Abstract
The kinetochore, which consists of centromere DNA and structural proteins, is essential for proper chromosome segregation in eukaryotes. In budding yeast, Sgt1 and Hsp90 are required for the binding of Skp1 to Ctf13 (a component of the core kinetochore complex CBF3) and therefore for the assembly of CBF3. We have previously shown that Sgt1 dimerization is important for this kinetochore assembly mechanism. In this study, we report that protein kinase CK2 phosphorylates Ser361 on Sgt1, and this phosphorylation inhibits Sgt1 dimerization.
The kinetochore is a structural protein complex located in the centromeric region of the chromosome coupled to spindle microtubules (1, 2). The kinetochore generates a signal to arrest cells during mitosis when it is not properly attached to microtubules, thereby preventing chromosome missegregation, which can lead to aneuploidy (3, 4). The molecular structure of the kinetochore complex of the budding yeast Saccharomyces cerevisiae has been well characterized; it is composed of more than 70 proteins, many of which are conserved in mammals (2).
The centromere DNA in the budding yeast is a 125-bp region that contains three conserved regions, CDEI, CDEII, and CDEIII (5, 6). CDEIII (25 bp) is essential for centromere function (7) and is bound to a key component of the centromere, the CBF3 complex. The CBF3 complex contains four proteins, Ndc10, Cep3, Ctf13 (8–15), and Skp1 (14, 15), all essential for viability. Mutations in any of the CBF3 proteins abolish the ability of CDEIII to bind to CBF3 (16, 17). All of the kinetochore proteins, except the CDEI-binding Cbf1 (18–20), localize to the kinetochores in a CBF3-dependent manner (2). Thus, CBF3 is a fundamental kinetochore complex, and its mechanism of assembly is of great interest.
We have previously found that Sgt1 and Skp1 activate Ctf13; thus, they are required for assembly of the CBF3 complex (21). The molecular chaperone Hsp90 is also required to form the active Ctf13-Skp1 complex (22). Sgt1 has two highly conserved motifs that are required for protein-protein interaction: the tetratricopeptide repeat (21) and the CHORD protein and Sgt1-specific motif. We and others have found that both domains are important for the interaction of Sgt1 with Hsp90 (23–26), which is required for assembly of the core kinetochore complex. This interaction is an initial step in kinetochore activation (24, 26, 27), which is conserved between yeast and humans (28, 29).
We have recently shown that Sgt1 dimerization is important for Sgt1-Skp1 binding and therefore for kinetochore assembly (30). In this study, we have found that protein kinase CK2 phosphorylates Sgt1 at Ser361, and this phosphorylation inhibits Sgt1 dimerization. Therefore, CK2 appears to regulate kinetochore assembly negatively in budding yeast.
EXPERIMENTAL PROCEDURES
Yeast Strains and Medium
Table 1 lists the genotypes of yeast strains used in this study. The medium for yeast growth and sporulation was prepared using previously described methods (31). Yeast transformation was done according to the method of Ito et al. (32). Strains that expressed tagged proteins were generated according to the procedure of Longtine et al. (33). Regions that encoded Myc tags were inserted at the 3′-end of the endogenous locus.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Referencea |
|---|---|---|
| YPH499 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 49 |
| Y1681 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 pRS414-3myc-SGT1 pRS416-3HA-SGT1 | |
| Y1682 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 pRS414-3myc-sgt1-S361A pRS416-3HA-sgt1-S361A | |
| Y1684 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 pRS414-3myc-sgt1-S361D pRS416-3HA-sgt1-S361D | |
| Y1686 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-S361A:LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | |
| Y1687 | Matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-S361D:LEU2 pRS416-SGT1 URA/CEN | |
| Y1734 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CKA1-myc | |
| Y1736 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cka1Δ ::His3MX6 PRS414-3HA-Sgt1 pRS415-3myc-Sgt1 | |
| Y26 | Matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 SGT1:LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | |
| CDY26 | MATaANB1-UB-R-lacI-4HA-SGT1 ::kan pACE1-UBR1 pACE1-ROX1 trp1-Δ 1 ura3-Δ 52 leu2 ::PET56 ade2-101 | 25 |
| YKK54 | Matα ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 sgt1-3:LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | 21 |
| Y1773 | Matα ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 cka1Δ ::His3MX6 CFIII(CEN3.L.YPH983) TRP1 SUP11 | |
| Y1775 | Mata ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 cka1Δ ::His3MX6 sgt1-3:LEU2 | |
| Y1870 | Matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1Δ ::HIS3-MX6 pRS415-3myc-Sgt1 | |
| Y1871 | Mat α ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1Δ ::HIS3-MX6 pRS415-3myc-sgt1-S361A | |
| Y1872 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 pRS415-3myc-Sgt1 | |
| Y1873 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cka1Δ ::His3MX6 pRS415-3myc-Sgt1 | |
| Y1874 | Mata ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cka1Δ ::His3MX6 pRS416-3HA-sgt1-S361A, pRS414-3myc-sgt1-S361A |
a Strains for which no reference is given were generated during this study.
Plasmid Construction and Primers
Table 2 lists the plasmids used in this study. Details about their construction (34) and primer sequences are available upon request.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Referencea |
|---|---|---|
| B1354 | pRS416-3HA-SGT1 URA CEN | 30 |
| B180 | pRS414-3HA-SGT1 TRP CEN | 30 |
| B1367 | pRS415-3myc-SGT1 LEU CEN | 30 |
| B940 | pDEST17-HIS-SGT1 | 30 |
| B942 | pDEST15-GST-SGT1 | |
| B1657 | pDEST15-GST-sgt1-S361A | |
| B824 | pDEST15-GST-hSGT1 | |
| BPB452 | pDEST15-GST-hsgt1A-S299A | |
| B191 | pRS414-3myc-SGT1 TRP CEN | 30 |
| B1374 | pRS414-3myc-sgt1-S361A TRP CEN | |
| B1375 | pRS414-3myc-sgt1-S361D TRP CEN | |
| B1377 | pRS416-3HA-sgt1-S361A URA CEN | |
| B1378 | pRS416-3HA-sgt1-S361D URA CEN | |
| B1699 | pRS415-3myc-sgt1-S361A LEU CEN |
a Plasmids for which no reference is given were generated during this study.
Antibodies
Anti-Skp1, anti-Sgt1, and anti-Hsp82 antibodies were used as previously described (21, 24, 35). Anti-hemagglutinin (HA2; Roche Applied Science), anti-Myc (Roche Applied Science), anti-GST (Abcam), and anti-His6 (Qiagen) antibodies were purchased.
Protein Expression and Immunoprecipitation
Immunoprecipitation using yeast lysates was performed as described previously (24). His6-Sgt1 and GST-Sgt1 proteins were expressed and purified according to the manufacturer's instructions, as previously described (24).
Two-dimensional Gel Electrophoresis
Myc-tagged Sgt1 was immunoprecipitated from yeast cell lysates using an anti-Myc antibody. Isoelectric focusing was performed with a 17-cm immobilized pH 3–10 gradient strips (Bio-Rad), following the manufacturer's instructions. Gel electrophoresis was performed in a Bio-Rad PROTEAN Plus Dodeca cell. After two-dimensional gel electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane, and immunoblotting was performed using anti-Myc antibodies.
CK2 Phosphorylation Assay
Phosphorylation of recombinant GST-Sgt1 or His-Sgt1 by human protein kinase CK2 was performed in a reaction mixture containing 100 ng to 1 μg of GST-Sgt1 or His-Sgt1 protein and 2 μCi of [γ-32P]ATP in 30 μl of reaction buffer (20 mm HEPES (pH 7.5), 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol, 1 mm CaCl2, and 0.2 mm ATP). The reactions were started by adding 250 units of CK2 (New England Biolabs) and incubated for 30 min at 30 °C. Reactions were stopped by adding 10 μl of 4× SDS buffer. Reaction samples were separated on a 4–15% SDS-polyacrylamide gel and stained with SYPRO Ruby protein stain (Molecular Probes). After images were taken, the gels were dried and autoradiographed. Phosphorylation signals were quantified by a PhosphorImager.
RESULTS
Phosphorylation of Ser361 on Sgt1
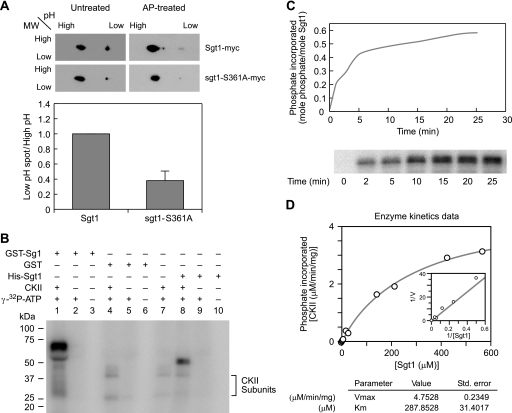
Matrix-assisted laser desorption time-of-flight mass spectrometry revealed that an oligopeptide, 348AGADPDTKRAMMKSF362, is phosphorylated in vivo (Fig. S1). Serine 361 is within the consensus site that is phosphorylated by CK2 (36), and this residue is conserved in humans (see Fig. 2B). Therefore, we tested by two-dimensional gel electrophoresis whether Ser361 is phosphorylated in vivo. Immunoprecipitated Sgt1-Myc appeared as two major spots on the two-dimensional gel (Fig. 1A, top left), and phosphatase treatment substantially diminished the low pH spot (Fig. 1A, top right), indicating that the low pH spot corresponds to phosphorylated forms. An unphosphorylated mutant sgt1-S361A protein showed a reduced low pH spot, indicating that Ser361 is phosphorylated in vivo (Fig. 1A, bottom left). Phosphatase treatment reduced the low pH spot of sgt1-S361A protein, suggesting that there are other phosphorylation sites.
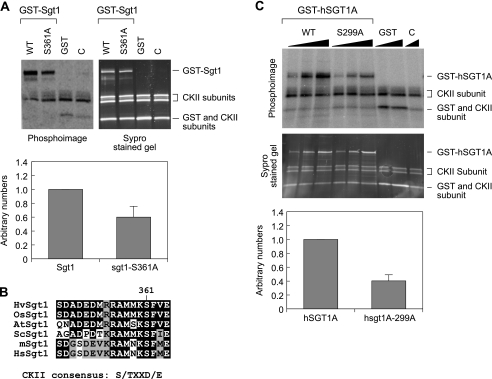
FIGURE 2.
A, CK2 phosphorylates Sgt1 at S361 in vitro. Top, 100 ng of recombinant GST-Sgt1 and GST-sgt1-S361A mutant proteins were used for the in vitro kinase assay (left, a phosphor image; right, a SYPRO Ruby-stained image). Bottom, phosphorylation signals were quantified by a PhosphorImager. Input Sgt1 (top bands on the gel) in the reaction was quantified from the SYPRO Ruby-stained gel image, and the phosphorylation signals were normalized. GST-Sgt1 was given the arbitrary value of 1. Numbers on the y axis are arbitrary. Signals were quantified from three independent experiments. C, a negative control without substrates. B, Sgt1 Ser361 is conserved from yeast to humans. A short peptide sequence around Ser361 in yeast Sgt1 is aligned with the amino acid sequence of the corresponding region in Sgt1 homologs of several other organisms, as indicated: barley (HvSgt1), rice (OsSgt1), Arabidopsis (AtSgt1), S. cerevisiae (ScSgt1), mouse (mSgt1), and human (HsSgt1). C, human SGT1A is phosphorylated by CK2 in vitro. Recombinant GST-hSGT1A, GST-hSGT1A-S299A (hSGT1A-S299 corresponds to yeast Sgt1-S361), and GST alone were used as substrate for the CK2-dependent in vitro kinase assay. WT, wild type.
FIGURE 1.
Sgt1 is phosphorylated at serine 361. A (top), Sgt1 is phosphorylated at Ser361 in vivo. The 3Myc-Sgt1 (Y1870) or 3Myc-sgt1-S361A (Y1871) was immunoprecipitated from cycling cells, and immunoprecipitates were treated with alkaline phosphatase (AP-treated). Immunoprecipitates were analyzed by two-dimensional gel electrophoresis. An anti-Myc antibody was used to detect Sgt1-Myc on the immunoblots. (bottom) The signal intensity was quantified, and the ratio of phosphorylated Sgt1 (toward low pH) to unphosphorylated Sgt1 (toward high pH) in untreated immunoprecipitates is shown (gray bars). B, recombinant human CK2 phosphorylates Sgt1 in vitro. Kinase activity was determined by assaying phosphorylation levels of GST-Sgt1 (lanes 1–3), GST only (lanes 4–6), or His-Sgt1 (lanes 8–10), incubated with [γ-32P]ATP, as described under “Experimental Procedures.” The arrowhead indicates autophosphorylated CK2. C, CK2-mediated phosphorylation of Sgt1. His-Sgt1 (1.6 pmol) was mixed in the reaction buffer with CK2 (710 pmol) and incubated with 0.5 μCi of [γ-32P]ATP for the indicated time periods at 30 °C. The protein was then analyzed by SDS-PAGE. Phosphorylated Sgt1 bands were quantified with a STORM 860 PhosphorImager and ImageQuant software (Amersham Biosciences), and data were converted into pmol units of phosphate relative to that of the control [γ-32P]ATP. The level of Sgt1 phosphorylation was calculated by dividing the pmol units of phosphate by the pmol units of Sgt1 used in the reaction. Bottom, the phosphor image of phosphorylated Sgt1. D, kinetics of Sgt1 phosphorylation by CK2. Reactions were performed in 15 μl of solution containing 20 mm HEPES (pH 7.5), 50 mm NaCl, 10 mm MgCl2, 0.2 mm ATP, 0.5 μCi of [γ-32P]ATP, 1 mm dithiothreitol, 1 mm CaCl2, 35 units of CK2, and varying concentrations of recombinant His-Sgt1 at 30 °C. Reactions were stopped after 10 min with SDS buffer, and proteins were resolved by SDS-PAGE. Phosphorylated Sgt1 was quantified using a STORM 860 PhosphorImager and ImageQuant software (Amersham Biosciences). Bottom, the kinetic properties Vmax and Km of Sgt1 phosphorylation were determined from Michaelis-Menten plots using GraFit (Erithacus Software Ltd.).
We also found that GST-Sgt1 and His6-Sgt1 were phosphorylated by human CK2 in vitro (Fig. 1B and Figs. S2 and S3). Sgt1 was phosphorylated with 0.6 mol of phosphate/mol of substrate (Fig. 1C). The Km and Vmax of CK2 for the phosphorylation of Sgt1 were 288 μm and 4.8 μmol/min/mg (Fig. 1D), respectively, which are within the same range as that for known CK2 substrates (37–39), indicating that Sgt1 is a suitable substrate for CK2 in vitro.
CK2 did not efficiently phosphorylate the sgt1-S361A mutant protein (Fig. 2A), indicating that the S361A on Sgt1 is a phosphorylation site for CK2 in vitro.
Serine 361 is within the consensus site that is phosphorylated by CK2 (Fig. 2B), and this residue is conserved in humans and corresponds to serine 299 in human SGT1A (Fig. 2B). Nowotny et al. (40) reported that an oligopeptide containing this serine residue, human SGT1A-(263–333), is phosphorylated by CK2 in vitro. A search at PhosphoBase (41) revealed that in human SGT1A-(263–333), residue Ser299 can be phosphorylated by CK2. Human SGT1A-(263–333) oligopeptide containing the E302K mutation was also efficiently phosphorylated, suggesting that the CK2 phosphorylation site might be different from that identified by computer prediction. However, whether Ser299 is a CK2 phosphorylation site has not been directly tested. Therefore, we made the human SGT1A-S299A protein and found that CK2 did not phosphorylate the mutant protein as efficiently as the wild-type protein (Fig. 2C). This finding strongly suggests that Ser299 is a CK2 phosphorylation site in vitro.
Inhibition of Sgt1 Dimerization by Phosphorylation of Ser361
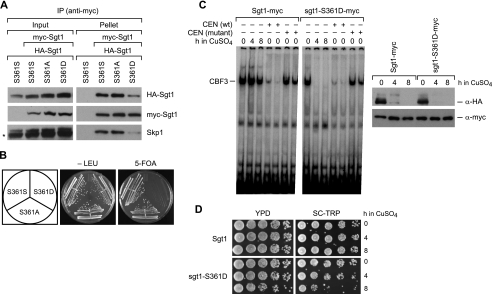
To examine the possible effect of phosphorylation of Ser361, we generated a nonphosphorylated mutant (S361A) strain and a phosphorylation-mimic mutant (S361D) strain. The S361A mutant protein bound to itself and Skp1 in vivo (Fig. 3A). However, the S361D protein did not bind to either itself or Skp1 efficiently, suggesting that Ser361-phosphorylated Sgt1 cannot form dimers effectively. Interestingly, the sgt1-S361D but not sgt1-S361A mutation was lethal when expressed in vivo as the sole version of Sgt1 (Fig. 3B). These results suggest that the phosphorylation of Ser361 inhibits Sgt1 dimerization, Sgt1 dimerization is essential for viability, and thus that CK2 negatively regulates Sgt1 dimerization.
FIGURE 3.
Phenotypes of the sgt1-S361D mutant. A, mimic of Sgt1 phosphorylation at Ser361 inhibits Sgt1-Sgt1 and Sgt1-Skp1 binding. Sgt1-S361S (wild type), Sgt1-S361A, or sgt1-S361D was expressed as HA-tagged and Myc-tagged proteins from two different plasmids (pRS416-HA and pRS414-Myc). Sgt1-S361S was expressed in the Y1681 strain, Sgt1-S361A in the Y1682 strain, and sgt1-S361D in the Y1684 strain. (All three strains were derived from the YPH499 strain.) TheYPH499 strain carrying only pRS416-HA-SGT1-S361S was used as a negative control (leftmost lane). B, mimic of Sgt1 phosphorylation at Ser361 is lethal. Yeast cells that were haploid for SGT1 (Y26), sgt1-S361A (Y1686), or sgt1-S361D carrying SGT1 on a URA/CEN plasmid (Y1687) were streaked onto plates that contained Sc+ 5-fluoroorotic acid (5-FOA) or Sc-Leu. Plates were incubated at 25 °C for 3 days. C (right), the addition of 0.5 mm CuSO4 to strain CDY26 (N-degron-Sgt1) induced the proteolysis of N-degron-4HA-Sgt1 after either a 4- or 8-h incubation with CuSO4, as determined by immunoblotting with anti-HA antibodies, but Myc-tagged Sgt1 or sgt1-S361D was not affected. Left, protein extracts of the indicated strains were collected at the indicated time points after adding CuSO4. Cell extracts (40 μg) were subjected to the band shift assay to examine the CBF3 assembly activity. Competitor CEN DNA (100 or 200 μg) or mutant CEN DNA (CCG motif at CDEIII region is mutated to CCC) was added to CuSO4-untreated extracts of each cell type. D, survival of cells after a 4- or 8-h incubation with 0.5 mm CuSO4, as determined by a dilution-spotting assay. The numbers of cells that were spotted onto the indicated plates were ∼1.25 × 106, 2.5 × 105, 5 × 104, 1 × 104, and 2 × 103. Plates were incubated at 30 °C for 2 days. IP, immunoprecipitation.
To confirm this hypothesis in vivo, we examined whether the CBF3 complex could be formed in sgt1-S361D mutant cells. Because the sgt1-S361D mutation is lethal, we used a conditional SGT1-null mutant by using N-degron-4HA-Sgt1 expressed under the control of a repressible promoter (25) to remove the N-degron-wild-type protein but not the Myc-tagged mutant protein from cells that were used for the CBF3 band shift assay (8). In this system, copper induces the expression of Ubr1 and Rox1. Ubr1 is a ubiquitin ligase that binds proteins containing an N-degron and facilitates their ubiquitination and subsequent destruction by the 26 S proteasome. Rox1 represses the transcription of the ANB1 promoter controlling the expression of N-degron-Sgt1. Thus, the addition of copper both blocks the expression of N-degron-Sgt1 and triggers its proteolysis. The levels of N-degron-Sgt1 decreased after CuSO4 was added and were substantially lower after a 4-h incubation, but the levels of Myc-tagged Sgt1 and Myc-tagged sgt1-S361D mutant protein did not change (Fig. 3C, right). The cells in which Sgt1-Myc or sgt1-S361D-Myc were expressed and N-degron-Sgt1 was depleted were used for the CBF3 band shift assay. Although the viability after an 8-h incubation was substantially reduced, that after a 4-h incubation appeared to be high enough to assess CBF3 activity (Fig. 3D). The CBF3 complex was not formed efficiently in cell lysates containing only sgt1-S361D-Myc (Fig. 3C, left), suggesting that the phosphorylation of Ser361 inhibits the formation of the CBF3 complex.
CK2 Phosphorylates Sgt1 in Vivo
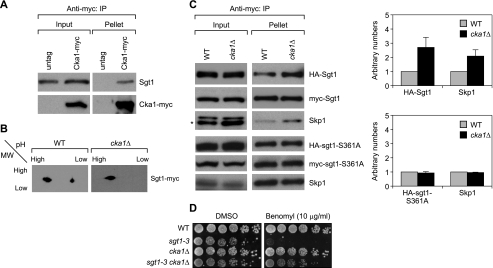
Next, we examined whether Sgt1 is phosphorylated by CK2. We found that Sgt1 interacted with Cka1 (a subunit of yeast CK2) in vivo (Fig. 4A) and that the low pH spot diminished in cka1Δ (Fig. 4B). These results strongly suggest that CK2 phosphorylates Sgt1 in vivo.
FIGURE 4.
CK2 phosphorylates Sgt1 in vivo. A, Cka1, a subunit of CK2, associates with Sgt1. Cell lysates obtained from cycling untagged wild-type (untag; strain YPH499) and Cka1-Myc (strain Y1734) cells were immunoprecipitated by using anti-Myc antibodies. Sgt1 was detected with anti-Sgt1 antibodies, and Cka1-Myc was detected with anti-Myc antibodies. An untagged strain was used as a negative control. B, phosphorylated Sgt1 diminished in cka1Δ cells. 3Myc-Sgt1 was immunoprecipitated from wild-type (WT; Y1872) cells or cka1Δ cells (cka1Δ; Y1873) harboring 3Myc-Sgt1. Immunoprecipitates were analyzed by two-dimensional gel electrophoresis, and immunoblotting was performed by using anti-Myc antibodies. C (top left panels), Sgt1 dimerization increases in cka1Δ cells. Immunoprecipitation was carried out on cell lysates from wild-type (WT; Y1681) cells or cka1Δ cells (cka1Δ; Y1736) harboring 3HA-Sgt1 and 3Myc-Sgt1. Bottom left panels, Sgt1 dimerization increases in cka1Δ cells, which is dependent on serine 361. Data are from experiments similar to those above, except that cell lysates were prepared from cells expressing HA-sgt1-S361A and Myc-sgt1-S361A in wild-type (Y1682) and cka1Δ cells (cka1Δ; Y1874). HA-tagged Sgt1 (wild type or mutant) was identified by anti-HA antibodies, and Myc-tagged Sgt1 (wild type or mutant) was identified by anti-Myc antibodies. Right panels, signals of HA-Sgt1 and Skp1 in Myc-Sgt1 immunoprecipitates were quantified from two independent experiments. Quantification was performed by giving an arbitrary value of 1 to HA-Sgt1 or Skp1 in the wild-type cells. D, the benomyl sensitivity of the sgt1–3 mutant was suppressed by the deletion of cka1. The indicated strains (wild type (YPH499), sgt1–3 (YKK54), cka1Δ (Y1773), and sgt1–3 cka1Δ (Y1775)) were grown on yeast extract-peptone-dextrose plates containing 10 μg/ml benomyl or DMSO. The numbers of cells that were spotted onto each plate (left to right) were ∼1.25 × 106, 2.5 × 105, 5 × 104, 1 × 104, 2 × 103, and 4 × 102. Plates were incubated at 30 °C for 3 days. IP, immunoprecipitation.
The phosphorylation of Ser361 appears to negatively regulate Sgt1 dimerization; thus, if Ser361 is phosphorylated by CK2 in vivo, then the amount of Sgt1 dimers should be greater in yeast casein-kinase mutants than in wild-type cells. Indeed, Sgt1 dimerization increased in cka1Δ mutant cells (Fig. 4C), and Skp1 binding also increased (Fig. 4C). Furthermore, sgt1-S361A dimerization or Skp1 binding did not increase in cka1Δ mutant cells, indicating that the stimulation of dimerization by reduction of CK2 activity is dependent on Ser361 phosphorylation (Fig. 4C). Consistent with these results, the deletion of cka1 suppressed the benomyl sensitivity of the sgt1–3 mutant (Fig. 4D) (21, 30). These results strongly suggest that CK2 phosphorylates Sgt1 at Ser361 and negatively regulates Sgt1 dimerization in vivo.
DISCUSSION
In this study, we have shown that Sgt1 dimerization, which is required for kinetochore assembly in budding yeast, is regulated negatively by CK2-mediated phosphorylation.
CK2 Negatively Regulates Sgt1 Dimerization
CK2 is a serine/threonine protein kinase that is ubiquitous in eukaryotes (42, 43). The enzyme is composed of a catalytic subunit a and a regulatory subunit b that form a native a2b2 holoenzyme (42, 43). CK2 of the budding yeast consists of two catalytic subunits, a and a′, and two regulatory subunits, b and b′, which are encoded by the CKA1, CKA2, CKB1, and CKB2 genes, respectively (44). Individual deletion of either CKA1 or CKA2 does not show any significant phenotypes, but their simultaneous deletion is lethal (45).
To demonstrate that Ser361 is phosphorylated by CK2 in vivo, we attempted to generate antibodies against phosphorylated Ser361. However, unfortunately, antibodies produced using a phosphorylated peptide were not specific for the phosphorylated form and recognized the unphosphorylated form even after affinity purification. Therefore, we used indirect approaches to answer this question. Two-dimensional gel analysis revealed that Sgt1 is phosphorylated by CK2. When CK2 activity was altered by deletion of Cka1 but not Cka2 or Ckb2, Sgt1 dimerization and Skp1 binding were stimulated (data not shown). It has been previously reported that different combinations of subunits exhibit properties typical for CK2 but differ in substrate specificity and sensitivity to inhibitors, which suggests that each CK2 isomer may regulate different processes or may have different mechanisms of regulation (46). Thus, the regulation of Sgt1 dimerization may be a specific function of Cka1.
Because the sgt1-S361D mutant is lethal, we had to perform coimmunoprecipitation experiments to test the dimerization activity of the sgt1 mutant proteins in the presence of endogenous wild-type Sgt1 (Fig. 3A). Since the sgt1-S361D protein presumably binds more efficiently to the wild-type Sgt1 than to sgt1-S361D, the dimerization activity detected by co-immunoprecipitation might be an underestimation.
The stimulation of Sgt1 dimerization by deletion of Cka1 was dependent on Ser361. The alteration of CK2 activity by deletion of Cka1 suppressed the benomyl sensitivity of the sgt1–3 mutant, and Sgt1 associated with Cka1 in vivo. Therefore, we conclude that CK2 negatively regulates Sgt1 dimerization by phosphorylating Ser361, which in turn represses kinetochore assembly. However, the biological significance of why Sgt1 dimerization needs to be negatively regulated is unknown and needs further analyses. A monomer form might have a different function.
Another interesting observation is that Ser361 is outside the Sgt1 dimerization domain but within the SGS domain (amino acids 307–395). Our binding analysis suggested that an inhibitory region may be present between amino acids 270 and 312 (30). The phosphorylation of Ser361 may enhance this internal inhibitory activity by affecting the structure of the SGS domain.
Highly Conserved Kinetochore Function of Sgt1
The human homolog of Sgt1 can rescue the yeast sgt1-null mutant (21), suggesting that the function of Sgt1 is conserved between humans and yeast. Sgt1 interacts with Hsp90 in yeast and human cells (28, 47). CENP-I, CENP-F, and Hec1, but not CENP-C, are absent from kinetochores in human cells depleted of SGT1 or HSP90 or treated with 17-AAG (an HSP90 inhibitor) (28, 29). This result strongly suggests that the human SGT1-HSP90 complex is crucial for assembly of the human kinetochore. Therefore, the kinetochore function of Sgt1 is evolutionarily conserved between humans and yeast.
However, Nyarko et al. (48) have shown by gel filtration chromatography that human SGT1A does not form dimers efficiently. Our recent analytical ultracentrifuge experiments have also revealed that human SGT1A exists mainly as a monomer (30). Further studies are required to determine whether human SGT1 is phosphorylated by CK2 in vivo and, if so, what role phosphorylation of this protein plays.
Supplementary Material
Acknowledgments
We thank V. Measday and R. Kitagawa for helpful comments, members of the Kitagawa laboratory for stimulating conversation and advice, V. Pagala and X. Ding for two-dimensional gel electrophoresis, D. Huang and J. Easton for kinetics experiments, and Vani Shanker for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Cancer Center Support Grant CA21765 and National Institutes of Health Grant GM68418. This work was also supported by the American Lebanese Syrian Association Charities.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- HA
- hemagglutinin
- GST
- glutathione S-transferase.
REFERENCES
- 1.Cheeseman I. M., Desai A. (2008) Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 2.Westermann S., Drubin D. G., Barnes G. (2007) Annu. Rev. Biochem. 76, 563–571 [DOI] [PubMed] [Google Scholar]
- 3.Sudakin V., Yen T. J. (2007) Biodrugs 21, 225–233 [DOI] [PubMed] [Google Scholar]
- 4.Weaver B. A., Silk A. D., Montagna C., Verdier-Pinard P., Cleveland D. W. (2007) Cancer Cell 11, 25–36 [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald-Hayes M., Clarke L., Carbon J. (1982) Cell 29, 235–244 [DOI] [PubMed] [Google Scholar]
- 6.Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. (1985) Cell 42, 913–921 [DOI] [PubMed] [Google Scholar]
- 7.Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P. (1988) Mol. Cell. Biol. 8, 2523–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lechner J., Carbon J. (1991) Cell 64, 717–725 [DOI] [PubMed] [Google Scholar]
- 9.Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., Hieter P. (1993) Cell 73, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh P. Y., Kilmartin J. V. (1993) J. Cell Biol. 121, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang W., Lechner J., Carbon J. (1993) J. Cell Biol. 121, 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lechner J. (1994) EMBO J. 13, 5203–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strunnikov A. V., Kingsbury J., Koshland D. (1995) J. Cell Biol. 128, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connelly C., Hieter P. (1996) Cell 86, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stemmann O., Lechner J. (1996) EMBO J. 15, 3611–3620 [PMC free article] [PubMed] [Google Scholar]
- 16.Sorger P. K., Doheny K. F., Hieter P., Kopski K. M., Huffaker T. C., Hyman A. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92, 12026–12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan K. B., Hyman A. A., Sorger P. K. (1997) Cell 91, 491–500 [DOI] [PubMed] [Google Scholar]
- 18.Baker R. E., Masison D. C. (1990) Mol. Cell. Biol. 10, 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai M., Davis R. W. (1990) Cell 61, 437–446 [DOI] [PubMed] [Google Scholar]
- 20.Cai M. J., Davis R. W. (1989) Mol. Cell. Biol. 9, 2544–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa K., Skowyra D., Elledge S. J., Harper J. W., Hieter P. (1999) Mol. Cell 4, 21–33 [DOI] [PubMed] [Google Scholar]
- 22.Stemmann O., Neidig A., Köcher T., Wilm M., Lechner J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azevedo C., Sadanandom A., Kitagawa K., Freialdenhoven A., Shirasu K., Schulze-Lefert P. (2002) Science 295, 2073–2076 [DOI] [PubMed] [Google Scholar]
- 24.Bansal P. K., Abdulle R., Kitagawa K. (2004) Mol. Cell. Biol. 24, 8069–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubacq C., Guerois R., Courbeyrette R., Kitagawa K., Mann C. (2002) Eukaryot. Cell 1, 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingelbach L. B., Kaplan K. B. (2004) Mol. Cell. Biol. 24, 8938–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catlett M. G., Kaplan K. B. (2006) J. Biol. Chem. 281, 33739–33748 [DOI] [PubMed] [Google Scholar]
- 28.Niikura Y., Ohta S., Vandenbeldt K. J., Abdulle R., McEwen B. F., Kitagawa K. (2006) Oncogene 25, 4133–4146 [DOI] [PubMed] [Google Scholar]
- 29.Steensgaard P., Garrè M., Muradore I., Transidico P., Nigg E. A., Kitagawa K., Earnshaw W. C., Faretta M., Musacchio A. (2004) EMBO Rep. 5, 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bansal P. K., Nourse A., Abdulle R., Kitagawa K. (2009) J. Biol. Chem. 284, 3586–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose M. D., Winston F., Hieter P. (1990) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 32.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 34.Kitagawa K., Abdulle R. (2002) BioTechniques 33, 288–292 [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa K., Abdulle R., Bansal P. K., Cagney G., Fields S., Hieter P. (2003) Mol. Cell 11, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 36.Pinna L. A. (1990) Biochim. Biophys. Acta 1054, 267–284 [DOI] [PubMed] [Google Scholar]
- 37.Carmichael D. F., Geahlen R. L., Allen S. M., Krebs E. G. (1982) J. Biol. Chem. 257, 10440–10445 [PubMed] [Google Scholar]
- 38.Fritz G., Kaina B. (1999) Oncogene 18, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 39.Mckenzie J. A., Strauss P. R. (2003) Anal. Biochem. 313, 9–16 [DOI] [PubMed] [Google Scholar]
- 40.Nowotny M., Spiechowicz M., Jastrzebska B., Filipek A., Kitagawa K., Kuznicki J. (2003) J. Biol. Chem. 278, 26923–26928 [DOI] [PubMed] [Google Scholar]
- 41.Kreegipuu A., Blom N., Brunak S. (1999) Nucleic Acids Res. 27, 237–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinna L. A., Meggio F. (1997) Prog. Cell Cycle Res. 3, 77–97 [DOI] [PubMed] [Google Scholar]
- 43.Poole A., Poore T., Bandhakavi S., McCann R. O., Hanna D. E., Glover C. V. (2005) Mol. Cell Biochem. 274, 163–170 [DOI] [PubMed] [Google Scholar]
- 44.Glover C. V., III (1998) Prog. Nucleic Acids Res. Mol. Biol. 59, 95–133 [DOI] [PubMed] [Google Scholar]
- 45.Padmanabha R., Chen-Wu J. L., Hanna D. E., Glover C. V. (1990) Mol. Cell. Biol. 10, 4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domañska K., Zieliñski R., Kubiñski K., Sajnaga E., Masłyk M., Bretner M., Szyszka R. (2005) Acta Biochim. Pol. 52, 947–951 [PubMed] [Google Scholar]
- 47.Lee Y. T., Jacob J., Michowski W., Nowotny M., Kuznicki J., Chazin W. J. (2004) J. Biol. Chem. 279, 16511–16517 [DOI] [PubMed] [Google Scholar]
- 48.Nyarko A., Mosbahi K., Rowe A. J., Leech A., Boter M., Shirasu K., Kleanthous C. (2007) Biochemistry 46, 11331–11341 [DOI] [PubMed] [Google Scholar]
- 49.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.