Abstract
Elucidating pore function at the 3-fold channels of 12-subunit, microbial Dps proteins is important in understanding their role in the management of iron/hydrogen peroxide. The Dps pores are called “ferritin-like” because of the structural resemblance to the 3-fold channels of 24-subunit ferritins used for iron entry and exit to and from the protein cage. In ferritins, negatively charged residues lining the pores generate a negative electrostatic gradient that guides iron ions toward the ferroxidase centers for catalysis with oxidant and destined for the mineralization cavity. To establish whether the set of three aspartate residues that line the pores in Listeria innocua Dps act in a similar fashion, D121N, D126N, D130N, and D121N/D126N/D130N proteins were produced; kinetics of iron uptake/release and the size distribution of the iron mineral in the protein cavity were compared. The results, discussed in the framework of crystal growth in a confined space, indicate that iron uses the hydrophilic 3-fold pores to traverse the protein shell. For the first time, the strength of the electrostatic potential is observed to modulate kinetic cooperativity in the iron uptake/release processes and accordingly the size distribution of the microcrystalline iron minerals in the Dps protein population.
The widely distributed bacterial Dps proteins (1, 2) belong to the ferritin superfamily and are characterized by strong similarities (3) but also distinctive differences with respect to “canonical” ferritins, the ubiquitous iron storage, and detoxification proteins found in biological systems. The structural resemblance is apparent in the overall molecular assemblage because both Dps proteins and ferritins are shell-like oligomers constructed from four-helix bundle monomers. However, Dps proteins are 12-mers of identical subunits that assemble with 23 symmetry, whereas ferritins are built by 24 highly similar or identical subunits related by 432 symmetry. The functional similarities consist in the common capacity to remove Fe(II) from cytoplasm, catalyze its oxidation, and store Fe(III) thus produced in the protein cavity, wherefrom the metal can be mobilized when required by the organism. However, ferritins use molecular oxygen as iron oxidant with the production of hydrogen peroxide, whereas Dps proteins prefer hydrogen peroxide, which is typically about 100-fold more efficient than molecular oxygen (1). This difference is of major importance because it renders Dps proteins capable of removing concomitantly Fe(II) and H2O2 whose combination leads to the production of reactive oxygen species via Fenton chemistry (4). This capacity confers H2O2 resistance and hence may be a virulence factor in certain pathogens (e.g. Campylobacter jejuni, Streptococcus mutans, and Porphyromonas gingivalis) because the H2O2 burst represents one of the first defense lines of the host during infection (5–7).
Key to a full understanding of the iron uptake and release processes at a molecular level is the route by which iron enters and exits the protein shell. In both 24-subunit ferritins and 12-subunit Dps proteins, the subunit assemblage creates pores across the protein shell that put the internal cavity in communication with the external medium. In ferritins there are two types of pores: largely hydrophobic ones along the axes with 4-fold symmetry and hydrophilic ones along the axes with 3-fold symmetry. The latter channels are funnel-shaped, with the smaller opening toward the protein cavity, and are lined with conserved glutamic and aspartic residues located in the narrow region of the funnel (8). These 3-fold pores were recognized to provide the route for iron entry into the protein cavity soon after resolution of the horse ferritin x-ray crystal structure (9). Later site-directed mutagenesis studies defined the role of specific residues (Asp131 and Glu134) that line the pore (10, 11), whereas electrostatic calculations related the passage of iron to the existence of a gradient that drives metal ions toward the protein interior cavity (12, 13). More recently, the 3-fold symmetry pores were shown to be involved also in the exit process of iron from the protein cavity. Thus, in H-frog ferritin used as model system, iron exit is affected by local protein unfolding promoted by site-specific mutagenesis of individual amino acid residues (14, 15), by the use of chaotropes (16), and by means of selected peptides designed to bind at these channels (17).
In Dps proteins, the protein shell is breached by two types of pores along the 3-fold axes, one type is formed by the N-terminal portion of the monomers and bears a strong similarity to the typical 3-fold channels of 24-subunit ferritins in that it is funnel-shaped, hydrophilic, and lined by conserved, negatively charged residues. It was therefore named “ferritin-like” and assumed to be involved in iron entry into the protein cavity upon resolution of the Listeria innocua x-ray crystal structure (18). The other type of pore is formed by the C-terminal ends of the monomers and was called “Dps type” because it is created at a subunit interface that is unique to Dps proteins. Although somewhat variable in length and in the size of the openings, the Dps type pore is mainly hydrophobic in nature (19).
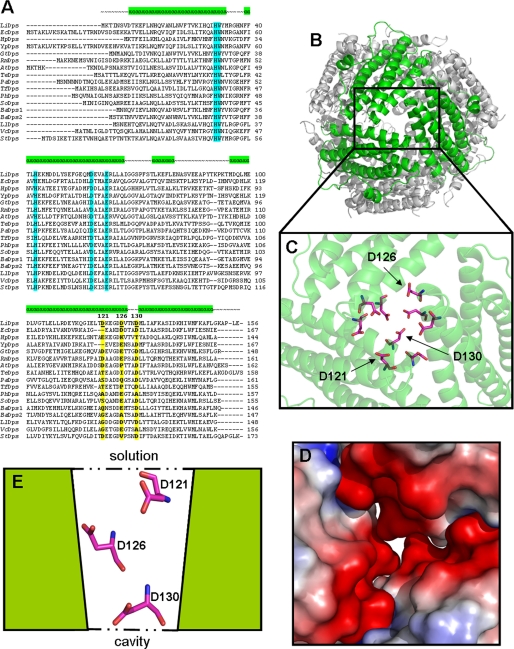
The present paper investigates the role of the ferritin-like pores in the iron uptake and release processes in Dps proteins using the well characterized L. innocua Dps (LiDps) as a model system (18, 20–22). In LiDps, the ferritin-like pores contain a set of three aspartate residues, Asp121, Asp126, and Asp130 that would be encountered in succession by a metal ion that is attracted by the electrostatic gradient they create and moves down the funnel-shaped pore toward the protein cavity (Fig. 1). Asp130, which is located in the narrowest part of the funnel, is conserved significantly among Dps proteins (∼ 80%), whereas Asp121 and Asp126 are less conserved (Fig. 1). Such considerations were used in the design of site-specific variants D121N, D126N, D130N, and D121N/D126N/D130N to elucidate the function of the ferritin-like pores in Dps proteins.
FIGURE 1.
Dps proteins sequences and conservation of the aspartate residues that line the 3-fold ferritin-like pores in L. innocua Dps. A, alignment of multiple Dps sequences from different bacteria: LiDps, non-heme iron-binding ferritin (L. innocua Clip11262]; EcDps, DNA-binding protein Dps (E. coli); HpDps, neutrophil-activating protein (Helicobacter pylori); YpDps, ferritin family protein (Yersinia pestis Angola); GtDps, DNA-protecting protein (Geobacillus thermodenitrificans NG80–2); RmDps, ferritin, and Dps (Ralstonia metallidurans CH34); AtDps, DNA protection during starvation conditions (Agrobacterium tumefaciens str. C58); TeDps Dps family DNA-binding stress response protein (Thermosynechococcus elongatus BP-1); PaDps, putative DNA-binding protein, Dps (Psychrobacter arcticus 273-4); TfDps, hypothetical protein Tfu_0799 (Thermobifida fusca YX); PhDps, DNA-binding DPS protein (Pseudoalteromonas haloplanktis TAC125); SoDps, Dps family protein (Shewanella oneidensis MR-1); BaDps1, general stress protein 20U (Bacillus anthracis str. Ames); BaDps2 general stress protein (B. anthracis str. Ames); LlDps, non-heme iron-binding ferritin (Lactococcus lactis subsp. lactis Il1403); VcDps, DPS family protein (Vibrio cholerae MZO-3); and StDps, DNA-binding ferritin-like protein (oxidative damage protectant) (Streptococcus thermophilus LMD-9). Residues forming the Dps catalytic center are highlighted in pale blue (His31, His43, Asp58, and Glu62 in LiDps); 3-fold pores aspartate residues are highlighted in yellow and marked in bold type. Alignment has been created with ClustalW2 (34). B, view of the junction of three monomers forming the 3-fold ferritin-like pore. C, Asp121, Asp126, and Asp130 aspartate residues comprised the pore area. D, three-dimensional view of the pore colored by charge. Red, negatively charged residues; blue, positively charged residues; white, uncharged residues. E, schematic representation of the vertical section of the pore. The images were created with PyMol (35).
The results demonstrate that iron uses the LiDps ferritin-like pores to enter and leave the protein shell and hence that these pores have the same role as the structurally similar 3-fold channels in 24-subunit ferritins. LiDps residue Asp130 is the most important determinant of the negative electrostatic gradient because of its location in the narrow part of the pores. Importantly, the data show for the first time that the electrostatic gradient at the pores modulates cooperativity in the iron uptake process and influences the size distribution of the iron core (23). The effect of the electrostatic gradient can be explained in terms of the electrostatic interaction effects between the fixed negative charges of the aspartate residues at the pores and the mobile positive charges of iron ions.
EXPERIMENTAL PROCEDURES
Mutagenesis
The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce the desired mutations. The single site-specific variants D121N, D126N, and D130N have been obtained using as template the pET-11a vector containing the L. innocua fri gene to introduce the mutations Asp121 → Asn, Asp126 → Asn, and Asp130 → Asn. The triple variant D121N/D126N/D130N has been obtained in two consecutive steps using as template the pET-11a vector containing the D121N gene and introducing the Asp130 → Asn mutation first and using this construct to introduce the Asp126 → Asn mutation. The oligonucleotides used in the PCRs are as follows: D121N_for, 5′-CGA ATA TAA ACA AGG CAT TGA GCT AAC TAA CAA AGA AGG CGA CG-3′; D121N_rev, 5′-CGT CGC CTT CTT TGT TAG TTA GCT CAA TGC CTT GTT TAT ATT CG-3′; D126N_for, 5′-GCTAACTAACAAAGAAGGCGACAATGTAACAAACAATATGC-3′; D126N_rev, 5′-GCA TAT TGT TTG TTA CAT TGT CGC CTT CTT TGT TAG TTA GC-3′; D130N_for, 5′-GGC GAC GAT GTA ACA AAC AAT ATG CTA ATT GCA TTT AAA GCT AGC-3′; and D130N_rev, 5′-GCT AGC TTT AAA TGC AAT TAG CAT ATT GTT TGT TAC ATC GTC GCC-3′.
Protein Expression and Purification
LiDps and the 3-fold ferritin-like pore variants D121N, D126N, D130N, and D121N/D126N/D130N were overexpressed in Escherichia coli strain BL21(DE3) (Novagen). Bacterial cultures were grown overnight at 37 °C in LB/Ampicillin 100 mg/ml, and then protein expression was induced with 1.0 mm isopropyl β-d-thiogalactopyranoside for 3 h at 37 °C. Bacterial cultures were centrifuged at 8.000 rpm in a Beckman centrifuge to obtain compact pellets. These pellets were frozen at −80 °C for 1 h and then thawed and resuspended in about 30.0 ml of 25.0 mm Bis-Tris-propane buffer, pH 7.5. Resuspended pellets were sonicated for 8 min (total time on) at 56% of amplitude (3.0 s time on and 9.0 s time off). Sonicated solutions were centrifuged for 2 h at 15,000 rpm to remove cellular debris. Supernatant solutions were heated for 8 min at 75 °C with stirring and then centrifuged for 30 min at 15,000 rpm at room temperature. A final 55% concentration of ammonium sulfate was added to the supernatant solutions, which were left for 30 min at room temperature and then overnight at 4 °C to obtain precipitation. After centrifugation for 30 min at 15,000 rpm, the supernatant solutions were applied by a peristaltic pump to a 5.0-ml MonoQ HiTrap HP column (Amersham Biosciences) equilibrated with 25.0 mm Bis-Tris-propane (BTP)4 buffer, pH 7.5. Chromatography was performed in an Amersham Biosciences chromatographic apparatus using a multi-steps program: 10 column volumes of washing with equilibration buffer, 25.0 mm BTP, pH 7.5; 10 column volumes wash of unspecific bound sample with 25.0 mm BTP, 100.0 mm NaCl, pH 7.5; 20 column volumes of linear gradient between 100 and 600 mm NaCl in 25.0 mm BTP, pH 7.5; and 10 column volumes with 25.0 mm BTP, 1.0 m NaCl, pH 7.5, followed by re-equilibration with 25.0 mm BTP, pH 7.5. All of the proteins eluted between 150 and 400 mm NaCl concentration. Fraction purity was assayed by SDS electrophoresis using 12% acrylamide gels. The final purity of proteins was tested also by native electrophoresis gel in Tris-glycine buffer, pH 8.5. The yield of purified protein ranged between 100 and 200 mg for liter of culture. Protein concentrations were determined spectrophotometrically using the molar absorptivity coefficient reported in Bozzi et al. (20); they are expressed on a dodecamer basis. The iron content, determined by the ferrozine assay (Fluka), was ∼1.0 Fe/dodecamer for all of the proteins studied.
Fe(II) Binding
Fe(II) binding was followed at 20 °C in an Applied Photophysics Π*-180 stopped flow instrument. The instrument syringes and the flow circuit were rendered anaerobic by extensive purging with degassed buffer. Intrinsic emission fluorescence was monitored after mixing equal volumes of a degassed 1.5 μm protein solution in 100 mm MOPS-NaOH buffer, pH 7.0, with a degassed 18.0 μm FeSO4 solution in 1.0 mm HCl. The excitation wavelength was 290 nm; the total intrinsic emission fluorescence was recorded using a filter at 320 nm; the entrance and exit slits were 4 nm. The half-times of the binding process were calculated by fitting the fluorescence quenching curves as a first order process. All of the fittings present χ2 values of >0.95.
Fe(II) Oxidation by Hydrogen Peroxide
Fe(II) oxidation by hydrogen peroxide was monitored at 20 °C in a Applied Photophysics Π*-180 stopped flow instrument recording changes in the absorbance value at 350 nm after mixing equal volumes of a 20.0 μm protein solution containing 240 μm H2O2 in 100 mm MOPS-NaOH buffer, pH 7.0, with a 240.0 μm FeSO4 solution in 1.0 mm HCl. The initial rates were determined by a linear fit of the data points from 0 to 6.0 ms (L. innocua Dps, D121N, and D126N) or from 0 to 30.0 ms (D130N, D121N/D126N/D130N, and control) after mixing, representing the first phase of the iron oxidation kinetic curves. The χ2 values for all of the fittings are >0.95.
Iron Mineralization
The kinetics of iron mineralization were monitored at 20 °C in a PerkinElmer Life Sciences spectrophotometer recording changes in the absorbance value at 350 nm after mixing solutions of LiDps and its pore variants in 100 mm MOPS-NaOH buffer, pH 7.0, with small volumes of FeSO4 solution in 1.0 mm HCl at 20 °C. The reaction was followed for 1 h in air while keeping the solutions under continuous stirring. The final concentrations in the assays were: 2.0 μm protein, 480.0 μm FeSO4 in 100.0 mm MOPS-NaOH buffer, pH 7.0. Because of the sigmoid shape of the progress curves under the conditions chosen, the half-times values are used for their comparison.
Analytical Ultracentrifugation
Size distribution of the iron core was assessed in sedimentation velocity experiments carried out on a Beckman XLI analytical ultracentrifuge using absorbance optics. The experiments were conducted at 30,000 rpm and 20 °C at a protein concentration of 2.0 μm in 100 mm MOPS-NaOH, pH 7.0. Radial absorbance scans were obtained at both 280 and 350 nm at a spacing of 0.003 cm with three averages in a continuous scan mode. Sedimentation coefficients were calculated using the software Sedfit (provided by Dr. P. Schuck, National Institutes of Health) and were reduced to water and 20 °C (s20,w) using standard procedures. The buffer density and viscosity were calculated by the software Sednterp.
Iron Release
The kinetics of iron release were studied using protein samples that had taken up ∼240 Fe(III)/molecule. LiDps and pore variants protein solutions in 50.0 mm MOPS-NaOH buffer, pH 7.0, were mixed with small volumes of reductants (25.0 mm NADH, 25.0 mm FMN or 25.0 mm sodium dithionite) and 25.0 mm bipyridyl. The final concentrations in the assay were: 0.2 μm proteins, 48.0 μm FeSO4, 2.5 mm reductants (NADH/FMN or sodium dithionite), and 2.5 mm bipyridyl in 50.0 mm MOPS-NaOH buffer, pH 7.0. The reaction was monitored for 20 min at 20 °C following the absorbance changes at 522 nm, indicative of the formation of [Fe+2-bipyridyl3] complex (E522 nm = 8430 m−1 cm−1). In the presence of NADH/FMN, the onset of iron release occurs about 3 min after mixing with the protein solution as indicated by the lack of absorbance changes at 522 nm. Initial rates were calculated fitting the linear portion of the kinetic curves within the first 20 s after the lag phase; all of the fittings present χ2 values of >0.95. The effect of chaotropes on the iron release process was assayed as described by Liu et al. (16).
RESULTS
To elucidate the role of the ferritin-like pores in the management of iron by Dps proteins, site-specific variants of the aspartate residues that line these pores in L. innocua Dps (LiDps) have been designed, and their behavior in the iron uptake and release processes has been investigated in comparison with the wild type protein. With regard to the iron uptake reaction, the kinetics of the various steps, namely Fe(II) binding, Fe(II) oxidation by hydrogen peroxide, and formation of the iron core, were followed; in addition the molecular mass (size) distribution of the iron core was determined by analytical ultracentrifugation in sedimentation velocity experiments.
Fe(II) Binding to the Ferroxidase Center
The ferroxidase centers of LiDps are located at the interface between 2-fold symmetry related monomers that contribute the iron ligands. Fe(II) binding to the ferroxidase center results in the quenching of the protein intrinsic fluorescence given the presence of a tryptophan residue (Trp32) at less than 5 Å (22).
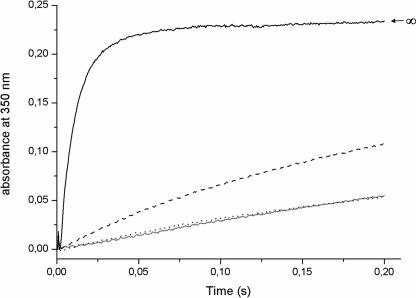
In LiDps, the stoichiometry of Fe(II) binding under anaerobic conditions corresponds to 12 Fe(II)/dodecamer (22). On this basis, the kinetics of Fe(II) binding was followed in anaerobic stopped flow experiments by measuring quenching of the intrinsic fluorescence upon addition of 12 Fe(II) to LiDps and its pore variants D121N, D126N, D130N, and D121N/D126N/D130N. In LiDps and in the D121N and D126N variants, the binding reaction is fast and is complete within the dead time of the instrument (≤3.0 ms). In contrast, Fe(II) binding to D130N and to the triple variant occurs at significantly slower rates and is characterized by a half-time of about 650 ms and 11 s, respectively (Fig. 2). The fact that the extent of quenching is the same for LiDps and all of the variants is in accord with the expectation that the site-specific mutations at the 3-fold ferritin-like pores have only a very localized structural effect.
FIGURE 2.
Aspartate residues at the L. innocua Dps ferritin-like pores control the rate of Fe(II) binding at the catalytic centers, measured as fluorescence quenching of the Trp32 residue. The Fe(II) binding to the catalytic centers was monitored after mixing a 1.5 μm protein solution with a 18.0 μm FeSO4 solution. Excitation wavelength was 290 nm, and the whole emission fluorescence signal up to 320 was recorded. The kinetic curves are representative of three to five experiments with two or three independent protein preparations. LiDps, D121N, and D126N binding kinetics are complete in the stopped flow mixing dead time (less than 3.0 ms). LiDps (solid line), D130N (dashed line), and D121N/D126N/D130N (dotted line). The D121N and D126N curves are superimposable to that of LiDps and have been omitted from the figure.
Fe(II) Oxidation by Hydrogen Peroxide
The kinetics of iron oxidation by hydrogen peroxide have been recorded by following the increase in absorbance at 350 nm because of Fe(III) formation. The iron/protein molar ratio was 12:1 to monitor exclusively ferroxidation at the catalytic centers. Under these conditions the rate of ferroxidation by molecular oxygen in the presence of LiDps is extremely slow (24). A 1:1 iron/hydrogen peroxide molar ratio was used to ensure full oxidation of the metal in the absence of protein (Fenton reaction stoichiometry).
When LiDps, D121N, and D126N are present, iron oxidation by hydrogen peroxide is about 70 times faster than the control reaction measured in the absence of protein. In contrast, the oxidation rate is increased only about 3-fold relative to control in the presence of the D130N variant and is unaffected when the triple variant D121N/D126N/D130N is present (Fig. 3 and Table 1).
FIGURE 3.
Aspartate 130, at the L. innocua Dps ferritin-like pores, is required for fast Fe(II) oxidation by hydrogen peroxide. Fe(II) oxidation by hydrogen peroxide was monitored at 350 nm after mixing 20.0 μm protein solutions containing 240 μm H2O2 in 100.0 mm MOPS buffer, pH 7.0, with a 240.0 μm FeSO4 solution in 1.0 mm HCl in a stopped flow instrument (mixing ratio, 1:1). The kinetic curves are representative of three to five experiments with two or three independent protein preparations (see Table 1 for the experimental variability). The gray line represents Fe(II) oxidation by hydrogen peroxide in absence of protein. LiDps (solid line), D130N (dashed line), and D121N/D126N/D130N (dotted line). The D121N and D126N curves are superimposable to that of LiDps and have been omitted from the figure.
TABLE 1.
Effect of 3-fold pore aspartate residues in the iron uptake process
| Proteina | Initial rate of Fe(II) oxidation by hydrogen peroxideb | t½ of Fe(III)OOH mineral formation in the presence of O2c |
|---|---|---|
| mm/s | min | |
| LiDps | 21.4 ± 2.1 | 5.4 ± 0.5 |
| D121N | 21.7 ± 1.1 | 4.9 ± 0.2 |
| D126N | 21.2 ± 2.6 | 3.6 ± 0.3 |
| D130N | 0.88 ± 0.07 | 11.5 ± 2.4 |
| D121N/D126N/D130N | 0.31 ± 0.06 | 17.4 ± 1.4 |
| Fe(II) + H2O2 (control) | 0.32 ± 0.02 |
a Recombinant L. innocua Dps proteins, produced by site-directed mutagenesis, expressed in E. coli strain as apoproteins (∼1 iron/molecule), were prepared in 100.0 mm MOPS, pH 7.0.
b Fe(II) oxidation by hydrogen peroxide was triggered by mixing 20.0 μm protein solutions containing 240 μm H2O2 in 100.0 mm MOPS buffer, pH 7.0, with a 240.0 μm FeSO4 solution in 1.0 mm HCl in a stopped flow instrument (mixing ratio, 1:1). Measurement of the change in absorbance, at 350 nm/s reflected the Fe(II) oxidation to the of ferric form. The initial rates were determined by a linear fit of the data points from 0 to 6.0 ms (LiDps, D121N, and D126N) or from 0 to 30.0 ms (D130N, D121N/D126N/D130N, and Fe(II) + H2O2) after mixing, which represents the first phase of iron oxidation kinetic curves; the χ2 values for all the fittings are >0.95. The data are averages of three to five measurements using two or three independent protein preparations. The error is presented as the standard deviation.
c The oxyhydroxide core was formed in the presence of oxygen by mixing the protein solution in 100 mm MOPS buffer, pH 7.0, with small volumes of a FeSO4 in 1.0 mm HCl solution. Final concentration in the assays was: 2.0 μm protein, 480.0 μm FeSO4 in 100.0 mm MOPS, pH 7.0. Half-times were calculated based on the final absorbance value at 350 nm reached after 1 h of reaction, corresponding to the >99% of iron converted in the oxidized form. The data are the averages of three to five measurements using two or three independent protein preparations. The error is presented as the standard deviation.
Formation of the Ferric Oxyhydroxide Mineral Core
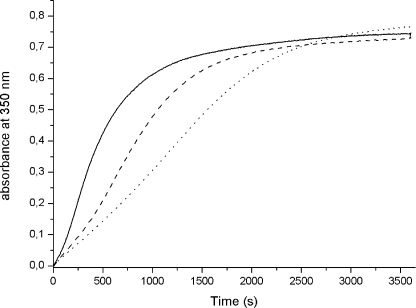
LiDps is able to store inside its cavity a microcrystalline ferric oxyhydroxide mineral core of about 500 iron atoms (20, 25). The kinetics of core formation by LiDps and the 3-fold ferritin-like pore variants have been monitored in air at 350 nm and room temperature according to the protocol employed currently for Dps proteins (19, 24, 26) and ferritin (27–30). The 240:1 iron/dodecamer molar ratio has been chosen so as to reach half the Dps storing capacity (26). For all of the proteins examined the kinetics of core formation is sigmoid, as expected for crystal formation that entails a slow nucleation step followed by fast, cooperative growth of the crystal. Notably, the absorbance values at the end of the reaction are alike, although the autocatalytic behavior displays significant variations (Fig. 4). Thus, the D121N and D126N are indistinguishable from LiDps, whereas kinetic cooperativity is less pronounced, and the half-time of the reaction is longer in the D130N variant and even more so in the triple variant relative to the wild type protein (Table 1).
FIGURE 4.
Iron mineralization kinetics in the presence of L. innocua Dps and its ferritin-like pores mutants. Fe(II) oxidation was monitored in air at 350 nm after mixing a 2.0 μm protein solution in 100.0 mm MOPS buffer, pH 7.0, with a small volumes of FeSO4 solution in 1.0 mm HCl. The kinetic curves presented are representative of three to five experiments with two or three independent protein preparations (see Table 1 for the experimental variability). LiDps (solid line), D130N (dashed line), and D121N/D126N/D130N (dotted line). The D121N and D126N curves are superimposable to that of LiDps and have been omitted from the figure.
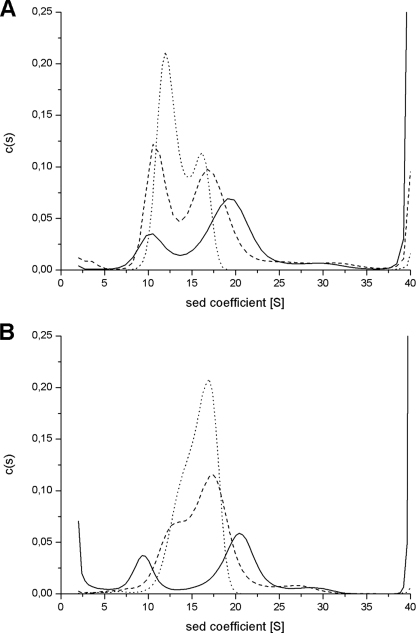
The difference in cooperativity of iron mineralization is reflected in the different size distribution of the mineralized iron in the sedimentation velocity patterns of LiDps and of the D130N and triple variants mineralized with 240 Fe(III)/dodecamer (Fig. 5A). The sedimentation profile of the wild type protein shows two clearly resolved peaks, one corresponding to the apoprotein (10 ± 0.5 S) and the other to heavily mineralized protein (∼19 S; 17, 21), as expected for a highly cooperative process. The sedimentation pattern of the D130N mutant likewise displays two peaks (∼10.5 and ∼16.5 S) attributable to iron-poor and iron-rich protein, in accord with the decreased cooperativity of the uptake process, which favors formation of partially filled protein molecules. This tendency is even more marked in the triple mutant where the iron-poor peak is centered at ∼12 S and the iron rich one at ∼16 S. A small heavier component (∼30 S) is also present that may be attributed to formation of a small amount of Fe(III) aggregates in solution (see below).
FIGURE 5.
Size distribution of the iron core in the protein cavity of L. innocua Dps and its D130N and D121N/D126N/D130N variants. The iron distribution in the proteins was assayed by analytical ultracentrifugation experiments. Mineralized LiDps, D130N, and D121N/D126N/D130N samples were prepared at 2.0 μm protein concentration by the addition of 480.0 μm FeSO4 (A; 240 Fe(III)/dodecamer) in one increment or 900.0 μm FeSO4 (B; 450 Fe(III)/dodecamer) in three increments, in MOPS, 100.0 mm, pH 7.0. Sodium citrate was added at 1.0 mm final concentration to samples containing 450 Fe(III)/dodecamer to remove Fe(III) aggregates. LiDps (solid lines), D130N (dashed lines), and D121N/D126N/D130N (dotted lines).
Wild type LiDps can be loaded fully with iron to yield a homogeneous population sedimenting at ∼21 S (20). Therefore, to better appreciate the effect of added iron on the mineral distribution in the LiDps wild type and mutant proteins, subsaturating amounts of 450 iron atoms were added to each, in three successive 150-atom aliquots (Fig. 5B). For LiDps, part of the protein remains iron-free and part becomes fully mineralized (∼20.5 S), whereas in the two mutants iron is distributed more homogeneously throughout the population of protein molecules. As a result, in the D130N mutant and in the D121N/D126N/D130N mutant, the iron-rich peak dominates (∼17 S). The sedimentation profiles of the proteins mineralized with 450 Fe(III)/dodecamer display an additional peak at around 30 S, even though the samples contain 1.0 mm citrate. This additional peak is significantly larger when citrate is omitted (data not shown), indicating that they are ferric oxyhydroxide particles that are formed outside the protein and are dissolved by citrate.
Release of Iron from the Ferric Oxyhydroxide Core
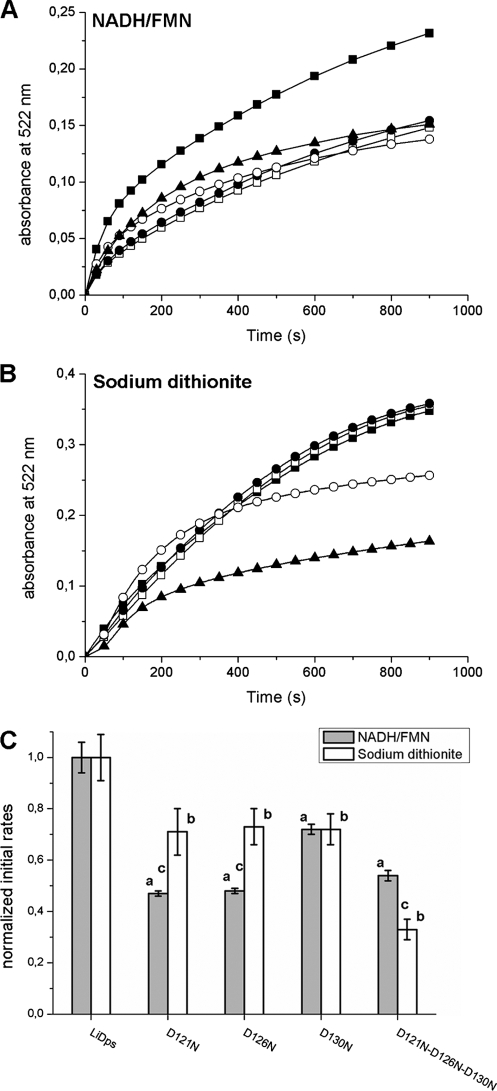
Samples of LiDps and the 3-fold ferritin-like pore variants containing 240 Fe(III)/dodecamer have been used to study iron release from the protein cavity. The release process was triggered by reduction of the ferric oxyhydroxide core with either a NADH/FMN mixture under aerobic conditions or a sodium dithionite solution under anaerobic conditions and was monitored as formation of the [Fe+2-bipyridyl3] complex at 522 nm (Fig. 6, A and B). The final concentrations in the assays were 0.2 μm protein, 48.0 μm FeSO4, 2.5 mm reductants (NADH/FMN or dithionite), and 2.5 mm bipyridyl in 50.0 mm MOPS-NaOH buffer, pH 7.0.
FIGURE 6.
Aspartate 121, 126, and 130 each contribute to iron removal from the hydrated ferric oxide mineral inside the L. innocua Dps protein. A and B, the iron release from LiDps and pore site-specific aspartate variants was monitored following the formation of the [Fe(II)-bipyridyl3] complex at 522 nm. Reduction and chelation were triggered by adding 2.5 mm of reductants (NADH/FMN (A) or sodium dithionite (B)) and 2.5 mm bipyridyl to solutions of protein with mineral (0.2 μm protein and 48.0 μm iron) in 100.0 mm MOPS, pH 7.0. ■, LiDps; □, D121N; ●, D126N; ○, D130N; ▲, D121N/D126N/D130N. C, initial rates of iron release calculated as described in Table 2. Each bar represents the value of the initial rate normalized for the LiDps value. The data are averages of five or six independent experiments, and the error is presented as standard deviation. a, D121N, D126N, D130N, and D121N/D126N/D130N variants are significantly different from LiDps (p < 0.0001) when iron release is triggered by NADH/FMN. b, D121N, D126N, D130N, and D121N/D126N/D130N variants are significantly different from LiDps (p ≤ 0.0002) when iron release is triggered by sodium dithionite. c, D121N, D126N, and D121N/D126N/D130N are significantly different (p ≤ 0.0002) when iron release is performed with NADH/FMN or sodium dithionite.
The progress curves describing iron release are complex, using both NADH/FMN and sodium dithionite (Fig. 6, A and B). When dithionite is used as reducing agent, the kinetic curves pertaining to LiDps and the D121N and D126N variants are almost superimposable and have a very slight sigmoid shape if any. In contrast, iron release from the D130N and the triple mutant is characterized by a markedly decreased rate and follows a sigmoid kinetics. When using the NADH/FMN mixture, the initial portion of the iron release curves (∼20 s) is linear and was used to determine the initial rates from the different proteins (Table 2 and Fig. 6C). The initial rate of iron release is fastest from LiDps as observed when sodium dithionite is the reducing agent. Notably, the amount of iron released after 2–3 h is similar for both reducing agents and ranges between 80 and 90% of the initial amount of stored iron in all of the proteins examined.
TABLE 2.
Effect of 3-fold pore aspartate residues in the iron release process
| Proteina | Initial rate of iron releaseb |
|
|---|---|---|
| NADH/FMN | Sodium dithionite | |
| μmols/s | ||
| LiDps | 0.177 ± 0.010 | 0.095 ± 0.009 |
| D121N | 0.084 ± 0.002 | 0.067 ± 0.009 |
| D126N | 0.084 ± 0.002 | 0.069 ± 0.007 |
| D130N | 0.127 ± 0.004 | 0.069 ± 0.006 |
| D121N/D126N/D130N | 0.095 ± 0.004 | 0.031 ± 0.004 |
a Recombinant L. innocua Dps proteins, produced by site-directed mutagenesis, expressed in E. coli strain as apoproteins (∼1 iron/molecule) were prepared in 100.0 mm MOPS, pH 7.0. Mineral was formed in vitro by the addition of FeSO4 in 1.0 mm HCl at the 240 irons/assembled protein ratio (see “Experimental Procedures”).
b Reduction and chelation were triggered by adding 2.5 mm reductants (FMN/NADH or sodium dithionite) and 2.5 mm bipyridyl to solutions of protein with mineral (0.2 μm protein and 48.0 μm iron). Measurement of the change in absorbance, at 522 nm/s (the absorbance maximum for the Fe(II)-bipyridyl complex), reflected the conversion of ferric mineral to Fe(II) in solution in the bipyridyl complex. The initial rates were determined by a linear fit of the data points from 0 to 20 s after mixing, which represents the first phase of iron release; the χ2 values for the fittings are all >0.95. The data are averages of five or six measurements using two or three independent protein preparations. The error is presented as the standard deviation. All of the differences between LiDps and the 3-fold pore variants are statistically significant (p values < 0.0003).
The effect of ionic strength, urea, and guanidine on the kinetics of iron release from LiDps was assayed as described in Liu et al. (16). No significant differences were apparent with respect to the kinetics recorded in the absence of chaotropes (data not shown).
DISCUSSION
The present study defines the role of the so-called ferritin-like pores in the iron oxidation/uptake and release processes by L. innocua Dps and in doing so shows that the striking structural similarity between these pores and the 3-fold channels of ferritins transfers to function. The conservation of the pore structure in all Dps proteins, especially of the aspartate residues that line the pores, in turn shows that their designation as ferritin-like is correct. From a mechanistic viewpoint, it is significant that substitution of the aspartate residues at the pores modulates cooperativity in the iron uptake process and thereby affects the size distribution of the microcrystalline iron core in the 12-subunit Dps proteins.
In 24-subunit ferritins, the 3-fold pore is shaped like a hydrophilic funnel that opens on the internal cavity with its narrow part and is contoured by a set of conserved aspartate residues, Asp131 and Glu134 in human H ferritin (8, 10). These localized charges generate a negative electrostatic gradient that guides iron from the external medium toward the interior of the protein as indicated by the calculations of Douglas and Ripoll (12) in the human H-chain homopolymer and of Takahashi and Kuyucak (13) in horse L-chain ferritin. As a result, the rate of iron oxidation decreases in the Glu134 → Ile or Phe variants of human H ferritin and even more so in the Asp131 → Ala mutant, because Asp131 is located in the tapered part of the funnel; moreover, the amount of iron incorporated decreases by about 50% (11). In LiDps the 3-fold ferritin-like pores are also funnel-shaped and lined by conserved aspartate residues that produce an electrostatic gradient with a maximum in the narrow part of the funnel where Asp130 is located (Fig. 1). The rate of iron uptake by LiDps was predicted, then, to decrease upon substitution of the ferritin-like pore aspartates because it reduces the electrostatic gradient at the pores. Further, the effects of substituting Asp130, Asp126, and Asp121 on the strength of the electrostatic gradient were predicted to reflect location. Indeed, removal of Asp130 results in a marked decrease in the rates of Fe(II) binding to the ferroxidase centers (Fig. 2) and of iron oxidation by hydrogen peroxide (Fig. 3 and Table 1), whereas substitution of Asp121 and Asp126 has practically no effect, in accord with their position in the wide part of the pores. However, the effects of Asp121 and Asp126 become apparent in the triple mutant D121N/D126N/D130N and add to that of Asp130 (Figs. 2 and 3). Thus, the half-time for Fe(II) binding to the ferroxidase centers, ≤3.0 ms in LiDps, increases to 650 ms in D130N and even further (∼11 s) in the triple mutant (Fig. 2). Moreover, protein-catalyzed iron oxidation by hydrogen peroxide, which is merely inhibited in the D130N variant, cannot be distinguished from the control reaction in the triple mutant, D121N/D126N/D130N (Table 1).
Iron mineralization by molecular oxygen, which takes place over minutes rather than milliseconds as oxidation by hydrogen peroxide, is also affected by the differential reduction of the negative electrostatic potential gradient because of substitution of Asp130, either singly or in combination with Asp121 and Asp126. The mineralization reaction is about 2–3-fold slower relative to LiDps in the D130N mutant and the triple variant, respectively. Importantly, the change in electrostatic potential also results in a progressive reduction of mineralization cooperativity (Fig. 4). This effect can be attributed to the modulation of kinetic cooperativity resulting from the interaction of mobile charges with clusters of fixed charge density (23), which, in turn, results in a significant difference in the size distribution of the iron core (Fig. 5). Thus, the strongly cooperative iron uptake process of LiDps leads to formation of two clearly defined homogeneous populations, namely iron-free protein (10 ± 0.5 S) and heavily mineralized protein (∼19 and ∼20.5 S, at the two molar ratios), whereas the metal is incorporated in all molecules of the D130N and triple mutants such that no iron-free protein peak is observed. This is especially evident when the mutant proteins are mineralized with 450 iron atoms/dodecamer; the triple mutant, for example, sediments as a single asymmetrical peak of ∼17 S. When 450 iron atoms were mixed with the variants of Asp130, some of the iron was oxidized outside the protein, based on the appearance of particles with high sedimentation coefficients that disappeared when citrate was added (Fig. 5). Oxidation of iron outside the protein likely explains the decrease in mineralized iron in the case of the corresponding 3-fold channel mutants of human H ferritin reported by Levi et al. (11).
The role of the different pore-specific aspartate residues in facilitating ferritin iron mineralization clearly matches their contribution to the negative electrostatic gradient at the pores. Does the relationship also apply to iron release from the mineral? In the case of 24-subunit ferritins, the kinetics of both iron mineralization and release can be explained in the framework of crystal formation/dissolution in a confined space (31). Such a model assumes that kinetics are controlled solely by the accessible core surface and describes the change in mineralization rate as a function of the number of iron atoms contained in the molecule and the shape of the release curves as a function of time. Importantly, because of the constraints imposed by the confined space, the model predicts that the crystal formation and dissolution rates depend on iron content such that both progress curves are sigmoidal when the content of hydrated iron oxide mineral in the protein cavity is low. Whereas this is a well known phenomenon in iron mineralization (32, 33) beyond the initial catalytic oxidation (3), relatively few data are available with regard to iron release (31). Indeed iron mineralization in the wild type and pore mutant proteins studied is autocatalytic (Fig. 4), although the progress curves of iron release differ in shape as the size of the cores differs in the various proteins studied (Fig. 5). Thus, the shape of the iron release curve by sodium dithionite (Fig. 6B) is not sigmoid in LiDps, because fully mineralized molecules are involved (s20,w = ∼19 S), but the curve is sigmoid in the case of the D130N and triple variants because it refers to partially filled molecules (s20,w = ∼17 S). Iron reduction using NADH/FMN under aerobic conditions (Fig. 6A), although more physiological, presents additional complexity because of the continuing change in the concentration of the reducing agents in the presence of oxygen but shows similar effects on the initial rates of the iron release process (Fig. 6C). Further, limited access of these reducing agents to the protein cavity may be limited by the pore size. If so mutation of the residues that correspond to those that unfold the pores in 24-subunit ferritins (3, 14, 17) should lead to a significant increase of the iron release rate.
Structural similarity between pores at the 3-fold symmetry axis of 12-subunit Dps protein cages and 24-subunit ferritins is now correlated to functional similarity. In addition, the linkage between the strength of the electrostatic gradient at the pores, cooperativity in the kinetics of iron uptake/release, and the size distribution of the iron mineral is now clear. Whether or not the hydrophobic residues in 24-subunit ferritin 3-fold pores that control pore unfolding and access to the mineral surface have the same function in Dps pores remains a subject for the future.
Acknowledgments
We thank Brie Fuqua for help at the Children's Hospital Oakland Research Institute.
This work was supported, in whole or in part, by National Institutes of Health Grant DK 20251 (to G. B. and E. C. T.).
- BTP
- Bis-Tris-propane
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Chiancone E., Ceci P., Ilari A., Ribacchi F., Stefanini S. (2004) Biometals 17, 197–202 [DOI] [PubMed] [Google Scholar]
- 2.Morikawa K., Ohniwa R. L., Kim J., Maruyama A., Ohta T., Takeyasu K. (2006) Genes Cells 11, 409–423 [DOI] [PubMed] [Google Scholar]
- 3.Liu X., Theil E. C. (2005) Acc. Chem. Res. 38, 167–175 [DOI] [PubMed] [Google Scholar]
- 4.Imlay J. A. (2003) Annu. Rev. Microbiol. 57, 395–418 [DOI] [PubMed] [Google Scholar]
- 5.Ishikawa T., Mizunoe Y., Kawabata S., Takade A., Harada M., Wai S. N., Yoshida S. (2003) J. Bacteriol. 185, 1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto Y., Poole L. B., Hantgan R. R., Kamio Y. (2002) J. Bacteriol. 184, 2931–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueshima J., Shoji M., Ratnayake D. B., Abe K., Yoshida S., Yamamoto K., Nakayama K. (2003) Infect. Immun. 71, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawson D. M., Artymiuk P. J., Yewdall S. J., Smith J. M., Livingstone J. C., Treffry A., Luzzago A., Levi S., Arosio P., Cesareni G., Thomas C. D., Shaw W. V., Harrison P. M. (1991) Nature 349, 541–544 [DOI] [PubMed] [Google Scholar]
- 9.Stefanini S., Desideri A., Vecchini P., Drakenberg T., Chiancone E. (1989) Biochemistry 28, 378–382 [DOI] [PubMed] [Google Scholar]
- 10.Treffry A., Bauminger E. R., Hechel D., Hodson N. W., Nowik I., Yewdall S. J., Harrison P. M. (1993) Biochem. J. 296, 721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levi S., Santambrogio P., Corsi B., Cozzi A., Arosio P. (1996) Biochem. J. 317, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas T., Ripoll D. R. (1998) Protein Sci. 7, 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi T., Kuyucak S. (2003) Biophys. J. 84, 2256–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagi H., Shi D., Ha Y., Allewell N. M., Theil E. C. (1998) J. Biol. Chem. 273, 18685–18688 [DOI] [PubMed] [Google Scholar]
- 15.Jin W., Takagi H., Pancorbo B., Theil E. C. (2001) Biochemistry 40, 7525–7532 [DOI] [PubMed] [Google Scholar]
- 16.Liu X., Jin W., Theil E. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3653–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X. S., Patterson L. D., Miller M. J., Theil E. C. (2007) J. Biol. Chem. 282, 31821–31825 [DOI] [PubMed] [Google Scholar]
- 18.Ilari A., Stefanini S., Chiancone E., Tsernoglou D. (2000) Nat. Struct. Biol. 7, 38–43 [DOI] [PubMed] [Google Scholar]
- 19.Franceschini S., Ceci P., Alaleona F., Chiancone E., Ilari A. (2006) FEBS J. 273, 4913–4928 [DOI] [PubMed] [Google Scholar]
- 20.Bozzi M., Mignogna G., Stefanini S., Barra D., Longhi C., Valenti P., Chiancone E. (1997) J. Biol. Chem. 272, 3259–3265 [DOI] [PubMed] [Google Scholar]
- 21.Chiaraluce R., Consalvi V., Cavallo S., Ilari A., Stefanini S., Chiancone E. (2000) Eur. J. Biochem. 267, 5733–5741 [DOI] [PubMed] [Google Scholar]
- 22.Su M., Cavallo S., Stefanini S., Chiancone E., Chasteen N. D. (2005) Biochemistry 44, 5572–5578 [DOI] [PubMed] [Google Scholar]
- 23.Ricard J., Kellershohn N., Mulliert G. (1989) Biophys. J. 56, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilari A., Latella M. C., Ceci P., Ribacchi F., Su M., Giangiacomo L., Stefanini S., Chasteen N. D., Chiancone E. (2005) Biochemistry 44, 5579–5587 [DOI] [PubMed] [Google Scholar]
- 25.Ilari A., Ceci P., Ferrari D., Rossi G. L., Chiancone E. (2002) J. Biol. Chem. 277, 37619–37623 [DOI] [PubMed] [Google Scholar]
- 26.Zhao G., Ceci P., Ilari A., Giangiacomo L., Laue T. M., Chiancone E., Chasteen N. D. (2002) J. Biol. Chem. 277, 27689–27696 [DOI] [PubMed] [Google Scholar]
- 27.Levi S., Luzzago A., Cesareni G., Cozzi A., Franceschinelli F., Albertini A., Arosio P. (1988) J. Biol. Chem. 263, 18086–18092 [PubMed] [Google Scholar]
- 28.Sun S., Arosio P., Levi S., Chasteen N. D. (1993) Biochemistry. 32, 9362–9369 [DOI] [PubMed] [Google Scholar]
- 29.Hilty S., Webb B., Frankel R. B., Watt G. D. (1994) J. Inorg. Biochem. 56, 173–185 [DOI] [PubMed] [Google Scholar]
- 30.Chasteen N. D., Harrison P. M. (1999) J. Struct. Biol. 126, 182–194 [DOI] [PubMed] [Google Scholar]
- 31.Courteix A., Bergel A. (1996) AIChE J. 42, 829–836 [Google Scholar]
- 32.Macara I. G., Hoy T. G., Harrison P. M. (1972) Biochem. J. 126, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanini S., Chiancone E., Vecchini P., Antonini E. (1976) Mol. Cell. Biochem. 13, 55–61 [DOI] [PubMed] [Google Scholar]
- 34.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 35.DeLano W. L. (2006) PyMOL, Version 0.99, DeLano Scientific LLC, San Carlos, CA [Google Scholar]