Abstract
Zinc is an essential nutrient and serves as a structural or catalytic cofactor for many proteins. Thus, cells need mechanisms to maintain zinc homeostasis when available zinc supplies decrease. In addition, cells require other mechanisms to adapt intracellular processes to suboptimal levels of zinc. By exploring the transcriptional responses to zinc deficiency, studies of the yeast Saccharomyces cerevisiae have revealed both homeostatic and adaptive responses to low zinc. The Zap1 zinc-responsive transcription factor regulates several genes in yeast, and the identity of these genes has led to new insights regarding how cells respond to the stress of zinc deficiency.
Zap1 Transcription Factor
In Saccharomyces cerevisiae, Zap1 is the central player in the response to zinc deficiency (1). This metal-responsive regulatory protein controls the expression of many genes in yeast. For most of its target genes, Zap1 acts as a transcriptional activator and increases expression when zinc levels are low. In a few other cases, Zap1 acts as a repressor. To perform these functions, Zap1 binds to zinc-responsive elements (ZREs)2 in the promoters of its target genes (2). The consensus sequence for a ZRE is ACCTTNAAGGT, although functional Zap1-binding sites can differ greatly from this consensus. Although some Zap1-regulated promoters contain multiple ZREs, most have only a single binding site (2, 3).
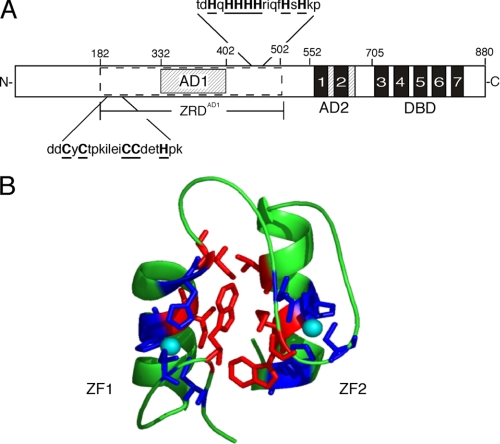
The Zap1 protein is 880 amino acids long (Fig. 1A). A DBD is found at its C terminus (4, 5). This domain is made up of five C2H2 zinc fingers. In addition, Zap1 contains two domains, AD1 and AD2, that activate transcription (6). Current evidence indicates that Zap1 is a direct sensor of zinc. The protein resides in the nucleus under all conditions of zinc status (6). When zinc levels rise, the metal binds to residues in the AD1 and AD2 regions of the protein, and this binding inhibits their ability to activate transcription (6–11). It is intriguing that Zap1 contains two ADs that are independently regulated by zinc. This feature is conserved among Zap1 orthologs from distantly related fungi, indicating that both ADs are critical for Zap1 function.
FIGURE 1.
Anatomy of the Zap1 transcription factor. In A, the zinc fingers of Zap1 are shown by the filled boxes, and AD1 and AD2 are indicated by the hatched boxes. The potential zinc ligands at the ends of ZRDAD1 are in boldface and underlined. In B, a model of the folded form of the ZF1/ZF2 finger pair is shown. Zinc atoms are cyan, the zinc ligands are blue, and the residues lining the hydrophobic interface between the fingers are red.
AD1 is located in the N-terminal half of the protein and is embedded within an ∼300-amino acid region required for its regulation by zinc (7). This region, designated ZRDAD1, contains no known zinc-binding motifs, but it binds multiple zinc ions in vitro. In addition, mutation of potential zinc ligands near the ends of ZRDAD1 renders AD1 constitutive. The Zap1 DBD has also been found to be required for shutting off AD1, suggesting that zinc binding promotes an interaction between AD1 and the DBD that masks AD1 function (6, 7).
AD2 is regulated by zinc binding to two additional C2H2 zinc fingers. These fingers, ZF1 and ZF2, bind zinc and fold into a novel finger pair structure in which the two fingers interact via a hydrophobic interface (Fig. 1B) (9). It is likely that this folded conformation prevents AD2 from recruiting coactivators to Zap1 target promoters. AD2 maps to the ZF2 domain, but both fingers are required for its zinc regulation (10). A remarkable feature of zinc binding to ZF1 and ZF2 is that it is very labile relative to structural sites (8, 9, 11). This observation suggests that regulating AD2 involves a constant cycle of zinc binding and release from ZF1 and ZF2, allowing rapid sampling of the cell's zinc status. Lability of zinc binding to AD1 has not been assessed.
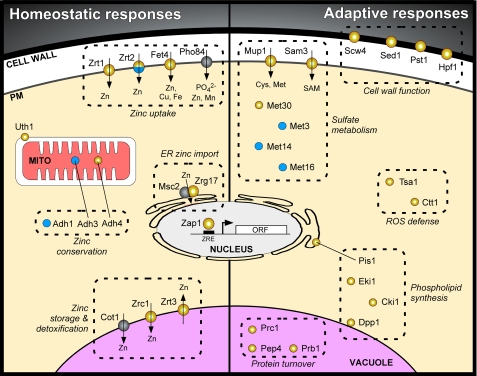
The genes that Zap1 regulates provide important insight into how cells respond to zinc-limiting conditions. Several studies have used genome-wide approaches to identify genes that are altered in expression in zinc-limited cells and those that are regulated directly by Zap1 (3, 12–15). Taken together, these studies suggest that >80 genes in yeast are direct targets of Zap1 activation. In addition, several genes are repressed in a Zap1-dependent manner, indicating that additional modes of regulation exist. The roles of many of these genes are depicted in Fig. 2 and discussed below.
FIGURE 2.
Transcriptional responses to zinc deficiency mediated by Zap1. The subcellular location and functional role of the protein products of known or likely Zap1 target genes are shown. Gene products shown in yellow represent those that are up-regulated by Zap1, and those in blue indicate down-regulated proteins. The symbol for Zrt2 is split because that gene is both activated and repressed by Zap1. Gray circles indicate gene products not regulated by zinc. PM, plasma membrane; MITO, mitochondria; ORF, open reading frame.
Homeostatic Responses to Zinc Deficiency
First among Zap1-mediated responses to zinc deficiency is the autoregulation of the ZAP1 gene itself (2). Zap1 activates its own transcription in zinc-limited cells, resulting in increased Zap1 protein levels. This increase may play an important role in the zinc responsiveness of other Zap1 target genes.
Zinc Uptake
A major response to Zap1 activation is the increased expression of transporters involved in zinc uptake. Several different proteins mediate zinc uptake in yeast. The ZRT1 and ZRT2 genes encode Zn2+-specific transporters, whereas Fet4 transports Fe2+, Cu+, and Zn2+ (16–18). Zap1 induces expression of ZRT1, ZRT2, and FET4 under zinc-limiting conditions. Zrt1 and Zrt2 are members of the ZIP family of transporters that play key roles in metal transport in bacteria, fungi, plants, and animals (19, 20). Fet4 homologs are found only in other fungi. Zrt1 has the highest affinity for zinc and plays the major role under zinc-limiting conditions. Zrt2 and Fet4 have lower affinity and contribute to zinc uptake under mildly low zinc conditions. The Pho84 phosphate transporter may also transport zinc, perhaps as a Zn·PO4 complex, when extracellular zinc levels are high (21). PHO84 expression is not altered by zinc status.
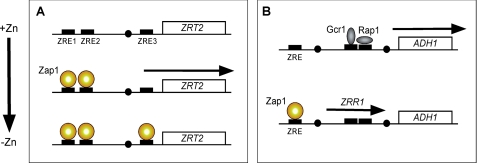
Surprisingly, the ZRT2 gene is both activated and repressed by Zap1 (22). ZRT2 expression increases under mild zinc deficiency but decreases to low levels under severe deficiency. This paradoxical regulation is due to the presence of three ZREs in the ZRT2 promoter (Fig. 3A). Two high affinity Zap1-binding sites, ZRE1 and ZRE2, are located upstream of the TATA box and mediate Zap1-dependent activation. The third ZRE, ZRE3, has a lower affinity for Zap1 binding and is located downstream of the TATA box near the transcription start site. ZRE3 is essential for repressing ZRT2 expression. Under moderate conditions of zinc deficiency, Zap1 binds to ZRE1 and ZRE2 and activates transcription. Under severe deficiency, when Zap1 levels rise due to autoregulation, Zap1 also binds to ZRE3 and blocks ZRT2 expression. This pattern of expression ensures that the low affinity Zrt2 transporter is not expressed at times when it is unable to contribute to zinc uptake.
FIGURE 3.
Mechanisms of ZRT2 (A) and ADH1 (B) repression mediated by Zap1. TATA elements (filled circles), transcription factor-binding sites (filled rectangles), and RNAs produced (arrows) are shown.
Vacuolar Zinc Storage
The vacuole is the major site of zinc storage in yeast. Under high zinc conditions, almost 109 atoms of zinc/cell can be stored in this organelle (23). This amount of zinc is sufficient to supply the needs of hundreds of progeny cells, so vacuolar zinc storage is likely to be of great utility to cells facing zinc-limiting conditions. Transport of zinc into the vacuole in zinc-replete cells is primarily the role of two transporters, Zrc1 and Cot1 (24). These proteins are also critical for zinc tolerance, indicating that the vacuole plays the major role in the resistance of yeast to excess zinc. Under zinc-limiting conditions, expression of the ZRT3 gene is up-regulated by Zap1 (24). ZRT3 encodes a ZIP protein related to Zrt1 and Zrt2 that pumps zinc out of the vacuole and into the cytoplasm, where it can then be utilized. Mutants defective for Zrt3 function hyperaccumulate zinc in the vacuole but mobilize that stored zinc very poorly.
Zinc Shock Tolerance
It was surprising to discover that ZRC1 is also up-regulated by Zap1 in zinc-limited cells (25, 26). This observation was unexpected given the role of Zrc1 in generating rather than mobilizing zinc stores. We have shown that ZRC1 induction is a novel “proactive” mechanism of zinc homeostasis (26). Because zinc-limited cells express high levels of zinc transporters like Zrt1, they are poised to accumulate substantial amounts of zinc should it become available. We refer to this condition as “zinc shock.” Zrc1 and its induction by Zap1 are critical for cells to withstand zinc shock by promoting the efficient transport of excess cytosolic zinc into the vacuole. Being proactive rather than reactive, this mechanism allows for more rapid resistance than would post-stress induction of zinc tolerance genes such as metallothioneins, as occurs in many other organisms.
Zinc Conservation
ADHs are among the most abundant zinc-binding proteins in the cell. Thus, it was intriguing to discover that two genes encoding ADH isozymes, ADH1 and ADH3, are repressed by Zap1 under low zinc conditions (3). The populations of Adh1 and Adh3 proteins together bind ∼1.5 × 106 atoms of zinc/cell under zinc-replete conditions (27, 28). This amount represents a sizeable portion of the total cellular zinc, so down-regulation of ADH1 and ADH3 expression would provide substantial amounts of the metal for other purposes. The mechanism Zap1 uses to repress ADH isozymes has recently been determined (Fig. 3B) (29). Zap1 represses ADH1 and ADH3 expression by means of intergenic transcripts that are activated by Zap1 and transcribed through the ADH1 and ADH3 promoters. These intergenic transcripts, designated ZRR1 and ZRR2, respectively, do not encode protein products themselves, but rather their synthesis through these promoter regions results in the transient displacement of transcription factors (Gcr1, Rap1) normally required for ADH1 and ADH3 expression. This displacement causes reduced expression of two of the most abundant zinc-binding proteins in the cell. Conversely, the ADH4 gene is induced by Zap1 (3, 14, 29), and this gene encodes an ADH similar to an iron-dependent isozyme from Zymomonas mobilis. Thus, by switching from zinc-dependent to zinc-independent ADH isozymes, the cell may conserve zinc for other uses. Alternatively, some evidence suggests that Adh4 may use zinc as its cofactor rather than iron (30). Adh4 is predicted to bind only one atom/monomer, whereas Adh1 and Adh3 each bind two. Thus, zinc conservation could occur under this scenario as well.
Other Homeostatic Responses
The Zap1-regulated genes just discussed have all been verified to be direct targets of Zap1. Several other genes have been implicated by expression studies to be Zap1 targets but have not yet been further tested. These genes may also play important roles in zinc homeostasis. One example is ZRG17. ZRG17 encodes a transporter protein involved in moving zinc into the ER for metalloproteins in that organelle (31). Zrg17 functions in a heteromeric complex with the Msc2 protein. Although MSC2 expression is not regulated by Zap1, ZRG17 expression probably is (3, 12, 14, 15). Thus, ER zinc transport and zinc homeostasis in that compartment may be controlled by Zap1 regulating ZRG17. Another potential target of Zap1 regulation is UTH1. Uth1 is a mitochondrial protein that is involved in the targeted degradation of mitochondria by autophagy (32). Mitochondrial autophagy involves the engulfment of mitochondria by the vacuole and their subsequent degradation by vacuolar enzymes. Induction of Uth1 may aid the degradation of mitochondria in zinc-limited cells, and these mitochondria could be a source of zinc for other purposes.
Adaptive Responses to Zinc Deficiency
The control of zinc uptake, vacuolar zinc storage, and isozyme switching among ADHs represent strategies of zinc homeostasis. It has also become clear that yeast cells use several strategies to adapt to conditions of zinc deficiency (Fig. 2). These include mechanisms to resist oxidative stress caused by zinc deficiency, changes in lipid synthesis pathways, and remodeling of sulfate assimilation. These adaptive responses can be as important as the homeostatic responses in allowing yeast to survive the stress of zinc deficiency.
Oxidative Stress Tolerance
Studies of many different organisms have shown that zinc deficiency can increase intracellular levels of ROS (33, 34). Increased ROS can then lead to lipid and protein oxidation and to DNA damage and mutations. Like other organisms, yeast cells experience increased oxidative stress when zinc-limited (35). Although the source of this increased ROS is unknown, we have discovered a mechanism that yeast cells use to protect themselves against it. Specifically, Zap1 activates expression of the TSA1 gene, which encodes the major cytosolic peroxiredoxin (35). Peroxiredoxins catalyze the degradation of hydrogen peroxide and organic hydroperoxides (36). Consistent with the regulation of TSA1 by Zap1, tsa1Δ mutants grow very poorly under zinc-limiting conditions. This indicates that Tsa1 plays a major role in protecting cells against ROS under low zinc conditions. Zap1 has also been implicated in the increased expression of CTT1, encoding cytosolic catalase, in low zinc (14). Thus, catalase may also degrade ROS that accumulates in zinc-limited cells. The source of oxidative stress in low zinc is under investigation.
Phospholipid Synthesis
The major phospholipids found in yeast membranes are PI, phosphatidylserine, PE, and PC. These phospholipids are synthesized from CDP-DAG via the CDP-DAG pathway (37). The CDP-DAG pathway is the primary mechanism of PE and PC synthesis when choline and ethanolamine are not supplied in the medium. However, these phospholipids can also be synthesized from ethanolamine and choline via the alternative Kennedy pathway. Intriguingly, Iwanyshyn et al. (38) noted that the activities of all of the enzymes of the CDP-DAG pathway are decreased in zinc-limited cells. Conversely, the activity of PI synthase, encoded by PIS1 and involved in converting CDP-DAG into PI, is increased. In addition, the activities of the Kennedy pathway enzymes ethanolamine kinase and choline kinase, encoded by EKI1 and CKI1, respectively, are increased in zinc-limited cells (39, 40). Thus, a metabolic remodeling occurs in low zinc such that CDP-DAG is diverted to PI synthesis and PE and PC synthesis switches from the CDP-DAG pathway to the Kennedy pathway.
Carman and co-workers (38) have determined the mechanism of this regulation. They found that repression of the CDP-DAG pathway enzymes occurs at the transcriptional level via the Opi1 repressor protein. Opi1 is a component of the regulatory machinery that regulates lipid synthesis in response to inositol. Opi1 is found in its inactive state bound to the nuclear/ER membrane. Under low zinc conditions, as under high inositol conditions, Opi1 is released and translocated into the nucleus. Opi1 is then recruited to the promoters of the CDP-DAG pathway enzymes (CHO1, PSD1, CHO2, OPI3), which are then repressed. In contrast, increased expression of PIS1 and the Kennedy pathway genes EKI1 and CKI1 is the result of transcriptional activation by Zap1 (39–41). The promoters of these genes contain ZREs that serve as Zap1-binding sites and are required for their induction in zinc-limited cells. In this way, zinc-limited cells increase their levels of PI and use alternative pathways to generate PE and PC. Whereas PE levels decrease by ∼50% in zinc-limited cells, PC levels are unaffected (38). It is interesting to note that the EKI1 and CKI1 promoters are also subject to Opi1 repression, suggesting that Zap1 activation overrides Opi1 control in zinc-limited cells.
An important remaining question is why this remodeling of lipid synthesis occurs in response to low zinc. Although the answer is still unclear, one possible explanation is suggested by our recent analysis of the regulation of sulfate assimilation by Zap1. As discussed further below, we have found that the assimilation of sulfate into sulfur-containing compounds such as methionine and SAM is repressed under zinc-limiting conditions. The conversion of PE to PC requires sequential methyltransferase reactions using SAM as the methyl donor. Repressed sulfate assimilation and reduced levels of SAM in zinc-limited cells may reduce the capacity of the CDP-DAG pathway for PC synthesis. Up-regulation of the Kennedy pathway would then help bypass that potential barrier to lipid synthesis.
Another gene involved in lipid metabolism that is regulated by Zap1 is DPP1, encoding DGPP phosphatase (42). Dpp1 is localized to the vacuole membrane and catalyzes the dephosphorylation of DGPP to PA and, subsequently, of PA to DAG. This results in a decrease in DGPP and PA levels in the vacuole membrane (43), but the importance of these effects in low zinc is unknown.
Sulfate Assimilation
Zap1 repression of ZRT2, ADH1, and ADH3 suggested that other genes may be repressed, either directly or indirectly, by Zap1 under zinc deficiency. An analysis of expression data obtained from previous microarray experiments indicated that >30 genes in the yeast genome show reduced expression in zinc-limited cells in a manner that is responsive to Zap1 activity (44). In addition to ADH1 and ADH3, the ADH2 zinc-dependent ADH was identified, indicating that it may also be repressed for zinc conservation.
The genes encoding the first three enzymes of the sulfate assimilation pathway, MET3, MET14, and MET16, are also repressed in a Zap1-dependent manner in low zinc. These enzymes convert SO42− into SO32−, which can then be converted into sulfur-containing compounds such as methionine, SAM, cysteine, and glutathione (45). Consistent with down-regulation of sulfate assimilation, the free pools of cysteine and methionine were greatly reduced in zinc-limited cells compared with zinc-replete cells (44).
We have recently determined the mechanism of this regulation. MET3, MET14, and MET16 transcription is mediated by the Met4 activator. When organic sulfur compounds (e.g. cysteine) are high, Met4 is ubiquitinated and inactivated by the SCFMet30 E3 ubiquitin ligase (46). In this way, transcription of the MET genes is decreased when the end products of the pathway accumulate. Zap1 commandeers this regulatory system in zinc-limited cells to repress sulfate assimilation regardless of the levels of organic sulfur compounds. To do this, Zap1 activates transcription of the MET30 gene via a ZRE in the MET30 promoter. Met30 is the rate-limiting subunit of SCFMet30, and its increased expression leads to increased SCFMet30 activity, Met4 ubiquitination, and decreased Met4 activation of its target genes.
Although the mechanism of this regulation is clear, how does this help zinc-limited cells? An attractive model is suggested by studies of oxidative stress tolerance. Sulfate assimilation requires large amounts of NADPH. Slekar et al. (47) demonstrated that using NADPH to combat oxidative stress, e.g. via NADPH-dependent glutathione and thioredoxin reductases, disrupts sulfate assimilation because of competition for limiting NADPH supplies. Conversely, we suspect that sulfate assimilation competes with oxidative stress responses for that same pool of NADPH. Thus, given that zinc-limited yeast cells have elevated ROS and an increased demand for NADPH, repression of sulfate assimilation would make more NADPH available for oxidative stress tolerance.
Other Adaptive Responses
As was the case for zinc homeostasis genes, there are several genes that may play important roles in the adaptation to zinc deficiency that have yet to be confirmed as direct Zap1 targets. Two of those genes, MUP1 and SAM3, could contribute to sulfate metabolism in zinc deficiency (14, 15). MUP1 encodes a cysteine and methionine uptake transporter protein (48). Similarly, Sam3 is a transporter that takes up exogenous SAM (49). Both of these genes are up-regulated in zinc-limited cells in a Zap1-dependent manner. Their induction makes sense in light of the decreased level of sulfate assimilation that was just discussed. In addition, several genes involved in cell wall function, including SCW4, SED1, PST1, and HPF1, are potential Zap1 targets, suggesting that cell wall remodeling occurs in low zinc. Furthermore, genes involved in vacuolar protein degradation (PEP4, PRB1, PRC1) are up-regulated perhaps to increase the rate of protein degradation. This could serve an adaptive role by aiding the removal of damaged proteins that could accumulate in zinc-limited cells. Alternatively, increased protein turnover could free up zinc for other uses. As a final example, two genes involved in protein modification in the secretory pathway, MCD4 and MNT2, are apparently up-regulated by Zap1 in low zinc. MNT2 encodes a mannosyltransferase. Mcd4 is a zinc-dependent enzyme involved in glycosylphosphatidylinositol anchor synthesis (50), so its increased expression may allow this enzyme to maintain its function under zinc deficiency.
Conclusions
The identification of Zap1 target genes has highlighted the strategies used by yeast to thrive under zinc-limiting growth conditions. Many of these responses are involved in zinc homeostasis and strive to maintain a consistent intracellular zinc environment while external conditions change. In addition, many other responses help the cell to adapt to conditions of zinc deficiency by altering metabolic processes that are compromised by that stress. In this minireview, I have discussed the roles of ∼30 of the >100 genes that are regulated positively or negatively by Zap1. Future studies addressing the function of other Zap1-regulated genes and their relevance to zinc-limited growth promise to provide new insights into how cells of all organisms respond to zinc deficiency.
This work was supported, in whole or in part, by National Institutes of Health Grants GM56285 and GM69786. This is the second article of five in the second Thematic Minireview Series on Metals in Biology. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
- ZRE
- zinc-responsive element
- DBD
- DNA-binding domain
- AD
- activation domain
- ZRDAD1
- zinc-responsive domain of AD1
- ZF
- zinc finger
- ADH
- alcohol dehydrogenase
- ER
- endoplasmic reticulum
- ROS
- reactive oxygen species
- PI
- phosphatidylinositol
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- DAG
- diacylglycerol
- SAM
- S-adenosylmethionine
- DGPP
- diacylglycerol-pyrophosphate
- PA
- phosphatidic acid.
REFERENCES
- 1.Zhao H., Eide D. J. (1997) Mol. Cell. Biol. 17, 5044–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao H., Butler E., Rodgers J., Spizzo T., Duesterhoeft S., Eide D. (1998) J. Biol. Chem. 273, 28713–28720 [DOI] [PubMed] [Google Scholar]
- 3.Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., Eide D. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97, 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans-Galea M. V., Blankman E., Myszka D. G., Bird A. J., Eide D. J., Winge D. R. (2003) Biochemistry 42, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 5.Bird A., Evans-Galea M. V., Blankman E., Zhao H., Luo H., Winge D. R., Eide D. J. (2000) J. Biol. Chem. 275, 16160–16166 [DOI] [PubMed] [Google Scholar]
- 6.Bird A. J., Zhao H., Luo H., Jensen L. T., Srinivasan C., Evans-Galea M., Winge D. R., Eide D. J. (2000) EMBO J. 19, 3704–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbig A., Bird A. J., Swierczek S., McCall K., Mooney M., Wu C. Y., Winge D. R., Eide D. J. (2005) Mol. Microbiol. 57, 834–846 [DOI] [PubMed] [Google Scholar]
- 8.Bird A. J., Swierczek S., Qiao W., Eide D. J., Winge D. R. (2006) J. Biol. Chem. 281, 25326–25335 [DOI] [PubMed] [Google Scholar]
- 9.Wang Z., Feng L. S., Matskevich V., Venkataraman K., Parasuram P., Laity J. H. (2006) J. Mol. Biol. 357, 1167–1183 [DOI] [PubMed] [Google Scholar]
- 10.Bird A. J., McCall K., Kramer M., Blankman E., Winge D. R., Eide D. J. (2003) EMBO J. 22, 5137–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao W., Mooney M., Bird A. J., Winge D. R., Eide D. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 8674–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan D. S. (2000) Genetics 156, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins V. J., Rogers P. J., Dawes I. W. (2003) Appl. Environ. Microbiol. 69, 7535–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C. Y., Bird A. J., Chung L. M., Newton M. A., Winge D. R., Eide D. J. (2008) BMC Genomics 9, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Nicola R., Hazelwood L. A., De Hulster E. A., Walsh M. C., Knijnenburg T. A., Reinders M. J., Walker G. M., Pronk J. T., Daran J. M., Daran-Lapujade P. (2007) Appl. Environ. Microbiol. 73, 7680–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H., Eide D. (1996) J. Biol. Chem. 271, 23203–23210 [DOI] [PubMed] [Google Scholar]
- 17.Zhao H., Eide D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waters B. M., Eide D. J. (2002) J. Biol. Chem. 277, 33749–33757 [DOI] [PubMed] [Google Scholar]
- 19.Guerinot M. L. (2000) Biochim. Biophys. Acta 1465, 190–198 [DOI] [PubMed] [Google Scholar]
- 20.Kambe T., Suzuki T., Nagao M., Yamaguchi-Iwai Y. (2006) Genomics Proteomics Bioinformatics 4, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen L. T., Ajua-Alemanji M., Culotta V. C. (2003) J. Biol. Chem. 278, 42036–42040 [DOI] [PubMed] [Google Scholar]
- 22.Bird A. J., Blankman E., Stillman D. J., Eide D. J., Winge D. R. (2004) EMBO J. 23, 1123–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simm C., Lahner B., Salt D., LeFurgey A., Ingram P., Yandell B., Eide D. J. (2007) Eukaryot. Cell 6, 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDiarmid C. W., Gaither L. A., Eide D. (2000) EMBO J. 19, 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyabe S., Izawa S., Inoue Y. (2000) Biochem. Biophys. Res. Commun. 276, 879–884 [DOI] [PubMed] [Google Scholar]
- 26.MacDiarmid C. W., Milanick M. A., Eide D. J. (2003) J. Biol. Chem. 278, 15065–15072 [DOI] [PubMed] [Google Scholar]
- 27.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 28.Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 29.Bird A. J., Gordon M., Eide D. J., Winge D. R. (2006) EMBO J. 25, 5726–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drewke C., Ciriacy M. (1988) Biochim. Biophys. Acta 950, 54–60 [DOI] [PubMed] [Google Scholar]
- 31.Ellis C. D., Macdiarmid C. W., Eide D. J. (2005) J. Biol. Chem. 280, 28811–28818 [DOI] [PubMed] [Google Scholar]
- 32.Kissová I., Deffieu M., Manon S., Camougrand N. (2004) J. Biol. Chem. 279, 39068–39074 [DOI] [PubMed] [Google Scholar]
- 33.Ho E. (2004) J. Nutr. Biochem. 15, 572–578 [DOI] [PubMed] [Google Scholar]
- 34.Powell S. R. (2000) J. Nutr. 130, Suppl. 5S, 1447S–1454S [DOI] [PubMed] [Google Scholar]
- 35.Wu C. Y., Bird A. J., Winge D. R., Eide D. J. (2007) J. Biol. Chem. 282, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 36.Rhee S. G., Chae H. Z., Kim K. (2005) Free Radic. Biol. Med. 38, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 37.Carman G. M., Han G. S. (2007) Biochim. Biophys. Acta 1771, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwanyshyn W. M., Han G. S., Carman G. M. (2004) J. Biol. Chem. 279, 21976–21983 [DOI] [PubMed] [Google Scholar]
- 39.Soto A., Carman G. M. (2008) J. Biol. Chem. 283, 10079–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersting M. C., Carman G. M. (2006) J. Biol. Chem. 281, 13110–13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han S. H., Han G. S., Iwanyshyn W. M., Carman G. M. (2005) J. Biol. Chem. 280, 29017–29024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han G. S., Johnston C. N., Chen X., Athenstaedt K., Daum G., Carman G. M. (2001) J. Biol. Chem. 276, 10126–10133 [DOI] [PubMed] [Google Scholar]
- 43.Han G. S., Johnston C. N., Carman G. M. (2004) J. Biol. Chem. 279, 5338–5345 [DOI] [PubMed] [Google Scholar]
- 44.Wu C. Y. (2008) The Zap1 Regulon in Yeast Saccharomyces Cerevisiae Ph.D. dissertation, University of Wisconsin-Madison, Madison, WI [Google Scholar]
- 45.Thomas D., Surdin-Kerjan Y. (1997) Microbiol. Mol. Biol. Rev. 61, 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser P., Su N. Y., Yen J. L., Ouni I., Flick K. (2006) Cell Div. 1, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slekar K. H., Kosman D. J., Culotta V. C. (1996) J. Biol. Chem. 271, 28831–28836 [DOI] [PubMed] [Google Scholar]
- 48.Kosugi A., Koizumi Y., Yanagida F., Udaka S. (2001) Biosci. Biotechnol. Biochem. 65, 728–731 [DOI] [PubMed] [Google Scholar]
- 49.Rouillon A., Surdin-Kerjan Y., Thomas D. (1999) J. Biol. Chem. 274, 28096–28105 [DOI] [PubMed] [Google Scholar]
- 50.Mann K. J., Sevlever D. (2001) Biochemistry 40, 1205–1213 [DOI] [PubMed] [Google Scholar]