Abstract
Clostridium perfringens enterotoxin (CPE) binds to the extracellular loop 2 of a subset of claudins, e.g. claudin-3. Here, the molecular mechanism of the CPE-claudin interaction was analyzed. Using peptide arrays, recombinant CPE-(116–319) bound to loop 2 peptides of mouse claudin-3, -6, -7, -9, and -14 but not of 1, 2, 4, 5, 8, 10–13, 15, 16, 18–20, and 22. Substitution peptide mapping identified the central motif 148NPL150VP, supposed to represent a turn region in the loop 2, as essential for the interaction between CPE and murine claudin-3 peptides. CPE-binding assays with claudin-3 mutant-transfected HEK293 cells or lysates thereof demonstrated the involvement of Asn148 and Leu150 of full-length claudin-3 in the binding. CPE-(116–319) and CPE-(194–319) bound to HEK293 cells expressing claudin-3, whereas CPE-(116–319) bound to claudin-5-expressing HEK293 cells, also. This binding was inhibited by substitutions T151A and Q156E in claudin-5. In contrast, removal of the aromatic side chains in the loop 2 of claudin-3 and -5, involved in trans-interaction between claudins, increased the amount of CPE-(116–319) bound. These findings and molecular modeling indicate different molecular mechanisms of claudin-claudin trans-interaction and claudin-CPE interaction. Confocal microscopy showed that CPE-(116–319) and CPE-(194–319) bind to claudin-3 at the plasma membrane, outside cell-cell contacts. Together, these findings demonstrate that CPE binds to the hydrophobic turn and flanking polar residues in the loop 2 of claudin-3 outside tight junctions. The data can be used for the specific design of CPE-based modulators of tight junctions, to improve drug delivery, and as chemotherapeutics for tumors overexpressing claudins.
The clinical use of many promising drug candidates is impeded by unacceptable pharmacokinetics (1). The ability of a drug to pass through tissue barriers is a major determinant for its delivery. In epithelia and endothelia, the paracellular route is blocked by tight junctions (TJ).4 Different approaches have been used to enhance transcellular drug delivery. These include the use of influx transporters, blocking of efflux transporters, or receptor-mediated endocytosis (2). Alternative approaches aim to enhance paracellular permeation of drugs by loosening the TJ (3, 4). This strategy has the advantage that it could improve the delivery of structurally unrelated drugs, and the drug itself does not have to be modified. Although different TJ modulators have been described, most of these are based on surfactants or chelators (3). These often have low tissue specificity and cause severe side effects, e.g. exfoliation of cells, which irreversibly compromise the barrier functions (5, 6). Fewer side effects may be obtained by more specific modulation of a molecular key component of the TJ (7).
TJ consist of transmembrane proteins, mainly the tetraspan proteins of the claudin family, as well as occludin and tricellulin (8). Other molecules associated with TJ include membrane-bound scaffolding and signaling proteins (9). However, claudins (Cld) are the major functional constituent of TJ (10). Claudins tighten the paracellular space, selectively for tissue, size, and charge. The tissue-specific combination of the claudin subtypes present in heteropolymers is assumed to determine the permeability properties of TJ (11). It was therefore proposed that tissue-specific drug delivery via the paracellular route would be possible by modulation of the barrier-function of claudins in a subtype-specific manner (7).
A subset of claudins, e.g. Cld3 and -4 but not -1 and -2, have been shown to be receptors for Clostridium perfringens enterotoxin (CPE) with high association constants of about 108 m−1 (12). CPE causes one of the most common food-borne diseases (13). It consists of two functional domains, an N-terminal region that mediates the cytotoxic effect and the C-terminal region (CPE-(184–319)), which binds to extracellular loop 2 (ECL2) of Cld3 but not of Cld1 nor to the ECL1 of Cld3 (12). Treatment of epithelial monolayers with non-cytotoxic CPE-(184–319) increases paracellular permeability (14). CPE-(184–319) enhanced drug absorption in rat jejunum 400-fold relative to sodium caprate, which is in clinical use (15). Thus, CPE is a promising tool to specifically modulate claudins, the key constituents of TJ, and thereby to enhance paracellular drug delivery. In addition, some studies have suggested the use of CPE for the chemotherapy of tumors overexpressing claudins (16–18).
Cld1 and -5 are potential targets for transepidermal and brain drug delivery, respectively (19, 20). However, it has been reported that these claudins do not interact with CPE (12). Modification of CPE could enhance and/or shift its claudin-subtype specificity. Therefore, the design of CPE-based TJ modulators could permit efficient claudin subtype-specific modulation, which would also be tissue-specific modulation of TJ. To achieve this, an understanding of the molecular mechanism of the CPE-claudin interaction is a necessary prerequisite. In this study, we identify the residues within the ECL2 of Cld3 that are involved in interaction with CPE.
EXPERIMENTAL PROCEDURES
Plasmids
For construction of plasmids encoding GST-CPE-(116–319), GST-CPE-(194–319), and GST-CPE-(290–319) fusion proteins, cDNA of CPE (kindly provided by Dr. Y. Horiguchi, Osaka, Japan) was amplified by PCR and cloned into pGEX-4T1 (GE Healthcare) using EcoRI and SalI; GST-CPE-(194–309) was generated by site-directed mutagenesis. Plasmids encoding Cld5wt-CFP, Cld5wt-YFP, and mutant fusion proteins have been described previously (21). A pECFP-N1-plasmid containing the Cld3 sequence with a stop codon ahead of CFP was generated by subcloning full-length Cld3 with SalI and BamHI from pSK-Cl-3, kindly provided by Dr. M. Furuse (Kyoto, Japan). Similarly, Cld5wt was subcloned from pGTCL-5 (Dr. M. Furuse, Kyoto, Japan) into pEYFP-N1 using EcoRI. To generate the in-frame fusion Cld3wt-CFP, the stop codon in pECFP-N1-Cld3, was removed by side-directed mutagenesis. Cld3wt-YFP was generated by subcloning Cld3wt-CFP in pECFP-N1 into pEYFP-N1 using SalI and BamHI. The plasmids encoding mutants of Cld3 (Y147A, N148D, L150A, E153V, A154N, and Q155E, N148D/L150A) were generated by site-directed mutagenesis of pECFP-N1-Cld3wt. pEGFP-Cld4 was kindly provided by Dr. W. Hunziker (Singapore).
Antibodies
Rabbit anti-Cld3, rabbit anti-Cld4, rabbit anti-Cld5, HRP- and Alexa Fluor 488-conjugated anti-rabbit antibodies, HRP-, Cy3-, and Alexa Fluor 647-conjugated anti-mouse antibodies were obtained from Invitrogen. Mouse anti-GST (GST-2) was from Sigma. To detect CFP and YFP fusion proteins, mouse anti-GFP antibodies (Clontech) were used for Western blots.
Expression and Purification of CPE Constructs
CPE-(116–319), CPE-(194–319), CPE-(194–309), and CPE-(290–319) with N-terminal GST fusions as well as GST (control) were expressed in Escherichia coli BL21. Bacteria were grown to A600 = 0.6–0.8, harvested by centrifugation, and lysed in PBS with 1% (v/v) Triton X-100, 0.1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, protease inhibitor mixture (Sigma) by using an EmulsiFlex-C3 homogenizer (Avestin, Mannheim, Germany). To remove insoluble cell debris, the lysates were centrifuged at 40,000 × g for 1 h at 4 °C. The recombinant proteins were purified from supernatants using glutathione-agarose (Sigma) and eluted proteins were dialyzed against PBS buffer with Ca2+ and Mg2+.
Surface Plasmon Resonance (SPR)
Measurements were performed using a Biacore 2000 instrument (Biacore AB, Uppsala, Sweden). Claudin3-ECL2 peptides (Cld3-(140–159), Cys-(6-aminocaproic acid)-NTIIRDFYNPLVPEAQKREM; and Cld3-(140–159)N148D/L150T, Cys-(6-aminocaproic acid)-NTIIRDFYDPTVPEAQKREM) were synthesized with N-terminal cysteine followed by a 6-aminocaproic acid linker. Peptides were immobilized on a CM5 chip using the thiol coupling procedure, according to the supplier's instructions (Biacore AB). To determine the binding affinities of GST-CPE-(116–319) and GST-CPE-(290–319), an amount of covalently coupled Cld3-(140–159) peptide, corresponding to a signal increase of ∼1600 resonance units was used. For comparison of Cld3-(140–159) and Cld3-(140–159)N148D/L150T, 500 and 250 resonance units of Cld3 peptides were immobilized, respectively. Binding experiments were performed as described earlier (22). The analytes GST-CPE-(116–319), GST-CPE-(290–319), and GST (control) were injected in concentrations between 0.01 and 20 μm in running buffer (PBS, pH 7.4, supplemented with 2 mm dithiothreitol). For regeneration 10 mm NaOH solution with 1 m NaCl was used. For every analyte concentration the maximal amount of GST-CPE-(116–319) and GST-CPE-(290–319) bound was calculated in femtomole (∼1000 resonance units = 1 ng of protein bound).
Peptide Arrays
Arrays were generated by spot-synthesis using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry as described (23). The AutoSpot-Robot ASS 222 (Intavis Bioanalytical Instruments AG, Köln, Germany) was used for automatic synthesis of 15–20-mer peptides on Whatman 50 cellulose membranes (C-terminal immobilization). The quality of synthesized peptides was evaluated by mass spectrometry of control spots. For binding experiments membranes were incubated with 10 μg/ml GST-CPE-(116–319) or GST in blocking buffer (Sigma) overnight at 8 °C, followed by incubations with mouse anti-GST and HRP-conjugated goat anti-mouse antibody in blocking buffer for 1.5 and 1 h at room temperature, respectively. The ECL Western blotting detection reagent (GE Healthcare) was applied and chemiluminescence intensities were measured as Boehringer light units (BLU) using a LumiImager (Roche Applied Science). Determined BLU values of GST-incubated SPOTs were subtracted from BLU values of corresponding GST-CPE-(116–319)-incubated spots (ΔBLU). Background corrected ΔBLU values were normalized to the binding of GST-CPE-(116–319) on Cld3-spots (%Cld3).
Cell Culture and Transfection
Caco-2 human intestinal carcinoma cells and HEK293 cells (HEK cells) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units ml−1 penicillin, 100 μg ml−1 streptomycin, and 1% l-alanyl-l-glutamine. Transient transfections of HEK cells were preformed with Lipofectamine 2000 (Invitrogen).
Electrical Cell Substrate Impedance Sensing (ECIS)
Caco-2 cells were seeded into the wells of ECISTM 8W10E+ electrode arrays for ECISTM model 1600R (Applied Biophysics) at a density of 5 × 105 cells per well and grown in a humidified CO2 incubator with 5% CO2. Junctional resistance was measured every 5 min at 400 Hz frequency, as in a previous report (24). When monolayers reached maximum resistance (∼1500 Ω) 10 μg/ml GST-CPE-(116–319) or GST were introduced into the medium.
Measurements of Transepithelial Electrical Resistance (TEER) and Paracellular Permeation
5 × 105 Caco-2 cells were seeded into a 24-well transwell filter (Millipore, Eschborn, Germany). TEER was determined with Endohm electrodes (Millipore). After reaching stable TEER values, cells were incubated with GST-CPE-(116–319) or GST for 16 h. For permeation studies cells were incubated with 25 μg/ml fluorescein (Sigma) in Hanks' balanced salt solution buffer on the apical side and Hanks' balanced salt solution on the basal side for 10 min. 100-μl samples were removed from the basal compartment and the fluorescence units were measured using a fluorescence plate reader (Tecan, Crailsheim, Germany) to calculate the permeation coefficient.
Immunocytochemistry
Immunocytochemistry was performed as described (25). Briefly, 2–3 days after transient transfection, HEK cells were incubated with 10 μg/ml GST-CPE-(116–319), GST-CPE-(194–319), or GST for 20 min, 1 h, or 14 h at 37 °C. Cells were fixed with acetone, incubated with anti-GST antibodies, followed by incubation with anti-Cld3 or anti-Cld5 antibodies, and incubated with 4′,6-diamidino-2-phenylindole, Cy3, or Alexa Fluor 647-conjugated goat anti-mouse and Alexa Fluor 488-conjugated goat anti-rabbit secondary antibodies. Cells were analyzed with the LSM 510 META confocal microscope (Zeiss, Jena, Germany).
Pull-down Assay
HEK cells seeded in 6-well plates were transiently transfected with Cld3wt, Cld3mutants, or GFP-Cld4. Two to 3 days after transfection, cells were washed with ice-cold PBS, with Ca2+ and Mg2+, scraped, harvested (300 × g, 5 min, 4 °C), and quick frozen in liquid nitrogen. The cells were lysed (1% Triton X-100 in PBS, EDTA-free protease inhibitor mixture (Roche), 0.1 mm phenylmethylsulfonyl fluoride), incubated on ice for 10 min, and centrifuged (5 min, 10,000 × g, 4 °C). The supernatants were incubated with GST, GST-CPE-(116–319), GST-CPE-(194–319), or GST-CPE-(194–309) bound to glutathione-Sepharose beads (GE Healthcare) in mini columns (MoBiTec, Göttingen, Germany) for 30 min on a shaker at 4 °C. Beads were centrifuged (2 min, 500 × g, 4 °C) and the flow-through (unbound fraction) collected. After washing three times with PBS, 0.5% Triton X-100, bound proteins were eluted with Laemmli buffer (eluate). Eluate and unbound fractions were analyzed by SDS-PAGE and Western blot as described previously (26).
Cellular CPE-binding Assay
Two to 3 days after transfection, HEK cells expressing Cld3wt, Cld3mutants, Cld3wt-CFP, Cld5wt, Cld5wt-YFP, or Cld5-YFPmutants were incubated with 1 or 10 μg/ml GST-CPE-(116–319), GST-CPE-(194–319), or GST for 1 h at 37 °C in 24- or 12-well plates. Cells were washed with ice-cold PBS supplemented with Ca2+ and Mg2+, scraped, and harvested (300 × g, 5 min, 4 °C). Cells were lysed with RIPA buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1.0 mm EDTA, 1.0% (v/v) Nonidet P-40, 0.5% (w/v) deoxycholate, 0.1% SDS, EDTA-free protease inhibitor mixture (Roche, Mannheim, Germany) for 10 min on ice. After centrifugation (10,000 × g, 5 min, 4 °C), supernatants were analyzed by SDS-PAGE and Western blot. For each claudin mutant the ratios of the BLU values of the GST-CPE and the claudin signal were calculated relative to the ratio for the claudinwt (internal standard).
Structural Bioinformatics and Molecular Modeling
The ECL2 of claudins was predicted by alignment of sequences of mouse (human when indicated) Cld-(1–23) with the GCG program package (GCG Wisconsin package, Accelrys Inc., San Diego, CA). A homology model for Cld3-ECL2-(134–164) was built on the model for murine Cld5-ECL2-(135–165) as described (21). The Cld5-ECL2 model was based on part of Protein Data Bank structure 2BDV (Phage-related Protein BB2244 from Bordetella bronchiseptica). This resulted in a helix-turn-helix motif for ECL2, where the helices of the ECL2 are extensions of the predicted transmembrane helices 3 and 4 of Cld3. All manual reciprocal dockings, manipulations, and optimizations of ECL2 models were performed with the program Sybyl 7.3 (Tripos Inc., St. Louis, MO). The models were energetically minimized, using the AMBER95 force field.
Statistics
Unless stated otherwise, results are shown as mean ± S.E. Statistical analyses were performed by one-way analysis of variance and followed by an unpaired Student's t test. p < 0.01 was taken as significantly different.
RESULTS
CPE-(116–319) Binds with Higher Affinity Than CPE-(290–319) to the ECL2 Peptide of Claudin-3
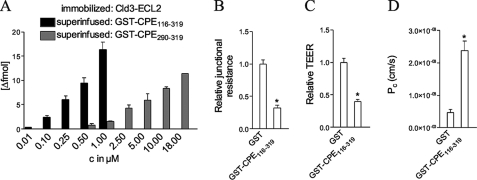
The C-terminal end of CPE-(290–319) is sufficient for interactions with Cld3 and Cld4 (27). In addition, further regions within CPE-(116–290) have been implicated in binding to Cld3/4 (28, 29). To determine whether CPE region 116–290 enhances the interaction of CPE with Cld3-ECL2, we compared binding of GST-CPE-(290–319) with the binding of GST-CPE-(116–319) to peptide Cld3-(140–159) (predicted ECL2, see “Experimental Procedures”). SPR spectroscopy using immobilized Cld3-ECL2-(140–159) showed that a 20-fold higher concentration of GST-CPE-(290–319) compared with GST-CPE-(116–319) was needed to obtain similar amounts of bound CPE (Fig. 1A). This demonstrates that GST-CPE-(116–319) binds much more strongly to Cld3-ECL2 than GST-CPE-(290–319). Thus, GST-CPE-(116–319) was used for further experiments. When confluent monolayers of Caco-2 cells were incubated with GST-CPE-(116–319), a decrease in junctional resistance, TEER, and an increase in paracellular fluorescein permeability compared with GST were detected (Fig. 1, B–D). Thus, GST-CPE-(116–319) binds to native claudins on living cells and influences paracellular tightness.
FIGURE 1.
A, CPE-(116–319) binds much more strongly to the ECL2 peptide of Cld3 than does CPE-(290–319). ECL2 peptide (amino acids 140–159) of mouse Cld3 was immobilized and superinfused with different concentrations of GST-CPE-(116–319) and GST-CPE-(290–319), respectively. The binding of GST was negligible (<1 fmol for <20 μm GST). The binding was analyzed by surface plasmon resonance spectroscopy. Mean ± S.E., n > 3 except for 18 μm GST-CPE-(290–319), n = 1. Incubation of Caco-2 cells with CPE-(116–319) reduces junctional resistance (B) and transepithelial electrical resistance, TEER (C), and increases the permeation coefficient (Pc) of fluorescein (D). For B, cells were incubated for 12 h with 10 μg/ml GST or GST-CPE-(116–319) and the junctional resistance relative to the value before GST or GST-CPE-(116–319) incubation was determined by electric cell-substrate impedance sensing. Mean ± S.E. (error bars); n ≥ 9; *, p < 0.001. For C and D, cells were incubated with 25 μg/ml GST or GST-CPE-(116–319) for 16 h and TEER determined relative to the value before incubation. Mean ± S.E. n = 6, *, p < 0.001.
Substitution Analysis Identified the Motif NPLVP in Peptides of the ECL2 of Claudin-3 as Essential for Binding to CPE-(116–319)
To compare the binding ability of GST-CPE-(116–319) to the ECL2 of different claudins, peptide arrays were used. Peptides corresponding to the predicted ECL2 (11) of Cld1–22, except -17 and -21 were synthesized on membranes and incubated with GST-CPE-(116–319) or GST as control. GST-CPE-(116–319) specifically bound to ECL2 peptides of Cld3, -6, -7, -9, and -14 but not Cld1, -2, -4, -5, -8, -10–13, -15, -16, -18–20, and -22 (Table 1).
TABLE 1.
GST-CPE-(116–319) binds to peptides corresponding to the extracellular loop two (ECL2) of claudin-3, -6, -7, -9, -14
20-mer peptides of the predicted ECL2 of the claudins (Cld1–16, 18, 19, 22, mouse; Cld20 human; 3rd column) were immobilized on a membrane and incubated with 10 μg/ml GST (1st column) or GST-CPE-(116–319) (2nd column). Bound CPE was visualized via mouse anti-GST and HRP-conjugated anti-mouse antibodies. Chemiluminescence intensities relative to that for Cld3 are shown (last column). Mean of n = 2. Representative membranes are shown. Underlined, region (NPLVP) highly conserved between claudins that bind CPE-(116–319). Bold, amino acids in this region that are identical to that of claudin-3.
For the identification of amino acids involved in the CPE-claudin interaction, additional peptide arrays were performed. Because GST-CPE-(116–319) did not bind to Cld5-ECL2 peptides (Table 1), we tested whether introduction of defined amino acids of Cld3-ECL2 into Cld5-ECL2 enables binding to CPE. In contrast to single or certain multiple substitutions including E146D, V154E, S155A, and Y158R, specifically the double substitution D149N/T151L in Cld5-ECL2 peptides facilitated the binding of GST-CPE-(116–319) (Table 2, upper part). Similarly, substitution of S149N in Cld2-ECL2 corresponding to substitution of D149N in Cld5 enabled binding of GST-CPE-(116–319) (Table 2, lower part). The additional substitution S155A in Cld2 further increased the binding. For Cld4, the double substitution M151L/A153P permitted binding of GST-CPE-(116–319) (Table 2, lower part). In addition to the peptide arrays, in surface plasmon resonance experiments N148D/L150T blocked binding of superinfused GST-CPE-(116–319) to immobilized CLd3-ECL2 peptides (0.75 μm GST-CPE-(116–319) resulted in binding of 10.3 ± 1.6 fmol of GST-CPE-(116–319) to Cld3wt and 0.0 ± 0.1 fmol of GST-CPE-(116–319) to Cld3-N148D/L150T, n = 4, mean ± S.E.). These data indicate that the positions corresponding to 148 and 150 in murine Cld3 are strongly involved in the claudin-CPE interaction.
TABLE 2.
Introduction of amino acids from the extracellular loop two (ECL2) of murine claudin-3 (Cld3wt) into the ECL2 of other claudins increased the binding of C. perfringens enterotoxin (CPE-116–319)
20-mer peptides were immobilized on a membrane and incubated with 10 μg/ml GST-CPE-(116–319). Bound CPE was visualized via mouse anti-GST and HRP-conjugated anti-mouse antibodies. Chemiluminescence intensities relative to that of Cld3, mean of n = 2, representative membranes are shown. Underlined, amino acid of Cld3 introduced in another claudin. Bold, amino acid substitution that enabled CPE binding.
To analyze the molecular determinants of the Cld3-CPE interaction in more detail, substitution mapping was performed. Here, peptide arrays were used that contained peptides with every possible single amino acid substitution for each position in Cld3-ECL2-(145–159) (Fig. 2). Cysteine was excluded because it leads to artificial covalent bonds during peptide synthesis. None of the substitutions at positions 145 to 147 had a considerable effect on the binding of GST-CPE-(116–319). In contrast, every substitution in the region 148NPLVP152 except V151I/L/R abolished binding of GST-CPE-(116–319). This indicates that the NPLVP motif is essential for the Cld3-CPE interaction. In addition, several substitutions at positions 154–158 also influenced the amount of GST-CPE-(116–319) bound. In particular, the introduction of negatively charged amino acids or proline at positions 154–157 strongly reduced the binding of GST-CPE-(116–319).
FIGURE 2.
Single amino acid substitution analysis of the ECL2 of Cld3 (Cld3-ECL2) identified the motif 148NPLVP152 as essential for the association of GST-CPE-(116–319). The C-terminal flank (Cld3-(154–157)) also contains amino acids involved in the interaction. 15-mer peptides of Cld3-ECL2 were immobilized on a membrane. In the left column, all spots contain the wild type sequence (wt); the letter and number give the position of the amino acid replaced in the same line of the second column. The membrane was incubated with 10 μg/ml GST-CPE-(116–319). Bound CPE was visualized via mouse anti-GST and HRP-conjugated anti-mouse antibodies. Binding was considered to be decreased or increased if the mean chemiluminescence was increased or decreased by a factor of 4, respectively, compared with the wt (n = 2). AA, amino acid; X, every amino acid except Cys and those mentioned as less or stronger. Dotted circles, wt sequence; dashed boxes, substitutions resulting in decreased binding. A representative membrane is shown.
Interaction between Full-length Claudin-3 and CPE-(116–319) Is Affected by Amino Acid Substitutions in ECL2
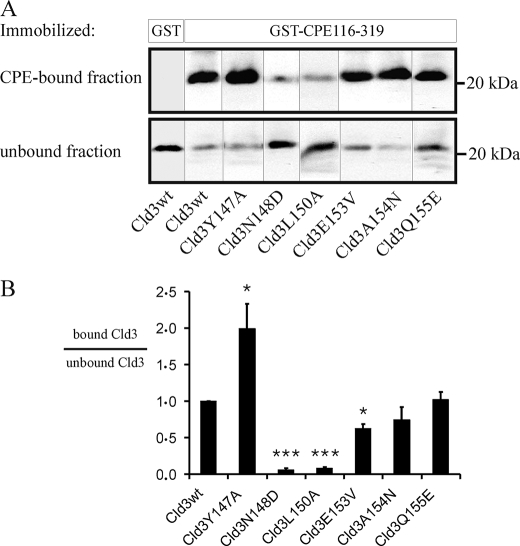
To determine whether amino acid substitutions in ECL2 affect binding of GST-CPE-(116–319) to full-length Cld3, transfections of Cld3 in TJ-free HEK cells (21) were performed. Lysates of the cells were used for pull-down assays with GST-CPE-(116–319) (Fig. 3). Cld3 bound specifically to GST-CPE-(116–319) but not GST. Substitutions N148D and L150A in the ECL2 of Cld3 caused a strong reduction in the amount of Cld3 that bound to GST-CPE-(116–319). In contrast, substitutions A154N and Q155E did not affect the interaction of full-length Cld3 with GST-CPE-(116–319). Substitution E153V used to mimic a sequence element in Cld5 slightly reduced the amount of bound Cld3. Removal of the aromatic side chain of Tyr147 (Y147A), which is involved in the trans-interaction between claudins (21), increased the amount of Cld3 bound to GST-CPE-(116–319).
FIGURE 3.
Binding of full-length Cld3 to CPE is affected by amino acid substitutions in the ECL2 of Cld3. HEK cells were transfected with Cld3wt or a Cld3mutant. Lysates thereof were used for pull-down assays with GST-CPE-(116–319) or GST immobilized on glutathione-Sepharose. Bound and unbound fractions were analyzed by SDS-PAGE and Western blot. A, representative blots are shown. Cld3wt bound specifically to GST-CPE-(116–319) but not to GST. The substitutions N148D and L150A reduced the binding strongly, Y147A increased the amount of bound Cld3. B, the intensity of the immunoreactive bands was quantified. For each mutant the ratio of intensity in the bound fraction to that in the unbound fraction relative to the ratio for Cld3wt (internal standard in each experiment) was determined (bound Cld3/unbound Cld3). Mean ± S.E. (error bars); n > 4; *, p < 0.05; ***, p < 0.001 to Cld3wt.
Binding of CPE to Claudin-3 on the Surface of Living Cells Is Affected by N148D/L150A Double Amino Acid Substitution
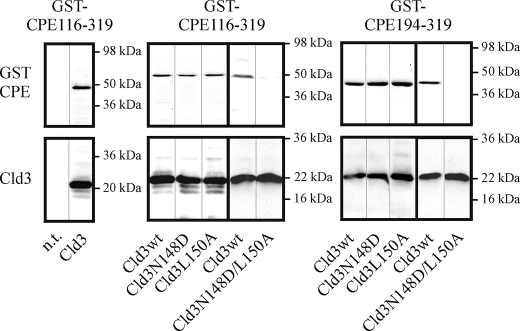
To analyze the binding of CPE to Cld3 in the plasma membrane of living cells, HEK cells were transfected with Cld3wt or Cld3mutants, incubated with GST-CPE-(116–319) or GST as a control, and the presence of bound GST-CPE in cell lysates tested by Western blots. GST-CPE-(116–319) bound specifically to Cld3-transfected but not to non-transfected cells (Fig. 4, left). This binding was not clearly affected by single amino acid substitutions N148D or L150A but strongly reduced by N148D/L150A double substitution in Cld3 (Fig. 4, middle). Similar results (Fig. 4, right) were obtained with GST-CPE-(194–319), which contains a CPE fragment of which the structure has been recently solved (30).
FIGURE 4.
Binding of GST-CPE to Cld3-transfected HEK cells. Three days after transfection, cells were incubated with 1 μg/ml GST-CPE-(116–319) (left, middle) or GST-CPE-(194–319) (right) for 1 h at 37 °C, washed with PBS, and the amount of bound GST-CPE and claudin in the lysate analyzed by SDS-PAGE and Western blot using anti-GST (top) and anti-Cld3 (bottom) antibodies. Left, GST-CPE-(116–319) bound to Cld3wt-transfected (Cld3) but not to non-transfected (n.t.) cells. Middle, GST-CPE-(116–319) bound to Cld3-N148D and Cld3-L150A to a similar extent as to Cld3wt. In contrast, binding to Cld3-N148D/L150A was strongly reduced. Right, similar results were obtained for GST-CPE-(194–319). Representative blots are shown.
CPE Binds to Cld3 on the Surface of Transfected HEK Cells Outside Tight Junctions
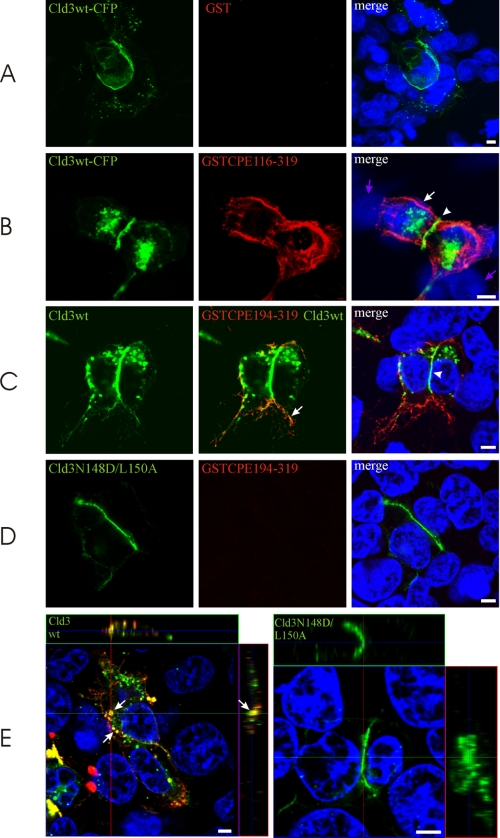
The binding of GST-CPE to Cld3-transfected HEK cells was also analyzed by confocal microscopy. Cld3 was detected in the plasma membrane and intracellular compartments (Fig. 5, A–C). Marked enrichment of Cld3 at contacts between two Cld3-expressing cells was observed. This enrichment is known to correspond to the formation of TJ strands (21). No binding of GST to either Cld3 expressing or non-expressing cells was detected (Fig. 5A). In contrast, GST-CPE-(116–319) bound specifically to Cld3 expressing cells and did not bind to non-expressing cells (Fig. 5B). Interestingly, almost no GST-CPE-(116–319) signals were detected at the contacts between two Cld3-expressing cells, where Cld3 was strongly enriched. However, strong GST-CPE-(116–319) signals were detected on the surface of the cells outside the contacts. Similar results were obtained with GST-CPE-(194–319) (Fig. 5C). Cld3-N148D/L150A was enriched at contacts similarly to Cld3wt (Fig. 5D). This indicates that the amino acid substitutions do not alter plasma membrane targeting and ability for trans-interaction of Cld3 (21). Instead, binding of GST-CPE-(194–319) was strongly reduced. In addition, incubation of transfected HEK cells with GST-CPE-(194–319) for 14 h resulted in internalization of CPE and Cld3 (Fig. 5E, left). This was blocked by the N148D/L150A substitution (Fig. 5E, right).
FIGURE 5.
Binding of GST-CPE-(116–319) and GST-CPE-(194–319) to Cld3 on the surface of transfected HEK cells. Cells were transfected with Cld3wt-CFP (A and B), Cld3wt (C and E), or Cld3-N148D/L150A (D and E), 3 days later, the cells were incubated with 10 μg/ml GST-CPE-(116–319), GST-CPE-(194–319), or GST for 20 min at 37 °C, washed with PBS, and fixed. GST (red) was stained using mouse anti-GST antibodies, nuclei (blue) with 4′,6-diamidino-2-phenylindole, and Cld3 (green) detected via CFP-fluorescence (A and B) or anti-Cld3 antibodies (C-E). Confocal images are shown. In contrast to GST (A, red), GST-CPE-(116–319) (B, red) and GST-CPE-(194–319) (C, red) bound to cells expressing Cld3 (green) but not to Cld3 negative/4′,6-diamidino-2-phenylindole positive cells (violet arrows). GST-CPE-(116–319) and GST-CPE-(194–319) bound to Cld3 on the cell surface (B and C, arrow) but not to Cld3 in the tight junction area at cell-cell contacts (B and C, arrowhead). For the detection of Cld3, high (C, left and middle) or low (C, right) detector gain was used to visualize colocalization of GST-CPE-(194–319) and Cld3 (arrow) or enrichment of Cld3 at contacts between two Cld3-expressing cells (arrowhead), respectively. Cld3-N148D/L150A (D) showed enrichment at contacts between two Cld3-expressing cells similar to Cld3wt. Binding of GST-CPE-(194–319) to Cld3-N148D/L150A-transfected cells was barely detected. E, 14 h incubation with 10 μg/ml GST-CPE-(194–319) led to internalization (arrows) of Cld3 (green) and GST-CPE-(194–319) (red) for Cld3wt, but not Cld3-N148D/L150A expressing cells. Z stacks, xy (quadrat), xz (top), and yz images (side) are shown and the position within the stack indicated by lines; 4′,6-diamidino-2-phenylindole is shown for the xy image only. Equal detector gains were used for A and B; C, right, and D; E, left; and E, right. Bar, 4 μm.
CPE-(116–319) but Not CPE-(194–319) Binds to Cld5-transfected HEK Cells
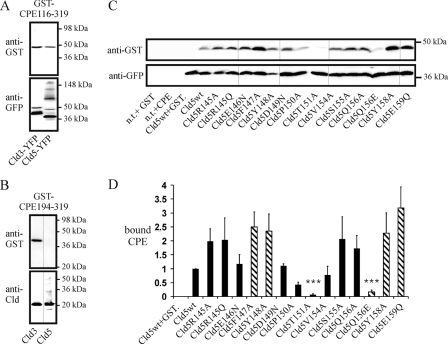
GST-CPE-(116–319) not only bound to Cld3- but also to Cld5-transfected cells (Fig. 6A). Cld3- and Cld5-YFP expression levels were detected via anti-YFP antibodies. Due to differences in the formation of SDS-resistant oligomers (31), the intensities of the Cld3- and Cld5-YFP bands were difficult to compare. Nevertheless, Cld5-YFP was expressed at least as high as Cld3-YFP, but gave a weaker signal for bound GST-CPE-(116–319). This indicates that GST-CPE-(116–319) binds more strongly to Cld3 than to Cld5. In contrast to GST-CPE-(116–319), GST-CPE-(194–319) bound to Cld3- but not to Cld5-transfected HEK cells (Fig. 6B).
FIGURE 6.
Binding of GST-CPE-(116–319) to Cld5-transfected HEK cells. Cells were incubated with 1 μg/ml GST-CPE-(116–319) (A) or GST-CPE-(194–319) (B) at 37 °C 3 days after transfection, washed with PBS, and the amount of bound GST-CPE and claudin in the lysate analyzed by SDS-PAGE and Western blot using anti-GST and anti-GFP (A) or anti-Cld3 and anti-Cld5 antibodies (B). Representative blots are shown. A, GST-CPE-(116–319) bound to Cld3-YFP- and Cld5-YFP-transfected cells. B, GST-CPE-(194–319) bound to Cld3- but not to Cld5-transfected cells. C, binding of GST-CPE-(116–319) to claudin-5 mutants. Transfected cells were incubated with 10 μg/ml GST-CPE-(116–319) for 1 h at 37 °C. Representative Western blots show that neither GST (n.t. + GST) nor GST-CPE-(116–319) (n.t. + GST-CPE116–319) bound to non-transfected cells. GST-CPE-(116–319) (Cld5wt) but not GST (Cld5wt+GST) bound to Cld5wt-YFP transfected cells. D, quantification of the binding. The ratio of GST-CPE-(116–319) intensity to Cld5-YFP intensity relative to the ratio for Cld5wt-YFP (internal standard) was determined for each mutant (bound CPE). Substitutions T151A and Q156E decreased whereas F147A, Y148A, Y158A, and E159Q increased the amount of GST-CPE-(116–319) bound. Striped columns, substitutions that block trans-interaction between claudins. Mean ± S.E. (error bars), n > 4. ***, p < 0.001 compared with Cld5wt.
Amino Acid Substitutions in ECL2 of Cld5 Affect the Binding of CPE-(116–319) to Cld5-transfected HEK Cells
To identify determinants in the ECL2 of Cld5 for the binding of CPE-(116–319), a set of Cld5-ECL2 mutants (21) was used. The plasma membrane targeting of these mutants was not affected. Transfection of most of the mutants resulted in binding of GST-CPE-(116–319) that was, at least, as strong as for Cld5wt-YFP (Fig. 6, C and D). In contrast, for mutants Cld5-T151A and Cld5-Q156E, the binding of GST-CPE-(116–319) was greatly reduced. In addition, Cld5-P150A showed reduced association with GST-CPE-(116–319). Interestingly, transfection of mutants F147A, Y148A, Y158A, E159Q, known to block the trans-interaction (21), resulted in increased binding of GST-CPE-(116–319). The binding of CPE-(116–319) to Cld5wt and a marked reduction of this binding by substitutions T151A and Q156E, but not Q156A and F147A, were also detected by immunostaining (supplemental Fig. S2).
Structural Determinants of Claudin-CPE Interaction
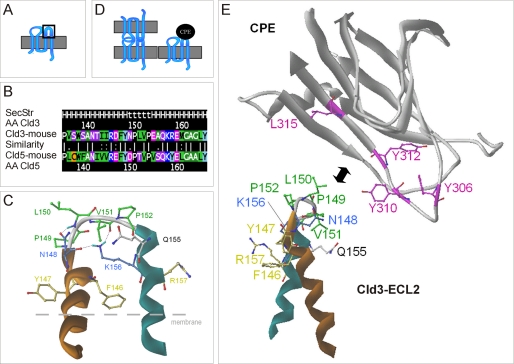
Our homologous helix-turn-helix model for the ECL2 of Cld3 shows a strong hydrophobic area at the turn (149PLVP, green in Fig. 7C) between the helices. This probably matches complementary with a hydrophobic pit on the surface of CPE formed by residues (Tyr306, Tyr310, Tyr312, Leu315, magenta, Fig. 7E). The Cld3 ECL2 residues Asn148 and Lys156 (blue in Fig. 7C) form loop-stabilizing hydrogen bridges. The residues (Phe146, Tyr147, and Arg157, yellow in Fig. 7E) point away from the potential CPE-binding motif and do not seem to be involved in CPE binding.
FIGURE 7.
Schemes of molecular interaction between CPE and the ECL2 of claudin-3. A, topology of claudins with ECL2 marked in a box. B, alignment of ECL2 of Cld3 and -5, with similarity of residues calculated with matrix blossom62 (dot = weak; colon = strong similarity; bar = identity). Secondary structure (SecStr) according to the model, helices (H), turn (t). C, homologous helix-turn-helix model for ECL2 of Cld3 based on the fragment of PDB code 2BDV in front view. N-terminal helix, orange; C-terminal helix, cyan. The hydrophobic residues 149PLVP (green) constituting the turn region of the loop are found to be important for CPE binding. The residues Asn148 and Lys156 (light blue) stabilize the turn-fold of the ECL2 by hydrogen bridges (hydrogens in cyan). Residues Phe146, Tyr147, and Arg157 (yellow) correspond to aromatic residues Phe147, Tyr148, and Tyr158 in Cld5, which were found to be important for trans-interaction. Heteroatoms are red (oxygen) and dark blue (nitrogen). D, according to the results, CPE cannot bind to trans-interacting claudins (left), probably due to steric hindrance, but needs a free ECL2 for binding (right). E, supposed spatial regions of interaction between binding sensitive residues (Tyr306, Tyr310, Tyr312, and Leu315 from literature (36)) visualized at the CPE-(194–319) x-ray structure (PDB code 2QUO) and the residues (148NPLVP, Gln155) identified in this study for Cld3.
DISCUSSION
This study identifies amino acid residues and a sequence motif in the ECL2 of claudin-3 involved in the interaction with CPE by the use of biochemical and cellular approaches. Together with molecular modeling and literature data the results allowed us to deduce determinants of the interaction.
In the recently solved x-ray structure of CPE-(194–319), region 290–319 is part of a nine-strand β sandwich that forms the claudin-binding domain (30). This explains our SPR results (Fig. 1) showing that the ECL2 of Cld3 interacts more weakly with GST-CPE-(290–319) than with GST-CPE-(116–319), because the latter includes the whole binding domain.
A Unique Pentapeptide Motif in Claudin-3 Is Important for Interaction with CPE
After incubation of peptide arrays with GST-CPE-(116–319), interactions with ECL2 of Cld3, -6, -7, -9, and -14 but not the 15 other claudins were found (Table 1). This is consistent with earlier reports on the interaction between full-length CPE and Cld3, -6, -7, and -14 but not Cld1, -2, -5, and -10 (12). In addition, for the first time, interaction of CPE-constructs with ECL2 peptides of Cld9, but not of Cld13, -15, -16, -18, -19, -20, and -22 has been demonstrated. All claudin subtype peptides that interacted with GST-CPE-(116–319) contain the consensus sequence NPL(V/L)(P/A). Introduction of Cld3-like residues in ECL2 peptides of Cld5 (D149N/T151L), Cld2 (S149N), or Cld4 (M151L/A153P) created the consensus sequence and enabled binding of GST-CPE-(116–319) (Table 2). Substitution mapping showed that a central region (mouse 148–152) and the C-terminal flank, but not the N-terminal flank, of the loop are involved in interaction of GST-CPE-(116–319) with Cld3-ECL2 peptides (Fig. 2). Negative charges, proline, and other residues at positions 154–157 block binding of GST-CPE-(116–319). But especially, the motif 148NPLVP152 was essential for the interaction, because any substitution other than V151I/L/R in this motif abolished CPE binding. In addition to the peptide arrays, SPR measurements confirmed the involvement of Asn148 and Leu150 in the interaction. Taken together, the data consistently demonstrate that the central motif 148NPLVP152 in the ECL2 of Cld3 is fundamental for the interaction with CPE.
Low and High Affinity Binding of CPE to Full-length Claudins
GST-CPE-(116–319) pull-down assays with lysates of Cld3-transfected HEK cells showed that substitutions N148D and L150A strongly inhibit the interaction of GST-CPE-(116–319) with full-length Cld3 (Fig. 3). This is comparable with findings obtained by ECL2-peptide arrays. Substitutions at other positions in full-length Cld3 did not inhibit the interaction to a similar extent.
Binding of GST-CPE-(116–319) and GST-CPE-(194–319) to Cld3 on the surface of living HEK cells was diminished by the double substitution N148D/L150A, but not by the single substitutions N148D and L150A (Figs. 4 and 5). This difference to the pull-down assay indicates that, in addition to amino acid substitutions, detergent solubilization decreases the affinity/avidity of the CPE-Cld3 interaction. This could be due to dissociation of multimeric claudin complexes (21) and/or destabilization of the claudin loop structure caused by solubilization.
Similar differences were found between membranous and non-membranous Cld5: GST-CPE-(116–319) bound to Cld5-expressing cells (Fig. 6 and supplemental Fig. S2) but not to solubilized Cld5 (data not shown) nor Cld5-ECL2 peptides (Table 2). Interestingly, GST-CPE-(194–319) did not bind to Cld5-transfected HEK cells (Fig. 6). In separate experiments, similar results were obtained with Cld5-transfected L-fibroblasts ruling out the possibility that this differential binding is due to unique properties of HEK cells. Preliminary experiments indicate that GST-CPE-(116–319) and GST-CPE-(194–319) as well as membranous and solubilized claudins differ in their oligomeric state. Hence, multivalent binding (32) between CPE-oligomers (33) and claudin-oligomers (34, 21) could enhance the presumably low affinity CPE-Cld5 interaction and promote binding between GST-CPE-(116–319) and Cld5 in the membrane, only.
In other studies, CPE-Cld5 interaction could not be detected (35). However, these authors used CPE-(184–319), a short fragment similar to CPE-(194–319), which, in contrast to CPE-(116–319), shows no binding to Cld5 in our study either. Fujita et al. (12) reported no high affinity binding of full-length CPE to Cld5. This does not rule out low affinity binding. Taken together, the data obtained with claudin peptides, full-length claudins, and from the literature, are consistent with the assumption of high affinity binding of CPE to Cld3 and low affinity binding of CPE to Cld5.
Previously, we characterized a set of Cld5-ECL2 mutants that were targeted to the plasma membrane in a similar manner to Cld5wt (21). To assess the CPE-Cld5 interaction in more detail, CPE binding to HEK293 cells transfected with these mutants was analyzed. In contrast to all other analyzed Cld5mutants, Cld5-T151A and Cld5-Q156E show strong reduction in binding of GST-CPE-(116–319) compared with Cld5wt. Interestingly, position Thr151 in Cld5 corresponds to Leu150 in Cld3, which is strongly involved in the interaction with CPE-(116–319). In addition, substitution T151L together with D149N enabled the binding of CPE-(116–319) to Cld5-ECL2 peptides. This underlines the involvement of position 150/151 in the CPE-Cld3/5 interaction. For Cld5, Q156E reduced but Q156A increased binding of CPE-(116–319) to transfected cells. The corresponding position in Cld3 peptides (Gln155) was the most sensitive one outside the NPLVP motif (Fig. 2). For example, Q155E or Q155D inhibited, whereas Q155A increased, binding of GST-CPE-(116–319) to Cld3 peptides. This indicates involvement of position Gln155 and Gln156 in the CPE-Cld3 and CPE-Cld5 interactions, respectively.
Structural Determinants Support an Interaction of the Helix-turn-helix Conformation of the ECL2 of Cld3 with CPE
The experimental binding data were combined with molecular homology modeling. Our previously generated helix-turn-helix homology model for the ECL2 of Cld5 indicates that mutations of exactly those residues that are supposed to stabilize the turn conformation led to folding defects (11). Because all of these stabilizing residues are identical (except a Asp → Asn exchange, which still preserves the same H-bonds) in Cld3, a comparable turn conformation could also be adapted for the ECL2 of Cld3 (Fig. 7). This is consistent with results of GST-CPE-(116–319) binding on ECL2 peptide arrays of Cld3 found in this study. For exactly those residues forming the turn (148NPLVP), almost any possible substitution abolished CPE binding (Fig. 2). Substitutions at Val151 that preserved CPE binding were mainly hydrophobic. This is consistent with the suggested turn conformation, because Val151 forms the hydrophobic core of the turn between the two helices and is itself not supposed to be involved in the CPE binding. The hydrophobic property on the surface of this turn portion of our helix-turn-helix model is formed by nPLvP.
In CPE, Tyr306, Tyr310, Tyr312, and Leu315 have been shown to be involved in the CPE-Cld4 interaction (36, 37). Similarly, we found that deletion of amino acids 310–319 diminished binding of Cld3 and Cld4 to GST-CPE (supplemental Fig. S1). On the basis of the complementary properties and shape, direct interaction of the hydrophobic ECL2-turn with a hydrophobic pit on the surface of CPE formed by Tyr306, Tyr310, Tyr312, or Leu315 (30) is very likely. This is supported by mutations Cld3-L150A, which strongly reduced the binding (Fig. 3), and additionally by the peptide array data (Fig. 2). Substitution of N148D, flanking the hydrophobic turn, strongly reduced CPE binding also (Fig. 3). This also indicates that the amino group of Cld3-N148 is involved in interaction with CPE. In addition, double substitution N148D/L150A in Cld3 diminished binding of GST-CPE-(116–319) and GST-CPE-(194–319) (Figs. 4 and 5). Consistently, Cld5 whose turn (DPTVP) contains no amino group and a smaller hydrophobic property than that of Cld3 (NPLVP) binds more weakly to CPE than does Cld3.
CPE has been shown to bind to full-length Cld4 and Cld8 (12, 14). Indeed, we found that GST-CPE-(116–319) and GST-CPE-(194–319) bind to full-length Cld4 (supplemental Fig. S1). In contrast, we did not observe an interaction between CPE and synthesized ECL2-peptides of Cld4 and Cld8. Similar results have been reported by others earlier. They found that CPE peptides do not bind to claudin-4 fragments without transmembrane segments (38). Strikingly, Cld4 and -8 do not contain the conserved second proline implicated in the stabilization of the loop structure (11). In addition, Cld4 is the only CPE-binding claudin that contains a methionine, instead of leucine (Leu150 in Cld3), in the turn region. It is possible, that due to lack of the corresponding leucine and proline, the conformation of ECL2 peptides of Cld4 and -8 on the peptide mapping membrane differs more markedly from the native structure than it does for CLd3, -6, -9, or -14. Indeed, substitutions of M150L/A153P in the Cld4 ECL2 peptide enabled binding to GST-CPE-(116–319) (Table 2). Our results with ECL2 peptides and full-length claudins, as well as other reports (38), are consistent with the idea that transmembrane segments stabilize the native conformation of the ECL2 and, thereby, increase the affinity of the CPE-claudin interaction.
CPE Binds to the Free ECL2 of Claudins Outside of Tight Junctions
Aromatic residues in CPE (Tyr306, Tyr310, Tyr312) are involved in interaction with claudins (36) and formation of an aromatic core within an ECL2-dimer is involved in the trans-interaction of claudins between two opposing cells (21). Others have speculated that aromatic residues in claudin could also interact with the aromatic residues in CPE (38). But interestingly, removal of the aromatic residues (F147A, Y148A, Y158A) in Cld5 resulted in an increase in binding of GST-CPE-(116–319) to transfected cells (Fig. 6). Similarly, substitution of Y147A in Cld3, which corresponds to Cld5-Y148A and also blocks trans-interaction, increased the amount of Cld3 pulled down with GST-CPE-(116–319) (Fig. 3). Moreover, in our model, the aromatic residues in the ECL2 do not sterically match the aromatic residues in CPE (Fig. 7). These data indicate that residues in the turn region of the loop, but not aromatic residues in the flanking helices, are involved in the claudin-CPE interaction.
It has to be stressed that the aforementioned substitutions (F147A, Y148A, and Y158A in Cld5; Y147A in Cld3) and E159Q in Cld5 block trans-interaction and strand formation of claudins (21) as well as increase binding of GST-CPE-(116–319). Consistently, confocal microscopy showed binding of GST-CPE-(116–319) and GST-CPE-(194–319) to Cld3 outside the tight junction area. These observations demonstrate that CPE binds to the free ECL2 of claudins that is not occupied by trans-interaction and not incorporated in TJ strands.
Treatment of epithelial cells with CPE increases paracellular permeability. To explain this, two mechanisms have been discussed (14). Either 1) CPE binding to TJ strands leads to direct depolymerization of the claudins, or 2) CPE sequesters claudins in the plasma membrane, preventing their incorporation in the strands during the dynamic assembly of strands. Our results strongly support the sequestration mechanism and this explains our and other findings (37) in which CPE treatment needs hours for the TJ to be opened, despite high affinity interaction with claudins (12).
In conclusion, we have shown that the molecular mechanism of the CPE-claudin-3 interaction is different from the trans-interaction between claudins. This improves the understanding of the molecular organization of TJ. In addition, the information gained is highly relevant for the design of CPE-based claudin modulators to improve drug delivery across tissue barriers (7) or treatment of tumors overexpressing claudins (16–18).
Supplementary Material
Acknowledgments
We thank Dr. Michael Beyermann for synthesis of ECL2 peptides and Angelika Ehrlich for excellent technical assistance.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- TJ
- tight junctions
- CPE
- Clostridium perfringens enterotoxin
- ECL
- extracellular loop
- Cld
- claudin
- SPR
- surface plasmon resonance
- BLU
- Boehringer light units
- GST
- glutathione S-transferase
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- HRP
- horseradish peroxidase
- PBS
- phosphate-buffered saline
- HEK
- human embryonic kidney
- TEER
- transepithelial electrical resistance.
REFERENCES
- 1.White R. E. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 133–157 [DOI] [PubMed] [Google Scholar]
- 2.Mizuno N., Niwa T., Yotsumoto Y., Sugiyama Y. (2003) Pharmacol. Rev. 55, 425–461 [DOI] [PubMed] [Google Scholar]
- 3.Aungst B. J. (2000) J. Pharm. Sci. 89, 429–442 [DOI] [PubMed] [Google Scholar]
- 4.Salama N. N., Eddington N. D., Fasano A. (2006) Adv. Drug Deliv. Rev. 58, 15–28 [DOI] [PubMed] [Google Scholar]
- 5.Hochman J., Artursson P. (1994) J. Control. Release 29, 253–267 [Google Scholar]
- 6.Yamamoto A., Uchiyama T., Nishikawa R., Fujita T., Muranishi S. (1996) J. Pharm. Pharmacol. 48, 1285–1289 [DOI] [PubMed] [Google Scholar]
- 7.Kondoh M., Takahashi A., Fujii M., Yagi K., Watanabe Y. (2006) Biol. Pharm. Bull. 29, 1783–1789 [DOI] [PubMed] [Google Scholar]
- 8.Chiba H., Osanai M., Murata M., Kojima T., Sawada N. (2008) Biochim. Biophys. Acta 1778, 588–600 [DOI] [PubMed] [Google Scholar]
- 9.González-Mariscal L., Betanzos A., Nava P., Jaramillo B. E. (2003) Progr. Biophys. Mol. Biol. 81, 1–44 [DOI] [PubMed] [Google Scholar]
- 10.Furuse M., Sasaki H., Fujimoto K., Tsukita S. (1998) J. Cell Biol. 143, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause G., Winkler L., Mueller S. L., Haseloff R. F., Piontek J., Blasig I. E. (2008) Biochim. Biophys. Acta 1778, 631–645 [DOI] [PubMed] [Google Scholar]
- 12.Fujita K., Katahira J., Horiguchi Y., Sonoda N., Furuse M., Tsukita S. (2000) FEBS Lett. 476, 258–261 [DOI] [PubMed] [Google Scholar]
- 13.Adak G. K., Long S. M., O'Brien S. J. (2002) Gut 51, 832–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoda N., Furuse M., Sasaki H., Yonemura S., Katahira J., Horiguchi Y., Tsukita S. (1999) J. Cell Biol. 147, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondoh M., Masuyama A., Takahashi A., Asano N., Mizuguchi H., Koizumi N., Fujii M., Hayakawa T., Horiguchi Y., Watanbe Y. (2005) Mol. Pharmacol. 67, 749–756 [DOI] [PubMed] [Google Scholar]
- 16.Kominsky S. L., Tyler B., Sosnowski J., Brady K., Doucet M., Nell D., Smedley J. G., 3rd, McClane B., Brem H., Sukumar S. (2007) Cancer Res. 67, 7977–7982 [DOI] [PubMed] [Google Scholar]
- 17.Santin A. D., Bellone S., Marizzoni M., Palmieri M., Siegel E. R., McKenney J. K., Hennings L., Comper F., Bandiera E., Pecorelli S. (2007) Cancer 109, 1312–1322 [DOI] [PubMed] [Google Scholar]
- 18.Morin P. J. (2007) Dis. Markers 23, 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuse M., Hata M., Furuse K., Yoshida Y., Haratake A., Sugitani Y., Noda T., Kubo A., Tsukita S. (2002) J. Cell Biol. 156, 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitta T., Hata M., Gotoh S., Seo Y., Sasaki H., Hashimoto N., Furuse M., Tsukita S. (2003) J. Cell Biol. 161, 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piontek J., Winkler L., Wolburg H., Müller S. L., Zuleger N., Piehl C., Wiesner B., Krause G., Blasig I. E. (2008) FASEB J. 22, 146–158 [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A., Utepbergenov D. I., Mueller S. L., Beyermann M., Schneider-Mergener J., Krause G., Blasig I. E. (2004) Cell. Mol. Life Sci. 61, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank R. (2002) J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 24.Schreibelt G., Kooij G., Reijerkerk A., van Doorn R., Gringhuis S. I., van der Pol S., Weksler B. B., Romero I. A., Couraud P. O., Piontek J., Blasig I. E., Dijkstra C. D., Ronken E., de Vries H. E. (2007) FASEB J. 21, 3666–3676 [DOI] [PubMed] [Google Scholar]
- 25.Blasig I. E., Winkler L., Lassowski B., Mueller S. L., Zuleger N., Krause E., Krause G., Gast K., Kolbe M., Piontek J. (2006) Cell. Mol. Life Sci. 63, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreeva A. Y., Krause E., Müller E. C., Blasig I. E., Utepbergenov D. I. (2001) J. Biol. Chem. 276, 38480–38486 [DOI] [PubMed] [Google Scholar]
- 27.Hanna P. C., Mietzner T. A., Schoolnik G. K., McClane B. A. (1991) J. Biol. Chem. 266, 11037–11043 [PubMed] [Google Scholar]
- 28.Kokai-Kun J. F., Benton K., Wieckowski E. U., McClane B. A. (1999) Infect. Immun. 67, 5634–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuyama A., Kondoh M., Seguchi H., Takahashi A., Harada M., Fujii M., Mizuguchi H., Horiguchi Y., Watanabe Y. (2005) J. Pharmacol. Exp. Ther. 314, 789–795 [DOI] [PubMed] [Google Scholar]
- 30.Van Itallie C. M., Betts L., Smedley J. G., 3rd, McClane B. A., Anderson J. M. (2008) J. Biol. Chem. 283, 268–274 [DOI] [PubMed] [Google Scholar]
- 31.Coyne C. B., Gambling T. M., Boucher R. C., Carson J. L., Johnson L. G. (2003) Am. J. Physiol. Cell. Mol. Physiol. 285, L1166–1178 [DOI] [PubMed] [Google Scholar]
- 32.Nishikawa K., Watanabe M., Kita E., Igai K., Omata K., Yaffe M. B., Natori Y. (2006) FASEB J. 20, 2597–2599 [DOI] [PubMed] [Google Scholar]
- 33.Robertson S. L., Smedley J. G., 3rd, Singh U., Chakrabarti G., Van Itallie C. M., Anderson J. M., McClane B. A. (2007) Cell. Microbiol. 9, 2734–2755 [DOI] [PubMed] [Google Scholar]
- 34.Mitic L. L., Unger V. M., Anderson J. M. (2003) Protein Sci. 218, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebihara C., Kondoh M., Hasuike N., Harada M., Mizuguchi H., Horiguchi Y., Fujii M., Watanabe Y. (2006) J. Pharmacol. Exp. Ther. 316, 255–260 [DOI] [PubMed] [Google Scholar]
- 36.Takahashi A., Komiya E., Kakutani H., Yoshida T., Fujii M., Horiguchi Y., Mizuguchi H., Tsutsumi Y., Tsunoda S., Koizumi N., Isoda K., Yagi K., Watanabe Y., Kondoh M. (2008) Biochem. Pharmacol. 75, 1639–1648 [DOI] [PubMed] [Google Scholar]
- 37.Harada M., Kondoh M., Ebihara C., Takahashi A., Komiya E., Fujii M., Mizuguchi H., Tsunoda S., Horiguchi Y., Yagi K., Watanabe Y. (2007) Biochem. Pharmacol. 73, 206–214 [DOI] [PubMed] [Google Scholar]
- 38.Ling J., Liao H., Clark R., Wong M. S., Lo D. D. (2008) J. Biol. Chem. 283, 30585–30595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.