Abstract
Vascular smooth muscle cells (VSMCs) maintain the ability to modulate their phenotype in response to changing environmental stimuli. This phenotype modulation plays a critical role in the development of most vascular disease states. In these studies, stimulation of cultured vascular smooth muscle cells with platelet-derived growth factor resulted in marked induction of c-jun expression, which was attenuated by protein kinase Cδ and calcium/calmodulin-dependent protein kinase inhibition. Given that these signaling pathways have been shown to relieve the repressive effects of class II histone deacetylases (HDACs) on myocyte enhancer factor (MEF) 2 proteins, we ectopically expressed HDAC4 and observed repression of c-jun expression. Congruently, suppression of HDAC4 by RNA interference resulted in enhanced c-jun expression. Consistent with these findings, mutation of the MEF2 cis-element in the c-jun promoter resulted in promoter activation during quiescent conditions, suggesting that the MEF2 cis-element functions as a repressor in this context. Furthermore, we demonstrate that protein kinase A attenuates c-Jun expression by promoting the formation of a MEF2·HDAC4 repressor complex by inhibiting salt-inducible kinase 1. Finally, we document a physical interaction between c-Jun and myocardin, and we document that forced expression of c-Jun represses the ability of myocardin to activate smooth muscle gene expression. Thus, MEF2 and HDAC4 act to repress c-Jun expression in quiescent VSMCs, protein kinase A enhances this repression, and platelet-derived growth factor derepresses c-Jun expression through calcium/calmodulin-dependent protein kinases and novel protein kinase Cs. Regulation of this molecular “switch” on the c-jun promoter may thus prove critical for toggling between the activated and quiescent VSMC phenotypes.
Vascular smooth muscle cells (VSMCs),2 unlike their skeletal and cardiac counterparts, do not terminally differentiate but can modulate their phenotype under conditions of growth or differentiation (1). Differentiated smooth muscle cells express high levels of contractile proteins and other muscle-specific genes, a phenotype that has been termed “quiescent” or “contractile.” However, in response to vascular injury, VSMCs down-regulate muscle-specific genes, increase their proliferation rate and migration capacity, and actively secrete matrix proteins. This proliferative phenotype has been called the “activated” or “synthetic” phenotype (1). Although proliferative VSMCs are undoubtedly required for vascular development and during vascular repair, this activated phenotype also plays a role in multiple smooth muscle diseases, such as atherosclerosis and restenosis following angioplasty (1). Therefore, the molecular mechanisms whereby VSMCs modulate their phenotype between the quiescent and activated states is of particular interest for our understanding of smooth muscle cell biology under physiological and pathological conditions.
The MADS box transcription factor, serum response factor (SRF), plays a critical role in smooth muscle phenotype modulation. SRF binds to a cognate cis-element termed the CArG box, which can be found in multiple copies in many smooth muscle structural genes (2). Conversely, SRF is also involved in smooth muscle proliferation by binding to a single CArG box in the proximal promoter of c-fos, a growth-responsive immediate-early gene (2). This dual role for SRF is largely regulated by recruiting co-activators, such as myocardin (3), to activate smooth muscle genes, or ternary complex factors, such as Elk-1, to activate immediate-early genes (4).
Another mammalian MADS box transcription factor, known as myocyte enhancer factor (MEF) 2 is functionally important in cardiac, skeletal, and smooth muscle cells. Recent studies have identified two smooth muscle marker genes that require a consensus MEF2-binding site in their respective promoter regions for expression in VSMCs in vivo. These genes encode myocardin, a master regulator of smooth muscle differentiation (5), and the histidine-rich calcium-binding protein (HRC), a sarcoplasmic reticulum protein expressed in skeletal, cardiac, and smooth muscle (6). In addition, gene targeting studies have revealed that MEF2C is required for proper vascular patterning and vascular smooth muscle differentiation (7). However, despite this emerging evidence supporting the role of MEF2 proteins in vascular smooth differentiation, MEF2 has also been associated with the activated, proliferative smooth muscle phenotype (8).
Analogous to SRF activation of the c-fos gene, MEF2 can increase the expression of the immediate-early gene, c-jun, which is known to act as a downstream target of the smooth muscle mitogen, platelet-derived growth factor (PDGF) (9, 10). It is currently not known whether PDGF induction of c-jun is mediated through MEF2; however, evidence from other cell lines suggests the involvement of MEF2 in the serum induction of c-jun (11). To date, very little is known regarding the role of MEF2 in smooth muscle phenotype modulation, but it appears that both SRF and MEF2 proteins have a regulatory role in smooth muscle proliferation and differentiation.
The transcriptional activity of MEF2 proteins is regulated by post-translational modifications, such as phosphorylation and sumoylation, and a number of interacting protein co-factors. The cellular consequences of the interaction between MEF2 and class II histone deacetylases (HDACs), and its regulation by calcium/calmodulin kinases (CaMK) and PKCδ/PKD signaling, have not thus far been elucidated in VSMCs. Interestingly, PDGF signaling is known to activate CaMKs and PKCδ/PKD during VSMC migration (12–14), and we have previously shown that the novel PKC isoforms, PKCδ and PKCϵ, can activate MEF2 proteins in HeLa and COS cells (15). Therefore, we speculated that PDGF induction of c-jun in VSMCs might be mediated by PKCδ- and CaMK-mediated derepression of MEF2.
Protein kinase A (PKA), the cyclic AMP-dependent protein kinase, potently inhibits vascular smooth muscle proliferation and may protect against vascular disease (16). In VSMCs, PKA is activated by prostaglandin I2 (prostacyclin) and β-adrenergic agonists. Interestingly, in humans, reduced production of prostaglandin I2 (prostacyclin) by cyclooxygenase II inhibition is associated with increased cardiovascular risk (17). One mechanism by which PKA has been shown to inhibit smooth muscle proliferation is to inhibit the expression of c-jun (18). In addition, recent evidence from our laboratory and from others suggests that PKA can promote HDAC4 repression of MEF2-dependent transcriptional activation in other cell types (19–21). Therefore, we evaluated the role of PKA signaling on MEF2-dependent c-jun expression in VSMCs.
In this report, we demonstrate that a MEF2 cis-element in the c-jun promoter serves as a repressor element in quiescent VSMCs and that this repression is largely abolished during conditions of cell growth. Consistent with this finding, HDAC4 is exported from the nuclear compartment during growth conditions or by exogenous expression of CaMK or PKD, whereas PDGF induction of c-jun is prevented by CaMK and PKCδ inhibition. In addition, gain and loss of function manipulation of HDAC4 levels reveal its involvement in regulation of c-jun expression in VSMCs, making this the first report to document that class II HDACs regulate immediate-early gene expression in connection with a proliferative phenotype in VSMCs. Furthermore, PKA promotes MEF2·HDAC4 repression of c-jun expression by inhibiting the activity of salt-inducible kinase 1 (SIK1). Finally, forced expression of c-Jun inhibits the ability of myocardin to activate smooth muscle gene expression, illustrating the fundamental importance of c-Jun regulation by MEF2 and HDAC4 during smooth muscle phenotype modulation.
EXPERIMENTAL PROCEDURES
Plasmids
MEF2 and c-Jun reporter constructs (pJC6, pJSX, and pJTX) in pGL3 and expression vectors for MEF2A, MEF2C, MEF2D, the MEF2A-VP16 fusion, the Gal4-MEF2A and Gal4-MEF2D fusions, and c-Jun have been described previously (15, 21, 22). Mouse CaMKIV was cloned by reverse transcription-PCR, and an activated construct was generated by truncation at amino acid 275. PCR products were ligated into the NotI-XbaI (CaMKIV) site of pcDNA3 for mammalian expression. An expression vector for rat CaMKII δB was kindly provided by A. Hudmon, and a constitutively active mutation was made by replacing threonine 287 with an aspartic acid residue by PCR-based mutagenesis. Expression vectors for the activated PKD and myocardin were generous gifts from E. Olson, and expression vectors for FLAG-tagged HDAC4 and HDAC5 were provided by S. Schreiber. The HDAC4-EGFP fusion and HDAC4 L175A vectors were kindly provided by X.-J. Yang. pSVL-SIK1 and pSVL-SIK1 S577A were kindly provided by H. Takemori. The Gal4-c-Jun fusion proteins were a kind gift from E. Yeh. The SM-MHC promoter was a gift from S. White, and the smooth muscle α-actin and calponin reporter genes were generously provided by J. Miano. The cardiac promoters for α-cardiac actin and α-myosin heavy chain were generously provided by M. Nemers, the PGC-1 promoter was purchased from Addgene, and the MMP-9 promoter was a gift from D. Boyd. The HRC promoter was provided by B. Black and subcloned into pGL4.10 (XhoI-HindIII). The 350-bp myocardin enhancer described by Creemers et al. (5), was PCR-amplified from mouse genomic DNA with KpnI and BglII restriction sites incorporated into the primers. The resulting DNA fragment was ligated with the c-fos minimal promoter (BglII-NcoI), described previously (22), into pGL4.10 (KpnI-NcoI). The MCP-1 luciferase construct was kindly provided by A. Garzino Demo. An expression vector containing the catalytic subunit of PKA (pFC-PKA) was purchased from Stratagene.
Cell Culture and Treatment of VSMCs
Rat A10 myoblasts (ATCC; CRL-1476) were maintained in growth medium consisting of 10% fetal bovine serum (FBS). Quiescence was obtained by refeeding the cells with either 1% or 0% FBS in Dulbecco's modified Eagle's medium overnight. C3H10T1/2 mouse embryonic fibroblasts (ATCC; CCL-226) and COS7 cells (ATCC) were maintained in standard Dulbecco's modified Eagle's medium with 10% FBS and re-fed in 5% horse serum to achieve quiescence. For conversion assays, C3H10T1/2 were grown to confluence and made quiescent for 4 days prior to harvesting.
Luciferase and β-Galactosidase Assays
Transient transfections of A10 and C3H10T1/2 cells were performed by a modified calcium phosphate-DNA precipitation with pCVM-β-galactosidase serving as an internal control for transfection efficiency (23). Luciferase and β-galactosidase activities were measured as described previously (24).
Immunoblot Analysis
Protein extractions were achieved using an Nonidet P-40 lysis buffer described previously (23). Protein concentrations were determined by Bradford assay, and 15 μg were resolved using SDS-PAGE and transfed to an Immobilon-P membrane (Millipore, Inc.). Immunoblotting was carried out using appropriate primary antibody in 5% powdered milk in PBS. Appropriate horseradish peroxidase-conjugated secondary antibody (Bio-Rad; 1:2000) was used in combination with chemiluminescence to visualize bands.
Nuclear/Cytosolic Fractionation
Nuclear and cytosolic fractions were obtained using a Pierce kit. The fractions were subjected to SDS-PAGE and immunoblotting as described above.
Immunofluorescence
A10 VSMCs, cultured as described in the figure legends, were fixed, permeabilized, and incubated with a primary HDAC4 antibody (Sigma) and TRITC-conjugated secondary antibody. Cells were visualized using standard fluorescence techniques or confocal microscopy.
siRNA Oligonucleotides
Sense and antisense siRNA oligonucleotides specific for mouse and rat HDAC4 (5′-GATCCACTGGTGCTTAACATTTGATTCAAGAGATCAAATGTTAAGCACCAGTTTTTTTGGAAA-3′) were purchased from Sigma Genosys, annealed, and ligated into pSilencer 3.0 H1 (Ambion). The siRNA for HDAC4 or a nonspecific scrambled control were transfected into A10 cells with Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. Transfected cells were enriched by puromycin selection (0.5 μg/ml) for 3 days prior to harvesting for protein extracts.
Carotid Injury of the MEF2 “Sensor” Mouse and Sprague-Dawley Rat
Wire injury of mouse carotid arteries and balloon injury of Sprague-Dawley rats were described previously (25, 26). Immunofluorescence and X-gal staining of mice harboring three tandem MEF2 consensus DNA-binding sites driving a LacZ reporter gene, which was described previously (27).
Human Aortic Tissues
Human abdominal aortic aneurysm segments were obtained from patients undergoing elective repair (n = 4, all men). The average age was 70.4 years. The average size of the aneurismal lesions estimated by CT scan and/or angiography was 6.95 cm. During graft replacement for abdominal aortic aneurysm, macroscopically normal adjacent normal aortic segments were carefully excised from four patients and used as controls. Immediately after procurement, segments were placed in sterile normal saline and transported to the laboratory. The protocol of this study was approved by the Clinical Research Ethics Committee at the St. Michael's Hospital and University of Toronto. Written informed consent was given by all patients. The samples were frozen in liquid nitrogen and stored for RNA analysis or were embedded in optimal cutting temperature compound, immediately frozen in liquid nitrogen, and stored at −80 °C.
Laser Capture Microdissection
Cryostat sections (∼8 μm) were mounted on membrane-based microdissection slides (Acutrus Engineering, Mountain View, CA) and fixed for 2 min with cold acetone. After washing twice 5 s each with diethyl pyrocarbonate-treated PBS, pH 7.6, the sections were incubated with fluorescein isothiocyanate-conjugated mouse anti-human smooth muscle (SM) α-actin antibody (Abcam Inc., Cambridge, MA; 1:20) for 5–8 min at room temperature. The sections were washed rapidly three times for 1 min each with diethyl pyrocarbonate-treated PBS followed by dehydration in graded ethanol solutions (70% one time for 1 min, 95% one time for 1 min, and 100% two times for 1 min each) and cleaned in xylene (two times for 5 min each). After air-drying for 5 min, laser capture microdissection was performed under direct microscopic visualization on the SM α-actin-positive stained areas. The Leica laser capture microdissection system (Leica Microsystem; Wetzlar GmbH) was set to the following parameters: laser diameter, 15 μm; speed, 1.5 ms; and amplitudes, 40 milliwatt. A total of 500–3000 target cells were captured for each sample.
Total RNA Isolation and Amplification
Total RNA from laser capture microdissection-captured cells was isolated by using the RNeasy micro RNA isolation kit (Qiagen). T7-based RNA amplification was performed by using the RiboAmp kit (Arcturus Engineering) according to the manufacturer's instructions.
Analysis of Gene Expression by Quantitative Real Time Reverse Transcription-PCR
Total RNA extracted either from laser-captured SMC or from alternating whole sections was reverse transcribed using Omniscript first-stand synthesis kit (Invitrogen) under conditions described by the supplier. cDNA was amplified by quantitative real time PCR (ABI prism 7700 sequence detection system; Applied Biosystems, Foster city, CA) using SYBR Green PCR Master Mix reagent (Qiagen). The primer pair sequences for each reaction was performed in duplicate by using equal amount of cDNA from each sample as template. The primer sequences of genes used in this study were: HDAC-4: forward, 5′-GGTTTGAGAGCAGGCAGAAC-3′, and reverse, 5′-CAGAGAATGAGGCCAAGGAG-3′; and GAPDH: forward, 5′-GAAGGTGAAGGTCGGAGTC-3′, and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. Thermal activation was initiated at 95 °C for 10 min, followed by 40 cycles of polymerase chain reaction (melting for 15 s at 95 °C and annealing/extension for 1 min at 60 °C). Relative quantitations of gene expression were calculated using standard curves and normalized to GAPDH in each sample.
Immunostaining Analysis of Aortic Tissue
Frozen segments from abdominal aortic aneurysm and adjacent NA tissues were sectioned in 10-μm-thick sections, briefly dried, and fixed in acetone. The sections were incubated in normal horse serum (Sigma) for 1 h, followed by a 1-h incubation with the primary antibody rabbit anti-human HDAC4 (1:200, Sigma). With intervening washes in PBS, the sections were then incubated for 30 min with biotin-conjugated horse anti-rabbit secondary antibody (1:200; Vector Laboratories, Burlingame, CA), followed by a 1-h incubation with Alexa fluor 488-conjugated streptavidin (1:200; Sigma). The sections were washed, mounted, and analyzed with confocal microscope (Leica Microsystem Inc, Exton, PA).
RESULTS
MEF2 Expression and Transcriptional Activation Following Carotid Injury
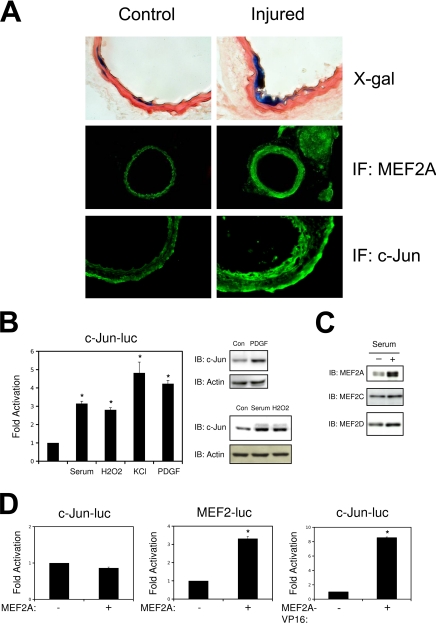
Previous studies have implicated MEF2 proteins in the activated smooth muscle response (8). Although MEF2 transcriptional activation following vascular injury has not, as yet, been reported, to this end, we utilized the MEF2 sensor mouse that we and others have previously used to evaluate MEF2 transcriptional activation during development (27, 28). As shown in Fig. 1A, carotid injury elicited a widespread increase in MEF2A expression, consistent with previous reports (8). MEF2 transcriptional activation, as indicated by X-gal staining of the MEF2 Lac Z-derived arteries, was observed at the site of injury (Fig. 1A). In addition, we observed an increased expression of the MEF2 target gene, c-jun, at the site of injury (Fig. 1A). Because the induction of MEF2A expression was not accompanied by a widespread increase in MEF2 transcriptional activation, we further studied the role of MEF2 proteins in the regulation of smooth phenotype. In particular, we analyzed the regulation of the c-jun promoter, a previously characterized MEF2 target gene that has been implicated as a key regulator of VSMC proliferation control. In the context of quiescent cultured smooth muscle cells, we found that the c-jun promoter, as predicted, was induced by serum stimulation, oxidative stress, depolarization, and PDGF treatment (Fig. 1B). These treatments resulted in corresponding increases in c-Jun protein expression, whereas treatment with transforming growth factor β1 had no effect (supplemental Fig. S1). To evaluate the role of MEF2 in c-jun expression, we first ectopically expressed MEF2 proteins with the c-jun reporter gene. Interestingly, and in contrast to other cell types, we found that MEF2 proteins were unable to activate c-jun expression in A10 smooth muscle cells (Fig. 1D and supplemental Fig. S1). However, MEF2 proteins were able to activate an artificial MEF2 reporter gene (MEF2-luc), a myocardin enhancer-based reporter gene, and the HRC promoter (HRC-luc) in this context (Fig. 1D and supplemental Fig. S1). In addition, a fusion protein consisting of the MEF2A DNA-binding domain fused to the VP16 transcriptional activation domain was able to activate the c-jun promoter (Fig. 1D). Collectively, these data suggest that MEF2 is capable of binding to both the c-jun and muscle-specific reporter regions in cultured smooth muscle cells, but the transcriptional responses of these target genes is divergent.
FIGURE 1.
MEF2 activity and expression in VSMCs in vivo and in vitro. Common carotid arteries of MEF2-LacZ mice were injured by inserting a 2-mm wire into the external carotid. Contralateral arteries were used as the control. A, X-gal staining and immunofluorescence (IF) for MEF2A and c-Jun 14 days following injury. B, A10 cells were transfected with the wild-type c-jun promoter (c-Jun-luc). Following recovery, the cells were serum-starved overnight and treated with 20% FBS, 100 μm H2O2, 60 mm KCl, or 10 ng/ml of PDGF for 4 h for luciferase extracts or 2 h for protein extracts. C, growth phase A10s in 10% FBS (+) or serum-free medium (−) were harvested for protein subjected to immunoblotting (IB) for MEF2A, MEF2C, and MEF2D. D, A10 cells were transfected with the c-jun or MEF2 reporter genes and MEF2A or MEF2A-VP16 (*, p < 0.05 was considered statistically significant).
c-Jun Expression Is Regulated by CaMK, PKCδ, and HDAC4 in Smooth Muscle Cells
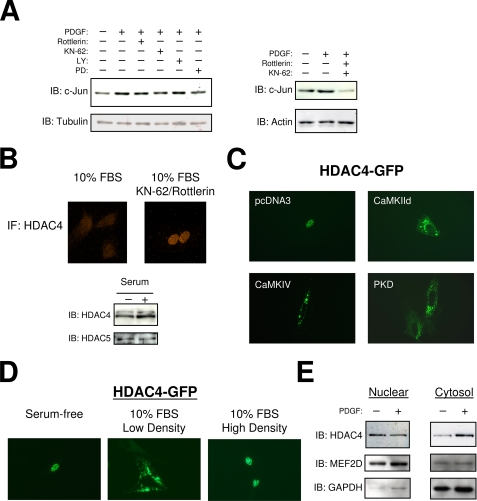
To examine the potential signaling pathways that regulate c-jun expression by PDGF, we utilized common pharmacological inhibitors in our culture model. As shown in Fig. 2A, inhibition of PKCδ by rottlerin, inhibition of CaMKII and IV by KN-62, or inhibition of MEK1 by PD98059 all resulted in a modest reduction in c-Jun protein, whereas inhibition of phosphatidylinositol 3-kinase by LY294002 had no effect. However, combination of rottlerin and KN-62 resulted in marked reduction in c-Jun, below levels observed in quiescent cells. In addition, the activation of CaMK or PKC signaling by A23187 or phorbol 12-myristate 13-acetate, respectively, also increased c-Jun protein expression (not shown). Given that previous studies have implicated the CaMKs and the novel PKCs in the regulation of class II HDACs, we next evaluated the role of KN-62 and rottlerin on the subcellular localization of HDAC4 (29, 30). Fig. 2B shows that HDAC4 is distributed throughout the cell during growth conditions, as determined by immunofluorescence. However, combined treatment with KN-62 and rottlerin resulted in nuclear accumulation of HDAC4. Furthermore, we utilized an HDAC4-GFP fusion protein and observed that it was primarily localized in the nucleus during serum-free quiescent conditions but was exported to the cytosol during low density growth conditions. This result was confirmed by nuclear and cytosolic fractionation studies demonstrating that PDGF treatment promotes nuclear export of HDAC4 (Fig. 2, D and E). Interestingly, when smooth muscle cultures were allowed to reach confluence, the HDAC4-GFP fusion protein was again primarily nuclear (Fig. 2D). Lastly, ectopic expression of activated CaMKs and activated PKD, a downstream HDAC kinase of PKCδ, resulted in a distribution of HDAC4-GFP to the cytosol (Fig. 2C). Together, these results indicate a growth-responsive role for HDAC4 that is regulated by PDGF activation of CaMKs and novel PKCs.
FIGURE 2.
PDGF induction of c-Jun is mediated by CaMK, PKCδ, and MEK. A, serum-starved A10 cells were treated with PDGF (10 ng/ml) for 2 h following 15 min pretreatment with rottlerin (5 μm), KN-62 (5 μm), LY294002 (10 μm), or PD98059 (10 μm). Protein extracts were immunoblotted (IB) with a c-Jun antibody (H79, Santa Cruz). B, growth phase VSMCs were treated with rottlerin (5 μm) and KN-62 (5 μm) followed by fixation with 4% paraformaldehyde. The fixed cells were then subjected to immunofluorescence (IF) with an HDAC4 primary antibody (Sigma). C, VSMCs were transfected with an EGFP fusion protein containing full-length human HDAC4 (HDAC4-GFP), and either activated CaMKII deltaB, CaMKIV, or PKD. Following serum starvation micrographs were obtained by standard fluorescent techniques. D, A10s were transfected with HDAC4-GFP. Micrographs were obtained in serum-free medium, low density growth medium (10% FBS), and high density growth medium (10% FBS). E, nuclear and cytosolic extracts were made from cultured VSMCs treated with 10 ng/ml PDGF for 2 h. The extracts were subjected to SDS-PAGE and immunoblotted for HDAC4, MEF2D, or GAPDH.
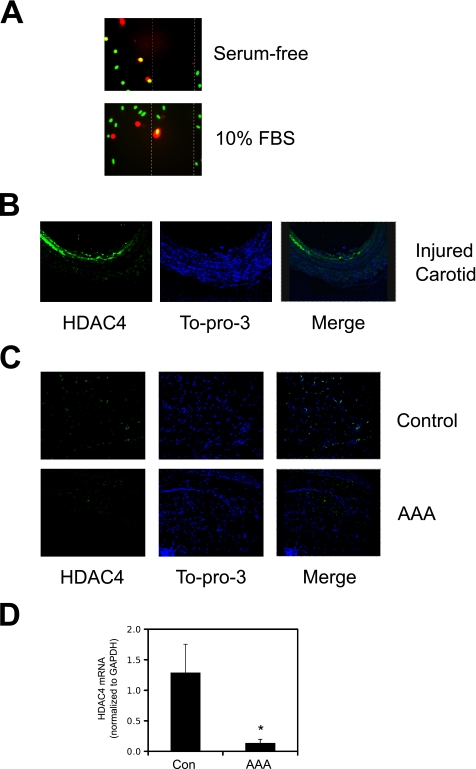
To validate this role for HDAC4 in vascular disease models, we utilized a scratch wound assay of VSMC migration. As shown in Fig. 3A, in confluent A10 cells that are positive for both HDAC4 and the 4′,6-diamidino-2-phenylindole nuclear stain, the HDAC4 signal is confined to the nuclear region. In contrast, in cells migrating into the wound, HDAC4 fluorescence is cytosolic. In addition, we utilized a rat model of carotid injury, because this animal model of vascular disease is more prone to neointimal formation than the mouse that harbors the MEF2-LacZ reporter gene (31, 32). Fig. 3B shows that HDAC4 staining is increased within the neointima of injured rat carotid arteries, where the HDAC4 immunofluorescence is more diffuse than the nuclear stain. This result is suggestive of a HDAC4 cytosolic distribution following vascular injury. Lastly, previous evidence has suggested a causal link between the c-Jun NH2-terminal kinase (JNK)-c-Jun pathway and the development of aneurysms (33). Therefore, we evaluated HDAC4 expression in human aortic aneurysms to evaluate whether this mechanism might be responsible for heightened c-Jun activity in an aneurysm. As shown in Fig. 3C, immunofluorescence of HDAC4 is reduced in abdominal human aneurysms. To validate that this reduction occurred in VSMCs, we utilized a technique of laser microdissection of smooth muscle α-actin-positive cells to purify RNA and perform quantitative PCR. Fig. 3D illustrates that HDAC4 expression is in fact reduced in VSMCs in human aortic aneurysms. However, we were unable to detect an increased c-Jun mRNA expression in this model (not shown). This finding is consistent with other reports indicating that c-Jun expression may not increase until rupture on an aneurysm (34). In this case, the down-regulation of HDAC4 may proceed an increase in c-Jun, which could occur with an appropriate rupture-induced stress signal. Together these results indicate that HDAC4 may be an important regulator of c-Jun expression in stenotic vascular diseases characterized by VSMC proliferation and migration; however, in arterial aneurysms, characterized by VSMC degeneration, down-regulation of HDAC4 is not sufficient to induce c-Jun expression.
FIGURE 3.
HDAC4 expression in models of vascular disease. A, A10 cells were grown to confluence and scraped with a standard 200-μl pipette tip. The cells were re-fed either serum-free medium or medium containing 10% FBS overnight and then fixed for immunofluorescence. Red, HDAC4; green, 4′,6-diamidino-2-phenylindole (i.e. nuclear). B, Sprague-Dawley rats were subjected to balloon-injury of the carotid artery. Following 14 days of recovery, the arteries were fixed and harvested for immunofluorescence. Green, HDAC4; blue, To-pro-3 (i.e. nuclear). C, human aortic aneurysms or a nondiseased control specimen were harvested during elective surgical reconstruction and fixed for immunofluorescence. Green, HDAC4; blue, To-pro-3 (i.e. nuclear). D, human control (Con) and aortic aneurysm sections were immunostained for smooth muscle α-actin and subjected to laser microdissection. Total RNA was isolated from collected cells and subjected to quantitative PCR for HDAC4 and GAPDH (n = 4; *, p < 0.05 was considered statistically significant).
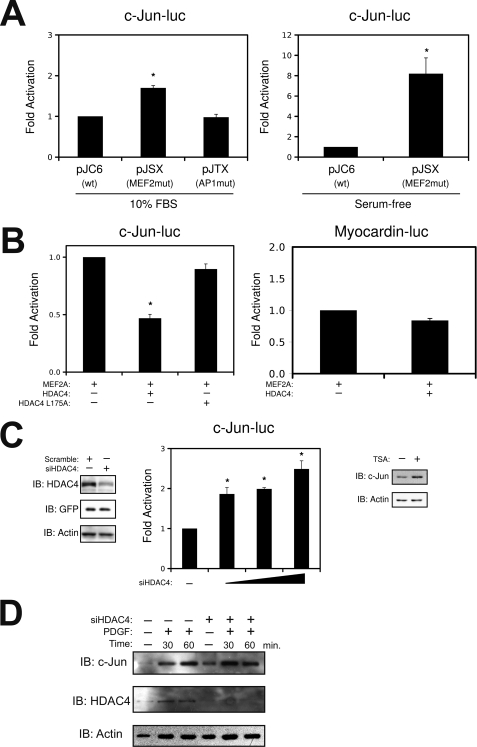
To dissect the function of the MEF2 cis-element within the c-jun promoter, we evaluated a c-jun reporter gene construct with a mutation in the MEF2-binding site under growth and quiescent conditions. As shown in Fig. 4A, mutation of the MEF2 cis-element site under growth conditions resulted in modest promoter activation, whereas mutation in the AP1 site had no effect. Interestingly, mutation in the MEF2 cis-element under quiescent conditions resulted in much greater promoter activation. These data suggest that the complex assembled at the MEF2 cis-element serves to repress c-jun expression under quiescent conditions. This was not the case for other MEF2-dependent reporter genes, because mutation of the MEF2 cis-element in the myocardin and HRC promoters did not result in activation (supplemental Fig. S2). Furthermore, ectopic expression of HDAC4 resulted in enhanced repression of the c-jun promoter, whereas ectopic expression of a mutant HDAC4 that cannot bind MEF2 proteins (HDAC4 L175A) or HDAC5 had no effect (Fig. 4B and supplemental Fig. S2). This repressive effect appears to be specific to c-jun, in that exogenous expression of HDAC4 had no effect on the myocardin and HRC promoters (Fig. 4 and supplemental Fig. S2). Consistent with these observations, suppression of HDAC4 expression by specific siRNA resulted in dose-dependent activation of the c-jun promoter, whereas treatment of quiescent smooth muscle cells with the deacetylase inhibitor, trichostatin A, resulted in an increase in c-Jun expression (Fig. 4C). Lastly, we evaluated the effect of the HDAC4 siRNA on endogenous c-Jun expression. Fig. 4D demonstrates a modest increase in c-Jun expression in quiescent VSMCs; however, when A10s cells were stimulated with PDGF, we observed and accelerated induction of c-Jun. Collectively, these data implicate MEF2, in conjunction with HDAC4, in the repression of the c-jun gene in quiescent conditions.
FIGURE 4.
The MEF2 cis-element in the c-jun promoter acts as a repressor element in quiescent VSMCs. A, A10 cells were transfected with a wild-type c-jun promoter (pJC6), a c-jun reporter with the MEF2 binding site mutated (pJSX), or a c-jun reporter gene with the AP1 site mutated (pJTX). The cells were harvested for luciferase under growth conditions (i.e. 10% FBS) or in serum-free Dulbecco's modified Eagle's medium. B, A10 cells were transfected with wild-type c-Jun-luc or myocardin-luc, with MEF2A, HDAC4, or HDAC4 L175A, as indicated. C, VSMCs were transfected with a specific siRNA targeted to HDAC4 (siHDAC4) or a scrambled nonspecific oligonucleotide in pSilencer H3 (Ambion). Following transfection, positive cells were selected using puromycin, followed by immunoblot analysis. For luciferase, increasing amounts of siHDAC4 were transfected with wild-type c-Jun-luc. Growth arrested A10 cells were treated with trichostatin A (1 μm, Sigma) for 2 h prior to harvesting. D, A10 cells were transfect siHDAC4 or scrambled control. Following recovery, positive cells were selected using puromycin, following by overnight quiescence in serum-free medium. The cells were stimulated with 20 ng/ml of PDGF as indicated and subjected to immunoblot (IB) analysis (*, p < 0.05 was considered statistically significant).
PKA Represses c-Jun Expression by Promoting the Nuclear Accumulation of HDAC4
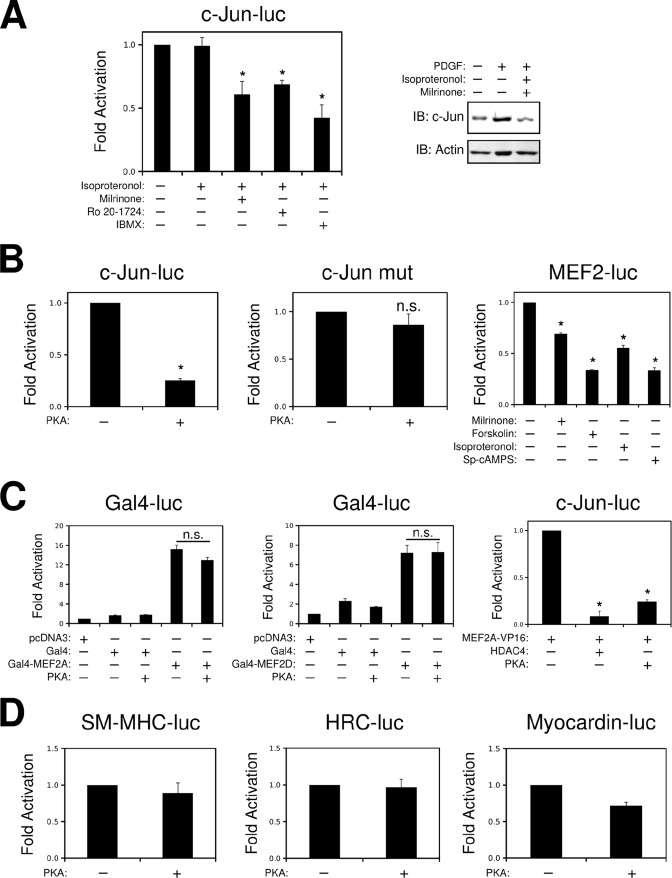
We have recently documented that PKA inhibits MEF2 transcriptional activity in skeletal muscle cells, in part by promoting the nuclear accumulation of class II HDACs (21). In addition, thrombin induction of c-Jun has been shown to be inhibited by cAMP in VSMCs, yet the mechanism for this phenomenon has not been completely elucidated (18). Therefore, we evaluated whether cAMP-mediated PKA activation could inhibit c-Jun induction by PDGF. As shown in Fig. 5A, the c-jun promoter is inhibited by combined treatment with the β-adrenergic agonist isoproteronol and phosphodiesterase (PDE) inhibitors. In addition, pretreatment with isoproteronol and the PDE3 inhibitor milrinone completely prevented the induction of c-Jun by PDGF in cultured VSMCs. This suppression of c-Jun expression could be rescued with the addition of PKA inhibitors Rp-cAMPS and H89 (supplemental Fig. S3); however, these pharmacological inhibitors were somewhat toxic in this cell line, similar to previously published work in A7r5 VSMC treated with the β-adrenergic receptor antagonist, propanolol (35). In addition, ectopic expression of the catalytic subunit of PKA reduced the expression of the wild-type c-jun reporter gene but not when the MEF2 cis-element was mutated (Fig. 5B). This effect was specific to c-jun, in that PKA failed to inhibit the expression of other smooth muscle marker genes, such as smooth muscle myosin heavy chain, myocardin, and HRC (Fig. 5D). Furthermore, Fig. 5B demonstrates that a MEF2-driven luciferase reporter is attenuated by a cAMP analog, milrinone, isoproteronol, and forskolin. Similar results were also obtained by ectopic expression of PKA (supplemental Fig. S3).
FIGURE 5.
PKA inhibits induction of the c-jun promoter through a MEF2-dependent mechanism. A, A10 cells transfected with c-Jun-luc were treated with isoproteronol (1 μm), milrinone (10 μm), Ro 20–1724 (10 μm), or IBMX (500 μm) as indicated overnight. Serum-starved A10s were preincubated with milrinone (10 μm) and isoproteronol (1 μm) for 15 min and then treated with PDGF (10 ng/ml), for 2 h. Protein extracts were prepared, and immunoblots (IB) were performed for c-Jun. B, A10 cells were transfected with wild-type c-Jun-luc or a construct with a mutation in the MEF2 cis-element (c-Jun mut) and the catalytic subunit of PKA (pFC-PKA, Stratagene), as indicated. A10 cells were transfected with a reporter containing a consensus MEF2-binding site (MEF2-luc). 24 h prior to harvesting, the cells were treated with 20 μm cAMP analog (Sp-cAMPS, Sigma), 10 μm milrinone, 10 μm forskolin, or 1 μm isoproteronol, as indicated. C, A10 cells were transfected with a Gal4-luciferase (Gal4-luc), a Gal4 DNA-binding domain (Gal4), and a Gal4-MEF2A or -MEF2D fusion containing the C terminus of MEF2A or -D, with or without PKA, as indicated. c-Jun-luc was transfected with the MEF2A-VP16 fusion with HDAC4, or PKA, as indicated. D, A10 cells were transfected with the smooth muscle myosin heavy chain (SM-MHC), HRC, or myocardin enhancer reporter genes and pFC-PKA, as indicated. The cells were harvested for luciferase 24 h after recovery (n.s., not significant; *, p < 0.05 was considered statistically significant).
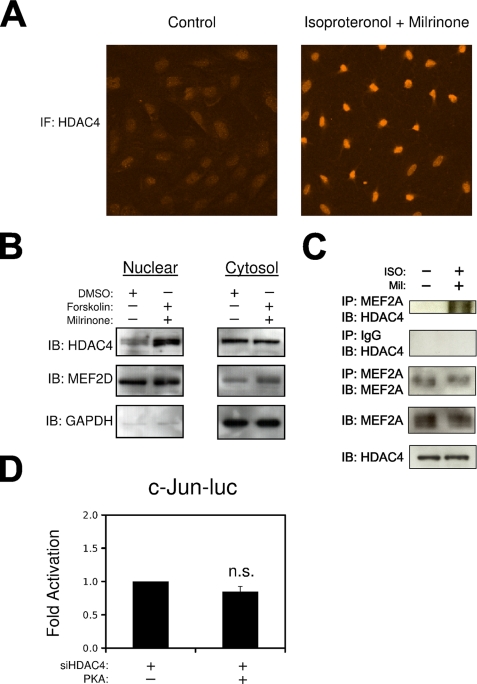
To identify a mechanism underlying PKA inhibition of MEF2-dependent c-jun regulation, we utilized Gal4 and VP16 fusions of MEF2A and MEF2D. As shown in Fig. 5C, PKA could not inhibit the Gal4-MEF2 fusion proteins that lack the N-terminal class II HDAC-binding domain but readily attenuated the activity of the MEF2A-VP16 fusion proteins that contain the class II HDAC-binding domain. In addition, Fig. 6 (A and B) demonstrates that activation of PKA increases the nuclear localization of HDAC4, as determined by immunofluorescence and nuclear/cytosolic fractionation. In addition, our previous work has shown that ectopic expression of PKA enhances the interaction between MEF2 and HDAC4, determined by co-immunoprecipitation in COS7 cells (21). Fig. 6C demonstrates that activation of endogenous PKA by treatment with isoproteronol, and milrinone increases the association of HDAC4 with MEF2A in A10 VSMCs. Lastly, Fig. 6D demonstrates that HDAC4 is required for PKA inhibition of the c-jun promoter, in that reduced expression of HDAC4 by siRNA targeting prevented the attenuation of the c-Jun reporter gene by the catalytic subunit of PKA.
FIGURE 6.
PKA inhibits c-jun expression through HDAC4. A, growth phase VSMCs were treated with milrinone (10 μm; Sigma) and isoproteronol (1 μm) for 2 h followed by fixation with 4% paraformaldehyde. Fixed cells were then subjected to immunofluorescence (IF) with an HDAC4 primary antibody (Sigma). B, A10 cells were serum-starved and pretreated with forskolin (10 μm) and milrinone (10 μm, Sigma), or Me2SO. Nuclear and cytosolic extractions were immunoblotted (IB) for HDAC4, MEF2D, and GAPDH. C, A10 cells were transfected with MEF2A and HDAC4 and treated for 2 h with isoproteronol and milrinone. Protein extracts were subjected to immunoprecipitation (IP) and immunoblotting as indicated. D, VSMCs were transfected with c-Jun-luc, siHDAC4, or PKA, as indicated (n.s., not significant).
PKA Enhances the Nuclear Accumulation of HDAC4 by Inhibiting the HDAC Kinase SIK1
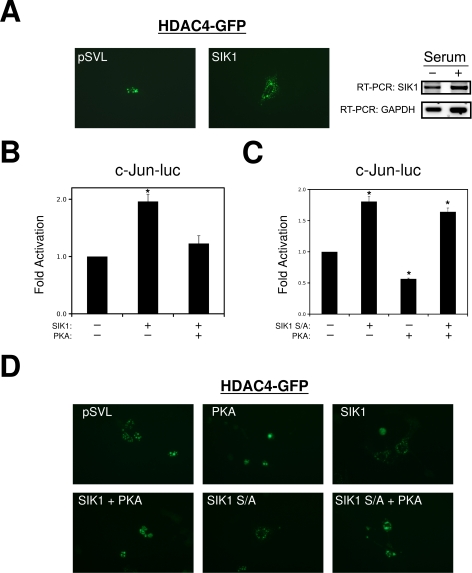
Recent studies in other cells types have identified the SIK1 as a potential PKA-regulated HDAC-kinase (20, 36). Therefore, we evaluated the role of SIK1 in MEF2-dependent c-jun expression in VSMCs. As shown in Fig. 7A, ectopic expression of SIK1 in quiescent VSMCs resulted in nuclear export of a HDAC4-GFP fusion protein. Furthermore, forced expression of SIK1 resulted in activation of the c-jun reporter gene (Fig. 7B). However, the addition of the catalytic subunit of PKA resulted in attenuation of SIK1 induction of c-jun. PKA has been shown to inhibit SIK1 by direct phosphorylation of serine 577, and a neutralizing mutation of this residue to alanine (SIK1 S/A) is sufficient to eliminate this effect (36). As shown is Fig. 7B, the SIK1 mutation is still capable of activating the c-jun reporter gene; however, PKA is not able to inhibit this mutated SIK1. Consistent with this finding, PKA could not inhibit the nuclear export of HDAC4-GFP by the mutated SIK1 in COS7 cells (Fig. 7D). Therefore, these data indicate that PKA inhibits c-jun expression in VSMCs by inhibiting SIK1 and promoting the nuclear accumulation of HDAC4.
FIGURE 7.
PKA inhibits HDAC4 nuclear export through SIK1. A, VSMCs were transfected with HDAC4-GFP and SIK1 or empty pSVL. The following serum starvation micrographs were obtained by standard fluorescent techniques. Growth phase A10s in 10% FBS (+) or serum-free medium (−) were harvested for total RNA and subjected to reverse transcription-PCR for SIK1 and GAPDH. B and C, A10 cells were transfected with c-Jun-luc, SIK1, SIK1 S577A, or PKA, as indicated. D, COS7 cells were transfected with HDAC4-GFP and SIK1, SIK1 S577A, or PKA, as indicated. The micrographs were obtained following 24 h of recovery.
Exogenous Expression of c-Jun Prevents Myocardin Induction of Smooth Muscle Marker Genes
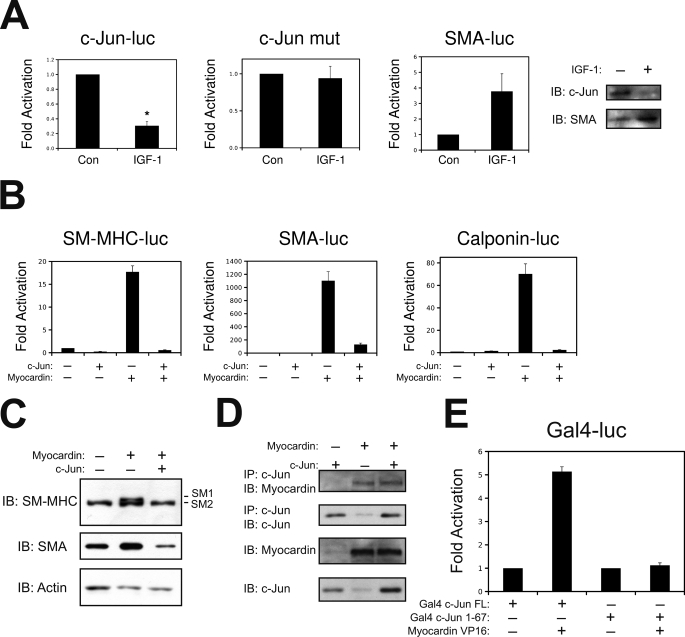
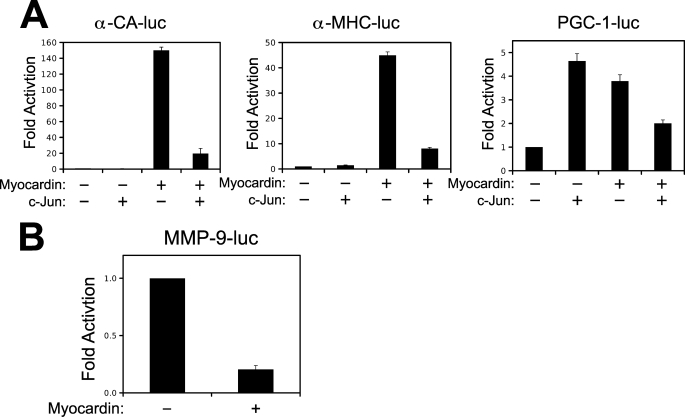
Although numerous studies have evaluated the role of c-Jun on the activated smooth muscle phenotype, to our knowledge, no reports exist evaluating the role of c-Jun on smooth muscle differentiation. A recent study has highlighted the role of insulin-like growth factor 1 (IGF-1) and phosphatidylinositol 3-kinase/AKT signaling in the promotion of smooth muscle differentiation by activating the transcriptional activity of myocardin (37). Therefore, we utilized this model of VSMC differentiation to evaluate c-jun expression. As shown in Fig. 8A, treatment of VSMCs with IGF-1 resulted in increased expression of smooth muscle α-actin (SMA). Interestingly, IGF-1 treatment simultaneously down-regulated c-jun, where this effect was dependent on the MEF2 cis-element. Therefore, we speculated that c-Jun could negatively modulate smooth muscle differentiation. In support of our hypothesis, constitutive expression of c-Jun attenuated myocardin induction of smooth muscle reporter genes for smooth muscle myosin heavy, smooth muscle α-actin, and calponin (Fig. 8B). In addition, we utilized a 10T1/2 conversion assay as a model to evaluate the role of c-Jun in smooth muscle differentiation. As shown in Fig. 8C, ectopic expression of the smooth muscle isoform of myocardin (myocardin 856) was sufficient to induce the endogenous expression of smooth muscle α-actin and smooth muscle myosin heavy chain, a definitive marker of the smooth muscle lineage (38). However, when c-Jun was co-expressed with myocardin, expression of these smooth muscle marker genes was attenuated. We hypothesized that c-Jun might attenuate the activation of myocardin by competing for a common co-activator. Previous studies have shown that both c-Jun and myocardin interact with the histone acetyltransferase and co-activator, p300 (39, 40). However, ectopic expression of p300 could not substantially rescue myocardin transcriptional activity once repressed by c-Jun (not shown). Therefore, we chose to evaluate whether c-Jun might inhibit myocardin through a physical interaction. This hypothesis seemed reasonable, given that c-Jun has previously been shown to physically interact and repress the transcriptional activation of other muscle-restricted transcription factors, like MyoD (41, 42). Fig. 8D demonstrates that myocardin immunoprecipitated with an antibody targeted to c-Jun when co-expressed in COS7 cells. The antibody to c-Jun resulted in a greater immunoprecipitation of myocardin that a control rabbit IgG (not shown). To validate this interaction between c-Jun and myocardin, we performed a mammalian two-hybrid assay in 10T1/2 cells using Gal4-c-Jun and myocardin-VP16 fusion proteins. As shown in Fig. 8E, myocardin-VP16 could activate the Gal4 fusion protein containing full-length c-Jun but not a Gal4 fusion protein containing the N-terminal transcriptional activation domain of c-Jun (1–67). This fusion protein lacks the B-zip domain of c-Jun, which has been shown to be critical for protein-protein interaction (43). Next, we speculated that if c-Jun can modulate the transcriptional activity of myocardin by physical interaction, myocardin might inhibit AP-1-dependent transcription. To evaluate this, we ectopically expressed myocardin with the AP-1-dependent promoter for matrix metalloprotease 9 (MMP-9) (44). Previous studies shown that MMP-9 is involved in both proliferative VSMC disease and degenerating disease, such as aneurysm (33, 45, 46). As shown in Fig. 9B, myocardin can repress the MMP-9 promoter in A10 VSMCs. Taken together, these data support the hypothesis that c-Jun and myocardin are mutual co-regulators that modulate VSMC phenotype in response to growth factor stimulation, such PDGF and IGF-1.
FIGURE 8.
Down-regulation of c-Jun is critical for VSMC differentiation. A, VSMCs were transfected with wild-type c-Jun-luc, c-Jun-luc containing a mutation in the MEF2 binding site (c-Jun mut), or a smooth muscle α-actin reporter gene (SMA-luc). Quiescent cells were treated with 50 ng/ml of IGF-1 overnight and harvested for luciferase extracts. Protein extracts from overnight treated IGF-1 A10 cells were subjected to immunoblotting for c-Jun or SMA (Sigma). B, 10T1/2 fibroblasts were transfected with smooth myosin heavy chain (SM-MHC-luc), smooth muscle α-actin (SMA-luc), or calponin (Calponin-luc) reporter genes with expression vectors for c-Jun and the smooth muscle isoform of myocardin (myocardin 856), as indicated. The cells were harvested for luciferase 24 h after recovery. C, 10T1/2 cells were transfected with myocardin 856 and c-Jun, as indicated. After a 24 h recovery, the cells were re-fed in 5% horse serum and allowed to differentiate for 4 days before harvesting for protein extracts and immunoblotting for SMA or SM-MHC (Biomedical Science). D, COS7 cells were transfected with c-Jun or myocardin 856, as indicated. Protein extracts were subjected to immunoprecipitation (IP) and immunoblotting (IB), as indicated. E, 10T1/2 cells were transfected with a Gal4 reporter gene, and Gal4-c-Jun fusion proteins containing full-length c-Jun (FL) or amino acids 1–67, with a myocardin-VP16 fusion protein, as indicated. The extracts were subject to luciferase assay. Con, control.
FIGURE 9.
Myocardin and c-Jun are mutual co-regulators. A, 10T1/2 cells were transfected with the cardiac α-actin promoter (α-CA-luc), the α-myosin heavy chain promoter (α-MHC-luc), or the PGC-1 promoter (PGC-1-luc), c-Jun, and the cardiac isoform of myocardin (myocardin 935), as indicated. The extracts were subjected to luciferase assay. B, 10T1/2 cells were transfected with the MMP-9 promoter (MMP-9-luc) and myocardin 856, as indicated. The extracts were subjected to luciferase assays.
Myocardin was originally identified as an activator of cardiac gene expression and has been shown to induce cardiac hypertrophy (47, 48). Interestingly, c-Jun expression can be induced by cardiac wall stress and hypertrophy in vitro and in vivo (49, 50). Therefore, we speculated that the interaction between c-Jun and myocardin might be an important regulator of myocardin-induced activation of cardiac gene expression. As shown in Fig. 9A, the cardiac isoform of myocardin (myocardin 935) potently activated the promoters for α-cardiac actin and α-myosin heavy chain. This induction was nearly completely attenuated by co-expression of c-Jun. The mitochondrial regulator PGC-1 is induced during cardiac hypertrophy but is thought to be down-regulated during the progression of heart failure (51). Interestingly, the PGC-1 promoter was induced by both c-Jun and myocardin 935, but co-expression of these transcription factors resulted in attenuation of the induction. Therefore, the interaction of c-Jun and myocardin may have implications to both vascular and cardiac disease.
DISCUSSION
Vascular diseases, such as atherosclerosis and restenosis, involve smooth muscle activation characterized by proliferation and migration to sites of injury. In the quiescent nonproliferating state, VSMCs are acted on by protective vasodilators, such as prostacyclin produced from the intact endothelium, and β2-adrenergic stimulation. Indeed, reduced prostacyclin production in humans by cyclooxygenase inhibition increases the risk of cardiovascular events (17). Prostanoids, like prostacyclin, activate PKA signaling and oppose growth factor-induced VSMC proliferation. However, vascular injury is known to increase the expression of PDEs, which may counteract PKA activation and allow growth factor-induced proliferation (16, 52). Thus, cAMP-dependent PKA activation may function as a signaling conduit controlling the phenotype of VSMCs. We report that these dilators can also function at the level of regulation of gene expression and demonstrate a novel role for PKA signaling in modulating MEF2-dependent repression of c-jun expression, a critical regulator of VSMC proliferation.
MEF2 proteins have been most extensively studied in striated muscle, where they are intimately involved in muscle development and various postnatal phenotypes (53). The role of MEF2 in vascular smooth muscle cells is less well characterized, although a role for MEF2C in VSMC differentiation and vascular ontogeny has been invoked (7). VSMCs represent an interesting model in which to study MEF2 site-directed gene expression, because VSMCs maintain the ability to modulate their postnatal phenotype in response to environmental stimuli, unlike other MEF2-dependent tissues such as striated muscle and neurons. Given the importance of the MEF2 target genes, myocardin, and c-jun to their respective quiescent and activated smooth muscle phenotypes, understanding the regulation of MEF2-dependent gene expression will be key in understanding smooth muscle phenotypic modulation in vascular disease. Like SRF, MEF2 activity is modulated by recruiting co-activators or co-repressors to promoter regions. Thus, it remains likely that site-directed transcriptional control of MEF2 is modulated by the unique combination of cis-elements present within these promoter regions that constitute a specific promoter architecture that serves to recruit a precise combination of co-factors and transcriptional regulators. Indeed, the regulation of MEF2 proteins by class II HDACs has not been established in VSMCs; however, HDAC5 has been shown to regulate the transcriptional activity of myocardin, and angiotensin-induced smooth hypertrophy is mediated through nuclear export of this histone deacetylase (39, 54).
In this report, we demonstrate that the MEF2 cis-element in the c-jun promoter acts as a repressor element in quiescent VSMCs, where growth factor-mediated activation of CaMK and PKC promotes nuclear export of HDAC4 to relieve MEF2 proteins from repression. This observation likely explains the absence of widespread MEF2 activation in vivo following vascular injury. Of the various CaMKs, CaMKIIδ appears to be the most likely kinase involved in c-Jun induction, given that recent evidence has demonstrated a critical role of this isoform during neointima formation, whereas CaMKIV has been implicated in VSMC differentiation (55, 56). In addition, we demonstrate that PKA can enhance the repression of c-jun by increasing the nuclear localization of HDAC4 through inhibition of SIK1. This repression of c-jun is of fundamental importance for VSMC differentiation, in that forced expression of c-Jun inhibits the ability of myocardin to activate smooth muscle-dependent gene expression.
PKA has been previously implicated in inhibition of VSMC proliferation and migration through inhibition of the MEK/ERK MAP kinase signaling pathway (16). In addition, PKA has been implicated in promoting VSMC differentiation and increasing the expression of smooth muscle marker genes, such as SM-MHC (57). Our data suggest that PKA does not directly increase the activity of smooth muscle promoters (Fig. 4) but promotes competence for smooth muscle differentiation through down-regulation of c-Jun.
PKA signaling is terminated by PDE enzymes that hydrolyze cyclic nucleotides to 5′ nucleotide monophosphates that do not activate PKA (52). Numerous studies have implicated PDE3 and PDE4 isoforms as the dominant cAMP metabolizing enzymes in VSMCs, and there is reported synergism between adenylate cyclase activators, PDE3 and/or PDE4 inhibitors in terms of VSMC relaxation, and inhibition of proliferation and migration (52). This is consistent with our data, in that combined treatment of isoproteronol and milrinone completely inhibited PDGF induction of c-Jun (Fig. 4).
PKA signaling has also been shown to have anti-inflammatory effects in VSMCs, where inhibition of PDE3 by cGMP signaling inhibits tumor necrosis factor-α-induced activation of NFκB-dependent gene expression (58). Interestingly, MEF2 proteins have been shown to play a role in VSMC inflammation through a consensus MEF2 cis-element in the promoter of the MCP-1 gene (59). Indeed, our preliminary evidence suggests that PKA inhibits the activity of a MCP-1 reporter gene (supplemental Fig. S3). Thus, it appears that PKA-mediated repression of MEF2-dependent gene expression will inhibit multiple components of the activated smooth muscle phenotype.
Interestingly, the phenotypic alterations mediated by PKA signaling differs between striated and VSMCs. Recent evidence from our laboratory has demonstrated that PKA can directly phosphorylate MEF2 proteins to inhibit skeletal muscle differentiation (21). In addition, transgenic mice expressing the catalytic subunit of PKA in the heart develop a dilated myopathy with down-regulation in MEF2-dependent cardiac marker genes (60). However, in VSMCs, PKA inhibits proliferation and, in contrast to striated muscle, may enhance smooth muscle differentiation. Therefore, based on our work, and the work of other laboratories, we propose that PKA-regulated inhibition of MEF2-dependent gene expression can result in different outcomes depending on the cellular context.
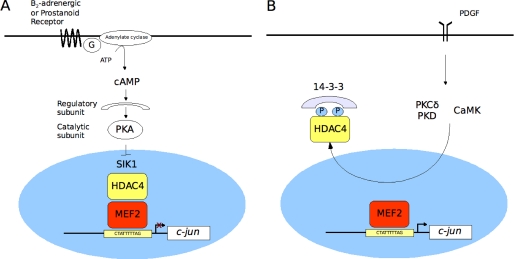
In summary, these studies support a novel link between MEF2 and the growth-responsive c-jun gene in quiescent VSMCs (Fig. 10), in which repression of c-jun expression is promoted by agents that elevate cellular cAMP such as prostacyclin or β2-adrenergic stimulation. This effect involves a mechanism in which PKA activation promotes the assembly of a MEF2·HDAC4 repressor complex. In view of the fundamental role of c-Jun as a modulator of VMSC differentiation, it will be important to determine whether MEF2 can mediate a protective effect of clinical relevance for vascular injury and disease.
FIGURE 10.
Model of c-jun regulation in VSMCs. A, in quiescent conditions, c-jun expression is repressed by a MEF2·HDAC4 complex, which is promoted by PKA-induced inhibition of SIK1. B, growth factor (i.e. PDGF) stimulation of VSMCs results in PKCδ/PKD- and CaMK-induced derepression through HDAC4 nuclear export and MEK/ERK-dependent activation of c-jun.
Supplementary Material
Acknowledgments
The Seneca College Office of Research and Innovation and Southlake Regional Health Centre are acknowledged for providing release time support for J. W. Gordon and J. J. Andreucci.
This work was supported in part by a postdoctoral fellowship from the Muscular Dystrophy Association of Canada and Canadian Institute of Health Research (to R. L. P.) and by a grant from the Canadian Institute of Health Research (to J. C. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- VSMC
- vascular smooth muscle cell
- MEF2
- myocyte enhancer factor 2
- HDAC
- histone deacetylase
- PKA
- protein kinase A
- PDGF
- platelet-derived growth factor
- PKC
- protein kinase C
- CaMK
- calcium/calmodulin-dependent protein kinase
- SIK
- salt-inducible kinase
- SRF
- serum response factor
- HRC
- histidine-rich calcium-binding protein
- PKD
- protein kinase D
- FBS
- fetal bovine serum
- TRITC
- tetramethylrhodamine isothiocyanate
- siRNA
- small interfering RNA
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- PBS
- phosphate-buffered saline
- SM
- smooth muscle
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PDE
- phosphodiesterase
- IGF
- insulin-like growth factor
- SMA
- smooth muscle α-actin
- MMP
- matrix metalloprotease
- MAP
- mitogen-activated protein
- ERK
- extracellular signal-regulated kinase
- MEK
- MAP kinase/ERK kinase
- MCP
- monocyte chemoattractant protein.
REFERENCES
- 1.Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 2.Miano J. M. (2003) J. Mol. Cell Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 3.Wang D. Z., Olson E. N. (2004) Curr. Opin. Genet. Dev. 14, 558–566 [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. (2004) Nature 428, 185–189 [DOI] [PubMed] [Google Scholar]
- 5.Creemers E. E., Sutherland L. B., McAnally J., Richardson J. A., Olson E. N. (2006) Development 133, 4245–4256 [DOI] [PubMed] [Google Scholar]
- 6.Anderson J. P., Dodou E., Heidt A. B., De Val S. J., Jaehnig E. J., Greene S. B., Olson E. N., Black B. L. (2004) Mol. Cell Biol. 24, 3757–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Q., Lu J., Yanagisawa H., Webb R., Lyons G. E., Richardson J. A., Olson E. N. (1998) Development 125, 4565–4574 [DOI] [PubMed] [Google Scholar]
- 8.Firulli A. B., Miano J. M., Bi W., Johnson A. D., Casscells W., Olson E. N., Schwarz J. J. (1996) Circ. Res. 78, 196–204 [DOI] [PubMed] [Google Scholar]
- 9.Zhan Y., Kim S., Yasumoto H., Namba M., Miyazaki H., Iwao H. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 82–88 [DOI] [PubMed] [Google Scholar]
- 10.Ioroi T., Yamamori M., Yagi K., Hirai M., Zhan Y., Kim S., Iwao H. (2003) J. Pharmacol. Sci. 91, 145–148 [DOI] [PubMed] [Google Scholar]
- 11.Han T. H., Prywes R. (1995) Mol. Cell Biol. 15, 2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilato C., Pauly R. R., Melillo G., Monticone R., Gorelick-Feldman D., Gluzband Y. A., Sollott S. J., Ziman B., Lakatta E. G., Crow M. T. (1995) J. Clin. Invest. 96, 1905–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zugaza J. L., Waldron R. T., Sinnett-Smith J., Rozengurt E. (1997) J. Biol. Chem. 272, 23952–23960 [DOI] [PubMed] [Google Scholar]
- 14.Markou T., Yong C. S., Sugden P. H., Clerk A. (2006) J. Biol. Chem. 281, 8321–8331 [DOI] [PubMed] [Google Scholar]
- 15.Ornatsky O. I., Cox D. M., Tangirala P., Andreucci J. J., Quinn Z. A., Wrana J. L., Prywes R., Yu Y. T., McDermott J. C. (1999) Nucleic Acids Res. 27, 2646–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bornfeldt K. E., Krebs E. G. (1999) Cell Signal 11, 465–477 [DOI] [PubMed] [Google Scholar]
- 17.Grosser T., Fries S., FitzGerald G. A. (2006) J. Clin. Invest. 116, 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao G. N., Runge M. S. (1996) J. Biol. Chem. 271, 20805–20810 [DOI] [PubMed] [Google Scholar]
- 19.Bolger T. A., Yao T. P. (2005) J. Neurosci. 25, 9544–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berdeaux R., Goebel N., Banaszynski L., Takemori H., Wandless T., Shelton G. D., Montminy M. (2007) Nat. Med. 13, 597–603 [DOI] [PubMed] [Google Scholar]
- 21.Du M., Perry R. L., Nowacki N. B., Gordon J. W., Salma J., Zhao J., Aziz A., Chan J., Siu K. W., McDermott J. C. (2008) Mol. Cell Biol. 28, 2952–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreucci J. J., Grant D., Cox D. M., Tomc L. K., Prywes R., Goldhamer D. J., Rodrigues N., Bédard P. A., McDermott J. C. (2002) J. Biol. Chem. 277, 16426–16432 [DOI] [PubMed] [Google Scholar]
- 23.Perry R. L., Parker M. H., Rudnicki M. A. (2001) Mol. Cell 8, 291–301 [DOI] [PubMed] [Google Scholar]
- 24.Kollias H. D., Perry R. L., Miyake T., Aziz A., McDermott J. C. (2006) Mol. Cell Biol. 26, 6248–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou G., Vogel W., Bendeck M. P. (2001) J. Clin. Invest. 107, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho B., Hou G., Pickering J. G., Hannigan G., Langille B. L., Bendeck M. P. (2008) Am. J. Pathol. 173, 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Angelis L., Zhao J., Andreucci J. J., Olson E. N., Cossu G., McDermott J. C. (2005) Dev. Biol. 283, 171–179 [DOI] [PubMed] [Google Scholar]
- 28.Naya F. J., Wu C., Richardson J. A., Overbeek P., Olson E. N. (1999) Development 126, 2045–2052 [DOI] [PubMed] [Google Scholar]
- 29.McKinsey T. A., Zhang C. L., Olson E. N. (2001) Curr. Opin. Genet. Dev. 11, 497–504 [DOI] [PubMed] [Google Scholar]
- 30.Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Mol. Cell Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhel D. G., Zhu B., Witte D. P., Hui D. Y. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 955–960 [DOI] [PubMed] [Google Scholar]
- 32.Assadnia S., Rapp J. P., Nestor A. L., Pringle T., Cerilli G. J., Gunning W. T., 3rd, Webb T. H., Kligman M., Allison D. C. (1999) Circ. Res. 84, 1252–1257 [DOI] [PubMed] [Google Scholar]
- 33.Yoshimura K., Aoki H., Ikeda Y., Fujii K., Akiyama N., Furutani A., Hoshii Y., Tanaka N., Ricci R., Ishihara T., Esato K., Hamano K., Matsuzaki M. (2005) Nat. Med. 11, 1330–1338 [DOI] [PubMed] [Google Scholar]
- 34.Laaksamo E., Tulamo R., Baumann M., Dashti R., Hernesniemi J., Juvela S., Niemelä M., Laakso A. (2008) Stroke 39, 886–892 [DOI] [PubMed] [Google Scholar]
- 35.Meier K. E., Gause K. C., Wisehart-Johnson A. E., Gore A. C., Finley E. L., Jones L. G., Bradshaw C. D., McNair A. F., Ella K. M. (1998) Cell Signal 10, 415–426 [DOI] [PubMed] [Google Scholar]
- 36.Takemori H., Katoh Y., Horike N., Doi J., Okamoto M. (2002) J. Biol. Chem. 277, 42334–42343 [DOI] [PubMed] [Google Scholar]
- 37.Liu Z. P., Wang Z., Yanagisawa H., Olson E. N. (2005) Dev. Cell 9, 261–270 [DOI] [PubMed] [Google Scholar]
- 38.Miano J. M., Cserjesi P., Ligon K. L., Periasamy M., Olson E. N. (1994) Circ. Res. 75, 803–812 [DOI] [PubMed] [Google Scholar]
- 39.Cao D., Wang Z., Zhang C. L., Oh J., Xing W., Li S., Richardson J. A., Wang D. Z., Olson E. N. (2005) Mol. Cell Biol. 25, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J. S., See R. H., Deng T., Shi Y. (1996) Mol. Cell Biol. 16, 4312–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L., Chambard J. C., Karin M., Olson E. N. (1992) Genes Dev. 6, 676–689 [DOI] [PubMed] [Google Scholar]
- 42.Bengal E., Ransone L., Scharfmann R., Dwarki V. J., Tapscott S. J., Weintraub H., Verma I. M. (1992) Cell 68, 507–519 [DOI] [PubMed] [Google Scholar]
- 43.Angel P., Karin M. (1991) Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 44.Gum R., Lengyel E., Juarez J., Chen J. H., Sato H., Seiki M., Boyd D. (1996) J. Biol. Chem. 271, 10672–10680 [DOI] [PubMed] [Google Scholar]
- 45.Bendeck M. P., Conte M., Zhang M., Nili N., Strauss B. H., Farwell S. M. (2002) Am. J. Pathol. 160, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courtman D. W., Franco C. D., Meng Q., Bendeck M. P.J. Vasc. Res. 41, 157–165 [DOI] [PubMed] [Google Scholar]
- 47.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 48.Xing W., Zhang T. C., Cao D., Wang Z., Antos C. L., Li S., Wang Y., Olson E. N., Wang D. Z. (2006) Circ. Res. 98, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 49.Sadoshima J., Jahn L., Takahashi T., Kulik T. J., Izumo S. (1992) J. Biol. Chem. 267, 10551–10560 [PubMed] [Google Scholar]
- 50.Schunkert H., Jahn L., Izumo S., Apstein C. S., Lorell B. H. (1991) Proc. Natl. Acad. Sci. U. S. A. 88, 11480–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finck B. N., Kelly D. P. (2007) Circulation 115, 2540–2548 [DOI] [PubMed] [Google Scholar]
- 52.Maurice D. H., Palmer D., Tilley D. G., Dunkerley H. A., Netherton S. J., Raymond D. R., Elbatarny H. S., Jimmo S. L. (2003) Mol. Pharmacol. 64, 533–546 [DOI] [PubMed] [Google Scholar]
- 53.Potthoff M. J., Olson E. N. (2007) Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 54.Xu X., Ha C. H., Wong C., Wang W., Hausser A., Pfizenmaier K., Olson E. N., McKinsey T. A., Jin Z. G. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 2355–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.House S. J., Singer H. A. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 441–447 [DOI] [PubMed] [Google Scholar]
- 56.Najwer I., Lilly B. (2005) Am. J. Physiol. Cell Physiol. 289, C785–793 [DOI] [PubMed] [Google Scholar]
- 57.Fetalvero K. M., Shyu M., Nomikos A. P., Chiu Y. F., Wagner R. J., Powell R. J., Hwa J., Martin K. A. (2006) Am. J. Physiol. Heart Circ. Physiol. 290, H1337–1346 [DOI] [PubMed] [Google Scholar]
- 58.Aizawa T., Wei H., Miano J. M., Abe J., Berk B. C., Yan C. (2003) Circ. Res. 93, 406–413 [DOI] [PubMed] [Google Scholar]
- 59.Suzuki E., Satonaka H., Nishimatsu H., Oba S., Takeda R., Omata M., Fujita T., Nagai R., Hirata Y. (2004) Circ. Res. 95, 42–49 [DOI] [PubMed] [Google Scholar]
- 60.Antos C. L., Frey N., Marx S. O., Reiken S., Gaburjakova M., Richardson J. A., Marks A. R., Olson E. N. (2001) Circ. Res. 89, 997–1004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.