Abstract
Kallikrein 4 (Klk4) is believed to play an essential role in enamel biomineralization, because defects in KLK4 cause hypomaturation amelogenesis imperfecta. We used gene targeting to generate a knockin mouse that replaces the Klk4 gene sequence, starting at the translation initiation site, with a lacZ reporter gene. Correct targeting of the transgene was confirmed by Southern blot and PCR analyses. Histochemical X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining demonstrated expression of β-galactosidase in maturation stage ameloblasts. No X-gal staining was observed in secretory stage ameloblasts or in odontoblasts. Retained enamel proteins were observed in the maturation stage enamel of the Klk4 null mouse, but not in the Klk4 heterozygous or wild-type mice. The enamel layer in the Klk4 null mouse was normal in thickness and contained decussating enamel rods but was rapidly abraded following weaning, despite the mice being maintained on soft chow. In function the enamel readily fractured within the initial rod and interrod enamel above the parallel enamel covering the dentino-enamel junction. Despite the lack of Klk4 and the retention of enamel proteins, significant levels of crystal maturation occurred (although delayed), and the enamel achieved a mineral density in some places greater than that detected in bone and dentin. An important finding was that individual enamel crystallites of erupted teeth failed to grow together, interlock, and function as a unit. Instead, individual crystallites seemed to spill out of the enamel when fractured. These results demonstrate that Klk4 is essential for the removal of enamel proteins and the proper maturation of enamel crystals.
Dental enamel is composed of highly ordered, very long crystals of calcium hydroxyapatite (Ca10(PO4)6(OH)2). Mature enamel crystallites are about 70 nm wide and 30 nm thick, but are of unmeasurable length (1), probably extending all the way from the dentin layer to the surface of the tooth (2). Enamel crystallites are organized into bundles called rods, with about 10,000 parallel crystals in a rod (3). Each enamel rod is the product of a single ameloblast, the cell type that forms a continuous sheet over the developing enamel and orchestrates its formation. Dental enamel of erupted teeth is ∼95% mineral (by weight) (4), with most of the non-mineral component being water. Protein comprises <1% of its weight. Forming enamel, however, is over 30% protein (5). Much of the protein is reabsorbed by ameloblasts and degraded in lysosomes (6, 7), but extracellular proteases also play a role in matrix protein removal (8–10).
Dental enamel formation is divided into secretory, transition, and maturation stages (11, 12). During the secretory stage, enamel crystals grow primarily in length. As the crystals extend, the enamel layer expands. Enamel crystallites lengthen along a mineralization front at the secretory surface of the ameloblast cell membrane. There, mineral deposits rapidly on the crystallite tips, and very slowly on their sides (3, 13, 14). By the end of the secretory stage the enamel crystals are full-length and the enamel layer as a whole is as thick as it will ever be, but it has only about 14% of the mineral as it will have when the tooth erupts (15). Following the secretory stage there is then a transition during which the ameloblasts greatly reduce their secretion of enamel proteins (16) and convert to maturation ameloblasts (17). During the maturation stage, mineral is deposited exclusively on the sides of pre-existing enamel crystallites (18), which grow in width and thickness until further growth is prevented by contact with adjacent crystals (19, 20). During early maturation the percentage protein by weight drops from 30 to 2% (5), and half of the total enamel mineral is deposited. The final 30–35% of mineral is deposited in the absence of significant protein and allows the crystals to grow firmly against one another and to mechanically interlock (15).
The major secretory stage enamel proteins are amelogenin (21, 22), ameloblastin (23–25), and enamelin (26, 27). These proteins function specifically during enamel formation, and the disease phenotypes exhibited by mice lacking these genes are confined to the developing teeth and include enamel agenesis (28–30). These genes are often deleted or are reduced to pseudogenes in vertebrates such as birds or baleen whales that evolved alternatives to developing teeth (31, 32). Although the enamel extracellular matrix proteins are critical for growing enamel crystals, they are not part of the final enamel product. Prior to tooth eruption, enamel proteins are digested by proteases and reabsorbed by ameloblasts. Two extracellular matrix proteases are involved in the cleavage of enamel proteins: matrix metalloproteinase 20 (Mmp-20)2 (33) and kallikrein 4 (Klk4) (34).
Mmp-20 is secreted along with amelogenin, ameloblastin, and enamelin by secretory stage ameloblasts (35–37). Mmp-20 activity can account for the range of cleavages observed in secretory stage enamel proteins (38) and appears to be the only protease secreted by ameloblasts during the secretory stage. Mmp20-null mice have enamel that is thinner and softer than normal, lacks enamel rod organization, and tends to chip off the crown surface (39, 40). Like the other secretory stage enamel proteins, Mmp20 expression appears to be restricted to developing teeth (41), as is the diseased phenotype when the human gene is defective (42–44).
Klk4 is a serine protease that is secreted by transition and maturation stage ameloblasts but is not expressed by secretory stage ameloblasts (45, 46). Klk4 might also be expressed by odontoblasts, the cells that form dentin (47). Klk4 has broad substrate specificity (48, 49) and is capable of activating other proteases (50–52) and protease activated receptors (53, 54). Unlike most proteins secreted by ameloblasts, Klk4 is expressed in other tissues, most notably the prostate (55) and endometrium (56). Much attention has been focused on the potential role of Klk4 in cancers. Klk4 is increased in breast cancer stromal cells (57), in prostate cancer cells (58–61), and ovarian cancer cells (62–65). Despite this focus on the potential role of Klk4 in tumors, very little is known about the normal expression and function of Klk4 in nondental tissues. A loss of function mutation in both human KLK4 alleles caused a hypomaturation enamel phenotype in the absence of any observable defects elsewhere in the body (66).
To gain insights into the role of Klk4 in normal dental enamel formation, and to better characterize the normal temporal and spatial patterns of Klk4 expression, we have used gene targeting to knock out normal Klk4 expression, while replacing the Klk4 code with lacZ, the bacterial gene encoding β-galactosidase reporter in mice. We demonstrate that Klk4 is not expressed by secretory stage ameloblasts, but is specifically expressed by ameloblasts later in enamel formation and is necessary for the proper removal of enamel proteins, the final thickening of enamel crystals, and ultimately, for hardening of the enamel layer.
EXPERIMENTAL PROCEDURES
Animal Protocol
All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committees (IACUC) at the University of Michigan.
Knockin Targeting Construct
The targeting construct was designed to precisely replace the Klk4 coding sequence with a lacZ (β-galactosidase) reporter containing a mouse nuclear localization signal (NLS) upstream of a phosphoglycerine kinase (PGK) promoter driving a neomycin (Neo) selection marker. The PGK-Neo coding sequence was flanked by loxP sites and later deleted by Cre recombinase (Cre). Proper integration of the targeting construct deleted the segment of the Klk4 gene starting with the translation initiation codon in exon 2 and ending downstream of exon 6, the last exon. The entire Klk4 coding region was included within the deleted segment. The targeting construct was assembled from the Klk4 5′ and 3′ homology arms and NLS-lacZ restriction fragments. The 5′ and 3′ homology arms and NLS-lacZ were each amplified by PCR, subcloned into pPCR, and characterized by DNA sequencing. The Klk4 5′ and 3′ homology arms were generated using C57BL/6 genomic DNA as template. These amplifications introduced appropriate restriction sites to facilitate genomic screening, and so the three components could be assembled into the final targeting vector.
The primers (5′-TGGCGCGCCCTCAAACCTGCCGTCTTAGCAC and 5′-TGGCGCGCCGCTAGCCTCCCTGCCTCCCCATGATCTC) for amplification of the 5′ homology arm introduced an NheI site (bold) that was later used in genomic screening of Southern blots using the 5′ probe (underlined sequences annealed to the template). The 5′ homology arm was 3596 bp in length. The primers (5′-TGCGGCCGCGGCGCGCCGATATCTCTAGAGCAGCCTGTGTCAGTGATGCTAA and 5′-TGCGGCCGCCCTGGGTATGATGAGAAGGTCAAA) for amplification of the 3′ homology arm introduced an EcoRV restriction site that was later used in genomic screening of Southern blots using the 3′ probe. The 3′ homology arm was 5019 bp in length.
The 5′ primer used to amplify the NLS-LacZ coding sequence included an NheI site, 76 nucleotides from the 3′ end of Klk4 intron 1, the 8 nucleotides from the 5′ end Klk4 exon 2 that precede the translation initiation codon, and the first 25 nucleotides of the NLS coding sequence (5′-TGCTAGCAAAGGCAAGGGAGAGAGATCATGGGGAGGCAGGGAGTAAGACTACCGTCCAGATTCATTGTTGTCTTCATTTCCAGGAGCCAACATGGCTCCCAAGAAGAAGAGGAAGG). The 3′ primer was: 5′-TGCTAGCTTAATTAATAAGATACATTGATGAGTTTGGACA). The 5′ homology arm was connected to NLS-lacZ at the introduced NheI site so that the NLS-lacZ initiation codon exactly replaced the Klk4 initiation codon in exon 2. The final targeting construct (Fig. 1A) was excised from the plasmid by digestion with NotI, and the 14-kbp linear construct was transfected into Bruce4 embryonic stem cells by electroporation at passage 24.
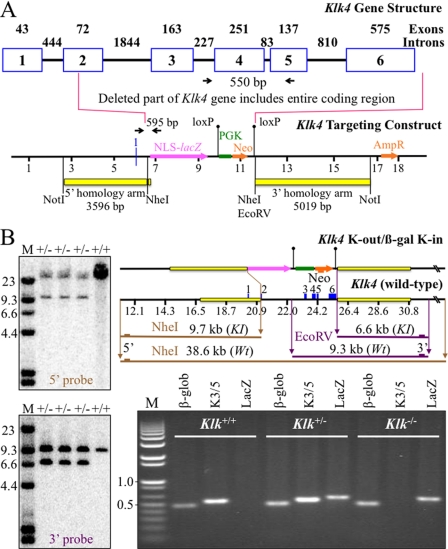
FIGURE 1.
Gene targeting strategy for the Klk4 mouse. A, depiction of the mouse Klk4 gene and the targeting construct. Numbered boxes indicate Klk4 exons (2 through 6 are coding). The number of nucleotides is shown above each exon and intron. The targeting construct was designed to replace the Klk4 coding region with NLS-lacZ so that the Klk4 promoter would drive reporter expression. The 5′ homology arm ended in exon 2 at the Klk4 translation initiation codon. The 3′ homology arm started downstream of exon 6, so that the entire Klk4 coding region was deleted. Downstream of the reporter gene was the selection gene (PGK-Neo) bracketed by loxP recombination signals that was later deleted by mating with mice expressing Cre recombinase. Black arrows mark the PCR primer annealing sites to amplify the wild-type Klk4 (550 bp) and the lacZ transgene (595 bp) sequences. B, Southern blots and PCR genotyping showing proper integration of the targeting construct. DNA isolated from tail biopsies of 3 putative Klk4+/lacZ mice were tested (lanes +/−) alongside the wild-type (+/+). Top gel, genomic DNA hybridized with the 5′ probe showing wild-type (38.6 kbp) and targeted allele (9.7 kbp) NheI bands. Bottom gel left, genomic DNA hybridized with the 3′ probe showing wild-type (9.3 kb) and the targeted allele (6.6 kb) EcoRV bands. Right, gene maps of the targeting construct and wild-type Klk4 gene show the predicted sizes of the relevant NheI and EcoRV restriction fragments and the hybridization sites for the 5′, 3′, and Neo probes. Bottom gel right, PCR genotyping to detect β-globin (β-glob, 494 bp), Klk4 intron 3 to exon 5 (K3/5, 550 bp), and Klk4-lacZ (lacZ, 595 bp).
Generation of KI Mice
Successful homologous recombination in the embryonic stem cells that survived G418/gancyclovier positive-negative selection was determined by Southern blot hybridization. Only one correctly targeted clone was identified (I_2D12) and injected into C57BL/6 blastocysts. The clone that produced germ line offspring was at passage 33 at the time of injection. The resultant male chimera was mated to C57BL/6 females, and the offspring were analyzed for germ line transmission by Southern blot hybridization (Fig. 1B) using external 5′ and 3′ probes. The 5′ probe (586 bp) was generated with primers 5′-GGCTCAGGTTGGGGAGAGAAAATG and 5′-GGGTGGTGGACAAAAAGGGAG and hybridized against NheI-digested genomic DNA. Both the wild-type (38.6 kbp) and the knockin (KI) (9.7 kbp) restriction fragments were observed. The 3′ probe (471 bp) was generated with primers 5′-TCTCCATCCCAGCCCCAAATAC and 5′-TCAATGACCCCAGAATCCTGC and hybridized against EcoRV-digested genomic DNA. Both the wild-type (9.3 kbp) and the knockin (6.6 kbp) restriction fragments were observed. A knockin-specific neomycin (Neo) probe (632 bp) was generated with primers 5′-GACTGGGCACAACAGACAATC and 5′-CCAAGCTCTTCAGCAATATCAC and hybridized against HindIII-digested genomic DNA to check for randomly integrated targeting vector. Only the expected band generated by the correctly targeted recombination (16.6 kbp) was observed (data not shown). Heterozygous offspring of the lone germ line chimera (A1_010) were mated with B6 Cre deleter mice to remove the PGK-Neomycin code. The presence or absence of Neo or Cre sequences was determined by PCR analyses using Neo (5′-AGACAATCGGCTGCTCTGAT and 5′-ATACTTTCTCGGCAGGAGCA; 261-bp amplicon) and Cre (5′-CGTACTGACGGTGGGAGAAT and 5′-TGCATGATCTCCGGTATTGA; 347-bp amplicon) primer pairs. Wild-type (Klk4+/+), heterozygous (Klk4+/lacZ or Klk4+/−), and homozygous (Klk4lacZ/lacZ or Klk4−/−) mice were distinguished by PCR genotyping of genomic DNA obtained by tail biopsy (Fig. 1) using three primer pairs: for β-globin (5′-CCAATCTGCTCACACAGGATAGAGAGGGCAGG and 5′-CCTTGAGGCTGTCCAAGTGATTCAGGCCATCG; 494-bp amplicon); for Klk4 intron 3 and exon 5 (5′-AACCTAAGGGACAGGGCAGT and 5′-TGAGGTGGTACACAGGGTCA; 550-bp amplicon), and Klk4-lacZ (5′-TGCCTCCAACCAGATAGGTC and 5′-GACAGTATCGGCCTCAGGAA; 595-bp amplicon). β-Globin was a positive control demonstrating adequate DNA template and reagents. A positive Klk4 band demonstrated the presence of at least one Klk4 allele. A positive Klk4-lacZ band demonstrated the presence of at least one knockin gene. The heterozygous (Klk4+/lacZ) mice were crossed with C57BL/6 to establish a breeding colony and backcrossed to Swiss Black mice for outbreeding. All mice were maintained on moistened chow and later switched to soft diet (DietGelTM R/L, ClearH2O, Portland, ME). Ozgene (Bentley DC, Western Australia, Australia) fabricated the targeting construct and generated the Klk4 knockout/lacZ knockin mice.
Physical Assessment and Blood Chemistry
The three mouse Klk4 genotypes (+/+, +/−, and −/−) were evaluated from birth for their appearance, physical activity, size discrepancy, rate of growth, food intake, and reproductive physiology. Peripheral blood samples were collected from 7-week-old mice for each of the three genotypes, and a comprehensive panel of blood chemistry tests was performed at the Animal Diagnostic Laboratory of the University of Michigan.
The β-Galactosidase Expression Assay
Mouse heads were collected from postnatal (PN) days 5, 6, 8, 12, and 14, fixed in 4% paraformaldehyde (PFA) overnight, and processed for β-galactosidase staining (67–69). To improve tissue fixation in mice 7 days old or later, the thorax and the right ventricle were opened, phosphate-buffered saline (PBS, 137 mm NaCl, 10 mm phosphate, 2.7 mm KCl, pH 7.4.) was slowly injected into the left ventricle to flush out circulatory fluid, and then 30 ml of 4% PFA, 0.1% glutaraldehyde in 0.1 m PBS, pH 7.4, was slowly injected. The mandibles were removed and immersed in fresh PFA-glutaraldehyde fixative solution for 90 min to overnight at 4 °C according to the age of mice and size of the tissue block, washed with PBS (3 × 15 min), then decalcified in 10% EDTA for up to 7 days according to the age of the mice (70). The decalcified tissues were washed in PBS (3 × 15 min) and embedded in Tissue-Tek® O.C.T. Compound (ProSciTech, Queensland, Australia). Tissue blocks were cryosectioned at 10-μm thickness, post-fixed for 5 min in 0.5% glutaraldehyde, washed in PBS (3 × 5 min), and incubated at 45 °C for 3 h in freshly prepared X-gal staining buffer, pH 8.0, containing 1 mg/ml X-gal, 100 mm HEPES, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 1 mm MgCl2, 2% Triton X-100, and 1 mm dithiothreitol (71). Tissue sections were rinsed and stored in PBS for counterstaining with hematoxylin then observed under a dissection (Nikon SMZ1000) or light microscope (Nikon Eclipse E600). All images were captured using a digital camera (Nikon DXM1200) and Act1 imaging software (Mager Scientific, Dexter, MI).
Immunohistochemistry
We used the Histostain-SP kit with the labeled-streptavidin-biotin detection system (Invitrogen). PN14 mice were perfused with ice-cold 4% PFA, 0.1% glutaraldehyde in PBS. The mouse heads were dissected and fixed in ice-cold 4% PFA, 0.1% glutaraldehyde in PBS overnight. PN14 maxillae and mandibles were demineralized using 10% EDTA for 5–7 days. The fixed maxillary and mandibular processes were dissected, embedded in paraffin, sectioned at 6 μm, spread in a water bath (52 °C), loaded on plus gold glass slides (Fisher), and left at room temperature overnight. The sections were deparaffinized 3× in xylene, rehydrated using 100%, 95%, 80%, and 70% ethanol, treated with PBS, and then treated with peroxidase quenching solution. The sections were incubated with serum blocking solution and then with amelogenin antibody (rM179 Ab 1:2000) at 4 °C overnight or enamelin antibody (mEnam223–336 Ab 1:100) at room temperature for 30 min, or with pre-immune serum. The slides were washed 3× with PBS, incubated with biotinylated secondary antibody, washed 3×, incubated with streptavidin-enzyme conjugate, washed 3×, incubated with substrate-chromogen mixture, washed 3×, counterstained with hematoxylin, and mounted with Aqueous Mounting Solution (Invitrogen) for microscopy and photography. The amelogenin antibodies were raised in rabbits against recombinant mouse amelogenin (72). The enamelin antibodies were raised in rabbits against the synthetic peptide (C)FEDFEKPKEKDPPK-NH2, (designated mEnam223–336 and containing amino acids Phe223–Lys236 of mouse enamelin (73)) conjugated to the KLH carrier protein (YenZym Antibodies, Burlingame, CA). Antibodies were generated using a protocol that included three immunizations, one test bleed, a fourth immunization, and a final bleed. Specific anti-peptide antibodies were purified from the final bleed using an affinity column containing the immobilized unconjugated mEnam223–336 peptide and were enzyme-linked immunosorbent assay-tested before being used for immunohistochemistry.
Radiography
Hemimandibles from 7-week-old Klk4+/+, Klk4+/−, and Klk4+/− mice were cleaned of soft tissues and radiographed using a FaxitronTM x-ray cabinet model MX-20 (Faxitron X-ray Corp., Wheeling, IL) operating at 30 kV. Hemimandibles from each of the three genotypes were evaluated and compared using digital micrographs obtained under identical Faxitron settings.
Micro-computed Tomography
Mouse hemimandibles were imaged using a cone-beam micro-computed tomography (μCT) system (eXplore Locus SP, Amersham Biosciences Pre-Clinical Imaging, London, Ontario, Canada) in the Orthopaedic Research Laboratory at the University of Michigan Department of Orthopaedic Surgery, as described previously (74). In brief, hemimandibles were exposed to polychromatic x-rays on a rotating stage. Measurements were taken at an operating voltage of 80 kV and 80 mA of current, with an exposure time of 1600 ms. The effective voxel size of the reconstructed image was 18 × 18 × 18 μm3.
SEM Evaluation of Molars and Incisors at 7 Weeks
SEM evaluations were performed at the University of Michigan Microscopy and Image-analysis Laboratory (Ann Arbor, MI). Ethanol-dehydrated, air-dried hemimandibles and mandibular incisors from 7-week-old Klk4+/+, Klk4+/−, and Klk4−/− mice were mounted on metallic stubs using conductive carbon cement, off-gassed in a vacuum desiccator overnight, and sputter-coated with an Au-Pd film to increase conductivity. The samples were imaged using an Amray EF 1910 scanning electron microscope operating at an accelerating voltage of 3–5 kV.
RESULTS
Generation of the Klk4-null/lacZ-knockin Mouse
We used gene targeting to generate a mouse strain carrying a null allele of kallikrein 4 that has a lacZ reporter gene inserted into the 5′ region of the Klk4 locus (Fig. 1A). Mice hosting the targeting construct in both Klk4 alleles are alternatively referred to as Klk4−/− or as Klk4lacZ/lacZ, to emphasize either the loss of Klk4 expression or the introduction of nuclear β-galactosidase expression within the context of the Klk4 gene. The lacZ coding sequence (encoding bacterial β-galactosidase) was modified to include a mouse NLS and inserted into the first coding exon (exon 2) of Klk4 so that its translation initiation codon (ATG) exactly replaced the translation initiation codon of Klk4. The Klk4 gene was deleted from the translation initiation codon until past the last exon (exon 6), so that none of the Klk4 coding sequence remained in the null mice. The coding sequences for the selection markers (PGK-Neo) downstream of the NLS-lacZ were bracketed with loxP sites and later removed by mating Klk4+/lacZ mice with Cre deleter mice. Correct targeting and integration were confirmed by Southern blot analyses and by reverse transcription-PCR genotyping using primer pairs specific for β-globin, Klk4, and the lacZ reporter (Fig. 1B). By replacing the Klk4 translation initiation site and 5′ code with the NLS-lacZ translation initiation site and code, we simultaneously knocked out Klk4 expression while knocking in NLS-lacZ expression in its place. Because the NLS-lacZ code is positioned in the same genomic context as wild-type Klk4, NLS-lacZ expression provides a sensitive reporter for native Klk4 expression.
Gross Tooth Morphology and Appearance
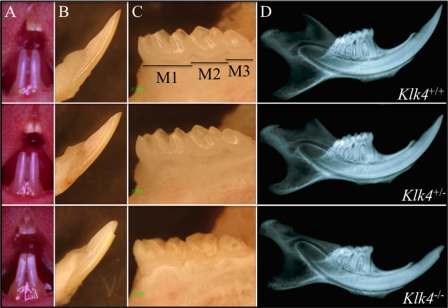
Photographic and radiographic evaluation at 7-weeks does not suggest any differences in the dentitions of wild-type (Klk4+/+) and heterozygous (Klk4+/−) mice (Fig. 2). Tooth shape, size, and color are indistinguishable. The Klk4 null mice, however, exhibit enamel defects that are obvious without magnification. The mandibular incisors of the Klk4−/− mice are chalky white in the erupted portion. The mandibular incisor enamel near the functional part of the incisal edge is always missing, being rapidly abraded during function and shows a prominent wear facet on its labial surface. The enamel of all teeth tends to fracture at points of contact with the opposing dentition. It appears as though the full thickness of enamel chips away, down to the surface of the underlying dentin.
FIGURE 2.
Photographic and radiographic examination at 7 weeks. Column A, oral photographs showing incisors. Column B, lateral view of mandibular incisors following soft tissue removal. Column C, lingual view of mandibular molars (M1–M3) following soft tissue removal. Column D, radiograph of hemimandible. No differences were detected between the wild-type (Klk4+/+, top row) and heterozygous (Klk4+/−, middle row) mice at this scale. The dental enamel of the null (Klk4−/−, bottom row) mice, however, is markedly different. It is chalky in color, and the crowns have chipped off in areas of occlusal contact.
Tissue-specific Expression of NLS-lacZ
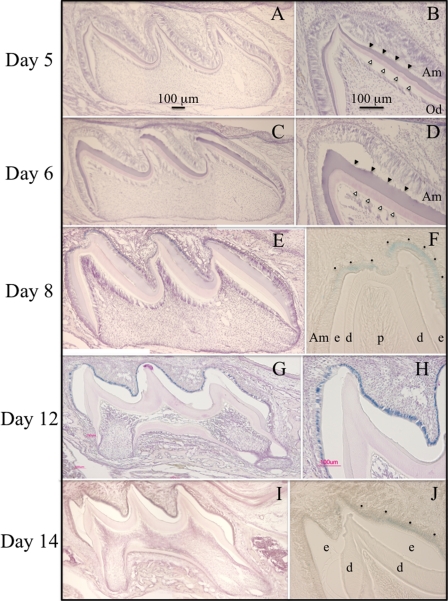
Replacing the Klk4 coding region with the NLS-lacZ code established the heterozygous mouse as a specific reporter for Klk4 expression. The NLS-lacZ is readily distinguished from native β-galactosidase activity because of its nuclear localization and because of its high pH optima. Mouse β-galactosidase is a lysosomal enzyme that is only marginally active at pH 7.5, whereas bacterial β-galactosidase can be strongly detected above pH 8 (75). X-gal is a substrate analogue that is cleaved by β-galactosidase to yield 5-bromo-4-chloro-3-hydroxyindole, an insoluble blue product. NLS-lacZ expression leaves a blue stain that localizes to the nucleus. We assayed developing dental structures in Klk4+/lacZ knockin mice for nuclear β-galactosidase expression by histochemistry at pH 8 (Fig. 3). X-gal staining was limited to ameloblasts starting at PN8 and continuing through PN14, the last day studied. Odontoblasts were negative. These results correlate well with previous studies of Klk4 expression in developing teeth by in situ hybridization (37, 46, 76) and support the conclusion that Klk4 is not expressed during the secretory stage (prior to PN7 in maxillary first molars), but is expressed in the transition stage (starting at PN8 in maxillary first molars) and thereafter. The close correlation between the lacZ staining in transition and maturation ameloblasts in the Klk4+/lacZ mice and Klk4 mRNA expression as determined by in situ hybridization provides additional confirmation that the knockin construct was correctly targeted.
FIGURE 3.
Histochemical detection of β-galacto-sidase activity in the maxillary first molars of heterozygous (Klk4+/lacZ) mice at postnatal days 5, 6, 8, 12, and 14. Mesial-distal sections through the buccal cusps are on the left. Higher magnification views of the distal cusp tips are on the right. A–D, no β-galactosidase histostaining was observed at days 5 and 6 when the ameloblasts are predominantly in the secretory stage. E and F, β-galactosidase histostaining was first observed at day 8, when ameloblasts near the cusp tip are in transition or early maturation stages. G and H, specific histostaining was strongest at day 12 and was restricted to the nuclei of ameloblasts, which are in the early maturation stage. I and J, histostaining in ameloblasts persisted at day 14, but was reduced in intensity. No histostaining was observed in odontoblasts at any stage. Am, ameloblasts; e, enamel; d, dentin; p, pulp; dots mark positive histostaining in F and J (which were not counterstained with hematoxylin); developing enamel (solid arrowheads) and dentin (outlined arrowheads) are marked in B and D.
Physicochemical Assessments
Our initial experience with the Klk4 null mice was that they often did not survive to maturity. We guessed that they were having difficult eating due to the loss of enamel from their teeth. We started weaning the Klk4 null mice later (at 28 versus 21 days) and provided a soft diet. The young null mice still appeared to be delayed in their growth, but eventually achieved a normal adult body size and survive normally. First generation litter sizes for the null mice crosses were low (5.8 pups/litter) but rebounded in the second generation crosses to the average for our wild-type colony (7.8 pups/litter). There were no statistically significant differences among the wild-type and Klk4+/− and Klk4−/− mice in a panel of blood chemical tests that included serum levels of calcium, phosphate, magnesium, alanine aminotransferase, and total protein (Table 1). The blood tests support the conclusion that the dental phenotype in the Klk4 null mice is not due to systemic alterations that affect calcium or phosphate homeostasis.
TABLE 1.
Blood chemistry of Klk4 mice
| Test | Age | Genotype | Results | Reference |
|---|---|---|---|---|
| Calcium | 7 weeks | +/+ | 9.6 | 5.9–9.4 mg/dl |
| +/− | 9.9 | |||
| −/− | 9.7 | |||
| Phosphorus | 7 weeks | +/+ | 8.2 | 6.1–10.1 mg/dl |
| +/− | 10.9 | |||
| −/− | 8.2 | |||
| Magnesium | 7 weeks | +/+ | 2.78 | 2.6–3.0 mm |
| +/− | 2.67 | |||
| −/− | 2.63 | |||
| Alanine aminotransferase | 7 weeks | +/+ | 79 | 28–132 units/liter |
| +/− | 130 | |||
| −/− | 51 | |||
| Total protein | 7 weeks | +/+ | 4.3 | 3.6–6.6 g/dl |
| +/− | 4.9 | |||
| −/− | 5.1 |
Immunohistochemistry
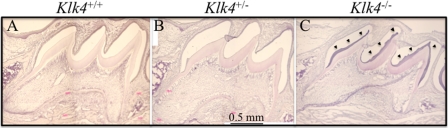
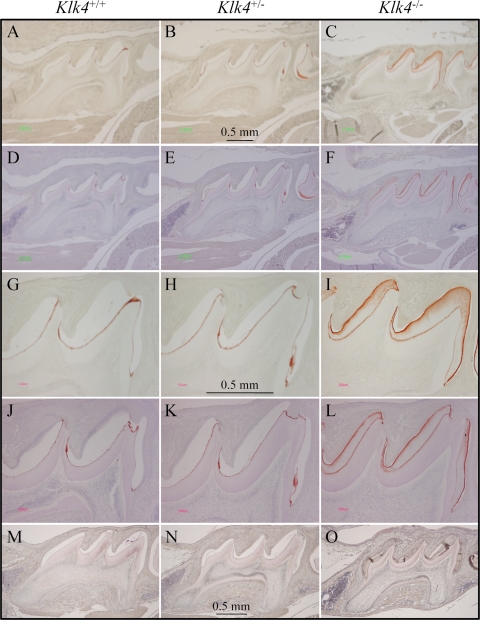
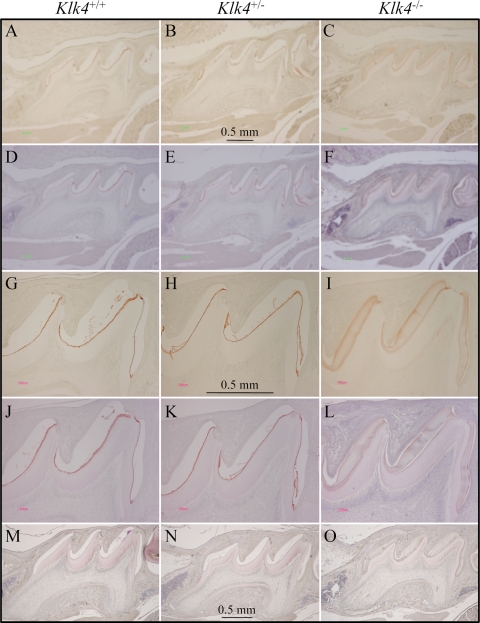
During our histological examination of cryosections of the developing teeth for X-gal histostaining we observed hematoxylin staining in the enamel space of the Klk4 null mouse that was not evident in either the wild-type or the Klk4 heterozygous mice (Fig. 4). While staining of the enamel extracellular space is normal during the secretory stage when enamel proteins comprise over 30% (by weight) of the matrix (5), such staining is not observed during the mid-maturation stage when the protein content usually falls to 1 or 2% of the matrix (15). In our cryostat sections the stained material seemed to be restricted to the deeper enamel near the DEJ, but later studies using paraffin sections showed that the residual material that stains with hematoxylin is distributed throughout the entire enamel space. To better characterize this material, we performed immunohistochemistry on PN14 paraffin sections using affinity-purified, anti-peptide antibodies specific for enamelin (Fig. 5) and polyclonal antibodies against recombinant mouse amelogenin (Fig. 6). In wild-type and heterozygous mice positive enamelin and amelogenin staining is only observed in molars during the maturation stage at the dentino-enamel junction (DEJ), where residual enamel protein is sometimes referred to as “enamel tufts” (77). The enamel space away from the DEJ does not immunostain with either antibody and does not counterstain significantly with hematoxylin, which is consistent with little residual enamel protein being present in maturing molar enamel of wild-type mice. In molars from Klk4 null mice, on the other hand, enamelin and amelogenin immunostaining is strongest at the enamel surface, diminishes with depth, and is strong again at the DEJ. These findings demonstrate that, in the absence of Klk4 expression, enamel proteins are not properly removed from the enamel space during the maturation stage and remain in the matrix.
FIGURE 4.
Residual protein in maturation stage enamel. Mesial-distal sections of mouse maxillary first molar cryosections at PN14. A and B, sections of maturation stage enamel are normally low in protein and appear clear in these hematoxylin-stained sections of day 14 wild-type and Klk4+/− molars. C, the enamel layer of the Klk4−/− molars stains purple (arrowheads) presumably due to the retention of processed enamel proteins.
FIGURE 5.
Immunohistochemistry of enamelin. A–F, lower magnification; G–L, higher magnification views of day 14 maxillary first molars in wild-type (Klk4+/+; left), heterozygous (Klk4+/−; middle) and null (Klk4−/−; right) mice immunostained using the mEnam223–336 Ab. M–O, negative control sections immunostained with pre-immune serum. Sections A–C and G–I are not counterstained. Sections D–F and J–L are hematoxylin counterstained. In the wild-type and heterozygous mice positive enamelin stain is only observed at the DEJ. In the Klk4−/− mice, enamelin immunostaining is strongest at the enamel surface, diminishes with depth, and is strong again at the DEJ.
FIGURE 6.
Immunohistochemistry of amelogenin. A–F, lower magnification; G–L, higher magnification views of day 14 maxillary first molars in wild-type (Klk4+/+, left), heterozygous (Klk4+/−, middle), and null (Klk4−/−, right) mice using the amelogenin (rM179) Ab. M–O, negative control sections immunostained with pre-immune serum. Sections A–C and G–I are not counterstained. Sections D–F and J–L are hematoxylin counterstained. In the wild-type and heterozygous mice positive amelogenin stain is only observed at the DEJ. In the Klk4−/− mice amelogenin immunostaining is strongest at the enamel surface, diminishes with depth, and is strong again at the DEJ. The immunostaining patterns for enamelin (Fig. 5) and amelogenin are essentially the same.
Scanning Electron Microscopy
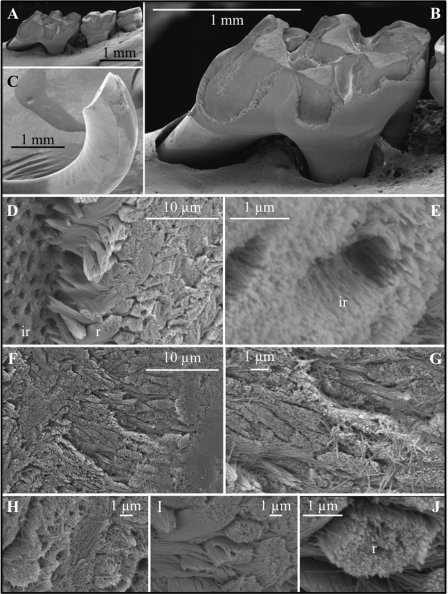
SEM of the dentition of the Klk4 null mouse at 7 weeks illustrates how enamel formed in the absence of Klk4 cannot withstand occlusal forces (Fig. 7). The enamel on all biting surfaces was abraded away (Fig. 7, A–C). At first glance, it appeared as though the breakage occurred at the DEJ, exposing the underlying dentin and dentinal tubules. Higher magnification of the chipped area near the tip of the incisor, however, showed that the break was in enamel. The broken surface contains numerous circular holes 1 to 2 μm in diameter. These holes do not appear to be exposed dentinal tubules based upon nanoscale anatomy of the crystallites lining the holes. They are also too numerous to be made by odontoblastic processes penetrating the enamel (enamel spindles), which are infrequent in mice. The mineralized wall abutting the floor of the break is comprised of enamel rods that appear to have separated and pulled out from the holes. If so, the enamel with the holes is interrod enamel (Fig. 7D). The tips of the rods near the holes are tapered, coming almost to a point, suggesting they may be the initial parts of the rods that formed near the DEJ. Higher magnification shows the holes are lined with what appear to be parallel enamel crystallites oriented toward the surface (Fig. 7E). This orientation suggests these crystallites belong to the initial interrod enamel covering the aprismatic enamel at the DEJ and are not dentin crytals lining dentinal tubules. Another unusual finding from the higher magnification SEM was that the enamel crystallites in the Klk4 mouse, even in teeth that have been erupted for a month, do not adhere to each other and individual crystallites are observed that have spilled out onto the broken enamel surface (Fig. 7, F–J).
FIGURE 7.
SEM of the mandibular molars (A and B) and mandibular incisor (C–J) of a Klk4 null mouse at 7 weeks. The enamel of all molars showed a significant loss of enamel from all working surfaces (buccal cusps, occlusal surface, and marginal ridges) (A and B). Similarly, the enamel layer was abraded at the working (buccal) surface of the mandibular incisor at its tip (C). Higher magnification of the chipped area near the tip of the incisor showed the break was in the enamel layer, close to, but not at the DEJ. The broken surface appears to be composed of interrod (ir) enamel with holes where enamel rods (r) had pulled out and separated (D) from the initial deposit of interrod enamel near the DEJ. The holes are too numerous to be made by odontoblastic processes penetrating the enamel (enamel spindles). The orientation of the crystallites on the walls of the holes is parallel to the direction of the tubular holes and to the crystallites between the holes (E). This is not the case for tubular and intertubular dentin. The enamel rods are well organized in the Klk4 null mouse (F–G and I–J), but the crystallites within a rod are not adherent and individual crystallites are seen spilled out onto the broken enamel surface (G and H).
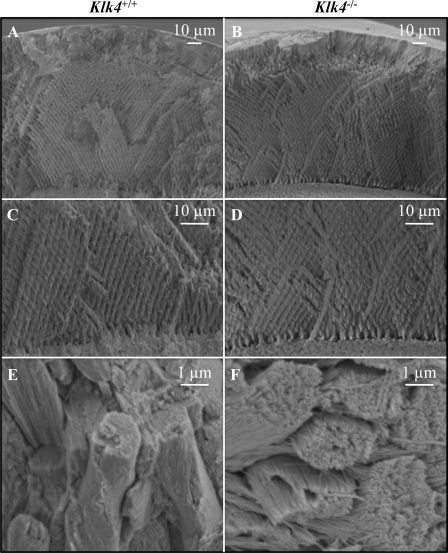
The enamel defects in the Klk4 mouse do not appear to have their origin in the secretory stage. The enamel in the Klk4 null mouse is the same thickness from the DEJ to the enamel surface as in the wild-type mouse (Fig. 8, A and B). It also shows a similar pattern of decussating enamel rods (Fig. 8, C and D). The thickness of the enamel layer as well as the decussating (X-like) pattern of enamel rods are both established during the secretory stage when the crystals are growing in length. The major difference between the enamel of the Klk4 null mouse and the wild type is that the enamel crystals in the null mouse persist as independent crystallites that can be distinguished from each other and can slide relative to its neighbors (Fig. 8, E and F). The enamel crystallites appear to have matured somewhat (grown in width and thickness) even in the absence of Klk4 activity. Individual crystallites in the rods are readily distinguished in the Klk4 null mouse enamel and conjure an impression of uncooked angel hair spaghetti. These images suggest that the fundamental failure in the Klk4 null condition is that the enamel crystallites fail to thicken and widen until they interlock. Because of this the crystals in the rods do not appear to function as a unit. This reduces the strength of the enamel and allows it to fracture in ways that wild-type enamel does not.
FIGURE 8.
Comparison of enamel from the wild-type and Klk4 null mice. A and B, SEM at the same scale showing mandibular incisor enamel from the DEJ (bottom) to the surface (top) that has been fractured in the erupted portion by pressing on it with a knife. There is no observable difference in the overall thickness of the enamel layer between the wild-type and Klk4 null mice. C and D, higher magnification showing the decussating patterns of enamel rods just above the DEJ. E and F, enamel rods in the wild-type mice have tightly packed crystallites that lose some aspect of their individuality. Enamel rods in the Klk4 null mice are compose of distinctly individual crystallites resembling angel hair spaghetti. Holes or vacancies in some rods give the impression that smaller bundles of crystallites broke at a slightly deeper level and slid out of the rod.
Micro-computed Tomography
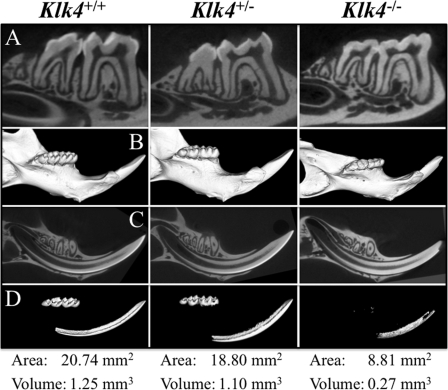
Hemimandibles from 7-week-old Klk4+/+, Klk4+/−, and Klk4−/− mice were scanned by micro-computed tomography and analyzed (Fig. 9). Sagittal sections through the mandibular first and second molars (Fig. 9A) and incisor (Fig. 9C) showed strong contrast between enamel and dentin in both the wild-type and Klk4 heterozygous mice. In the Klk4 null mice the enamel was partially abraded from the molars and at the tip of the incisor. In the retained enamel, which was most evident on the null incisor, there was no contrast between the density of enamel and that of dentin. The three-dimensional micro-computed tomography reconstructions of the hemimandibles using a low arbitrary density demarcation displayed the entire mandible (Fig. 9B). Similar reconstructions were made using an arbitrary density demarcation that eliminated all of the dentin and bone of the wild-type hemimandible but displayed the entire enamel layers of the molars and the maturation stage enamel of the incisor (Fig. 9C). Using this same threshold, most of the enamel in the heterozygous mouse was displayed, with about a 10% drop in area and volume relative to the wild type. In the heterozygous teeth, the enamel near the cemento-enamel junction dropped below threshold, especially in the first molar (Fig. 9D). So did the enamel at the base of the incisor, suggesting a delay in the onset of maturation. In the null mouse, virtually all of the molar enamel was either abraded away or below threshold. The incisor enamel reached threshold even later (closer to the erupted region) than the heterozygous mouse, suggesting that some maturation of the enamel mineral occurred even in the null mouse, but that it was delayed and never reached the mineral density of the wild type.
FIGURE 9.
Micro-computed tomography of hemimandibles at 7 weeks. Row A, 18-μm section through the mandibular first and second molars. The enamel layers of the wild-type and Klk4 heterozygous mouse molars have similar intensities, which sharply contrast with dentin. The enamel is largely abraded from the molars of the null mouse. Row B, three-dimensional micro-computed tomography reconstruction of the hemimandibles. Row C, 18-μm sections through the mandibular incisor. As in the molars, the enamel layers of the wild-type and Klk4 heterozygous mouse have similar intensities that sharply contrast with dentin. The incisal edges are sharp and unbroken. The incisor enamel of the Klk4 null mouse does not contrast well with dentin and is abraded at the incisal edge. Row D, three-dimensional micro-computed tomography reconstruction of the hemimandibles as in row B, but using a threshold cutoff so that only highly mineralized structures are seen. The enamel crowns of the three molars and the incisor in the wild-type hemimandible are uniquely above the threshold. Dentin and bone are both below threshold. The area and volume of the supra-threshold signal (enamel) of Klk4 heterozygous mouse was reduced by ∼10% from the wild type. In the molar region of the heterozygote, only the thin enamel near the cemento-enamel junction of the first molar appeared to drop below threshold, whereas virtually the entire enamel layer of the molars from the null mouse dropped below threshold.
DISCUSSION
The Klk4 knockout/lacZ knockin mouse confirms that Klk4 is not expressed by secretory stage ameloblasts. Klk4 expression began in PN8 maxillary molars, which is consistent with previous in situ hybridization analyses showing the onset of Klk4 expression by ameloblasts is at the beginning of the transition stage. The enamel layer in the Klk4 null mouse achieves normal thickness and is composed of decussating enamel rods. No Klk4 expression by odontoblasts was observed. These findings lead us to the conclusions that Klk4 does not function during the secretory stage of amelogenesis and that the observed enamel defects in the Klk4 mice are due to the lack of secretion and activity of Klk4 in the transition and maturation stages.
The fundamental process of biomineralization occurring during the transition/maturation stages of amelogenesis is the growth in width and thickness of highly elongated pre-existing enamel crystals. This is a biologically complicated process that involves the cycling of ameloblasts between smooth and ruffle-ended phases. These modulations occur about every 8 h (79). How this cycling is controlled is completely unexplored. During the ruffle-ended part of the cycle, there is an increase in the flux of calcium and phosphate into the matrix with increased mineral deposition and a drop in matrix pH (80, 81). The ruffled ends of the ameloblasts are due to membrane infoldings that appear to be involved in the endocytosis of degraded enamel proteins. In early maturation they are associated with electron-dense material (enamel proteins?) and disappear half-way through the maturation stage when little or no enamel proteins remain (15). The smooth ended phase is associated with neutralizing the acid released by mineralization. This involves the production of bicarbonate by carbonic anhydrase II (82) and its transport into the matrix by anion exchanger II (83) possibly in association with the cystic fibrosis transmembrane conductance regulator (84).
The complete maturation of enamel crystals requires the removal of residual enamel proteins from the matrix. It is believed that enamel proteins bind to the sides of enamel crystals, possible at kinks or similar growth sites, and inhibit the deposition of ions on the crystal surface (85). When maturation-stage enamel crystals are incubated in supersaturated solutions of calcium phosphate, no crystal growth is observed unless the maturation-stage enamel is first pretreated with either 8 m urea or sodium hypochlorite to remove residual protein (86). Serine proteinases facilitate enamel crystal growth in vitro (87), and the binding affinity of recombinant amelogenins for synthetic calcium hydroxyapatite is reduced by proteolysis with MMP-20 and Klk4 (88). Despite this consistent experimental support, in vivo studies and this report suggest that the importance of this mechanism for proper enamel maturation may be overemphasized.
Analyses of strip dissections of rat incisors clearly show that as much as a third of enamel maturation is deposited in the maturation stage prior to the removal of enamel proteins (15). This study shows that, although enamel crystallites do not completely mature in the Klk4 null mouse, they greatly increase in width and thickness during the maturation stage despite the pathological retention of enamel proteins. The influx of calcium and phosphate ions may displace enamel proteins from the mineral, much as phosphate buffer elutes proteins from a hydroxyapatite column. There is, however, a delay in enamel maturation in the Klk4 heterozygous and null mice based upon the incisal shift in the position where the enamel surpasses an arbitrary density demarcation in the micro-computed tomography images of the mouse incisors (Fig. 9D). This delay is not critical in the heterozygous mouse, because the enamel ultimately mineralizes to a level necessary for function.
The ultimate structural defect in the enamel of the Klk4 null mouse appears to be the failure of the crystals to mature fully, that is, so the individual crystallites grow into contact with adjacent crystals, interlock, and behave as a structural unit. This final bit of the maturation process might be blocked by the physical presence of residual enamel proteins, which occupy the spaces between crystallites. The removal of enamel proteins, specifically amelogenins, has been proposed to be the principle mechanism for providing the physical space for pre-existing crystals to expand within the defined volume of the inner secretory stage enamel (89). The enamel phenotype of the Klk4 null mouse is consistent with the perspective that the proline-rich enamel protein matrix, especially after the removal of the charged amelogenin C terminus, is a relatively inert packing material that supports and separates enamel crystals by occupying the space between them. The main role of Klk4 is to catalyze the proteolytic degradation of the residual matrix facilitating its removal (by degradation and reabsorption into ameloblasts) to provide space for the expansion of pre-existing enamel crystals.
Prior to this study the only evidence of a critical role for Klk4 in dental enamel formation was the identification of a missense mutation in a codon for an active site residue in both KLK4 alleles of a family with autosomal recessive hypomaturation amelogenesis imperfecta (66). We have generated a Klk4 null mouse that causes a similar dental phenotype and permits experimental analyses of its pathogenesis. Our analyses have focused mainly on the extracellular mineralizing matrix, but it is conceivable that Klk4 plays additional roles, perhaps in helping to regulate the modulations of maturation stage ameloblasts through the stimulation of protease-activated receptors.
Although no phenotype outside of the dentition was observed in the Klk4 null mice, Klk4 presumably also plays roles in other tissues or in disease processes, such as cancer. The Klk4 null mouse should prove a useful experimental model for further discoveries of the role of Klk4 in normal and pathological conditions.
Acknowledgments
We thank Dr. Dorothy R. Sorenson of the University of Michigan Microscopy and Image-analysis laboratory for her help and expertise in SEM, Dr. Jaclynn M. Kreider of the University of Michigan, Orthopaedic Research Laboratories for her help and expertise in micro-computed tomography, Dr. Thomas Saunders of the University of Michigan Transgenic Animal Model Core and Dr. Ashok Kulkarni of the NIDCR, National Institutes of Health for their insightful consultations and technical support related to gene targeting, and the team of scientists at Ozgene Ltd., Australia, who fabricated the Klk4 knockin construct and established the mouse founders.
This work was supported, in whole or in part, by National Institutes of Health Grant DE 15846 from NIDCR.
- Mmp-20
- matrix metalloproteinase 20
- Klk4
- kallikrein 4
- Cre
- Cre recombinase
- DEJ
- dentino-enamel junction
- KI
- knockin
- Neo
- neomycin
- NLS
- nuclear localization signal
- PBS
- phosphate-buffered saline
- PFA
- paraformaldehyde
- PGK
- phosphoglycerine kinase
- PN
- postnatal
- SEM
- scanning electron microscopy
- β-gal
- β-galactosidase.
REFERENCES
- 1.Warshawsky H., Nanci A. (1982) J. Dent. Res. Spec. No., 1504–1514 [PubMed] [Google Scholar]
- 2.Daculsi G., Menanteau J., Kerebel L. M., Mitre D. (1984) Calcif. Tissue Int. 36, 550–555 [DOI] [PubMed] [Google Scholar]
- 3.Daculsi G., Kerebel B. (1978) J. Ultrastruct. Res. 65, 163–172 [DOI] [PubMed] [Google Scholar]
- 4.Deakins M., Volker J. F. (1941) J. Dent. Res. 20, 117–121 [Google Scholar]
- 5.Fukae M., Shimizu M. (1974) Arch. Oral Biol. 19, 381–386 [DOI] [PubMed] [Google Scholar]
- 6.Sasaki T. (1984) Histochemistry 80, 263–268 [DOI] [PubMed] [Google Scholar]
- 7.Smid J. R., Young W. G., Monsour P. A. (2001) Eur. J. Oral Sci. 109, 260–266 [DOI] [PubMed] [Google Scholar]
- 8.Smith C. E., Pompura J. R., Borenstein S., Fazel A., Nanci A. (1989) Anat. Rec. 224, 292–316 [DOI] [PubMed] [Google Scholar]
- 9.Bartlett J. D., Simmer J. P. (1999) Crit. Rev. Oral Biol. Med. 10, 425–441 [DOI] [PubMed] [Google Scholar]
- 10.Simmer J. P., Hu J. C. (2002) Connect. Tissue Res. 43, 441–449 [DOI] [PubMed] [Google Scholar]
- 11.Reith E. J. (1970) J. Ultrastruct. Res. 30, 111–151 [DOI] [PubMed] [Google Scholar]
- 12.Kallenbach E. (1971) J. Ultrastruct. Res. 35, 508–531 [DOI] [PubMed] [Google Scholar]
- 13.Nylen M. U., Eanes E. D., Omnell K. A. (1963) J. Cell Biol. 18, 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miake Y., Shimoda S., Fukae M., Aoba T. (1993) Calcif. Tissue Int. 53, 249–256 [DOI] [PubMed] [Google Scholar]
- 15.Smith C. E. (1998) Crit. Rev. Oral Biol. Med. 9, 128–161 [DOI] [PubMed] [Google Scholar]
- 16.Wakida K., Amizuka N., Murakami C., Satoda T., Fukae M., Simmer J. P., Ozawa H., Uchida T. (1999) Histochem. Cell Biol. 111, 297–303 [DOI] [PubMed] [Google Scholar]
- 17.Kallenbach E. (1974) Tissue Cell 6, 173–190 [DOI] [PubMed] [Google Scholar]
- 18.Kerebel B., Daculsi G., Kerebel L. M. (1979) J. Dent. Res. 58, 844–851 [DOI] [PubMed] [Google Scholar]
- 19.Mjor I., Fejerskov O. (eds) (1986) Human Oral Embryology and Histology, pp. 50–3; 89, Munksgaard, Copenhagen [Google Scholar]
- 20.Provenza D. V. (1988) Fundamentals of Oral Histology and Embryology, 2nd ed., p. 151, Lea & Febiger, Philadelphia [Google Scholar]
- 21.Snead M. L., Zeichner-David M., Chandra T., Robson K. J., Woo S. L., Slavkin H. C. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7254–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snead M. L., Lau E. C., Zeichner-David M., Fincham A. G., Woo S. L., Slavkin H. C. (1985) Biochem. Biophys. Res. Commun. 129, 812–818 [DOI] [PubMed] [Google Scholar]
- 23.Cerny R., Slaby I., Hammarstrom L., Wurtz T. (1996) J. Bone Miner. Res. 11, 883–891 [DOI] [PubMed] [Google Scholar]
- 24.Krebsbach P. H., Lee S. K., Matsuki Y., Kozak C. A., Yamada K. M., Yamada Y. (1996) J. Biol. Chem. 271, 4431–4435 [DOI] [PubMed] [Google Scholar]
- 25.Hu C. C., Fukae M., Uchida T., Qian Q., Zhang C. H., Ryu O. H., Tanabe T., Yamakoshi Y., Murakami C., Dohi N., Shimizu M., Simmer J. P. (1997) J. Dent. Res. 76, 648–657 [DOI] [PubMed] [Google Scholar]
- 26.Hu C. C., Fukae M., Uchida T., Qian Q., Zhang C. H., Ryu O. H., Tanabe T., Yamakoshi Y., Murakami C., Dohi N., Shimizu M., Simmer J. P. (1997) J. Dent. Res. 76, 1720–1729 [DOI] [PubMed] [Google Scholar]
- 27.Hu C. C., Hart T. C., Dupont B. R., Chen J. J., Sun X., Qian Q., Zhang C. H., Jiang H., Mattern V. L., Wright J. T., Simmer J. P. (2000) J. Dent. Res. 79, 912–919 [DOI] [PubMed] [Google Scholar]
- 28.Gibson C. W., Yuan Z. A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J. T., Decker S., Piddington R., Harrison G., Kulkarni A. B. (2001) J. Biol. Chem. 276, 31871–31875 [DOI] [PubMed] [Google Scholar]
- 29.Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., Krebsbach P. H., Nanci A., Kulkarni A. B., Yamada Y. (2004) J. Cell Biol. 167, 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J. C., Hu Y., Smith C. E., McKee M. D., Wright J. T., Yamakoshi Y., Papagerakis P., Hunter G. K., Feng J. Q., Yamakoshi F., Simmer J. P. (2008) J. Biol. Chem. 283, 10858–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sire J. Y., Delgado S. C., Girondot M. (2008) BMC Evol. Biol. 8, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deméré T. A., McGowen M. R., Berta A., Gatesy J. (2008) Syst. Biol. 57, 15–37 [DOI] [PubMed] [Google Scholar]
- 33.Bartlett J. C. (2004) in Handbook of Proteolytic Enzymes (Barrett A., Rawlings N., Woessner J. eds) pp. 561–563, Academic Press, Amsterdam [Google Scholar]
- 34.Simmer J. P. (2004) in Handbook of Proteolytic Enzymes (Barrett A., Rawlings N., Woessner J. eds) pp. 1612–1613, Academic Press, Amsterdam [Google Scholar]
- 35.Bartlett J. D., Simmer J. P., Xue J., Margolis H. C., Moreno E. C. (1996) Gene 183, 123–128 [DOI] [PubMed] [Google Scholar]
- 36.Bègue-Kirn C., Krebsbach P. H., Bartlett J. D., Butler W. T. (1998) Eur. J. Oral Sci. 106, 963–970 [DOI] [PubMed] [Google Scholar]
- 37.Hu J. C., Sun X., Liu S., Zhang C., Bartlett J. D., Simmer J. P. (2002) Eur. J. Oral Sci. 110, 307–315 [DOI] [PubMed] [Google Scholar]
- 38.Ryu O. H., Fincham A. G., Hu C. C., Zhang C., Qian Q., Bartlett J. D., Simmer J. P. (1999) J. Dent. Res. 78, 743–750 [DOI] [PubMed] [Google Scholar]
- 39.Caterina J. J., Skobe Z., Shi J., Ding Y., Simmer J. P., Birkedal-Hansen H., Bartlett J. D. (2002) J. Biol. Chem. 277, 49598–49604 [DOI] [PubMed] [Google Scholar]
- 40.Bartlett J. D., Beniash E., Lee D. H., Smith C. E. (2004) J. Dent. Res. 83, 909–913 [DOI] [PubMed] [Google Scholar]
- 41.Turk B. E., Lee D. H., Yamakoshi Y., Klingenhoff A., Reichenberger E., Wright J. T., Simmer J. P., Komisarof J. A., Cantley L. C., Bartlett J. D. (2006) Biochemistry 45, 3863–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J. W., Simmer J. P., Hart T. C., Hart P. S., Ramaswami M. D., Bartlett J. D., Hu J. C. (2005) J. Med. Genet 42, 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozdemir D., Hart P. S., Ryu O. H., Choi S. J., Ozdemir-Karatas M., Firatli E., Piesco N., Hart T. C. (2005) J. Dent. Res. 84, 1031–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papagerakis P., Lin H. K., Lee K. Y., Hu Y., Simmer J. P., Bartlett J. D., Hu J. C. (2008) J. Dent. Res. 87, 56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmer J. P., Fukae M., Tanabe T., Yamakoshi Y., Uchida T., Xue J., Margolis H. C., Shimizu M., DeHart B. C., Hu C. C., Bartlett J. D. (1998) J. Dent. Res. 77, 377–386 [DOI] [PubMed] [Google Scholar]
- 46.Hu J. C., Ryu O. H., Chen J. J., Uchida T., Wakida K., Murakami C., Jiang H., Qian Q., Zhang C., Ottmers V., Bartlett J. D., Simmer J. P. (2000) J. Dent. Res. 79, 70–76 [DOI] [PubMed] [Google Scholar]
- 47.Fukae M., Tanabe T., Nagano T., Ando H., Yamakoshi Y., Yamada M., Simmer J. P., Oida S. (2002) J. Dent. Res. 81, 668–672 [DOI] [PubMed] [Google Scholar]
- 48.Ryu O., Hu J. C., Yamakoshi Y., Villemain J. L., Cao X., Zhang C., Bartlett J. D., Simmer J. P. (2002) Eur. J. Oral Sci. 110, 358–365 [DOI] [PubMed] [Google Scholar]
- 49.Matsumura M., Bhatt A. S., Andress D., Clegg N., Takayama T. K., Craik C. S., Nelson P. S. (2005) Prostate 62, 1–13 [DOI] [PubMed] [Google Scholar]
- 50.Takayama T. K., McMullen B. A., Nelson P. S., Matsumura M., Fujikawa K. (2001) Biochemistry 40, 15341–15348 [DOI] [PubMed] [Google Scholar]
- 51.Borgoño C. A., Gavigan J. A., Alves J., Bowles B., Harris J. L., Sotiropoulou G., Diamandis E. P. (2007) Biol. Chem. 388, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 52.Yoon H., Laxmikanthan G., Lee J., Blaber S. I., Rodriguez A., Kogot J. M., Scarisbrick I. A., Blaber M. (2007) J. Biol. Chem. 282, 31852–31864 [DOI] [PubMed] [Google Scholar]
- 53.Ramsay A. J., Dong Y., Hunt M. L., Linn M., Samaratunga H., Clements J. A., Hooper J. D. (2008) J. Biol. Chem. 283, 12293–12304 [DOI] [PubMed] [Google Scholar]
- 54.Mize G. J., Wang W., Takayama T. K. (2008) Mol. Cancer Res. 6, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 55.Nelson P. S., Gan L., Ferguson C., Moss P., Gelinas R., Hood L., Wang K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3114–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernando S. C., Buck J. S., Ashworth M. D., Ross J. W., Geisert R. D., DeSilva U. (2006) Reproduction 132, 939–947 [DOI] [PubMed] [Google Scholar]
- 57.Mangé A., Desmetz C., Berthes M. L., Maudelonde T., Solassol J. (2008) Biochem. Biophys. Res. Commun. 375, 107–112 [DOI] [PubMed] [Google Scholar]
- 58.Day C. H., Fanger G. R., Retter M. W., Hylander B. L., Penetrante R. B., Houghton R. L., Zhang X., McNeill P. D., Filho A. M., Nolasco M., Badaro R., Cheever M. A., Reed S. G., Dillon D. C., Watanabe Y. (2002) Oncogene 21, 7114–7120 [DOI] [PubMed] [Google Scholar]
- 59.Obiezu C. V., Soosaipillai A., Jung K., Stephan C., Scorilas A., Howarth D. H., Diamandis E. P. (2002) Clin. Chem. 48, 1232–1240 [PubMed] [Google Scholar]
- 60.Klokk T. I., Kilander A., Xi Z., Waehre H., Risberg B., Danielsen H. E., Saatcioglu F. (2007) Cancer Res. 67, 5221–5230 [DOI] [PubMed] [Google Scholar]
- 61.Gao J., Collard R. L., Bui L., Herington A. C., Nicol D. L., Clements J. A. (2007) Prostate 67, 348–360 [DOI] [PubMed] [Google Scholar]
- 62.Dong Y., Kaushal A., Bui L., Chu S., Fuller P. J., Nicklin J., Samaratunga H., Clements J. A. (2001) Clin. Cancer Res. 7, 2363–2371 [PubMed] [Google Scholar]
- 63.Obiezu C. V., Scorilas A., Katsaros D., Massobrio M., Yousef G. M., Fracchioli S., Rigault de la Longrais I. A., Arisio R., Diamandis E. P. (2001) Clin. Cancer Res. 7, 2380–2386 [PubMed] [Google Scholar]
- 64.Xi Z., Kaern J., Davidson B., Klokk T. I., Risberg B., Tropé C., Saatcioglu F. (2004) Gynecol. Oncol. 94, 80–85 [DOI] [PubMed] [Google Scholar]
- 65.Prezas P., Arlt M. J., Viktorov P., Soosaipillai A., Holzscheiter L., Schmitt M., Talieri M., Diamandis E. P., Krüger A., Magdolen V. (2006) Biol. Chem. 387, 807–811 [DOI] [PubMed] [Google Scholar]
- 66.Hart P. S., Hart T. C., Michalec M. D., Ryu O. H., Simmons D., Hong S., Wright J. T. (2004) J. Med. Genet. 41, 545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lobe C. G., Koop K. E., Kreppner W., Lomeli H., Gertsenstein M., Nagy A. (1999) Dev. Biol. 208, 281–292 [DOI] [PubMed] [Google Scholar]
- 68.Chai Y., Jiang X., Ito Y., Bringas P., Jr., Han J., Rowitch D. H., Soriano P., McMahon A. P., Sucov H. M. (2000) Development 127, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z., Song Y., Zhao X., Zhang X., Fermin C., Chen Y. (2002) Development 129, 4135–4146 [DOI] [PubMed] [Google Scholar]
- 70.Rosen D. (1981) Neuropathol. Appl. Neurobiol. 7, 331–340 [DOI] [PubMed] [Google Scholar]
- 71.Young D. C., Kingsley S. D., Ryan K. A., Dutko F. J. (1993) Anal. Biochem. 215, 24–30 [DOI] [PubMed] [Google Scholar]
- 72.Simmer J. P., Lau E. C., Hu C. C., Aoba T., Lacey M., Nelson D., Zeichner-David M., Snead M. L., Slavkin H. C., Fincham A. G. (1994) Calcif. Tissue Int. 54, 312–319 [DOI] [PubMed] [Google Scholar]
- 73.Hu C. C., Simmer J. P., Bartlett J. D., Qian Q., Zhang C., Ryu O. H., Xue J., Fukae M., Uchida T., MacDougall M. J. (1998) Connect. Tissue Res. 39, 47–61 [DOI] [PubMed] [Google Scholar]
- 74.Park C. H., Abramson Z. R., Taba M., Jr., Jin Q., Chang J., Kreider J. M., Goldstein S. A., Giannobile W. V. (2007) J. Periodontol. 78, 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss D. J., Liggitt D., Clark J. G. (1999) Histochem. J. 31, 231–236 [DOI] [PubMed] [Google Scholar]
- 76.Simmer J. P., Sun X., Yamada Y., Zhang C. H., Bartlett J. D., Hu J. C. (2004) in Biomineralization, Formation, Diversity, Evolution and Application (Kobayashi I., Ozawa H. eds) pp. 348–352, Tokai University Press, Hadano, Japan [Google Scholar]
- 77.Sognnaes R. F. (1949) J. Dent. Res. 28, 549–557 [DOI] [PubMed] [Google Scholar]
- 78.Boyde A. (1989) in Handbook of Microscopic Anatomy, Teeth (Oksche A., Vollrath L. eds) pp. 309–473, Springer-Verlag, Berlin [Google Scholar]
- 79.Smith C. E., McKee M. D., Nanci A. (1987) Adv. Dent. Res. 1, 162–175 [DOI] [PubMed] [Google Scholar]
- 80.Sasaki S., Takagi T., Suzuki M. (1991) Arch. Oral Biol. 36, 227–231 [DOI] [PubMed] [Google Scholar]
- 81.Smith C. E., Issid M., Margolis H. C., Moreno E. C. (1996) Adv. Dent. Res. 10, 159–169 [DOI] [PubMed] [Google Scholar]
- 82.Toyosawa S., Ogawa Y., Inagaki T., Ijuhin N. (1996) Cell Tissue Res. 285, 217–225 [DOI] [PubMed] [Google Scholar]
- 83.Lyaruu D. M., Bronckers A. L., Mulder L., Mardones P., Medina J. F., Kellokumpu S., Oude Elferink R. P., Everts V. (2008) Matrix Biol. 27, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright J. T., Kiefer C. L., Hall K. I., Grubb B. R. (1996) J. Dent. Res. 75, 966–973 [DOI] [PubMed] [Google Scholar]
- 85.Simmer J. P., Fincham A. G. (1995) Crit. Rev. Oral Biol. Med. 6, 84–108 [DOI] [PubMed] [Google Scholar]
- 86.Robinson C., Kirkham J., Stonehouse N. J., Shore R. C. (1989) Connect. Tissue Res. 22, 139–145 [PubMed] [Google Scholar]
- 87.Robinson C., Hallsworth A. S., Shore R. C., Kirkham J. (1990) Caries Res. 24, 226–230 [DOI] [PubMed] [Google Scholar]
- 88.Sun Z., Fan D., Fan Y., Du C., Moradian-Oldak J. (2008) J. Dent. Res. 87, 1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukae M., Yamamoto R., Karakida T., Shimoda S., Tanabe T. (2007) J. Dent. Res. 86, 758–763 [DOI] [PubMed] [Google Scholar]