Abstract
Upon DNA replication stress, stalled DNA replication forks serve as a platform to recruit many signaling proteins, leading to the activation of the DNA replication checkpoint. Activation of Rad53, a key effector kinase in the budding yeast Saccharomyces cerevisiae, is essential for stabilizing DNA replication forks during replication stress. Using an activity-based assay for Rad53, we found that Mrc1, a replication fork-associated protein, cooperates with Mec1 to activate Rad53 directly. Reconstitution of Rad53 activation using purified Mec1 and Mrc1 showed that the addition of Mrc1 stimulated a more than 70-fold increase in the ability of Mec1 to activate Rad53. Instead of increasing the catalytic activity of Mec1, Mrc1 was found to facilitate the phosphorylation of Rad53 by Mec1 via promotion of a stronger enzyme-substrate interaction between them. Further, the conserved C-terminal domain of Mrc1 was found to be required for Rad53 activation. These results thus provide insights into the role of the adaptor protein Mrc1 in activating Rad53 in the DNA replication checkpoint.
Faithful replication of the genome is important for the survival of all organisms. During DNA replication, replication stress can arise from a variety of situations, including intrinsic errors made by DNA polymerases, difficulties in replicating repeated DNA sequences, and failures to repair damaged DNA caused by either endogenous oxidative agents or exogenous mutagens such as UV light and DNA-damaging chemicals (1–3). In eukaryotes, there is an evolutionarily conserved DNA replication checkpoint that becomes activated in response to DNA replication stress. It helps to stabilize DNA replication forks, block late replication origin firing, and delay mitosis and ultimately helps recovery from stalled replication forks after DNA repair (4–7). Defects in the DNA replication checkpoint could result in elevated genomic instabilities, cancer development, or cell death (8, 9).
Aside from replicating the genome, the DNA replication forks also provide a platform to assemble many signaling proteins that function in the DNA replication checkpoint. In the budding yeast Saccharomyces cerevisiae, Mec1, an ortholog of human ATR,2 is a phosphoinositide 3-kinase-like kinase (PIKK) involved in sensing stalled DNA replication forks. Mec1 forms a protein complex with Ddc2 (ortholog of human ATRIP). The Mec1-Ddc2 complex is recruited to stalled replication forks through replication protein A (RPA)-coated single-stranded DNA (10, 11). The Mec3-Rad17-Ddc1 complex, a proliferating cell nuclear antigen (PCNA)-like checkpoint clamp and ortholog of the human 9-1-1 complex, was shown to be loaded onto the single- and double-stranded DNA junction of the stalled replication forks by the clamp loader Rad24-RFC complex (12). Once loaded, the Mec3-Rad17-Ddc1 complex stimulates Mec1 kinase activity (13). Dbp11 and its homolog TopBP1 in vertebrates are known components of the replication machinery (14). In addition to regulating the initiation of DNA replication, they were found to play a role in the DNA replication checkpoint (15–17). They interact with the 9-1-1 complex and directly stimulate Mec1/ATR activity in vitro (18–20). Thus, the assembly of multiple protein complexes at stalled DNA replication forks appears to facilitate activation of the DNA replication checkpoint (13, 18).
Mrc1 (for mediator of replication checkpoint) was originally identified to be important for cells to respond to hydroxyurea in S. cerevisiae and Schizosaccharomyces pombe (21, 22). Mrc1 is a component of the DNA replisome and travels with the replication forks along chromosome during DNA synthesis (23–25). Deletion of MRC1 causes defects in DNA replication, indicating its role in the normal progression of DNA replication (23). Interestingly, when DNA replication is blocked by hydroxyurea, Mrc1 undergoes Mec1- and Rad3 (S. pombe ortholog of Mec1)-dependent phosphorylation (21, 22). In S. cerevisiae, mutations of Mrc1 at the (S/T)Q sites, which are consensus phosphorylation sites of the Mec1/ATR family kinases, abolishes hydroxyurea-induced Mrc1 phosphorylation in vivo, suggesting a direct phosphorylation of Mrc1 by Mec1 (21, 22).
Rad53 and Cds1, homologs of human Chk2, are the major effector kinases in the DNA replication checkpoints in S. cerevisiae and S. pombe, respectively. Activation of Rad53 is a hallmark of DNA replication checkpoint activation and is important for the maintenance of DNA replication forks in response to DNA replication stress (5, 6). Thus, it is important to understand how Rad53 activity is controlled. Interestingly, mutation of all the (S/T)Q sites of Mrc1 not only abolishes the phosphorylation of Mrc1 by Mec1 but also compromises hydroxyurea-induced Rad53 activation in S. cerevisiae (21). Similarly, mutation of the TQ sites of Mrc1 in S. pombe was shown to abolish the binding between Cds1 and Mrc1 as well as Cds1 activation (22). Further, mutation of specific TQ sites of Mrc1 in S. pombe abolishes its binding to Cds1 in vitro and the activation of Cds1 in vivo (26). Thus, Mec1/Rad3-dependent phosphorylation of Mrc1 is responsible for Mrc1 binding to Rad53/Cds1, which is essential for Rad53/Cds1 activation.
An intriguing property of the Chk2 family kinases is their ability to undergo autophosphorylation and activation in the absence of other proteins in vitro (27, 28). First, autophosphorylation of a conserved threonine residue in the activation loop of Chk2 family kinase was found to be an essential part of their activation processes (26, 29–31). Second, a direct and trans-phosphorylation of the N-terminal TQ sites of the Chk2 family kinases by the Mec1/ATR family kinases is also important for their activation in vivo. Analogous to the requirement of N-terminal TQ site phosphorylation of Chk2 by ATR in human (32), the activation of Rad53/Cds1 in vivo requires phosphorylation of TQ sites in their N termini by Mec1/Rad3 (33, 34).
Considering that Mec1, Mrc1, and many other proteins are recruited at stalled DNA replication forks and have been shown to be involved in DNA replication checkpoint activation, a key question remains unresolved: what is the minimal system that is capable of activating Rad53 directly? Given the direct physical interaction between Mrc1 and Rad53 and the requirement of Mrc1 and Mec1 in vivo, it is likely that they both play a role in Rad53 activation. Furthermore, what is the molecular mechanism of Rad53 activation by its upstream activators? To address these questions, a faithful reconstitution of the activation of Rad53 using purified proteins is necessary. In this study, we developed an activity-based assay consisting of the Dun1 kinase, a downstream substrate of Rad53, and Sml1, as a substrate of Dun1, to quantitatively measure the activity of Rad53. Using this coupled kinase assay from Rad53 to Dun1 and then to Sml1, we screened for Mrc1 and its associated factors to see whether they could directly activate Rad53 in vitro. Our results showed that Mec1 and Mrc1 collaborate to constitute a minimal system in direct activation of Rad53.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
The plasmids used are summarized in supplemental Table S1. Yeast strains are summarized in supplemental Table S2. MRC1 was first cloned into the pFA6a-3XHA-Kan plasmid using PacI and AscI followed by mutagenesis. Various mutations of MRC1 were then introduced into yeast cells via homologous recombination (35). All plasmids and mutations introduced into yeast cells were confirmed by DNA sequencing.
Partial Purification of Mrc1 and Mass Spectrometry Analysis
All purification steps were performed at 4 °C. 2 liters of yeast cells (SCY216, SCY152, and SCY230 for mec1Δ, rad53Δ, and Mrc1-TAF/rad53Δ, respectively) were grown in YPD (yeast extract, peptone, and dextrose) medium to log phase (A600 ∼ 0.8). Spheroplasts were prepared as described (36). The extract was prepared from spheroplasts in 10 ml of buffer A (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm DTT, 0.1% Tween 20, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1.5 μm pepstatin A, 1 μm leupeptin, 0.2 mm benzamidine, 5 mm β-glycerophosphate, 5 mm sodium fluoride) by sonication and clarification at 70,000 rpm in a MLA80 rotor for 10 min. 200 μl of anti-FLAG M2 affinity resin (Sigma) was added to 10 ml of the extract (∼30 μg/μl protein), and the bead/extract mix was rotated for 3 h. The anti-FLAG resins were then washed with 15 bead volumes of TBSD (50 mm Tris-HCl, pH 7.5, 150 mm sodium chloride, 1 mm DTT, 0.1% Tween 20), and proteins were eluted by incubation with 1 bead volume of TBSD containing 10% glycerol and 200 μg/ml 3× FLAG peptide (Sigma) for 1 h at room temperature.
Elution equivalent to 0.5 liter of yeast culture was prepared for mass spectrometry analysis as follows. Elution was denatured with 1% SDS followed by reduction and alkylation with 10 mm DTT and 30 mm iodoacetamide, respectively. Proteins were then precipitated and washed with 50% ethanol, 50% acetone, and 0.1% acetic acid. The precipitated proteins were then resuspended and subjected to trypsin (Roche Applied Science) digestion. Mass spectrometry analysis was similar as described previously (37) except a Thermo Finnigan (Thermo Scientific, Waltham, MA) LTQ mass spectrometer was used for sample analysis.
Tandem Affinity Purification of Epitope-tagged Endogenous Rad53 and Dun1
2 liters of yeast cells were grown in YPD medium to log phase and broken in an ice-cooled bead beater (Hamilton Beach/Proctor-Silex, Inc.) in 40 ml of buffer A. Crude extracts were clarified by centrifugation at 15,000 rpm in a JA-25.50 rotor for 30 min and added to ∼100 μl of anti-FLAG M2 resins and IgG resins (IgG-Sepharose 6 Fast Flow, GE Healthcare) for the immunoprecipitation of Rad53 and Dun1, respectively. The bead/extract mix was rotated for 3 h, and the resins were washed with 5 bead volumes of TBSD supplemented with 1 m sodium chloride followed by washing with 15 bead volumes of TBSD (standard wash). Rad53 was eluted by 2 bead volumes of TBSD containing 200 μg/ml 3× FLAG peptide and then bound to 20 μl of Ni-NTA resin (Qiagen) for 2 h. The Ni-NTA resins were washed, and Rad53 was eluted by 4 bead volumes of TBSD containing 10% glycerol and 200 mm imidazole. Dun1 was eluted by incubation with 2 bead volumes TBSD containing 10 unit of tobacco etch virus protease for 2 h at 30 °C, and the supernatant was added to 20 μl of anti-FLAG M2 resin for 2 h. The anti-FLAG M2 column was washed, and Dun1 was eluted by incubation with 4 bead volumes of TBSD containing 10% glycerol and 200 μg/ml 3× FLAG peptide for 1 h at room temperature. Both Rad53 and Dun1 purifications yielded a protein concentration of ∼20 ng/μl. Concentrations of Rad53 and Dun1 were determined by comparison with a known bovine serum albumin standard using 10% SDS-PAGE analysis and silver staining.
Tandem Affinity Purification of Recombinant Rad53 and Mrc1
BL21 cells were used to overexpress Rad53 and Rad53 kinase-dead protein (Rad53KD) using plasmids HZE1452 and HZE1446, respectively. Extract was prepared from 2 liters of cells in 20 ml of buffer A by sonication and clarification at 30,000 rpm in a JA-25.50 rotor for 30 min. 250 μl of IgG resins were added to the extract, the bead/extract mix was rotated for 2 h, and the IgG resins were washed. For the purification of active Rad53, Rad53 was eluted using 3 bead volumes of TBSD containing 10 units of PreScission protease (Amersham Biosciences); and the supernatant was bound to 100 μl of Ni-NTA resins for 2 h. The Ni-NTA resins were washed, and Rad53 was eluted by 2 bead volumes of TBSD containing 200 mm imidazole. Eluted Rad53 was dialyzed in TBSD supplemented with 10% glycerol. The final Rad53 concentration was 500 ng/μl. For the purification of inactive Rad53, all the steps were the same except for an additional inaction step after the IgG binding of Rad53. To inactive/dephosphorylate Rad53, IgG-bound Rad53 was incubated with 1000 units of Lambda phosphatase (New England BioLabs) in 1 bead volume of TBSD supplemented with 5 mm magnesium chloride and 2% glycerol for 12 h at room temperature. Following the wash to remove Lambda phosphatase, Rad53 was eluted by PreScission protease.
BL21 cells were used to overexpress Mrc1 WT and various mutant proteins using the plasmids HZE1516-1521. The purification of these recombinant proteins was similar to that described above for Rad53. The introduction of a His6 tag at the C terminus of Mrc1 incidentally introduced a point mutation in the tag, resulting in a sequence of GGGSSSSSS in the C termini of all Mrc1 proteins used. All of these Mrc1 proteins were still purified using the Ni-NTA resin. Concentration of Mrc1 was determined by comparing with a known bovine serum albumin standard using 10% SDS-PAGE analysis and Coomassie staining.
Overexpression and Purification of Dun1 and Mec1-Ddc2 Complex
pYES-PP-Dun1 was transformed in SCY152. 100 ml of cells were grown in CSM-Ura 2% glucose to an A600 of 1.5. Cells were pelleted, resuspended in 100 ml of medium containing CSM-Ura, 2% galactose, and 0.1% glucose, and grown for 12 h. An extract was prepared by breaking the cells in 10 ml of buffer A using a vortex (Vortex-Genie 2, Scientific Industries), and centrifugation at 13,200 rpm in a F45-24-11 rotor for 10 min. 100 μl of IgG resins were added to the extract, and the bead/extract mix was rotated for 2 h. After the washing of IgG resins, Dun1 was eluted by incubating it with 2 bead volumes of TBSD containing 10% glycerol and 5 units of PreScission protease overnight. The concentration of purified Dun1 was 500 ng/μl.
S. cerevisiae strain SCY001, transformed with plasmid pBL504 (Mec1/GST-Ddc2), was grown and induced with galactose under the same conditions as described above. 12 liters of cells were broken in an ice-cooled bead beater in 300 ml of buffer B (buffer A plus 0.01% Nonidet P-40) and clarified by centrifugation at 15,000 rpm in a JA-25.50 rotor for 30 min. Proteins were precipitated using ammonium sulfate to 55% saturation. The precipitate was collected after centrifugation at 15,000 rpm for 30 min. The protein pellet was resuspended in 30 ml of buffer B and then incubated with 3 ml of anti-FLAG M2 resin for 4 h. After washing, proteins were eluted with 3 bead volumes of TBSD containing 100 μg/ml 3× FLAG peptide. 300 μl of glutathione-Sepharose 4 Fast Flow resin (Amersham Biosciences) was added to the FLAG-eluted sample, and the sample was rotated overnight. The glutathione resins were washed, and Mec1-Ddc2 was eluted by incubating it with 2 bead volumes of TBSD containing 50 units of PreScission protease for 4 h. The eluted sample was loaded onto a 1-ml heparin-agarose column (GE Healthcare), washed with 10 ml of TBSD, and eluted with TBSD containing 500 mm sodium chloride. Finally, eluted Mec1-Ddc2 was dialyzed in TBSD supplemented with 10% glycerol. The final Mec1-Ddc2 concentration was 100 ng/μl.
Standard Kinase Reaction
In a typical kinase assay, 20 mm Tris, pH 7.5, 50 mm NaCl, 0.2 mm ATP, 1 mm MgCl2, 1 mm DTT, and 10 μCi of [γ32P]ATP were used. A standard kinase reaction for 30 min at 30 °C was used unless stated otherwise.
In Vitro Rad53 FHA1 Domain Binding Assay
The binding assay was similar to that described previously (37). Briefly, GST fusion proteins of the wild type and R70A mutant FHA1 domains of Rad53 were first purified using glutathione resins (Promega). The GST-FHA1 domain bound glutathione resins were then incubated with either unphosphorylated or phosphorylated recombinant Mrc1-FLAG for 3 h (recombinant Mrc1-FLAG was purified using the same protocol as described for the purification of recombinant Mrc1, except anti-FLAG resins were used during the second step of purification). After washing, the FHA1-bound proteins were eluted by boiling with SDS sample buffer containing 10 mm DTT and then analyzed by 10% SDS-PAGE. This was followed by an anti-FLAG Western blot to detect the presence of Mrc1-FLAG. To phosphorylate Mrc1-Flag by Mec1, 100 μl of kinase reaction containing 80 nm recombinant Mrc1-FLAG with or without 0.6 nm Mec1 was incubated for 2 h at 30 °C. After the kinase reaction, the reaction mixture was incubated with the WT and R70A mutant GST-FHA1 domains of Rad53 (bound to glutathione resins) as described above.
RESULTS
Biochemical Screen for Rad53 Activators Identified Mec1 and Mrc1
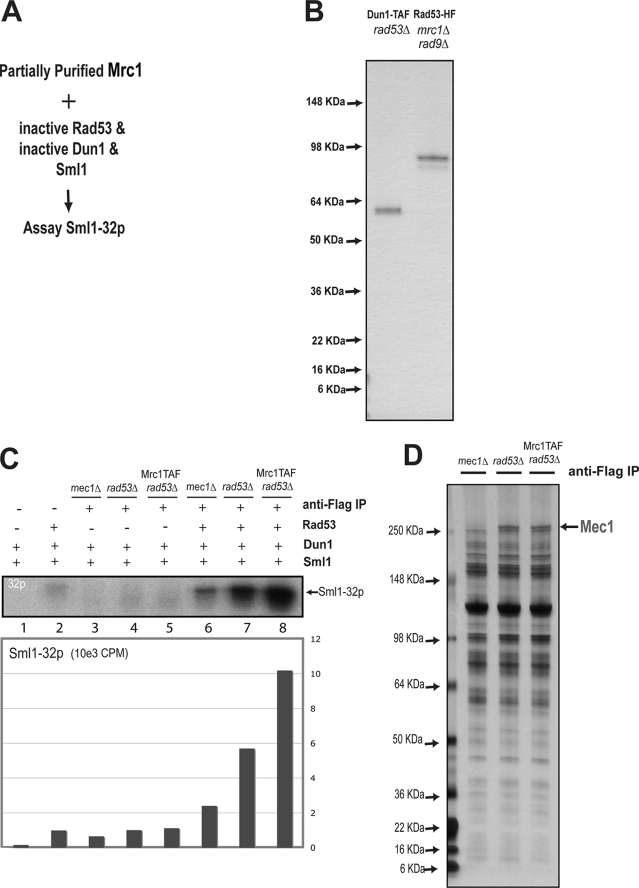
To identify factors that may activate Rad53 directly, we developed a Rad53 activity-based assay consisting of inactive Rad53, inactive Dun1, and recombinant Sml1 (Fig. 1, A and 1B). As shown previously (29), Dun1 activation requires Rad53 phosphorylation, and only activated Dun1 can hyperphosphorylate Sml1. We therefore used this inactive Rad53-Dun1-Sml1 (RDS) system as a reporter to identify potential activator(s) of Rad53 (Fig. 1A). As discussed above, Mrc1 is known to bind to Rad53 after its phosphorylation by Mec1 (21, 37). To address whether Mrc1 and its associated proteins could directly activate Rad53 in vitro, we immunoprecipitated epitope-tagged Mrc1-TAF in rad53Δ cells using immobilized anti-FLAG antibody. As controls, parallel anti-FLAG immunoprecipitation experiments were performed using cell extracts from non-epitope-tagged rad53Δ cells and mec1Δ cells. Each immunoprecipitate was then added to the inactive RDS system to perform a kinase reaction with [γ32P]ATP (Fig. 1, A and C). The amount of hyperphosphorylated Sml1 was quantified using scintillation counting. As shown in Fig. 1C, the highest amount of Sml1 phosphorylation was observed using the immunoprecipitated sample from Mrc1-TAF in rad53Δ cells (lane 8). The amount of Sml1 phosphorylation was reduced 10-fold to the basal level when inactive Rad53 was omitted in the kinase assay, indicating that Rad53 is required for Sml1 phosphorylation (see Fig. 1C, lane 5). In contrast, the anti-FLAG-immunoprecipitated sample from either mec1Δ cells or rad53Δ cells was less potent, showing a 2- or 5-fold increase in Sml1 phosphorylation compared with the basal level, respectively. Reproducible results were obtained in repeated experiments leading to the following conclusions. First, Mrc1 facilitates Rad53 activation (Fig. 1C, compare lanes 7 and 8). Second, there are unknown factors that co-purify with Mrc1 to help Rad53 activation (Fig. 1C, compare lane 6 with lanes 7 and 8). Third, these unknown factors are Mec1-dependent and Rad53-independent (Fig. 1C, compare lanes 6 and 7) despite the fact that no epitope-tagged gene was present in mec1Δ and rad53Δ cells.
FIGURE 1.
Identification of Mrc1 and Mec1, which cooperate in Rad53 activation. A, schematic of the RDS assay and its use to screen Rad53 activators. B, silver staining of inactive Rad53 and Dun1, purified from endogenously tagged Rad53-His6FLAG/rad9Δmrc1Δ cells and Dun1-TAF/rad53Δ cells, respectively. C, anti-FLAG immunoprecipitates from Mrc1-TAF/rad53Δ and rad53Δ cells stimulate the activation of Rad53. Anti-FLAG immunoprecipitates from mec1Δ, rad53Δ, and Mrc1-TAF/rad53Δ cells were added to the inactive RDS system to activate Rad53. The inactive RDS system consists of the following: 0.5 nm Rad53, 0.85 nm Dun1, and 3 μm GST-Sml1. Typically 1% of the anti-FLAG immunoprecipitate from two liters of yeast cell culture was used. Phosphorylated Sml1 was visualized using autoradiography and quantified using scintillation counting (see “Experimental Procedures” for details). These results are representative of three independent experiments. D, silver staining of 10% of the anti-FLAG immunoprecipitates from two liters of mec1Δ, rad53Δ, and Mrc1-TAF/rad53Δ cells. The band corresponding to Mec1 was identified by mass spectrometry.
To identify these unknown factors, the immunoprecipitated samples from these cells were analyzed using silver staining (Fig. 1D). Although most of the protein bands are common to all three samples, a distinct band with a molecular weight of more than 250 kDa is present only in lanes 2 and 3 and is absent in lane 1, where mec1Δ cells is used (Fig. 1D). This band was excised from the gel and identified as Mec1 by mass spectrometry. To identify additional specific proteins in these immunoprecipitated samples that were not visualized by silver staining, we performed in-solution trypsin digestion and mass spectrometry analysis. Again, Mec1 and Ddc2 were identified in both samples immunoprecipitated from rad53Δ and Mrc1-TAF/rad53Δ cells but not from mec1Δ cells. In addition, Mrc1 was found only in the sample immunoprecipitated from Mrc1-TAF/rad53Δ cells. These results suggest that Mec1 and Mrc1 may be sufficient and act together to promote Rad53 activation (Fig. 1C), whereas the lack of either Mec1 in the mec1Δ sample or Mrc1 in the rad53Δ sample compromises the activation of Rad53 and thus the hyperphosphorylation of Sml1.
Characterization of the RDS System
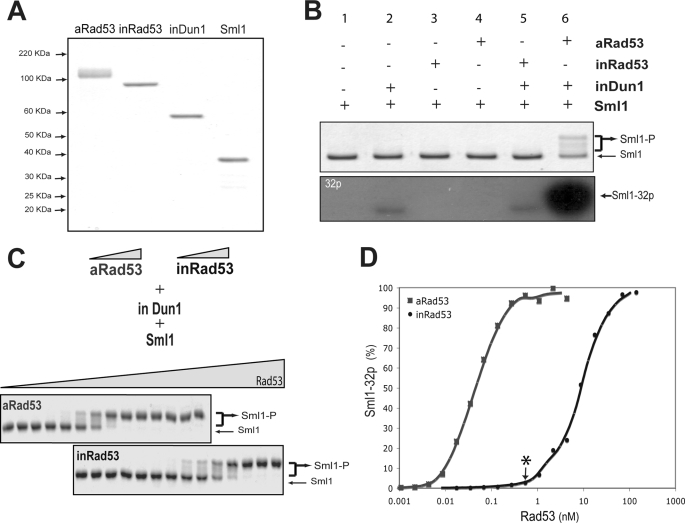
The above observation that Mec1 and Mrc1 might act together to activate Rad53 prompted us to examine their effects further. To this end, it is necessary to have sufficient amounts of inactive Rad53 and inactive Dun1. We chose to purify recombinant Rad53 from Escherichia coli, which is known to be active (29). As shown in Fig. 2A, following an extended Lambda phosphatase treatment, recombinant Rad53 is completely dephosphorylated and shows a faster migration in the gel (Fig. 2A). This Lambda phosphatase-dephosphorylated Rad53 has an activity similar to the endogenous Rad53 purified from rad9Δ mrc1Δ cells. Longer Lambda phosphatase treatment had no additional effect on its activity. Thus, it is considered inactive.3 To prepare inactive Dun1, Dun1 was overexpressed and purified from rad53Δ cells (Fig. 2A). The activities of these purified kinases were then analyzed using Sml1 as a substrate. Only when active Rad53 and inactive Dun1 and Sml1 were used was a characteristic gel shift of the hyperphosphorylated Sml1 observed, whereas the same amount of inactive Rad53 did not cause appreciable hyperphosphorylation of Sml1 (Fig. 2B). Further characterization of the activities of active Rad53 with inactive Dun1 and Sml1 was carried out to determine the concentration ranges of active Rad53 to be used so that changes in its activity could be better detected (see supplemental Fig. S1).
FIGURE 2.
Characterization of the Rad53-Dun1-Sml1 system. A, Coomassie staining of purified proteins: active Rad53 (aRad53), inactive Rad53 (inRad53), inactive Dun1 (inDun1), and GST-Sml1 (see “Experimental Procedures” for details). B, specificity of Sml1 hyperphosphorylation. Only when active Rad53 and Dun1 are both present, is significant Sml1 hyperphosphorylation detected. The following proteins were used in the assay: 17 pm active Rad53 or inactive Rad53, 8.5 nm inactive Dun1, and 3 μm GST-Sml1. C, comparison of inactive Rad53 and active Rad53 activities. 2-fold titrations of inactive Rad53 (ranging from 17 pm to 140 nm) and active Rad53 (ranging from 0.5 pm to 4.4 nm) were used in the kinase reaction. Dun1 and Sml1 concentrations and kinase reaction conditions are the same as in B unless noted otherwise. D, quantification of phosphorylated Sml1 from C. Rad53 concentration is plotted using log10 scale, whereas the amount of phosphorylated Sml1 is normalized to the fully phosphorylated Sml1 (using 2.2 nm active Rad53). For later experiments, 0.5 nm inactive Rad53, indicated by an asterisk, 8.5 nm inactive Dun1, and 3 μm GST-Sml1 were chosen to constitute the inactive Rad53-Dun1-Sml1 (RDS) system.
To quantify the difference in activity between active and inactive Rad53, increasing amounts of Rad53 were used to produce Dun1-dependent Sml1 hyperphosphorylation using either active or inactive Rad53. As shown in Fig. 2C, from left to right there is a 2-fold increase in the amount of active or inactive Rad53 in adjacent lanes. Quantification of the hyperphosphorylated Sml1 reveals an ∼250-fold difference in the concentration between active and inactive Rad53 to achieve the same level of Sml1 hyperphosphorylation (Fig. 2D). This allowed us to choose a concentration of inactive Rad53, i.e. 0.5 nm, for an inactive RDS system so that the effects of Mec1 and Mrc1 on Rad53 activation could be studied. As indicated in Fig. 2D, active Rad53 at this concentration (0.5 nm) is capable of producing an almost complete Sml1 hyperphosphorylation, whereas inactive Rad53 is not. Thus, there is a wide dynamic range to detect any increase in Rad53 activity.
Reconstitution of Rad53 Activation Using Purified Mec1-Ddc2 Complex and Mrc1
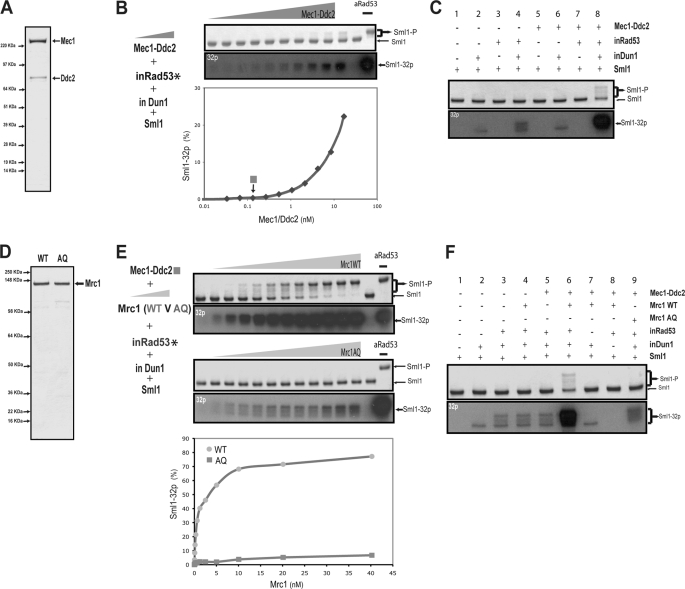
We first examined the ability of Mec1-Ddc2 complex to activate Rad53. The Mec1-Ddc2 complex was purified to near homogeneity (Fig. 3A; see “Experimental Procedures” for details). The addition of increasing amounts of this purified Mec1-Ddc2 complex to the inactive RDS system leads to a gradual increase of Sml1 hyperphosphorylation (Fig. 3B). However, even at the highest concentration of Mec1-Ddc2 complex used (17 nm), the amount of hyperphosphorylated Sml1 is still much less than the use of active Rad53 (0.5 nm). Thus, the Mec1-Ddc2 complex alone does activate the inactive RDS system, albeit with a relatively poor efficiency. Nevertheless, this effect of Mec1 depends on the presence of both inactive Rad53 and inactive Dun1, thus confirming the specificity of Rad53 activation by Mec1 (Fig. 3C).
FIGURE 3.
Reconstitution of Rad53 activation using purified Mec1-Ddc2 complex and Mrc1. A, Coomassie staining of purified Mec1-Ddc2 complex. B, effect of increasing amounts of Mec1 on Rad53 activity, assayed using phosphorylated Sml1, as shown by both Coomassie staining (upper panel) and autoradiography (lower panel). 2-Fold titrations of the Mec1-Ddc2 complex (ranging from 33 pm to 17 nm) were added to the inactive RDS system. 0.13 nm Mec1, indicated by a square, was chosen for the later kinase reactions unless noted otherwise. C, specificity of Rad53 activation by Mec1. The amounts of proteins and kinase reaction conditions are the same as for the inactive RDS system. Only when Mec1, Rad53, and Dun1 were all present was significant Sml1 phosphorylation observed, as shown by both Coomassie staining (upper panel) and autoradiography (lower panel). D, Coomassie staining of purified wild-type and AQ mutant proteins of Mrc1 from E. coli. E, effect of Mrc1 on Rad53 activation by Mec1. 2-Fold titrations of both wild-type and AQ mutant proteins of Mrc1 (ranging from 40 pm to 40 nm) were used together with Mec1 and the inactive RDS system. The effect of the WT and AQ mutant proteins of Mrc1 was quantified using phosphorylated Sml1, which is shown by both Coomassie staining (upper panel) and autoradiography (lower panel). F, specificity of the reconstituted phosphorylation cascade from Mec1 to Sml1. 0.6 nm Mrc1 (wild type and AQ mutant) was used, and other components are the same as mentioned in E. Sml1 phosphorylation is shown the same way as described in E.
To examine the effect of Mrc1, we chose a low concentration of Mec1-Ddc2 (0.13 nm) that causes no appreciable hyperphosphorylation of Sml1 (see arrow in Fig. 3B). Recombinant Mrc1, either the WT or the AQ mutant (21) protein, was purified from E. coli to near homogeneity (Fig. 3D). As shown previously, the mrc1-AQ mutant is defective in checkpoint activation in vivo (21). We examined the effect of adding increasing amounts of either the WT or AQ mutant protein of Mrc1 to the inactive RDS in the presence of 0.13 nm Mec1. As shown in Fig. 3E, the addition of the Mrc1-AQ mutant protein leads to a relatively small increase of Sml1 hyperphosphorylation in this reconstituted system. In contrast, the addition of the same amount of Mrc1 WT protein leads to a more drastic increase in Sml1 hyperphosphorylation as compared with Mrc1-AQ. At the highest concentration of WT Mrc1 used (>30 nm), a similar amount of hyperphosphorylated Sml1 was obtained compared with the use of active Rad53 (0.5 nm) as a positive control (see Fig. 3E, rightmost lane). Compared with the absence of Mrc1, WT Mrc1 leads to a >70-fold increase in the amount of Sml1 phosphorylation. Further, significant hyperphosphorylation of Sml1 was detected only in the kinase reaction containing Mec1, WT Mrc1, inactive Rad53, and inactive Dun1. Omission of any one of these components, or replacing WT Mrc1 by Mrc1-AQ, severely reduces Sml1 hyperphosphorylation (Fig. 3F). Therefore the phosphorylation cascade from Mec1 to Rad53, Dun1, and Sml1 is faithfully reconstituted in vitro, and Mrc1 greatly facilitates this phosphorylation cascade from Mec1 to Sml1.
Phosphorylation Status of Mrc1, Rad53, and Dun1 in the Reconstituted System
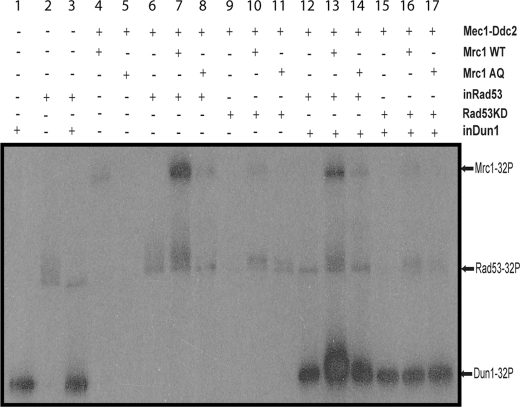
The phosphorylation of Mrc1, Rad53, and Dun1 in the reconstituted system was examined more closely. As shown in Fig. 4, inactive Rad53 alone does autophosphorylate itself, leading to a gel-shifted and phosphorylated Rad53 (see Fig. 4, lane 2). The addition of inactive Dun1 appears to suppress this Rad53 autophosphorylation, and Dun1 becomes more phosphorylated in the same reaction (Fig. 4, compare lanes 2 and 3 for Rad53 phosphorylation and lanes 1 and 3 for Dun1 phosphorylation). Interestingly, the addition of inactive Dun1 again suppresses the phosphorylation of both Mrc1 and Rad53 (Fig. 4, compare lanes 7 and 13), and Dun1 in the same reaction becomes phosphorylated. Thus, Dun1 appears to suppress the phosphorylation of Mrc1 by Rad53 and direct Rad53 phosphorylation to Dun1 in the reconstituted phosphorylation cascade. This effect of Dun1 is likely related to the direct interaction between Dun1 and Rad53 (38).
FIGURE 4.
Phosphorylation status of Mrc1, Rad53, and Dun1 in the reconstituted system. The same kinase reaction condition as used in Fig. 3F except 8 nm Mrc1 and 0.5 nm Rad53KD were used here. Each kinase reaction (50 μl) was analyzed by 10% SDS-PAGE and autoradiography.
A comparison of lanes 4, 7, and 10 in Fig. 4 reveals a much higher level of Mrc1 phosphorylation in the presence of inactive Rad53 but not the Rad53KD mutant protein. Thus, Rad53 appears to phosphorylate Mrc1 after it becomes activated. As expected, Mrc1-dependent activation and hyperphosphorylation of Rad53 is also observed by comparing lanes 6 and 7 in Fig. 4. To ask whether Mrc1 could specifically promote trans-phosphorylation of Rad53 by Mec1, the phosphorylation of Rad53KD in the presence or absence of Mrc1 was examined (Fig. 4, compare lanes 9 and 10). The results show that Mrc1 increases the phosphorylation of Rad53KD by Mec1. In all cases, the use of the Mrc1-AQ mutant protein greatly compromises not only its own phosphorylation but also the phosphorylation of Rad53 and Dun1. Thus, phosphorylation of Mrc1 by Mec1 is essential for the induced phosphorylation and activation of Rad53 by Mec1.
Mrc1 Promotes Enzyme-Substrate Association between Mec1 and Rad53
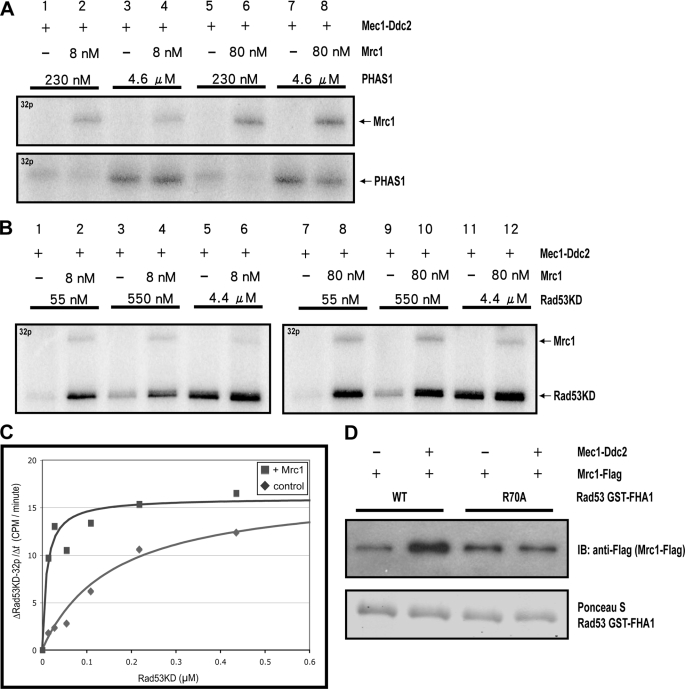
To stimulate Rad53 activation, Mrc1 can either stimulate the catalytic activity of Mec1 or promote association between Mec1 and Rad53. To examine these possibilities, PHAS-1 was used as an artificial substrate to measure the catalytic activity of Mec1. As shown in Fig. 5A, the addition of Mrc1 does not promote more PHAS-1 phosphorylation by Mec1, and thus the catalytic activity of Mec1 is not affected by Mrc1. Interestingly, when Rad53KD was used as a substrate for Mec1, the presence of Mrc1 strongly stimulated Rad53KD phosphorylation by Mec1. The degree of stimulation appears to be higher when the concentration of Rad53KD is lower (Fig. 5B, compare lanes 1 and 2 with lanes 5 and 6). The same trend is observed when the concentration of Mrc1 is 10-fold higher (Fig. 5B, compare lanes 1–6 with lanes 7–12).
FIGURE 5.
Mrc1 promotes enzyme-substrate association between Mec1-Ddc2 and Rad53KD. A, Mrc1 does not stimulate Mec1 catalytic activity toward PHAS-1. PHAS-1 was used as a substrate for Mec1 in the absence or presence of Mrc1 with the indicated concentrations. 0.13 nm Mec1 was used in all experiments shown in this figure. B, Rad53KD phosphorylation by Mec1 is stimulated by Mrc1. The concentration of each protein is shown. C, rate of Rad53KD phosphorylation by Mec1-Ddc2 as a function of Rad53KD concentration in the absence (♦) or presence (■) of 80 nm Mrc1. D, binding between Rad53 WT and R70A mutant FHA1 domains to Mrc1 with or without prior Mec1 phosphorylation (see “Experimental Procedures” for details).
To further characterize the effect of Mrc1 on Rad53KD phosphorylation by Mec1, the kinetics of Rad53KD phosphorylation by Mec1 was measured. As shown in Fig. 5C, the initial rate of Rad53KD phosphorylation by Mec1 was plotted as a function of Rad53KD concentration (see supplemental Fig. S2 for details). In the absence of Mrc1, as the concentration of Rad53KD increases, there was a gradual increase in the rate of Rad53KD phosphorylation by Mec1. Even at the highest concentration of Rad53KD used (>0.4 μm), we did not observe a saturation of Rad53KD phosphorylation, which would correspond to a maximum velocity for Mec1 to phosphorylate Rad53KD. In contrast, in the presence of Mrc1, the rate of Rad53KD phosphorylation reached an apparent maximum at a much lower concentration of Rad53KD and became independent of Rad53KD concentration thereafter (Fig. 5C). This kinetic analysis shows that Mrc1 appears to reduce the apparent Km of the phosphorylation of Rad53KD by Mec1 significantly. Again, Mrc1 did not appreciably change the maximum velocity of this reaction, suggesting that Mrc1 does not alter the catalytic activity of Mec1 significantly. Accordingly, Mrc1 appears to promote enzyme-substrate association between Rad53 and Mec1, thus increasing Rad53 phosphorylation by Mec1.
It was shown previously that the Rad53 FHA1 domain binds to phosphorylated Mrc1 after DNA damage (37). To further examine this binding, we tested whether the Rad53 FHA1 domain binds to purified Mrc1 and, if so, whether this binding is mediated by the phosphorylation of Mrc1 by Mec1. As shown in Fig. 5D, the wild-type Rad53 FHA1 domain appears to have a tighter binding to phosphorylated Mrc1 as compared with unphosphorylated Mrc1. Further, regardless of the phosphorylation status of Mrc1, the FHA1(R70A) mutant protein binds to Mrc1, which is similar to the binding between wild-type FHA1 and unphosphorylated Mrc1 (Fig. 5D). The R70A mutation of the FHA1 domain is expected to compromise FHA binding to phosphorylated threonine residue within a phosphorylated peptide or protein (39). This basal level of binding between the FHA1 domain and unphosphorylated Mrc1 is not unexpected because FHA domains are known to recognize phosphorylated threonine residues and their surrounding amino acid residues in phosphorylated peptides (39). This result thus shows that the binding between phosphorylated Mrc1 and Rad53 is direct and is promoted by Mec1 phosphorylation of Mrc1.
The Conserved Mrc1 C-terminal Domain Is Required for Its Phosphorylation by Mec1 and Rad53 Activation
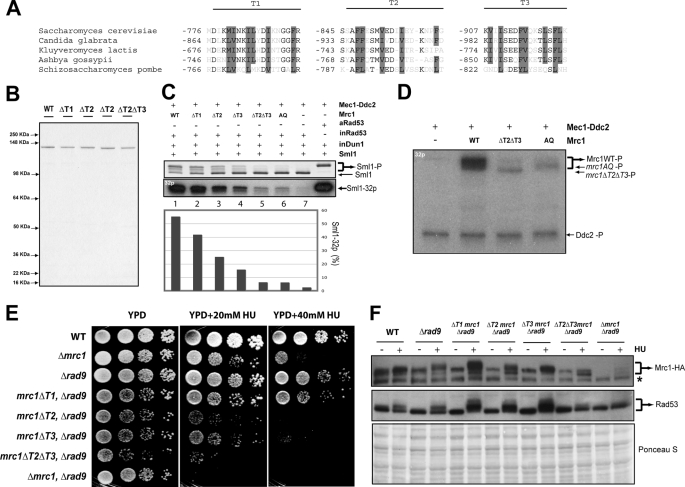
We performed a sequence alignment of the yeast Mrc1 orthologs and identified a conserved C-terminal domain containing blocks of hydrophobic residues (Fig. 6A). Secondary structure prediction suggests that these blocks likely form a coiled-coil domain. To examine its role in Rad53 activation, short stretches of 16–19 amino acid residues within this C-terminal region of Mrc1 were deleted from Mrc1 (indicated as T1, T2, and T3 in Fig. 6A). These internal deletion mutant proteins of Mrc1, purified from E. coli, were tested for their ability to promote Rad53 activation by Mec1 (Fig. 6B). Comparing the use of WT and various C-terminal deletion mutant proteins of Mrc1, i.e. mutations to remove the T1, T2, or T3 region of Mrc1, a partial loss of Sml1 hyperphosphorylation was observed. Interestingly, deletion of both the T2 and T3 regions of Mrc1 virtually eliminated its ability to promote Sml1 hyperphosphorylation (Fig. 6C).
FIGURE 6.
Mrc1 C-terminal domain is required for its phosphorylation by Mec1 and Rad53 activation. A, alignment of three conserved blocks, T1-(776–794), T2-(845–864), and T3-(907–923), from the C-terminal domain of Mrc1 with the corresponding sequences of four related yeast species including S. pombe. Conserved residues with similar properties are marked with black, and hydrophobic residues are shown with a gray background. B, Coomassie staining of purified Mrc1 mutant proteins. C, mutations of the Mrc1 C-terminal domain compromises Rad53 activation in vitro. 8 nm Mrc1 wild-type and mutant (ΔT1, ΔT2, ΔT3, ΔT2ΔT3, and AQ) proteins were tested using the inactive RDS system and Mec1 (same conditions as described for Fig. 4). D, phosphorylation of Mrc1 mutants (ΔT2ΔT3 and AQ) by Mec1 is diminished in vitro. The protein concentrations in the kinase reactions when present are the following: 17 nm Mec1, 160 nm Mrc1 WT, ΔT2ΔT3 mutant, and AQ mutant proteins. E, Mrc1 C-terminal domain mutations cause increased sensitivity to hydroxyurea. 5-Fold dilutions of logarithmically growing cells of each strain indicated were spotted on YPD plates and on YPD plates containing 20 and 40 mm hydroxyurea, and the plates were incubated at 30 °C for 3 days. F, Mrc1 and Rad53 phosphorylation in vivo depends on an intact Mrc1 C-terminal domain. Wild-type or mutant cells were incubated in the absence (−) or presence (+) of 100 mm hydroxyurea for 2 h at 30 °C. The gel mobility of Mrc1–3XHA (top panel) and Rad53 (middle panel) were analyzed by SDS-PAGE followed by Western blotting using anti-hemagglutinin (HA) (Roche Applied Science) and anti-Rad53 (Santa Cruz Biotechnology) antibodies, respectively. *, a common contaminating band. The bottom panel shows Ponceau S staining of the same membrane.
To further address the defect of the C-terminal mutation of Mrc1, the phosphorylation of Mrc1 mutant proteins by Mec1 was examined. As shown in Fig. 6D, although WT Mrc1 is efficiently phosphorylated by Mec1 (see lane 2), the internal deletion of both the T2 and T3 regions of Mrc1 greatly compromises its phosphorylation by Mec1, which is reduced to a level comparable with that of the nonspecific phosphorylation of the Mrc1-AQ mutant protein by Mec1. Thus, the C-terminal region of Mrc1 appears to be required for Mec1 to phosphorylate Mrc1. That phosphorylation of Mrc1 by Mec1 is required for Rad53 binding explains the defects of Mrc1 C-terminal mutants in Rad53 activation in vitro (see Fig. 6C). To address the in vivo relevance of the conserved C-terminal domain of Mrc1, these mutations were introduced to the endogenous MRC1 locus. Because Rad9 is known to act redundantly with Mrc1 for Rad53 activation, various C-terminal mutants of MRC1 were introduced into the rad9Δ background. As shown in Fig. 6E, in the rad9Δ background, mutations in the C-terminal domain of Mrc1 lead to a higher sensitivity to hydroxyurea (HU). The mrc1(ΔT2 ΔT3)rad9Δ mutant is almost as sensitive as the rad9Δ mrc1Δ mutant.
Finally, we asked whether HU-induced phosphorylation of Mrc1 and Rad53 is affected by these internal deletion mutations in the Mrc1 C terminus. As shown in Fig. 6F, HU-induced gel shifts of both Mrc1 and Rad53 are not appreciably perturbed by mutations to remove T1, T2, or T3 individually, whereas deletion of both the T2 and T3 regions of Mrc1 greatly diminishes the gel shift of both Mrc1 and Rad53, indicating that HU-induced Rad53 activation in vivo is compromised. These findings are consistent with the effects of various Mrc1 mutant proteins in Rad53 activation in vitro. In conclusion, the conserved C-terminal domain of Mrc1 is required for its ability to mediate Rad53 activation by Mec1 in vitro and in vivo. Specifically, the C-terminal domain helps to promote Mrc1 phosphorylation by Mec1.
DISCUSSION
Activation of the Rad53 and the Chk2 family kinases is a hallmark of DNA replication checkpoint activation. Extensive studies have identified the role of Mec1 in the phosphorylation of Rad53 and a key role of Mrc1 in the recruitment of Rad53 after Mrc1 is phosphorylated by Mec1 for Rad53 activation (21, 33). Both Mec1 and Mrc1 are known to be recruited to the DNA replication forks via distinct mechanisms; thus they are poised to exert their activities on Rad53, which is known to help maintain DNA replication forks in response to genotoxic stresses (4, 6). Although many proteins are also present in the DNA replication forks (24), it is satisfying to find that the Mec1-Ddc2 complex and Mrc1 constitute a minimal in vitro system to directly activate Rad53 through a biochemical screen to identify Rad53 activators.
Our previous study suggested that by using Dun1 and its substrate, Sml1, it is possible to establish an activity-based assay for Rad53 that is not only sensitive but, more importantly, specific and quantitative (29). To reach the same level of Sml1 phosphorylation, the concentration of active Rad53 was found to be almost 250-fold lower than inactive Rad53. Using an inactive RDS system, the ability of Mec1 to activate Rad53 directly was confirmed. Importantly, this experiment also revealed that Mec1 by itself is a relatively poor Rad53 activator in vitro. Remarkably, addition of purified WT Mrc1 leads to an over 70-fold stimulation of Rad53 activation, as measured by Sml1 phosphorylation. This Mrc1 effect is specific, as omission of any one of the components in the reconstitution reaction eliminates the effect entirely (Fig. 3F). Moreover, when Mrc1 is replaced by the Mrc1-AQ mutant protein, the effect of Mrc1 is greatly diminished. In summary, these biochemical findings from our reconstituted system faithfully recapitulated the known genetic and biochemical results from previous studies. Thus, this study has established the first reconstituted kinase cascade of the DNA replication checkpoint.
The reconstitution of Rad53 activation by Mec1 and Rad53 further allowed us to address the role of Mrc1 in Rad53 activation. Although autophosphorylation is a general property of the Rad53 and Chk2 family kinases and is undoubtedly involved in Rad53 activation, the presence of upstream activators became essential when Rad53 was present at a limiting concentration in vivo or in vitro in our assay. Several studies have found that the catalytic activity of the Mec1/ATR family kinases is regulated by its association with various DNA replication-associated proteins, including Dpb11 and the 9-1-1 complex. Interestingly, Mrc1 appears to act through a different mechanism. Instead of increasing the catalytic activity of Mec1, the primary role of Mrc1 appears to be promoting an enzyme-substrate association between Mec1 and Rad53. Two features of Mrc1 were found to be required for this role. First, the (S/T)Q sites of Mrc1 that are phosphorylated by Mec1 are essential. This (S/T)Q phosphorylation of Mrc1 is believed to be responsible for Rad53 binding in vivo. Using purified proteins, phosphorylated Mrc1 showed a tighter binding to the Rad53 FHA1 domain (Fig. 5D). Second, the conserved C-terminal coiled-coiled domain of Mrc1 was found to be required for Mrc1 to become better phosphorylated by Mec1, and thus Rad53 activation, even though the C-terminal truncation mutations do not affect (S/T)Q sites of Mrc1 (Fig. 6). Although we cannot exclude the possibility that some of these C-terminal mutations may affect the folding of Mrc1, the expression of various Mrc1 mutants in both bacteria and yeast are similar to that of WT Mrc1. Further, the addition of the purified Mrc1 C-terminal domain alone or of synthetic T2 and T3 peptides had no appreciable effect on the ability of wild-type Mrc1 to stimulate Rad53 activation in a competition experiment.3 Future studies are needed to address the role of the conserved C-terminal domain of Mrc1, which is clearly required for Rad53 activation by Mec1.
The role of Mrc1 in promoting Rad53 activation is also reminiscent of another adaptor protein, Rad9, in Rad53 activation. As shown previously, Rad9 stimulates Mec1 to phosphorylate Rad53KD in vitro (40). Thus, this property of adaptor proteins including Mrc1 and Rad9 appears to be a common mechanism by which the downstream checkpoint kinase such as Rad53 is activated by upstream kinases such as Mec1. As indicated by our detailed kinetic analysis, the role of adaptor protein Mrc1 appears to be in promoting a better enzyme-substrate association between these kinases.
The addition of Mrc1 significantly reduces the apparent Km of the phosphorylation of Rad53KD by Mec1 (Fig. 5). We have not been able to measure the affinity between Mec1 and Mrc1 due to the nature of their transient interaction and a lack of sufficient concentration of these purified proteins. Such transient interaction between an enzyme and its substrate is not unexpected. Nevertheless, the kinetic results suggest a tighter association between them (see Fig. 5).
The importance of a specific protein-protein association in DNA damage checkpoint signaling is further revealed by the observations that a forced assembly of signaling proteins to the same location in the genome could activate Rad53 without introducing DNA damage signals (41, 42). In addition, many proteins are known to localize to the site of DNA damage in cells (43). Thus, an understanding of the protein-protein interactions for DNA damage and replication checkpoint adaptor proteins is needed to understand the specificity of DNA damage and replication checkpoint signaling in vivo. Although Mec1 and Mrc1 constitute a minimal system for Rad53 activation, it should be emphasized that additional DNA replication fork-associated proteins may further promote the ability of these proteins to activate Rad53. For example, replication protein A-coated single-stranded DNA is known to be responsible for the recruitment of the Mec1-Ddc2 complex to the DNA replication forks. On the other hand, the fission yeast Mrc1 has been suggested to bind to specific DNA structures (44). Finally, additional replication fork-associated proteins, including Tof1, Dpb11, and various alternative replication factor C (RFC) complexes, may also contribute to the activation of Rad53. Given the direct association of Mrc1 with Rad53 and the genetic requirement of Mrc1 for Rad53 activation, it is likely that additional factors act through Mrc1. Thus, an interesting future direction will be to use this reconstitution system to study how additional components of the DNA replication forks may contribute to Rad53 activation.
Acknowledgments
We are grateful to Jean Y. J. Wang, Christopher Putnam, Samantha G. Zeitlin, Hans Hombauer, and Jerzy Majka for advice and encouragement, to Peter Burgers for pBL504 plasmid, and to Richard D. Kolodner and Samantha G. Zeitlin for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM080469 (to H. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
S. Chen and H. Zhou, unpublished observations.
- ATR
- ataxia telangiectasia-mutated and Rad3-related
- DTT
- dithiothreitol
- Ni-NTA
- nickel-nitrilotriacetic acid
- WT
- wild type
- RDS
- Rad53-Dun1-Sml1
- Rad53KD
- Rad53 kinase-dead protein
- HU
- hydroxyurea
- GST
- glutathione S-transferase
- FHA
- forkhead associated.
REFERENCES
- 1.Samadashwily G. M., Raca G., Mirkin S. M. (1997) Nat. Genet. 17, 298–304 [DOI] [PubMed] [Google Scholar]
- 2.Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. (2002) Annu. Rev. Genet. 36, 617–656 [DOI] [PubMed] [Google Scholar]
- 3.Kolodner R. D., Putnam C. D., Myung K. (2002) Science 297, 552–557 [DOI] [PubMed] [Google Scholar]
- 4.Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. (2001) Nature 412, 557–561 [DOI] [PubMed] [Google Scholar]
- 5.Santocanale C., Diffley J. F. (1998) Nature 395, 615–618 [DOI] [PubMed] [Google Scholar]
- 6.Tercero J. A., Diffley J. F. (2001) Nature 412, 553–557 [DOI] [PubMed] [Google Scholar]
- 7.Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J. (1998) Genes Dev. 12, 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myung K., Datta A., Kolodner R. D. (2001) Cell 104, 397–408 [DOI] [PubMed] [Google Scholar]
- 9.Hartwell L. H., Kastan M. B. (1994) Science 266, 1821–1828 [DOI] [PubMed] [Google Scholar]
- 10.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 11.Nakada D., Hirano Y., Tanaka Y., Sugimoto K. (2005) Mol. Biol. Cell 16, 5227–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majka J., Burgers P. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majka J., Niedziela-Majka A., Burgers P. M. (2006) Mol. Cell 24, 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia V., Furuya K., Carr A. M. (2005) DNA Repair 4, 1227–1239 [DOI] [PubMed] [Google Scholar]
- 15.Araki H., Leem S. H., Phongdara A., Sugino A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11791–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamimura Y., Masumoto H., Sugino A., Araki H. (1998) Mol. Cell. Biol. 18, 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Elledge S. J. (2002) Genetics 160, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. (2006) Cell 124, 943–955 [DOI] [PubMed] [Google Scholar]
- 19.Mordes D. A., Nam E. A., Cortez D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18730–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navadgi-Patil V. M., Burgers P. M. (2008) J. Biol. Chem. 283, 35853–35859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., Bousset K., Furuya K., Diffley J. F., Carr A. M., Elledge S. J. (2001) Nat. Cell Biol. 3, 958–965 [DOI] [PubMed] [Google Scholar]
- 22.Tanaka K., Russell P. (2001) Nat. Cell Biol. 3, 966–972 [DOI] [PubMed] [Google Scholar]
- 23.Szyjka S. J., Viggiani C. J., Aparicio O. M. (2005) Mol. Cell 19, 691–697 [DOI] [PubMed] [Google Scholar]
- 24.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 25.Lou H., Komata M., Katou Y., Guan Z., Reis C. C., Budd M., Shirahige K., Campbell J. L. (2008) Mol. Cell 32, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y. J., Davenport M., Kelly T. J. (2006) Genes Dev. 20, 990–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert C. S., Green C. M., Lowndes N. F. (2001) Mol. Cell 8, 129–136 [DOI] [PubMed] [Google Scholar]
- 28.Xu X., Tsvetkov L. M., Stern D. F. (2002) Mol. Cell. Biol. 22, 4419–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S. H., Smolka M. B., Zhou H. (2007) J. Biol. Chem. 282, 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn J. Y., Li X., Davis H. L., Canman C. E. (2002) J. Biol. Chem. 277, 19389–19395 [DOI] [PubMed] [Google Scholar]
- 31.Usui T., Petrini J. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2797–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S. J., Schwartz M. F., Duong J. K., Stern D. F. (2003) Mol. Cell. Biol. 23, 6300–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka K., Boddy M. N., Chen X. B., McGowan C. H., Russell P. (2001) Mol. Cell. Biol. 21, 3398–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 36.Pasero P., Duncker B. P., Gasser S. M. (1999) Methods 18, 368–376, 323 [DOI] [PubMed] [Google Scholar]
- 37.Smolka M. B., Chen S. H., Maddox P. S., Enserink J. M., Albuquerque C. P., Wei X. X., Desai A., Kolodner R. D., Zhou H. (2006) J. Cell Biol. 175, 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H., Yuan C., Hammet A., Mahajan A., Chen E. S., Wu M. R., Su M. I., Heierhorst J., Tsai M. D. (2008) Mol. Cell 30, 767–778 [DOI] [PubMed] [Google Scholar]
- 39.Durocher D., Taylor I. A., Sarbassova D., Haire L. F., Westcott S. L., Jackson S. P., Smerdon S. J., Yaffe M. B. (2000) Mol. Cell 6, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 40.Sweeney F. D., Yang F., Chi A., Shabanowitz J., Hunt D. F., Durocher D. (2005) Curr. Biol. 15, 1364–1375 [DOI] [PubMed] [Google Scholar]
- 41.Bonilla C. Y., Melo J. A., Toczyski D. P. (2008) Mol. Cell 30, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S. J., Duong J. K., Stern D. F. (2004) Mol. Biol. Cell 15, 5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004) Cell 118, 699–713 [DOI] [PubMed] [Google Scholar]
- 44.Zhao H., Russell P. (2004) J. Biol. Chem. 279, 53023–53027 [DOI] [PubMed] [Google Scholar]