Abstract
Methionine oxidation leads to the formation of S- and R-diastereomers of methionine sulfoxide (MetSO), which are reduced back to methionine by methionine sulfoxide reductases (MSRs) A and B, respectively. MSRBs are classified in two groups depending on the conservation of one or two redox-active Cys; 2-Cys MSRBs possess a catalytic Cys-reducing MetSO and a resolving Cys, allowing regeneration by thioredoxins. The second type, 1-Cys MSRBs, possess only the catalytic Cys. The biochemical mechanisms involved in activity regeneration of 1-Cys MSRBs remain largely elusive. In the present work we used recombinant plastidial Arabidopsis thaliana MSRB1 and MSRB2 as models for 1-Cys and 2-Cys MSRBs, respectively, to delineate the Trx- and glutaredoxin-dependent reduction mechanisms. Activity assays carried out using a series of cysteine mutants and various reductants combined with measurements of free thiols under distinct oxidation conditions and mass spectrometry experiments show that the 2-Cys MSRB2 is reduced by Trx through a dithiol-disulfide exchange involving both redox-active Cys of the two partners. Regarding 1-Cys MSRB1, oxidation of the enzyme after substrate reduction leads to the formation of a stable sulfenic acid on the catalytic Cys, which is subsequently glutathionylated. The deglutathionylation of MSRB1 is achieved by both mono- and dithiol glutaredoxins and involves only their N-terminal conserved catalytic Cys. This study proposes a detailed mechanism of the regeneration of 1-Cys MSRB activity by glutaredoxins, which likely constitute physiological reductants for this type of MSR.
Proteins are prone to oxidative modifications due to the action of reactive oxygen species. Methionine (Met), one of the most susceptible amino acids to oxidation (1), is converted into methionine sulfoxide (MetSO),3 resulting in altered conformation and activity for many proteins (1). Methionine sulfoxide reductases (MSRs), which catalyze the reduction of MetSO back to Met, are present in most living organisms. MSRA, the first MSR isolated (2), is specific of the MetSO S-diastereomer and participates in protection against oxidative stress (3). A second MSR type, MSRB, which catalytically reduces the MetSO R-diastereomer, was identified later (4). MSRA and MSRB are monomeric enzymes that display no sequence or structural homologies but share a similar three-step catalytic mechanism, (i) reduction of MetSO by MSR and formation of a sulfenic acid intermediate on the “catalytic” cysteine (Cys), (ii) formation of a disulfide bond between catalytic and “resolving” Cys and release of H2O, and (iii) reduction of the disulfide bond by a reductant (5, 6). Thioredoxins (Trxs) have been proposed to be the biological reductant for MSRs (2, 7). Trxs are small and ubiquitous disulfide reductases with a WC(G/P)PC active site. They function as electron donors and play essential roles in many processes through control of protein conformation and activity by supplying the reducing power needed to reduce disulfide bonds in target proteins.
Most MSRBs, named 2-Cys MSRBs, possess two conserved Cys and are actually reduced by Trxs (7, 8). However, in a second class of MSRBs, termed 1-Cys MSRBs and representing ∼40% of known MSRBs, the resolving Cys residue corresponding to Cys-63 in Escherichia coli is replaced by Thr or Ser (8, 9). Although some of these MSRBs possess another potential resolving Cys (9), most 1-Cys MSRBs do not have any additional Cys, indicating that an alternative mechanism, which does not involve the formation of an intramolecular disulfide reduction, is needed for their regeneration (7). Contrasting data concerning the role of Trxs in providing electrons to these MSRBs have been reported. Several studies showed that cytosolic Trx is not an efficient reductant for human 1-Cys MSRBs (10–12), whereas mitochondrial Trxs were recently reported to efficiently regenerate mitochondrial 1-Cys MSRBs (13). It has been proposed that regeneration of mammalian and plant 1-Cys MSRBs could involve direct reduction of the cysteine sulfenic acid form generated during catalysis (10, 13–15).
Arabidopsis thaliana possesses two plastidial MSRBs referred to as MSRB1 and MSRB2 and related to 1-Cys and 2-Cys MSRB types, respectively. MSRB2 possesses two CXXC motifs potentially implicated in the coordination of a zinc atom, a Cys in position 187 corresponding to the catalytic Cys-117 of E. coli MSRB, a potential resolving Cys in position 134, and an additional Cys in position 115. MSRB1 also contains the four Cys residues potentially coordinating zinc, the potential catalytic Cys-186, and a Cys in position 116, whereas the potential resolving Cys is replaced by a Thr in position 132. Previously, we showed that various types of canonical Trxs are efficient electron suppliers to MSRB2, whereas MSRB1 can only be reduced by the peculiar Trx CDSP32 (chloroplastic drought-induced stress protein of 32 kDa) and by Grxs (15–17). Grxs are oxidoreductases of the Trx superfamily possessing either a monothiol CXXS or a dithiol CXXC active site and are generally reduced by glutathione (18). Grxs are able to reduce protein disulfides, but also glutathione-mixed disulfides, a reaction termed deglutathionylation, for which Trxs are not efficient catalysts (19, 20). Classical dithiol Grxs can reduce disulfide bonds using both active site Cys residues, as shown for E. coli ribonucleotide reductase, but can also reduce glutathione-mixed disulfides through a monothiol mechanism that requires only the N-terminal active site Cys (21). CXXS-type Grxs catalyze deglutathionylation either through a monothiol mechanism, as recently shown for chloroplastic GrxS12 (CSYS active site) (22), or through a dithiol mechanism as suggested for Grxs with a CGFS active site (20, 23).
We reported recently the involvement of Grxs in the regeneration of MSRB activity (15). Nevertheless, the precise biochemical mechanism underlying regeneration by Grxs remains unknown. In this study we performed a detailed analysis of the roles of redox-active Cys in reductants (Trxs and Grxs) and in acceptors (plastidial Arabidopsis MSRBs). We provide evidence that reduction of MSRB2 by Trxs is achieved through a classical dithiol-disulfide exchange. The data on MSRB1 reveal that 1-Cys MSRBs are regenerated by Grxs through a glutathionylation step of the catalytic Cys.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Nucleotide substitutions at specific positions in MSRB1 and MSRB2 were performed using the QuikChange mutagenesis method (Stratagene, La Jolla, CA). The pQE-30 expression vector (Qiagen, Valencia, CA) carrying the coding sequence of MSRB2 (17) was used as a template for mutagenic PCR. The MSRB1 coding sequence was cloned into the BamHI and PstI restriction sites of the pQE-30 expression vector and used to generate mutated forms. Primers for site-directed mutagenesis contained a modified restriction enzyme site allowing screening (supplemental Table 1).
Expression and Purification of Recombinant Proteins
Arabidopsis NADPH thioredoxin reductase B, wild-type (WT), and mutated forms of poplar Trx h1, GrxC4, and GrxS12 were purified as described (22, 24–27). Recombinant WT and mutant forms of MSRB1 and MSRB2 proteins fused to an N-terminal His6 tag were produced in M15rep4 E. coli strains and purified as previously described (15). After elution, proteins were desalted in 30 mm Tris-HCl, pH 8.0, using HiTrapTM desalting column (GE Healthcare). Proteins were reduced using 10 mm DTT for 30 min at room temperature, and excess DTT was removed by desalting using IllustraTM NAP-5 Sephadex G-25 column (GE Healthcare) in the same buffer. Protein concentrations were determined using the bicinchoninic acid assay (BC Assay Reagent, Interchim, Montluçon, France) or spectrophotometrically using molar extinction coefficients at 280 nm of 26,930 m−1·cm−1 for WT MSRB1, C116S MSRB1, T132A MSRB1, T132C MSRB1, and C189S MSRB1 and of 16,960 m−1·cm−1 for WT MSRB2 and C134T MSRB2. Protein purity was verified using SDS-PAGE gels stained with ImperialTM Protein Stain (Pierce).
MSR Activity Assay
The activity of recombinant MSRB proteins was determined by monitoring reduction of the synthetic substrate, dabsyl-MetSO, in the presence or absence of 20 mm DTE or Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) using a method modified from Ref. 15. Dabsyl-Met and dabsyl-MetSO were separated by high performance liquid chromatography (HPLC) using a C18 reverse phase column, SunFireTM 3.5 μm, 3.0 × 50 mm (Waters, Milford, MA) and 29 mm acetate buffer, pH 4.16, and acetonitrile as solvents. Km for substrate (KDabsyl-MetSO) was determined using substrate concentrations ranging from 7.8 μm to 1 mm and nonlinear regression (SigmaPlot 10.0, Systat Software, San Jose, CA). Alternatively, the MSRB activity was measured after NADPH oxidation at 340 nm in the presence of a Trx-reducing system (either 200 or 400 μm NADPH, 2 μm Arabidopsis NADPH thioredoxin reductase B, and a saturating concentration of poplar Trx h1 or C42S Trx h1 (40–100 μm)) or of a Grx-reducing system (400 μm NADPH, 0.5 unit yeast glutathione reductase (Sigma), 10 mm GSH, and 0.13 to 50 μm Grx) using a saturating concentration of N-acetyl-MetSO (2 mm) and 1–6 μm MSRB. The reaction was carried out at 25 °C in a 500-μl reaction volume. MSRB activity was calculated from the slope due to NADPH consumption after subtracting the background activity in the absence of enzyme, considering that 1 mol of oxidized NADPH corresponds to 1 mol of Met formed.
Thiol Titration
Free thiols were determined using the 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) procedure (28). Pre-reduced proteins (20–50 μm) were incubated in 30 mm Tris-HCl, pH 8.0, in the presence or not of 0.5 mm N-acetyl-MetSO. After 15 min of incubation at room temperature, 100 μm DTNB was added, and the absorbance at 412 nm was measured after 30 min of incubation in the dark. The free thiol content was estimated using a molar extinction coefficient of 14,150 m−1.cm−1 for thiobis-2-nitrobenzoic acid.
Mass Spectrometry Analyses
Recombinant Arabidopsis MSRB1 and MSRB2 were reduced using 10 mm DTT for 1 h at room temperature and subsequently subjected to desalting on NAP-5 columns equilibrated with 50 mm HEPES-NaOH, pH 7.2. Pre-reduced proteins were treated for 15 min at room temperature with 2 mm N-acetyl-MetSO or for 30 min with 2 mm N-acetyl-MetSO and 2 mm GSH and subjected to desalting as described above. MALDI-TOF mass spectrometry analyses were performed before and after treatment in the presence of 10 mm DTT for 30 min at room temperature. Mass determination of MSRB1 and MSRB2 proteins was carried out on whole proteins or after tryptic digestion as described in Augusto et al. (29).
RESULTS
MetSO Reduction by WT and Mutated MSRBs
Arabidopsis plastidial MSRBs mainly differ in the presence or absence of a potential resolving Cys. MSRB2 possesses a conserved Cys in position 134, whereas MSRB1 contains a Thr at the corresponding position 132. To investigate the consequences for activity of this substitution, we assayed the MetSO reduction activity of WT and mutated MSRBs using DTE as a chemical reductant, an HPLC-based method, and dabsylated MetSO as a substrate. We measured a turnover number (kcat) of 0.075 s−1 for MSRB1 (Table 1). The replacement of Thr-132 by an Ala or a Cys in MSRB1 led to 6- and 14-fold activity decreases, respectively, compared with that of the WT form. Using mutated C186S MSRB1, no activity was detected (data not shown), confirming that Cys-186 is the catalytic residue absolutely required for MetSO reduction. We also tested the activity of mutated MSRB1 in which Cys-116 was changed into Ser. This mutant form was still able to reduce substrate with a turnover number representing two-thirds that of the WT form (Table 1). This result indicates that Cys-116 is dispensable for MetSO reduction.
TABLE 1.
Catalytic parameters of WT and mutated MSRBs
Turnover numbers were determined using an HPLC-based method for the reduction of dabsyl-MetSO to dabsyl-Met with 1 μm MSRB and 0.5 mm dabsyl-MetSO in the presence of 20 mm DTE or TCEP. Measurements of stoichiometry were carried out using 5 μm pre-reduced MSRB and 0.1 mm dabsyl-MetSO. Data presented are the means ± S.D. (n = 3). Considering that only the R-diastereomer is reduced by MSRBs, KDabsyl-MetSO values were divided by two.
|
kcat |
Stoichiometry | KDabsyl-MetSO | ||
|---|---|---|---|---|
| +DTE | +TCEP | |||
| s−1 | Mol of Met formed·mol of MSRB−1 | μm | ||
| MSRB1 | 0.075 ± 0.018 | 0.002 ± 0.000 | 1.0 ± 0.1 | 2.2 ± 0.6 |
| C116S MSRB1 | 0.047 ± 0.009 | 0.9 ± 0.1 | 7.9 ± 2.2 | |
| T132A MSRB1 | 0.012 ± 0.001 | 0.7 ± 0.1 | ||
| T132C MSRB1 | 0.005 ± 0.000 | 0.9 ± 0.1 | 5.2 ± 1.1 | |
| MSRB2 | 0.028 ± 0.007 | 0.014 ± 0.002 | 0.9 ± 0.1 | 2.1 ± 0.6 |
| C134T MSRB2 | 0.064 ± 0.012 | 1.3 ± 0.1 | 18.1 ± 3.2 | |
A turnover number of 0.028 s−1 was determined for MSRB2 (Table 1). This value, 3-fold lower than that of MSRB1, is in agreement with previous results (15). Interestingly, activity of mutated MSRB2, in which Cys-134 was replaced by a Thr, was higher than that of WT MSRB2 and in the same range of that recorded for MSRB1. A similar value was recorded for C134S MSRB2 (data not shown). To determine whether the different levels of activity observed for these proteins were due to alterations in substrate reduction or in regeneration of catalytic Cys by reductant, we carried out assays in the absence of DTE to calculate the stoichiometry of the reactions and the Km for dabsyl-MetSO (KDabsyl-MetSO) (Table 1). MSRB1 and MSRB2 possess comparable KDabsyl-MetSO for the substrate, in the range of 2 μm. For all WT and mutated MSRBs, an expected stoichiometry of nearly 1 mol of Met formed per mol of enzyme was measured. The T132C MSRB1 mutation resulted in a 2.5-fold increase in the KDabsyl-MetSO value compared with that of WT MSRB1. This indicates that the mutation does not significantly affect the capacity of MetSO reduction and suggests that the very low activity observed in the presence of DTE likely originates from a modification affecting the reduction of the catalytic Cys. With regard to C134T MSRB2, the KDabsyl-MetSO is 9-fold higher than that of WT MSRB2. Nevertheless, C134T MSRB2 exhibits a higher activity than the WT enzyme in the presence of DTE, suggesting that the mutation favors the regeneration of the catalytic Cys. These results demonstrate that MSRB1 and MSRB2 share a similar capacity of MetSO reduction and indicate that they very likely possess distinct features regarding the regeneration of their activity.
Formation of a Disulfide Bond in MSRB2 after MetSO Reduction
To delineate the mechanisms of reduction of plastidial MSRBs, we assayed the capacity of TCEP, a specific non-thiol based reductant of disulfide bonds (30, 31), to regenerate the activity of MSRBs (Table 1). A very low turnover number, representing only 3% that of the value measured in the presence of DTE, was recorded for MSRB1. This result reveals that TCEP is not an efficient reductant for MSRB1. In contrast, a kcat of 0.014 s−1 was recorded for MSRB2. This value corresponds to nearly 50% that of the activity determined using DTE and shows that TCEP regenerates MSRB2 activity. As TCEP specifically reduces disulfide bridges, these results argue for the formation of disulfide bond in MSRB2 but not in MSRB1.
The free thiol content of pre-reduced WT and mutated MSRBs was then measured in the presence or absence of N-acetyl-MetSO under non-denaturing conditions to avoid titration of the four zinc-coordinating Cys (Table 2). Reduced MSRB1 was found to contain 2.4 thiols, consistent with the expected 2 free thiols, Cys-186 and Cys-116. After the addition of N-acetyl-MetSO, 1.6 thiols were measured, corresponding to a decrease of 0.8 free thiols (Table 2). These results are consistent with the oxidation of one single Cys, very likely Cys-186, to a sulfenic acid form. Note that the formation of an intermolecular disulfide can be excluded, as no MSRB1 dimer was observed after oxidation of WT or mutated forms, even in the presence of a large amount of diamide (data not shown). Thiol titration experiments were also achieved using T132C MSRB1. The reduced form of T132C MSRB1 contains about 3 thiols, likely corresponding to Cys-116, Cys-186, and the Cys replacing Thr in position 132 (Table 2). The decrease of ∼1 thiol after oxidation by substrate is consistent with the oxidation of only the catalytic Cys-186, with Cys-116 and Cys-132 still being free to react with DTNB. These data indicate that no disulfide bridge is formed in WT or mutated MSRB1.
TABLE 2.
Free thiol content in WT and mutated MSRB proteins in the absence or presence of N-acetyl-MetSO
The content of free thiols in pre-reduced MSRBs was titrated using a standard DTNB assay in the absence or presence of a saturating concentration of N-acetyl-MetSO (0.5 mm). Data are expressed in mol of SH·mol of enzyme−1. Data presented are the means ± S.D. (n = 3).
| Number of free Cys | Number of free thiol measured |
|||
|---|---|---|---|---|
| Before N-acetyl-MetSO treatment | After N-acetyl-MetSO treatment | Decrease in free thiols (before to after) | ||
| MSRB1 | 2 | 2.4 ± 0.1 | 1.6 ± 0.1 | 0.8 ± 0.1 |
| T132C MSRB1 | 3 | 3.3 ± 0.5 | 2.4 ± 0.4 | 0.9 ± 0.1 |
| MSRB2 | 3 | 3.2 ± 0.1 | 1.3 ± 0.1 | 1.9 ± 0.1 |
| C134T MSRB2 | 2 | 1.9 ± 0.1 | 1.2 ± 0.2 | 0.7 ± 0.1 |
In the case of reduced MSRB2, 3.2 thiol groups were titrated, likely corresponding to the two redox-active Cys (Cys-134 and Cys-187) and the additional free Cys (Cys-115). On the other hand, only 1.3 thiols were titrated after incubation with substrate (Table 2). The calculated decrease of 1.9 units of free thiols is consistent with the formation of an intramolecular disulfide bridge. Reduced C134T MSRB2 was found to contain ∼2 thiols/mol. A decrease of 0.7 units of thiol was observed after incubation with N-acetyl-MetSO, likely due to oxidation of one unique Cys, Cys-187, to a sulfenic acid intermediate after reaction with substrate, with the Cys-115 still free to react with DTNB. Thiol titration carried out using C134S MSRB2 gave similar values (data not shown). Consistent with TCEP reduction assays, the thiol titration data led us to conclude that a disulfide bond is formed between Cys-134 and Cys-187 of MSRB2 after MetSO reduction.
Reduction of MSRBs by Trx
To determine the regeneration mechanism of MSRB2 by Trxs, we then monitored the activity of MSRB2 and C134T MSRB2 in the presence of a Trx-reducing system composed of NADPH, Arabidopsis NADPH thioredoxin reductase B, and poplar Trx h1. The replacement of the potential resolving Cys, Cys-134, by Thr allowed us to mimic the MSRB1 sequence. In this assay WT MSRB2 exhibited a turnover number of 1.28 s−1, whereas no significant consumption of NADPH was observed for C134T MSRB2, indicating that the mutation compromises MSRB recycling by Trx (Table 3). Similar results were obtained with a mutated MSRB2, in which Cys-134 was replaced by Ser (data not shown). These results are consistent with the absence of recycling by Trx previously reported for MSRB1, which naturally possess a Thr at the position corresponding to the resolving Cys-134 (15). To gain further insight into the reduction mechanism, we assayed the capacity of C42S Trx h1, a mutant form lacking the second active site Cys, to supply reducing power to MSRB2. No activity was detected using this mutant, indicating that the resolving Cys-42 of Trx h1 is required for MSRB2 reduction (data not shown). Then, we tested whether T132C MSRB1 could be reduced by Trx h1. Despite the presence of Cys instead of Thr in position 132, corresponding to the potential resolving Cys of MSRB2, we observed that this mutated MSRB1 does not use canonical Trxs as electron donors (data not shown). This could be due to an inappropriate distance between the sulfhydryl group of the introduced Cys and the one of the catalytic Cys and/or to an orientation not compatible with the formation of a disulfide bond with the catalytic Cys.
TABLE 3.
Turnover numbers of WT and mutated MSRBs using Trx h1, GrxC4, and GrxS12 as electron donors
Measurements were carried out under steady-state conditions after NADPH oxidation at 340 nm as described under “Experimental Procedures.” Data are represented as the means ± S.D. (n = 3). ND, not detected.
|
kcat |
|||
|---|---|---|---|
| Trx h1 | GrxC4 | GrxS12 | |
| s−1 | |||
| MSRB1 | ND | 0.48 ± 0.02 | 0.58 ± 0.01 |
| C116S MSRB1 | 0.34 ± 0.12 | 0.50 ± 0.02 | |
| T132A MSRB1 | 0.37 ± 0.06 | 0.31 ± 0.04 | |
| T132C MSRB1 | ND | ND | ND |
| MSRB2 | 1.28 ± 0.03 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| C134T MSRB2 | ND | 0.30 ± 0.01 | 0.36 ± 0.06 |
Reduction of MSRBs by Grxs
We investigated the capacity of Grxs to reduce WT and mutated MSRBs. First, we determined MSR activity using HPLC quantification of dabsyl-MetSO reduction. The addition of either Grx or GSH alone in the reaction mixture did not significantly increase dabsyl-Met production, unlike the combined presence of GSH and either GrxC4 or GrxS12 (data not shown). These results indicate that both GSH and Grxs are required for MSRB1 regeneration. Then activities were determined using the NADPH coupled system (Table 3). In the presence of 50 μm GrxC4 or GrxS12, MSRB1 was efficiently reduced, as it exhibited activities nearly 10-fold higher than the basal activity measured when adding all components except Grx (kcat of 0.48 and 0.58 s−1 with GrxC4 and GrxS12, respectively) (Table 3). Turnover numbers of 0.37 and 0.31 s−1 were recorded using T132A MSRB1 with GrxC4 and GrxS12, respectively. These data indicate that this mutation only slightly affects regeneration by Grxs. In contrast, no significant activity could be detected when using T132C MSRB1 (data not shown). This result is consistent with DTE regeneration assays, which indicated that changing Thr-132 in Cys likely modifies the MSRB1 regeneration capacity. In the case of C116S MSRB1, the activity values measured when using GrxC4 and GrxS12 as electron donors were 0.34 and 0.50 s−1, respectively (Table 3). These values, relatively comparable with those measured for the WT form, show that Cys-116 is not involved in the regeneration of MSRB1 activity by Grxs. Thus, because no other Cys can act as a resolving residue, these data indicate that regeneration of MSRB1 activity involves only the catalytic Cys-186. Regarding MSRB2, no activity was recorded using Grxs as electron donors (Table 3) (15). But very interestingly, in the case of C134T MSRB2, substantial turnover numbers (0.30 and 0.36 s−1) were recorded using GrxC4 and GrxS12, respectively, and were in the same range as those obtained for MSRB1. These results demonstrate that the replacement of the resolving Cys by a Thr in 2-Cys MSRB2 is sufficient to allow regeneration by Grxs.
Reduction of MSRB1 by Mutated Grxs
To delineate the mechanism used by Grxs for regenerating MSRB1 activity, we compared the ability of WT and mutated forms of GrxC4, possessing a classical dithiol CPYC active site but no extra-Cys, and of GrxS12, possessing an atypical monothiol CSYS active site and a Cys in the C-terminal part (Cys-87), to supply electrons to MSRB1. On one hand, C27S GrxC4 and C29S GrxS12, in which the first redox active Cys was changed to Ser, lost their ability to supply electrons to MSRB1. On the other hand, mutations to Ser of the other Cys, Cys-30 in GrxC4, or Cys-87 in GrxS12 did not prevent the capacity of Grxs to reduce MSRB1 (Table 4). Regarding kinetic parameters, the saturation curves obtained when varying Grx concentrations were found to follow Michaelis-Menten kinetics (data not shown), in agreement with previous data (15). The apparent Km between WT MSRB1 and Grxs (Km Grx), measured in steady-state conditions, were found to be in the low μm range, with a value higher for GrxC4 than for GrxS12 (6.8 and 1.3 μm, respectively), whereas comparable turnover numbers, about 0.5 s−1, were calculated for both Grxs. These differences resulted in a 6-fold higher catalytic efficiency (kcat/Km Grx) in the presence of GrxS12 (Table 4). The C30S mutation in GrxC4 leads to a 7-fold increase of Km Grx and a 2-fold increase of kcat, resulting in a 4-fold decrease in catalytic efficiency consistent with previous observations (26). For GrxS12, the C87S mutation did not significantly alter its catalytic parameters. This is consistent with the recent biochemical characterization of GrxS12 revealing that Cys-87 is dispensable for activity and does not form a disulfide bridge with the catalytic Cys-29 (22). Altogether, these results demonstrate that both GrxC4 and GrxS12 use only the N-terminal active site Cys to regenerate MSRB1 activity and that GSH is absolutely required for this process.
TABLE 4.
Kinetic parameters of MSRB1 using WT and mutated GrxC4 and GrxS12 as electron donors
Measurements were carried out after NADPH oxidation at 340 nm as described under “Experimental Procedures.” Data are represented as the means ± S.D. (n = 3).
| kcat | KmGrx | kcat/KmGrx | |
|---|---|---|---|
| s−1 | μm | m−1·s−1 | |
| GrxC4 | 0.48 ± 0.02 | 6.8 ± 0.8 | 73,400 ± 10,800 |
| C27S GrxC4 | 0.07 ± 0.01 | ||
| C30S GrxC4 | 0.83 ± 0.07 | 47.1 ± 8.5 | 18,300 ± 4,300 |
| GrxS12 | 0.58 ± 0.01 | 1.3 ± 0.1 | 444,000 ± 42,600 |
| C29S GrxS12 | 0.05 ± 0.01 | ||
| C87S GrxS12 | 0.48 ± 0.01 | 1.5 ± 0.1 | 330,600 ± 26,300 |
Glutathionylation of MSRB1 after MetSO Reduction
We used MALDI-TOF mass spectrometry to determine the masses of MSRBs in the reduced form or after oxidation by MetSO. The mass measured for reduced MSRB2 was very close to that expected (−1.1 Da), whereas the experimental mass of reduced MSRB1 was slightly inferior (−9.0 Da) to that theoretically deduced from the sequence but inconsistent with an amino acid cleavage (Table 5, Fig.1A). After oxidation in the presence of 2 mm N-acetyl-MetSO, the masses of MSRB1 and MSRB2 were found to increase by 15.3 and 4.3 Da, respectively (Table 5, Fig. 1B). While the increase is not significant in the case of MSRB2, the increase observed for MSRB1 is significantly higher than the experimental error (±0.05%, ∼±8 Da) and affects most of the protein pool. The mass increase is in the range of 16 Da and corresponds to the addition of an oxygen atom in the protein (Fig. 1). These results argue for the formation of a stable sulfenic acid on Cys-186 of MSRB1 after oxidation by the substrate.
TABLE 5.
Molecular masses of MSRB1 and MSRB2 after reducing and oxidizing treatments
Molecular masses were determined using MALDI-TOF mass spectrometry. Masses correspond to major peaks and are in daltons. The differences were calculated between the measured masses of treated and untreated proteins. Accuracy of the measurement is ±8 Da.
| Reduced |
+N-Acetyl-MetSO |
+GSH |
+N-Acetyl-MetSO + GSH |
|||||
|---|---|---|---|---|---|---|---|---|
| Theoretical | Measured | Measured | Difference | Measured | Difference | Measured | Difference | |
| Da | Da | Da | Da | |||||
| MSRB1 | 16,623.6 | 16,614.6 | 16,629.9 | +15.3 | 16,615.4 | +0.8 | 16,919.4 | +304.8 |
| MSRB2 | 16,859.8 | 16,858.9 | 16,863.2 | +4.3 | 16,866.4 | +7.5 | 16,859.2 | +0.3 |
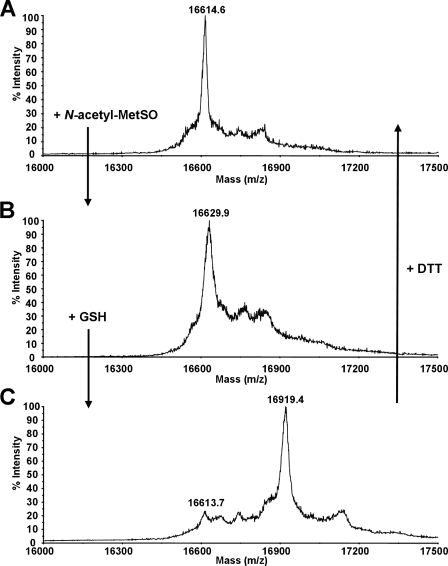
FIGURE 1.
MALDI-TOF mass spectrometry of MSRB1 incubated with N-acetyl-MetSO and GSH. Mass spectra of reduced MSRB1 (A) and of MSRB1 incubated with either N-acetyl-MetSO (B) or N-acetyl-MetSO and GSH (C). After N-acetyl-MetSO treatment, an increase of 15.3 Da was recorded (B), whereas an increase of 304.8 Da was observed after treatment with N-acetyl-MetSO in the presence of GSH (C). The shift observed in C was fully reversed by DTT treatment. Accuracy of the measurement is ±8 Da.
Then we performed mass analyses to determine whether the catalytic Cys-186 of MSRB1 could undergo glutathionylation after MetSO reduction. After incubation in the presence of N-acetyl-MetSO and GSH, a shift in molecular mass was observed for MSRB1 but not for MSRB2 (Fig. 1C, Table 5). This shift of 304.8 Da is perfectly consistent with the presence of one glutathione adduct per molecule of MSRB1 (theoretical additional mass, 305 Da) and affects most of the protein pool. Moreover, after DTT treatment of MetSO/GSH-treated MSRB1, the molecular mass of the protein shifted back to the mass of the reduced form, showing that glutathionylation of the protein is fully reversible. In contrast, no mass increase was observed after treatment with GSH alone. These results suggest that after the formation of the sulfenic acid upon MetSO reduction, Cys-186 is the residue that undergoes glutathionylation. This hypothesis was validated by peptide mass fingerprinting after tryptic digestion of glutathionylated MSRB1 (Fig. 2). The peptide profile of glutathionylated MSRB1 reveals an additional peak compared with the profile of the reduced protein. This peak corresponds to the peptide Arg-184—Lys-193, containing the catalytic Cys-186, shifted by 305 Da. These data unambiguously reveal that the mass increase is due to the formation of a mixed disulfide between GSH and the thiol of Cys-186. The modification appeared to be reversible as the additional peak was no longer detected after DTT treatment.
FIGURE 2.
MALDI-TOF mass spectrometry of MSRB1 tryptic peptides incubated with N-acetyl-MetSO and GSH. MALDI-TOF spectra of tryptic peptides before (A) or after (B) N-acetyl-MetSO + GSH treatment. After the treatment, the molecular mass of the peptide 184RYCLNSAALK193, containing the catalytic Cys-186, is partially shifted by 305 Da. The shift was fully reversed by DTT treatment.
DISCUSSION
The goal of the present study was to delineate the mechanisms used by Grxs for the reduction of the Arabidopsis plastidial 1-Cys MSRB1 and to compare it to the Trx-dependent reduction of the 2-Cys MSRB2. Despite the distinct number of redox-active Cys, MSRB1 and MSRB2 display very similar biochemical capacities and an expected stoichiometry of 1 mol of Met formed per mol of enzyme. Using DTE as a reductant, MSRB1 was found to be 3-fold more efficient than MSRB2 (Table 1). Interestingly, the mutation of Cys-134 to Thr in MSRB2 led to an increase (2.3-fold) in activity up to the value range recorded for MSRB1 (Table 1). This has been observed previously, for example with the MSRB domain of the Neisseria meningitidis PILB protein (5), for which the Cys to Ser mutation of the resolving Cys resulted in a 12-fold increase of the activity. As the reduction of the disulfide bond formed between catalytic Cys and resolving Cys has been demonstrated to be the rate-limiting step in MSRs (6), the higher activity recorded with C134T and C134S MSRB2 compared with that of the WT form (Table 1 and data not shown) could originate from a shorter catalytic cycle. Indeed, in the case of mutated MSRB2 and of MSRB1, the cycle is very likely composed of only two steps, reduction of MetSO and direct reduction by DTE of the sulfenic acid formed on catalytic Cys. Interestingly, most MSRB proteins from bacteria and animal cells belonging to the 1-Cys type display a Thr or a Ser in place of the resolving Cys (7, 10). The substitution of Thr-132 by Cys or Ala in MSRB1 resulted in a substantial loss of activity. In agreement with these data, Kim and Gladyshev (10) reported that replacement of Ser by Cys in mammalian MSRB3 led to a strong decrease in the MetSO reductase activity when using DTT as an electron donor. However, T132A MSRB1, but not T132C MSRB1, was still efficiently reduced by Grxs. These results point out that Thr could be an important residue at this position regarding catalytic and/or regeneration mechanisms of MSRB1. However, the analysis of the known three-dimensional structures of MSRB possessing a Thr or a Ser at this position (Protein Data Base accession codes 2K8D (Methanobacterium MSRB) and 2KAO (mouse MSRB1)) reveals that these hydroxylated residues are far from the active site or from any residue involved in the catalytic mechanism. But as MSRs are very flexible enzymes displaying considerable changes between reduced and oxidized forms,4 only a dynamic and/or structural characterization of MSRB1 would help to uncover a potential role of Thr-132.
Using site-directed mutagenesis, we investigated the role of Cys-134 in the Trx-mediated recycling process of MSRB2. Activity measurements combined with the titration of free thiol groups in various oxidation states (Table 3) and mass spectrometry analyses (Table 5) led us to propose that the sulfenic acid formed on Cys-187 is reduced by the nucleophilic attack of the sulfur atom of Cys-134, resulting in the formation of an intramolecular disulfide bridge. This hypothesis has been validated by the fact that TCEP, a specific reductant of disulfide bonds (30–31), regenerates MSRB2 activity. Moreover, the modeling of MSRB2 three-dimensional structure indicates that the distance between the two sulfur atoms (3.6 Å) is compatible with the formation of an intramolecular disulfide bond (data not shown). This bond is reduced subsequently by Trx through a classical dithiol-disulfide exchange involving both catalytic and resolving Cys of the two partners. The proposed mechanism of MSRB2 reduction by canonical Trxs is in agreement with those described for other 2-Cys MSRBs (5, 7) that possess two redox-active Cys or a catalytic selenocysteine and a resolving Cys (10). Our data indicate that this mechanism is likely conserved in 2-Cys MSRBs of higher plants and green algae, which are very similar to Arabidopsis MSRB2 (8).
Titration of free thiol groups and reduction assays using TCEP showed that only one Cys is oxidized by the substrate in MSRB1 and in C134T MSRB2 (Tables 1 and 2). In other respects, mutated MSRB1, in which the Cys-116 was replaced by a Ser, was found to be almost as active as WT MSRB1 and to retain its capacity to be reduced by Grxs. This demonstrates that Cys-116 cannot act as a resolving Cys and that MSRB1 uses only Cys-186 for the catalytic activity and the Grx-dependent regeneration mechanism. Moreover, the fact that the replacement of resolving Cys-134 of MSRB2 by a Thr allows the generation of an “MSRB1-like” enzyme regenerated by Grxs leads us to propose that the distinct regeneration mechanisms for MSRB1 and MSRB2 originate from the presence or not of a stable sulfenic acid intermediate, which is linked to the absence or presence of a resolving Cys. Hence, in MSRB2, the resolving Cys-134 very likely rapidly attacks the sulfenic acid formed after MetSO reduction, whereas in MSRB1 and in C134T MSRB2, the sulfenic acid form of catalytic Cys is much more stable due to the absence of a resolving one. The mass spectrometry results validate this hypothesis. Indeed, after incubation with an excess of N-acetyl-MetSO, an increase of nearly 16 Da was observed for most of the MSRB1 protein pool assayed. In comparison, no significant mass difference was recorded for MSRB2. The formation of a transient sulfenic acid was shown in Drosophila 2-Cys MSRB after MetSO reduction; mass spectrometry of dimedone-treated WT 2-Cys MSRB after MetSO reduction revealed a small peak with a +138 Da mass shift, suggesting the presence of a dimedone-reacted sulfenic acid, but the major fraction of the enzyme was converted by MetSO to an oxidized form not able to bind dimedone, suggesting a very fast reduction of the sulfenic acid intermediate by the resolving Cys (7). A similar result was observed for E. coli MSRA (32). In contrast, our data reveal the presence of a stable sulfenic acid intermediate in 1-Cys MSRB after MetSO reduction.
The mass spectrometry results acquired on MSRB1 incubated with N-acetyl-MetSO and GSH showed that the sulfenic acid formed on Cys-186 is attacked by glutathione. The fact that GSH is absolutely required for the Grx-dependent MSRB1 regeneration, as shown by activity assays, is also consistent with the observation that Grxs alone are unable to reduce the sulfenic acid on MSRB1. The use of monothiol and dithiol Grxs and of Cys to Ser-mutated forms showed that only the catalytic Cys is required to provide MSRB1 with electrons (Table 4). These results indicate that reduction of MSRB1 by Grxs is very likely performed through a monothiol mechanism involving only the N-terminal active site Cys. Based on our data, we propose a model (Fig. 3) in which the catalytic mechanism for 1-Cys MSRB1 involves (i) the formation of a sulfenic acid on Cys-186 due to MetSO reduction, (ii) the glutathionylation of Cys-186 through reaction of the sulfenic acid with GSH, and (iii) the regeneration of reduced MSRB1 through deglutathionylation mediated by Grxs. Then the thiol group of an external reduced glutathione would reduce glutathionylated Grx, as it is well established that Grxs can catalyze protein deglutathionylation using only the N-terminal active site Cys (21, 22, 33).
FIGURE 3.
Model for the regeneration of MSRB1 by Grxs. The first step consists of MetSO reduction with the concomitant release of 1 mol of Met and the formation of a stable sulfenic acid intermediate on catalytic Cys-186 (1). The sulfenic acid is then attacked by GSH leading to the liberation of one molecule of water and to the formation of a glutathione adduct (2), which is subsequently solved by Grxs through the N-terminal Cys of the active site (3).
Our results show for the first time a glutathionylation step in the regeneration of the activity of MSRB enzymes. Interestingly, these results are related to those reported on plant type II peroxiredoxins (Prxs). These are thiol-dependent peroxidases which also use the sulfenic acid chemistry and the GSH/Grx system for their regeneration. Note that these type II Prxs accept the Trx system as an alternative reducing system (34). The study of the Grx-mediated Prx regeneration mechanism also showed that only the N-terminal catalytic Cys of Grxs is required. However, the order in which GSH and Grx are involved in this process is still unclear because covalent heterodimers can be formed between Prx and Grx in the absence of GSH (34). Interestingly, the regeneration mechanism of human 1-Cys Prx by glutathione S-transferase has been shown to implicate glutathionylation of the oxidized catalytic Cys (35, 36). The data acquired in the present work extend the participation of glutathionylation in activity regeneration of another type of enzyme involved in protection against oxidative modification, i.e. 1-Cys methionine sulfoxide reductases B. In other respects, several studies proposed that reversible glutathionylation of Cys could be a protective mechanism during oxidative stress. For instance, Zaffagnini et al. (37) reported that plant chloroplastic glyceraldehyde-3-phosphate dehydrogenase is transiently inactivated in vitro by glutathionylation of the catalytic Cys after an oxidative treatment. Similar results were recently reported for the 20 S proteasome of yeast (38). In conclusion, glutathionylation appears to fulfill at least two distinct roles during oxidative stress, (i) regeneration of the activity of stress-specific enzymes such as MSRs and Prxs and (ii) transient inactivation and protection of metabolic enzymes. In addition, as glutathionylation has also been shown to alter, either positively or negatively, the activity of many signaling proteins, including several members of the NF-κB pathway and protein-tyrosine phosphatases, to cite only few examples, it is tempting to speculate that glutathionylation also participates to the signaling pathways in response to oxidative stress (39–41).
From an evolutionary point of view, the presence of a single MSRB gene in most prokaryotes suggests that a prototypic enzyme exists and that the various types of enzymes evolved from this ancestor. Regardless of which enzyme was the ancestor, the 1-Cys or the 2-Cys MSRB, it is worth mentioning that the absence of resolving Cys leads to a drastic change in the regeneration system, as 1-Cys MSRBs are reduced by the GSH/Grx system. But it is worth mentioning that this system is not the only possible reducing system. Indeed, some mammalian Trxs have been shown to reduce 1-Cys MSRB2 and MSRB3 (13), and we previously reported that the peculiar plant Trx CDSP32, which participates in the tolerance to oxidative stress and interacts with MSRB1 in plant extracts (42, 43), regenerates the activity of this 1-Cys MSRB (14, 15) without the addition of GSH or of another thiol compound (14). This process might involve direct reduction of the sulfenic acid and the formation of a heterodimeric disulfide complex (13). Several other compounds have been proposed as potential reducing agents for 1-Cys MSRBs. Indeed, Sagher et al. reported that selenocystamine, selenocysteine (11), and thionein, the reduced apoprotein of zinc-metallothionein (12), are able to reduce human 1-Cys MSRB2 and MSRB3. However, the in vivo significance of these data remains largely unclear. Our data give credence to a function of GSH, the major low molecular weight thiol in cells, and Grxs in the physiological reducing system for 1-Cys MSRBs. This is in agreement with the roles of GSH in protection against oxidative stress and redox homeostasis that are clearly established in plants (for review, see Ref. 18) as well as in other organisms (for review, see Ref. 44). Preliminary results acquired with Arabidopsis plants lacking both MSRB1 and MSRB2 genes and identification of their potential targets reveal that the two plastidial MSRBs could have redundant functions in protecting plants against oxidative damage (data not shown). In contrast, the specificity observed for their reductants reveals a difference in electron supply (NADPH for the GSH/Grx system, ferredoxin for the Trx system), which could be associated with a preservation of the plastidial MSRB capacity under various environmental conditions.
Supplementary Material
Acknowledgments
We are very grateful to Patricia Henri (Commissariat à l'Energie Atomique (Cadarache, France) (CEA), Institut de Biologie Environmentale et Biotechnology (IBEB), Service de Biologie Végétale et Microbiologie Environnementales (SBVME), Laboratoire d'Ecophysiologie Moléculaire des Plantes (LEMP)) for technical assistance, Noelle Becuwe (CEA, IBEB, SBVME, LEMP) for mutagenesis, and Stéphan Cuiné (CEA, IBEB, SBVME, LB3M) and Rémy Puppo (CEA, IBEB, SBVME, LEMP) for helpful advice in protein purification and HPLC experiments.
This work was supported by Agence Nationale de la Recherche, ANR-Génoplante Grants GNP05010G (to P. R., E. L., and N. R.) and ANR-JC45751 (to M. Z. and S. D. L.). Financial support (to L. T.) from Région Provence-Alpes-Côtes-d'Azur is acknowledged.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
F. Favier, personal communication.
- MetSO
- methionine sulfoxide
- MSR
- methionine sulfoxide reductases
- Trx
- thioredoxin
- Grx
- glutaredoxin
- GSH
- reduced glutathione
- DTT
- reduced dithiothreitol
- DTE
- reduced dithioerythritol
- DTNB
- 5,5′-dithiobis-2-nitrobenzoic acid
- TCEP
- Tris(2-carboxyethyl) phosphine hydrochloride
- Prx
- peroxiredoxins
- WT
- wild type
- HPLC
- high performance liquid chromatography
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight.
REFERENCES
- 1.Davies M. J. (2005) Biochim. Biophys. Acta 1703, 93–109 [DOI] [PubMed] [Google Scholar]
- 2.Brot N., Weissbach L., Werth J., Weissbach H. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 2155–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moskovitz J., Berlett B. S., Poston J. M., Stadtman E. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimaud R., Ezraty B., Mitchell J. K., Lafitte D., Briand C., Derrick P. J., Barras F. (2001) J. Biol. Chem. 276, 48915–48920 [DOI] [PubMed] [Google Scholar]
- 5.Olry A., Boschi-Muller S., Marraud M., Sanglier-Cianferani S., Van Dorsselear A., Branlant G. (2002) J. Biol. Chem. 277, 12016–12022 [DOI] [PubMed] [Google Scholar]
- 6.Boschi-Muller S., Gand A., Branlant G. (2008) Arch. Biochem. Biophys. 474, 266–273 [DOI] [PubMed] [Google Scholar]
- 7.Kumar R. A., Koc A., Cerny R. L., Gladyshev V. N. (2002) J. Biol. Chem. 277, 37527–37535 [DOI] [PubMed] [Google Scholar]
- 8.Tarrago L., Laugier E., Rey P. (2009) Mol. Plant 2, 202–217 [DOI] [PubMed] [Google Scholar]
- 9.Neiers F., Kriznik A., Boschi-Muller S., Branlant G. (2004) J. Biol. Chem. 279, 42462–42468 [DOI] [PubMed] [Google Scholar]
- 10.Kim H. Y., Gladyshev V. N. (2005) PLoS Biol. 3, e375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagher D., Brunell D., Brot N., Vallee B. L., Weissbach H. (2006) J. Biol. Chem. 281, 31184–31187 [DOI] [PubMed] [Google Scholar]
- 12.Sagher D., Brunell D., Hejtmancik J. F., Kantorow M., Brot N., Weissbach H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8656–8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H. Y., Kim J. R. (2008) Biochem. Biophys. Res. Commun. 371, 490–494 [DOI] [PubMed] [Google Scholar]
- 14.Ding D., Sagher D., Laugier E., Rey P., Weissbach H., Zhang X. H. (2007) Biochem. Biophys. Res. Commun. 361, 629–633 [DOI] [PubMed] [Google Scholar]
- 15.Vieira Dos Santos C., Laugier E., Tarrago L., Massot V., Issakidis-Bourguet E., Rouhier N., Rey P. (2007) FEBS Lett. 581, 4371–4376 [DOI] [PubMed] [Google Scholar]
- 16.Rey P., Pruvot G., Becuwe N., Eymery F., Rumeau D., Peltier G. (1998) Plant J. 13, 97–107 [DOI] [PubMed] [Google Scholar]
- 17.Vieira Dos Santos C., Cuiné S., Rouhier N., Rey P. (2005) Plant Physiol. 138, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouhier N., Lemaire S. D., Jacquot J. P. (2008) Annu. Rev. Plant Biol. 59, 143–166 [DOI] [PubMed] [Google Scholar]
- 19.Jung C. H., Thomas J. A. (1996) Arch. Biochem. Biophys. 335, 61–72 [DOI] [PubMed] [Google Scholar]
- 20.Zaffagnini M., Michelet L., Massot V., Trost P., Lemaire S. D. (2008) J. Biol. Chem. 283, 8868–8876 [DOI] [PubMed] [Google Scholar]
- 21.Bushweller J. H., Aslund F., Wüthrich K., Holmgren A. (1992) Biochemistry 31, 9288–9293 [DOI] [PubMed] [Google Scholar]
- 22.Couturier J., Koh C. S., Zaffagnini M., Winger A. M., Gualberto J. M., Corbier C., Decottignies P., Jacquot J. P., Lemaire S. D., Didierjean C., Rouhier N. (2009) J. Biol. Chem. 284, 9299–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamarit J., Belli G., Cabiscol E., Herrero E., Ros J. (2003) J. Biol. Chem. 278, 25745–25751 [DOI] [PubMed] [Google Scholar]
- 24.Behm M., Jacquot J. P. (2000) Plant Physiol. Biochem. 38, 363–369 [Google Scholar]
- 25.Jacquot J. P., Rivera-Madrid R., Marinho P., Kollarova M., Le Maréchal P., Miginiac-Maslow M., Meyer Y. (1994) J. Mol. Biol. 235, 1357–1363 [DOI] [PubMed] [Google Scholar]
- 26.Rouhier N., Gelhaye E., Jacquot J. P. (2002) FEBS Lett. 511, 145–149 [DOI] [PubMed] [Google Scholar]
- 27.Rouhier N., Kauffmann B., Tete-Favier F., Palladino P., Gans P., Branlant G., Jacquot J. P., Boschi-Muller S. (2007) J. Biol. Chem. 282, 3367–3378 [DOI] [PubMed] [Google Scholar]
- 28.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 29.Augusto L. A., Decottignies P., Synguelakis M., Nicaise M., Le Maréchal P., Chaby R. (2003) Biochemistry 42, 3929–3938 [DOI] [PubMed] [Google Scholar]
- 30.Rüegg U. T., Rudinger J. (1977) Methods Enzymol. 47, 111–126 [DOI] [PubMed] [Google Scholar]
- 31.Getz E. B., Xiao M., Chakrabarty T., Cooke R., Selvin P. R. (1999) Anal. Biochem. 273, 73–80 [DOI] [PubMed] [Google Scholar]
- 32.Boschi-Muller S., Azza S., Sanglier-Cianferani S., Talfournier F., Van Dorsselear A., Branlant G. (2000) J. Biol. Chem. 275, 35908–35913 [DOI] [PubMed] [Google Scholar]
- 33.Gallogly M. M., Starke D. W., Mieyal J. J. (2009) Antioxid. Redox Signal. 11, 1059–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouhier N., Gelhaye E., Jacquot J. P. (2002) J. Biol. Chem. 277, 13609–13614 [DOI] [PubMed] [Google Scholar]
- 35.Manevich Y., Feinstein S. I., Fisher A. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3780–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralat L. A., Manevich Y., Fisher A. B., Colman R. F. (2006) Biochemistry 45, 360–372 [DOI] [PubMed] [Google Scholar]
- 37.Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., Miginiac-Maslow M., Noctor G., Trost P., Lemaire S. D. (2007) FEBS J. 274, 212–226 [DOI] [PubMed] [Google Scholar]
- 38.Silva G. M., Netto L. E., Discola K. F., Piassa-Filho G. M., Pimenta D. C., Bárcena J. A., Demasi M. (2008) FEBS J. 275, 2942–2955 [DOI] [PubMed] [Google Scholar]
- 39.Dixon D. P., Fordham-Skelton A. P., Edwards R. (2005) Biochemistry 44, 7696–7703 [DOI] [PubMed] [Google Scholar]
- 40.Dalle-Donne I., Rossi R., Giustarini D., Colombo R., Milzani A. (2007) Free Radic. Biol. Med. 43, 883–898 [DOI] [PubMed] [Google Scholar]
- 41.Shelton M. D., Mieyal J. J. (2008) Mol. Cells 25, 332–346 [PMC free article] [PubMed] [Google Scholar]
- 42.Broin M., Cuiné S., Eymery F., Rey P. (2002) Plant Cell 14, 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rey P., Cuiné S., Eymery F., Garin J., Court M., Jacquot J. P., Rouhier N., Broin M. (2005) Plant J. 41, 31–42 [DOI] [PubMed] [Google Scholar]
- 44.Holmgren A., Johansson C., Berndt C., Lönn M. E., Hudemann C., Lillig C. H. (2005) Biochem. Soc. Trans. 33, 1375–1377 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.