10 mRNA was significantly reduced in SYF cells compared with SYF cells in which c-Src has been introduced (23) (Fig. 5, A and B).
FIGURE 5.
SV-stimulated gene expression of IFN-β and IP10 is decreased in cells devoid of c-Src. A and B, fibroblasts from c-Src family kinase triple null (SYF) mice and triple null fibroblasts in which c-Src expression has been restored (Src) were infected with SV (300 HAU/ml) for various times prior to total RNA isolation and measurement of IFN-β (A) and IP10 (B) mRNA by qRT-PCR. C, Src and SYF cells were infected with SV (300 HAU/ml) for 6 h and stained intracellularly for IRF-3 (Alexa546). A total of 300 cells from each cell line were counted, and average percentages of cells with nuclear IRF-3 are shown in the inset.
Phosphorylation of IRF-3 leads to its nuclear translocation (24). The localization of IRF-3 was examined by immunofluorescent staining of endogenous IRF-3 using an anti-IRF-3 antibody in SV-infected SYF cells and SYF cells expressing c-Src. SV-stimulated IRF-3 nuclear translocation was significantly reduced in c-Src lacking SYF fibroblasts compared with cells that express c-Src (Fig. 5C). These data corroborate results obtained with kinase-inactive c-Src and c-Src siRNA, providing further evidence that c-Src is implicated in RIG-I-elicited antiviral responses.
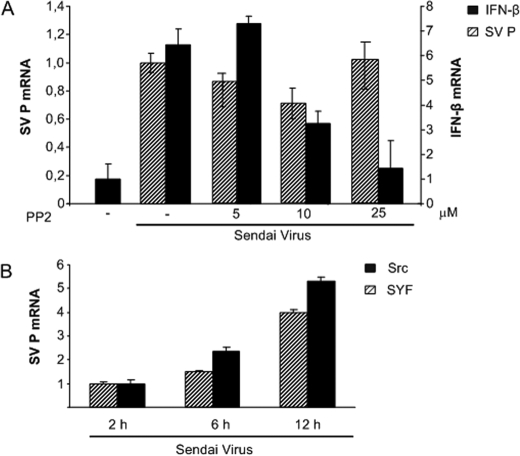
The reduced SV-stimulated IRF-3 activation and IFN-β production observed after c-Src inhibition (Figs. 2–5) could be due to effects of c-Src inhibition on viral entry or replication. To investigate this possibility, we examined mRNA levels of the SV P-gene in cells treated with the c-Src inhibitor PP2. HEK293 cells were pretreated with various concentrations of PP2 prior to infection with SV for 6 h. Using qRT-PCR analysis, we found that levels of SV P-gene mRNA were largely unaffected by treatment with PP2 (Fig. 6A). In contrast, IFN-β production in the same RNA samples was strongly reduced by PP2 (Fig. 6A). As shown in Fig. 5, we found reduced IRF-3 nuclear translocation and decreased mRNA levels of IFN-β and IP10 in fibroblasts derived from SYF triple-deficient mice relative to c-Src-expressing fibroblasts. Again, to examine if this effect was due to inhibition of SV infectivity in c-Src-deficient cells, we analyzed the presence of SV P-protein in Src and SYF fibroblasts. Analysis by qRT-PCR showed that SV P-protein mRNA increased with increased duration of infection in both SYF fibroblasts and c-Src-expressing fibroblasts and that mRNA levels of SV P-protein were only moderately reduced in SYF cells compared with in c-Src-expressing cells (Fig. 6B). Conversely, SV-stimulated IFN-β and IP10 production were significantly reduced in SYF cells relative to c-Src-expressing cells (Fig. 5, A and B). We also found that PP2 failed to inhibit SV-induced IL-8 mRNA levels.3 Taken together, these results suggest that the reduced IFN-β production observed after c-Src inhibition is primarily caused by effects on signaling after viral recognition rather than effects on SV entry and replication.
FIGURE 6.
Effect of c-Src inhibition on mRNA levels of SV P-protein. A, HEK293 cells were pretreated with the c-Src inhibitor PP2 for 45 min prior to SV infection for 6 h. Total RNA was isolated, and the expression of SV P mRNA or cellular IFN-β was examined. SV P-protein and IFN-β mRNA levels were compared with GAPDH mRNA, as described under “Experimental Procedures.” SV P mRNA in cells treated with various concentrations of PP2 prior to infection with SV is compared with SV P mRNA in cells infected with SV in medium. B, Src and SYF fibroblasts were infected with SV (300 HAU/ml) for 2, 6, or 12 h prior to total RNA isolation and determination of mRNA expression of SV P-protein by qRT-PCR. SV P mRNA levels were compared with GAPDH mRNA, as described under “Experimental Procedures,” and expressed relative to SV P mRNA levels measured at 2 h of infection.
Molecular Components of the RIG-I-signaling Pathway Induce IRF-3-dependent Transcription That Involves c-Src Activity
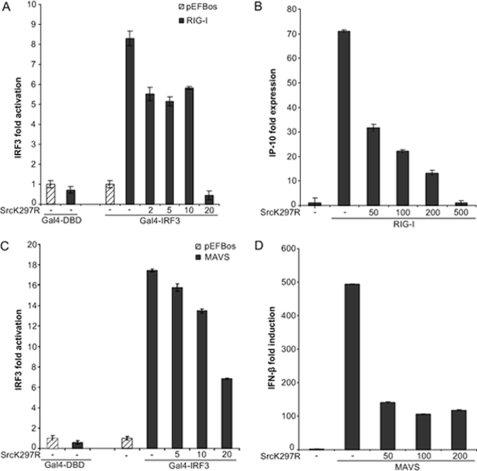
SV has previously been shown to activate IRF-3 and induce IFN-β transcription via RIG-I and MAVS in HEK293 cells (9, 19). Moreover, ectopic expression of RIG-I and MAVS stimulates IRF-3 activation in the absence of viral nucleic acid (9). To more accurately position the level at which c-Src operates in SV-stimulated responses, we first examined the effect of overexpression of kinase-inactive c-Src on RIG-I- and MAVS-induced IRF-3 activation. IRF-3 activation following RIG-I or MAVS transfection occurs independently of signaling elicited from SV and bypasses effects dependent on viral characteristics (e.g. cellular infectivity). HEK293 cells were cotransfected with RIG-I or MAVS together with kinase-inactive c-Src and the Gal4-based IRF-3 luciferase reporter. As shown in Fig. 7, A and B, kinase-inactive c-Src attenuated RIG-I- and MAVS-stimulated IRF-3 activation. Also, kinase-inactive c-Src suppressed RIG- and MAVS-induced transcription from endogenous IP10 and IFN-β promoters, as determined by qRT-PCR (Fig. 7, C and D). These results imply that c-Src participates downstream of the cytoplasmic RNA helicase RIG-I and its mitochondrial adapter protein MAVS in antiviral responses.
FIGURE 7.
Ectopic expression of dominant negative c-Src impairs RIG-I- and MAVS-elicited IRF-3-dependent gene expression/antiviral responses. A and C, HEK293 cells were transfected with 20 ng of RIG-I (A) or MAVS (C) and cotransfected with various concentrations (ng) of kinase inactive c-Src (K297R) or the corresponding empty vector together with an IRF3-dependent Gal4-luciferase vector. 24 h posttransfection cells were lysed, and IRF-3-dependent luciferase activity was determined. B and D, HEK293 cells were transfected with 0.5 μg of RIG-I (C) or MAVS (D) and cotransfected with various concentrations (ng) of kinase-inactive c-Src (K297R) or the corresponding empty vector prior to total RNA isolation and assessment of endogenously expressed IFN-β and IP10 mRNA by qRT-PCR.
Interaction of c-Src with Signaling Components of the RIG-I-mediated Pathway Enhances Antiviral Responses
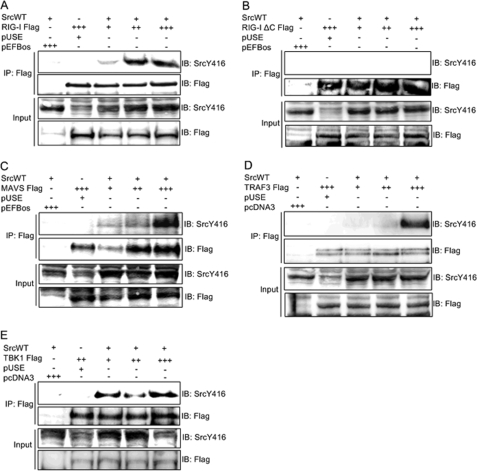
To further examine the involvement of c-Src in RIG-I-initiated antiviral signaling, we investigated if c-Src physically interacts with components mediating RIG-I-elicited signaling. Hence, c-Src was coexpressed with various concentrations of FLAG-tagged RIG-I, MAVS, TRAF3, or TBK1 in HEK293 cells prior to lysis, precipitation with anti-FLAG, and immunoblotting for activated c-Src (SrcpY416). We found that c-Src associated with full-length RIG-I, MAVS, TRAF3, and TBK1 proteins (Fig. 8). In contrast, c-Src failed to associate with a RIG-I mutant lacking the two N-terminal CARDs (Fig. 8B). As a control, we checked that this RIG-I mutant lacking the two N-terminal CARDs was indeed immunoprecipitated. Moreover, we verified that c-Src was overexpressed in the lysates used for immunoprecipitation. Hence, these results indicate that the CARD motifs, previously demonstrated to be critical for activation of downstream signaling pathways (2), mediate the interaction between RIG-I and c-Src.
FIGURE 8.
The tyrosine kinase c-Src interacts with molecular components of RIG-I-induced antiviral responses. HEK293 cells were transfected with 0.6 μg of c-Src and 0.75, 1.0, or 1.5 μg of plasmids expressing either of the FLAG-tagged proteins: RIG-I (A), a RIG-I mutant lacking the CARD motifs (RIG-I ΔC) (B), MAVS (C), TRAF3 (D), or TBK1 (E), or corresponding concentrations of empty vectors. Lysates were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblotting (IB) with anti-SrcpY416. Membranes were reprobed with anti-FLAG. Input lysates were analyzed by immunoblotting using the indicated antibodies.
Collectively, these results suggest that c-Src contributes to RIG-I-mediated signaling through association, either directly or indirectly, with signaling complexes containing RIG-I, MAVS, TRAF3, and TBK1.
TRAF3 Association with c-Src Occurs through the TRAF3 RING Domain
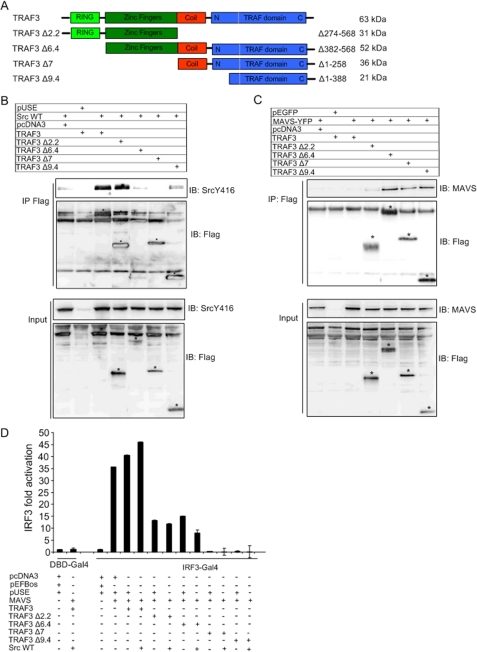
Our previous and present data indicate that c-Src modulates signaling from TLR3 and RIG-I. TRAF3 associates with the PRR adapter proteins TRIF (TLR3), MyD88 (TLR9), and MAVS (RIG-I), and IRF-3 activation induced downstream of these components converges on TRAF3 (14, 15, 25). We found that c-Src associated with TRAF3 (Fig. 8). Therefore, we further characterized the role of c-Src in relation to TRAF3. TRAFs constitute a family of cytoplasmic adapter proteins that link pathogen-recognizing or cytokine receptors with downstream signaling. Members of the TRAF family contain an N-terminal RING domain followed by one or more zinc fingers, a coiled-coil region, and a C-terminal TRAF homology domain. To map regions of TRAF3 that are implicated in the interaction between c-Src and TRAF3, we performed coimmunoprecipitation assays for c-Src and various N- and C-terminal TRAF3 deletion mutants (schematically depicted in Fig. 9A). HEK293 cells were transiently transfected with c-Src and FLAG-tagged TRAF3 mutants. In this assay, c-Src coimmunoprecipitated with wild-type TRAF3 and a mutant containing the RING and zinc finger domains (TRAF3 Δ2.2) but showed no significant interaction with TRAF3 deletion mutants containing the TRAF domain (TRAF3 Δ9.4) or the coiled-coil region and the TRAF domain (TRAF3 Δ7; Fig. 9B). Likewise, the zinc fingers of TRAF3 do not appear to be required for c-Src association, since the mutant TRAF3 Δ6.4, which encompasses the TRAF domain and the zinc fingers (Fig. 9A), failed to interact with c-Src. This indicates that c-Src interacts with the RING domain of TRAF3. However, an obstacle to using protein truncation mutants is that the proteins may fail to be expressed or to fold properly. Therefore, we first compared the expression levels of these mutants compared with wild-type TRAF3 in input lysates. The TRAF3 mutants were expressed to similar extents as wild-type TRAF3 (Fig. 9, B and C, lower panel). Additionally, to further confirm the biological functionality of the TRAF3 truncation mutants, we examined their ability to interact with the RIG-I adapter protein MAVS, since MAVS has previously been found to interact with TRAF3 (25). Fig. 9C shows that MAVS associated with N-terminal deletion mutants containing the zinc fingers and the TRAF domain (TRAF3 Δ6.4, TRAF3 Δ7, and TRAF3 Δ9.4) but failed to interact with a mutant containing only the RING and the zinc fingers (TRAF3 Δ2.2). Hence, the TRAF domain of TRAF3 mediates interaction with MAVS. Of note, we found that it was occasionally difficult to detect expression of wild-type TRAF3 (Fig. 9C), which may reflect that the protein is subject to proteolytic degradation mechanisms. Nevertheless, collectively, these results suggest that c-Src interacts with TRAF3 (directly or indirectly) through the RING domain, whereas the TRAF3 TRAF domain mediates MAVS-TRAF3 association.
FIGURE 9.
Distinct TRAF3 domains mediate association with c-Src and MAVS. A, schematic diagram of wild type (wt) form and deletion mutants of TRAF3. B and C, FLAG-tagged TRAF3 or deletion mutants (5.0 μg) were cotransfected with c-Src (3.0 μg) (B) or MAVS-YFP (3.0 μg) (C) in HEK293 cells. The physical interaction of c-Src and MAVS with different TRAF3 mutants was determined by co-immunoprecipitation assays using an anti-FLAG antibody for immunoprecipitation and detecting coprecipitated c-Src (B) and MAVS (C) with anti-phospho-c-Src or anti-MAVS, respectively. Immunoblots were reprobed with anti-FLAG. The expression of c-Src and MAVS in input lysates was monitored by immunoblotting. D, HEK293 cells were transfected with an IRF-3-dependent Gal4-based luciferase reporter and MAVS (20 ng), TRAF3, or TRAF3 deletion mutants (40 ng) and c-Src (20 ng) in the indicated combinations. Luciferase activity was determined 24 h posttransfection.
To assess the functional consequences of interrupting the binding of TRAF3 to c-Src or MAVS, we coexpressed MAVS, c-Src, and the TRAF3 deletion mutants together with the Gal4-IRF-3 reporter. Although simultaneous expression of wild-type TRAF3 and c-Src augmented MAVS-stimulated IRF-3 activation, all of the different TRAF3 truncation mutants examined abrogated IRF-3 activation induced by MAVS and c-Src (Fig. 9D). These results are in accordance with previous findings showing that both successive N- and C-terminal TRAF3 truncation mutants are unable to rescue antiviral responses in TRAF3 knock-out cells (25). The authors thus suggested that antiviral activity requires a structurally intact TRAF3 molecule.
DISCUSSION
Detection of pathogenic nucleic acids by the innate immune system is brought about by several components, including transmembrane TLRs, the interferon-induced enzyme RNA-activated protein kinase R, the cytoplasmic RNA helicase RIG-I, and the DNA-binding sensor DAI (26). In this study, we show that impairment of c-Src function by either pharmacological inhibition, expression of a kinase-inactive c-Src mutant, or genetic knock-out strategies effectively suppress SV-stimulated, RIG-I-dependent transcriptional activation of IRF-3 and lead to reduced expression of the IRF-3 target genes IFN-β and IP10. This suggests that c-Src participation in antiviral immune mechanisms is of biological significance, regulating IFN-β and the chemokine IP10 that control activation and migration of immune cells during viral infections. In conjunction with our previous results (6), these results show that c-Src contributes to distinct antiviral signaling pathways emanating from different pathogen recognition receptors, which suggests that c-Src may play a broader role in innate defense mechanisms against diverse viruses.
Concerning the location of c-Src in the signaling pathway from RIG-I to IRF-3, we found that enforced expression of RIG-I and its adapter protein MAVS (to bypass RIG-I activation) induced IRF-3-dependent gene expression that was effectively suppressed by impaired c-Src function (following c-Src siRNA depletion or expression of kinase-defective c-Src). Hence, these results further suggest that c-Src acts downstream of RIG-I and MAVS. Recently, TRAF3 was found to be a critical intermediary molecule that integrates TLR-dependent and -independent IFN production and antiviral responses (14, 15). Since our previous (6) and present data showed that c-Src contributes to both TLR-dependent and -independent signaling and associates with TRAF3 upon infection with SV, we sought to gain mechanistic insight into molecular determinants of the TRAF3-c-Src association. We found that c-Src associated with TRAF3 mutants that contained the N-terminal RING and zinc finger domains (lacking the coiled-coil and TRAF domains), whereas it failed to interact with different N-terminal truncation mutants that lacked the RING domain (while encompassing the coiled-coil, zinc finger, and TRAF motifs). This suggests that the RING domain of TRAF3 directs interaction with c-Src. Conversely, we found that the TRAF domain was sufficient for binding of TRAF3 to MAVS (Fig. 9C). These results corroborate previous data demonstrating that MAVS contains a TRAF interaction motif that governs association to the TRAF domain of TRAF3 (25). Interestingly, these results demonstrate functional specificity of different TRAF3 domains in antiviral signaling mediated by MAVS and c-Src.
Numerous studies have indicated that an important step downstream of PRRs is the assembly of multicomponent signaling complexes that activate critical kinases. These signaling complexes serve as structural platforms to assemble downstream signaling molecules. The complexes have been shown to contain TRIF or MAVS (depending pathogenic agent), TANK (TRAF family member-associated NF-κB activator), and TRAF3 and have been suggested to stimulate the IRF-3-phosphorylating kinase TBK1 (27, 28). We found that c-Src physically associated with MAVS, TRAF3, and TBK1, suggesting that c-Src is organized into IRF-3-activating signaling complexes. Moreover, wild-type c-Src acted synergistically with MAVS and TRAF3 in IRF-3 activation, whereas kinase-inactive c-Src had no effect on MAVS- and TRAF3-stimulated IRF-3. In addition, activating phosphorylation of c-Src was enhanced by co-transfection of MAVS and TRAF3. Taken together, this indicates a functional relationship between MAVS, TRAF3, and c-Src in that IRF-3-activating complexes entail MAVS, TRAF3, and c-Src and that the presence of MAVS and TRAF3 augments c-Src activation. Concerning the functional outcome of c-Src involvement, it is possible that c-Src-mediated phosphorylation might modulate protein-protein interactions through distinct modular domains in target proteins that may promote recruitment of antiviral signaling components to regulate the activity of TBK1 (directly or indirectly). It has also been suggested that alternative kinases are implicated in IRF-3 activation through phosphorylation of IRF-3 residues other than those targeted by TBK1 (29, 30). Hence, c-Src may modulate activation of such kinases. In this regard, it could be mentioned that we have previously found that c-Src mediates TLR3-elicited Akt activation but not p38 and p42/44 MAP kinase or c-Jun N-terminal kinase activation (6). This demonstrates that c-Src activates distinct pathways downstream of TLRs. Interestingly, the scaffold protein TANK was recently shown to be subject to phosphorylation and lysine 63-linked polyubiquitination (31), thus illustrating the complexity of IRF-3 regulatory mechanisms.
Regarding assembly of multiprotein complexes involving TRAF3 and c-Src, it is presently not clear how TRAF3 is activated and conveys signaling to IRF-3. Nevertheless, TRAF3 was recently reported to become ubiquitinated, presumably through lysine 63-linked polyubiquitin chains that recruit the kinase TBK1 (32). It is generally believed that ligand binding to TRAF-associating receptors induces TRAF oligomerization that leads to lysine 63-linked ubiquitination of target proteins. This promotes protein-protein interactions and activation of the kinase TAK1 in the NF-κB signaling pathway and recruits TBK1 to TRAF3 in the IRF-3 pathway (32, 33). Interestingly, TRAF3 has been suggested to function as a regulator of protein kinases that control IFN-producing pathways (34). Specifically, TRAF3 interacts with several nonrelated kinases: the IRF-3 phosphorylating kinases TBK1 and IKKϵ; IRAK1, which acts in MyD88-dependent TLR pathways; NIK, which activates the noncanonical NF-κB pathway; and protein kinase R, which binds viral double-stranded RNA. Although NIK has been shown to interact with TRAF3 through the TRAF domain (35), the underlying mechanisms responsible for TRAF3 binding to TBK1, IKKϵ, IRAK1, and protein kinase R are not known. In this study, we present novel results showing that c-Src binds to TRAF3 through the TRAF3 RING domain, thus illustrating that different modular domains link NIK and c-Src to TRAF3 function. Hence, TRAF3 may function as a scaffold protein that recruits kinases (e.g. c-Src), which mediates phosphorylation-dependent posttranslational modifications of proteins critical for IRF-3 activation, such as ubiquitination. In this connection, phosphorylation and ubiquitination have been found to mutually regulate each other through several distinct mechanisms (36).
In conclusion, our results provide further insight into the role of c-Src in sensing of pathogenic RNA and contribute to increased understanding of the IRF-3 regulatory pathway. As such, our results may have implications for antiviral strategies.
Acknowledgments
We thank Veslemøy Malm Landsem and Mari Sæther for excellent technical assistance.
This work was supported by the National Programme for Research in Functional Genomics in Norway (FUGE) (to M. W. A.), FUGE Mid-Norway (to M. W. A.), the Research Council of Norway (to M. W. A.), the Faculty of Medicine, Norwegian University of Science and Technology (to I. B. J. and M. W. A.), and the Cancer Fund at St. Olavs Hospital (to M. W. A.).
T. T. Nguyen, I. B. Johnsen, C. Knetter, E. Lien, K. A. Fitzgerald, F. Drabløs, and M. W. Anthonsen, manuscript in preparation.
- PRR
- pattern recognition receptor
- IFN
- interferon
- TLR
- Toll-like receptor
- SV
- Sendai virus
- CARD
- caspase recruitment domain
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- CAT
- chloramphenicol acetyltransferase
- IRF
- interferon regulatory factor
- siRNA
- small interfering RNA
- SYF
- Src/Yes/Fyn
- qRT
- quantitative real time
- HAU
- hemagglutination units.
REFERENCES
- 1.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 3.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 4.Gitlin L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R. A., Diamond M. S., Colonna M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. (2004) Nat. Immunol. 5, 190–198 [DOI] [PubMed] [Google Scholar]
- 6.Johnsen I. B., Nguyen T. T., Ringdal M., Tryggestad A. M., Bakke O., Lien E., Espevik T., Anthonsen M. W. (2006) EMBO J. 25, 3335–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. (2005) Immunity 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 8.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 9.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 11.Ishii K. J., Coban C., Kato H., Takahashi K., Torii Y., Takeshita F., Ludwig H., Sutter G., Suzuki K., Hemmi H., Sato S., Yamamoto M., Uematsu S., Kawai T., Takeuchi O., Akira S. (2006) Nat. Immunol. 7, 40–48 [DOI] [PubMed] [Google Scholar]
- 12.Wathelet M. G., Lin C. H., Parekh B. S., Ronco L. V., Howley P. M., Maniatis T. (1998) Mol. Cell 1, 507–518 [DOI] [PubMed] [Google Scholar]
- 13.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 14.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., Mann M., Karin M. (2006) Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 15.Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 16.Force W. R., Cheung T. C., Ware C. F. (1997) J. Biol. Chem. 272, 30835–30840 [DOI] [PubMed] [Google Scholar]
- 17.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 18.Wagner A. M., Loganbill J. K., Besselsen D. G. (2003) Comp. Med. 53, 173–177 [PubMed] [Google Scholar]
- 19.Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B. G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K. A. (2005) J. Immunol. 175, 5260–5268 [DOI] [PubMed] [Google Scholar]
- 20.Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 21.Toshchakov V., Jones B. W., Perera P. Y., Thomas K., Cody M. J., Zhang S., Williams B. R., Major J., Hamilton T. A., Fenton M. J., Vogel S. N. (2002) Nat. Immunol. 3, 392–398 [DOI] [PubMed] [Google Scholar]
- 22.Cai W., He J. C., Zhu L., Lu C., Vlassara H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13801–13806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinghoffer R. A., Sachsenmaier C., Cooper J. A., Soriano P. (1999) EMBO J. 18, 2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin R., Heylbroeck C., Pitha P. M., Hiscott J. (1998) Mol. Cell Biol. 18, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha S. K., Pietras E. M., He J. Q., Kang J. R., Liu S. Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T. L., Wang Y., Cheng G. (2006) EMBO J. 25, 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. (2007) Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 27.Guo B., Cheng G. (2007) J. Biol. Chem. 282, 11817–11826 [DOI] [PubMed] [Google Scholar]
- 28.Ryzhakov G., Randow F. (2007) EMBO J. 26, 3180–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K. P., Yamaoka S. (2006) Nat. Immunol. 7, 598–605 [DOI] [PubMed] [Google Scholar]
- 30.Clément J. F., Bibeau-Poirier A., Gravel S. P., Grandvaux N., Bonneil E., Thibault P., Meloche S., Servant M. J. (2008) J. Virol. 82, 3984–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatot J. S., Gioia R., Chau T. L., Patrascu F., Warnier M., Close P., Chapelle J. P., Muraille E., Brown K., Siebenlist U., Piette J., Dejardin E., Chariot A. (2007) J. Biol. Chem. 282, 31131–31146 [DOI] [PubMed] [Google Scholar]
- 32.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., Dixit V. M. (2007) Science 318, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 33.Gao M., Karin M. (2005) Mol. Cell 19, 581–593 [DOI] [PubMed] [Google Scholar]
- 34.Schneider K., Benedict C. A., Ware C. F. (2006) Nat. Immunol. 7, 15–16 [DOI] [PubMed] [Google Scholar]
- 35.Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 36.Hunter T. (2007) Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]