Abstract
Transcription is essential for neurite and axon outgrowth during development. Recent work points to the involvement of nuclear factor of activated T cells (NFAT) in the regulation of genes important for axon growth and guidance. However, NFAT has not been reported to directly control the transcription of axon outgrowth-related genes. To identify transcriptional targets, we performed an in silico promoter analysis and found a putative NFAT site within the GAP-43 promoter. Using in vitro and in vivo experiments, we demonstrated that NFAT-3 regulates GAP-43, but unexpectedly, does not promote but represses the expression of GAP-43 in neurons and in the developing brain. Specifically, in neuron-like PC-12 cells and in cultured cortical neurons, the overexpression of NFAT-3 represses GAP-43 activation mediated by neurotrophin signaling. Using chromatin immunoprecipitation assays, we also show that prior to neurotrophin activation, endogenous NFAT-3 occupies the GAP-43 promoter in PC-12 cells, in cultured neurons, and in the mouse brain. Finally, we observe that NFAT-3 is required to repress the physiological expression of GAP-43 and other pro-axon outgrowth genes in specific developmental windows in the mouse brain. Taken together, our data reveal an unexpected role for NFAT-3 as a direct transcriptional repressor of GAP-43 expression and suggest a more general role for NFAT-3 in the control of the neuronal outgrowth program.
Transcription involves many protein-protein and protein-DNA interactions. This allows for the integration of multiple signaling pathways by a limited set of transcription factors that work in combination to either activate or repress genes relevant to the current cellular signaling context (1, 2). These diverse “inputs” are integrated by the binding of transcriptional activators/repressors along with their coactivators/repressors and the modification of chromatin itself to result in the final “output” of a unique nucleoprotein complex capable of either inducing or repressing transcription (3). Transcription is therefore a key regulation point as it allows for the integration of diverse and subtle cellular context during neural development (4, 5).
Not surprisingly, axon sprouting and outgrowth are under tight transcriptional control, and the expression of pro-axon growth genes is limited to appropriate spatial and temporal stages of neural development. Therefore, an examination of how changing developmental cues are integrated at the level of transcription might reveal novel mechanisms that regulate axon sprouting and outgrowth.
One gene involved in axon outgrowth and guidance is GAP-43 (growth-associated protein 43), a neurotrophin-dependent membrane-bound phosphoprotein highly expressed during the development of the nervous system (6–8). It is found localized to the axon and growth cones of developing neurons and shows preferential expression in the forebrain and in highly plastic central nervous system regions such as the olfactory bulb, hippocampus, dorsal root ganglia, and ascending sensory pathways in the spinal cord (9, 10). It is also significantly up-regulated in regenerating neurons subsequent to axon lesion (11, 12). Studies examining the transcriptional control of GAP-43 have identified a ∼1000-bp promoter region upstream of the protein-coding region that is sufficient to respond to neurotrophin signaling and to determine neuron-specific expression (13–16). Thus, the GAP-43 proximal promoter provides a relatively compact and neuron-specific model to investigate the transcriptional machinery and chromatin context required for axon outgrowth in developing and regenerating neurons.
We have recently characterized a novel role for the transcription factor and tumor suppressor protein p53 in both axon growth and physiological nerve regeneration, where it functions as a transcriptional activator of several neuronal pro-axon outgrowth and pro-regeneration genes, including GAP-43 (17, 18). Specifically, p53 promotes GAP-43 expression through a novel binding site within the 5′ promoter region. The in silico promoter analysis also revealed a putative binding site for the transcription factor nuclear factor of activated T cells (NFAT)2 adjacent to the p53 site.
The NFAT family has been shown to play a role in the developing and possibly in the adult nervous system. Transgenic mice containing an NFAT reporter showed that NFAT transcriptional activity is highest in the brain (19, 20), and NFAT-3 is specifically expressed in the spinal cord and the brain, with high levels found in the olfactory bulb, cerebellum, and certain regions of the cortex (21–24). NFAT activity is important in neuronal growth and guidance during vertebrate development and appears to be downstream of neurotrophin and netrin signaling pathways (25–27).
There are five NFAT family members named NFAT-1/p/c2, NFAT-2/c/c1, NFAT-3/c4, NFAT-4/c3/x, and NFAT-5/TonEBP (28–30). However, only NFAT1–4 contain the Ca2+ sensor/translocation domain (31, 32) and are thus dependent upon intracellular Ca2+ activation of the phosphatase calcineurin (33–35). Dephosphorylation of several residues in the Ca2+ sensor/translocation domain by calcineurin results in exposure of a nuclear localization sequence and nuclear import (36, 37). In fact, the drugs cyclosporin A (CsA) and FK506, which are potent and specific inhibitors of calcineurin, are often used as inhibitors of NFAT transcriptional activity (38). Although additional post-translational modifications affect transcriptional activity, the nuclear localization of NFAT1–4 and the cooperative binding with other transcription factors appear to be a major regulatory mechanism for the transcriptional activity of NFAT complexes (30, 39–41).
In this study, we examine whether NFAT-3 is a direct transcriptional regulator of GAP-43 expression within neurotrophin signaling and whether this implies a role in neuronal outgrowth program during development. Unexpectedly, our data suggest that NFAT-3 is a negative and not a positive regulator of GAP-43 during neuronal outgrowth and maturation.
EXPERIMENTAL PROCEDURES
Cell Cultures
Cell Lines
Pheochromocytoma (PC-12) cell lines were grown in Dulbecco's modified medium supplemented with 10% horse serum, 5% fetal bovine serum, and 100 units/ml penicillin and 100 mg/ml streptomycin (Invitrogen) in a humidified atmosphere of 5% CO2 in air at 37 °C.
Primary Cortical Neurons
Rat cortical neurons or mouse cortical neurons were derived from wild-type adult rat (Sprague-Dawley) or mice (C57BL/6) and NFAT-3 null mice (19). Cortices were extracted, dissociated, and cultured as reported previously (18). Briefly, cortices were minced in Hanks' balanced salt solutions buffer supplemented with 2.5 μg/ml amphotericin B (Sigma, A2942), 100 units/ml penicillin, and 100 mg/ml streptomycin, and then 1,800 units/ml trypsin was added. The cells were incubated at 37 °C for 20 min, Dulbecco's modified medium (supplemented with 10% fetal bovine serum) was added, and the cells were dissociated by triturating. The partially dissociated tissue was allowed to settle for 5–10 min, the supernatant collected, and the remaining tissue pellet was retitrated an additional four times. The combined supernatants were filtered through a 40-μm cell strainer (BD Biosciences) and plated at the appropriate density. After 1 h, medium was aspirated off with any dead cells, and Neurobasal medium supplemented with B27, Glutamax, penicillin, and streptomycin was added. (All reagents were from Invitrogen except where indicated.) Embryonic day 13 (E13) mouse brains from wild-type and NFAT-3 null mice were also extracted, dissociated, and plated in the presence of Neurobasal medium plus retinoic acid and forskolin to induce neuronal differentiation.
In Silico Transcription Factor Binding Sites Analysis
A large region (∼10,000 bp) of the 5′-untranslated region of the GAP-43 gene was analyzed using MatInspector, a search algorithm within a suite of software programs provided by Genomatix that identifies transcription binding sites using a large library of weight matrices. An additional algorithm, P-MATCH, was also used to identify potential NFAT transcription binding sites for SCG-10, CAP-23, and L1cam.
Luciferase Assays
Reporter and Expression Plasmid Construct
The region of the GAP-43 promoter containing a putative NFAT binding site (see Fig. 1A) was cloned by PCR and ligated into the HindIII and XhoI restriction sites of a luciferase reporter derived from the NFAT/AP-1 3× luciferase reporter (Addgene, plasmid 11783). The pEGFP-NFAT-3 expression plasmid was also obtained from Addgene (plasmid 10961).
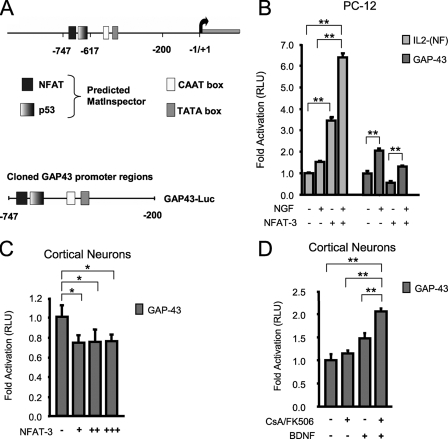
FIGURE 1.
NFAT-3 represses GAP-43 expression in PC-12 and cortical neurons. A, a depiction of the GAP-43 promoter region with the putative NFAT binding site. B, NFAT-3 represses GAP-43 transcription in PC-12 cells. Transient transfection on PC-12 cells with control IL-2-Luc, GAP-43-Luc, and EGFP-NFAT-3 expression plasmid. Cells were allowed to express EGFP-NFAT-3 for 24 h and then stimulated with 100 ng/ml NGF for an additional 24 h (see “Experimental Procedures” for DNA amounts used). RLU, relative light units. C, NFAT-3 represses GAP-43 transcription in neurons. Transient transfection was performed on cultured E16 rat cortical neurons with GAP-43-Luc reporter and increasing EGFP-NFAT-3 expression plasmid. (+, 125 ng; ++, 250 ng; +++, 350 ng) D, inhibition of the calcineurin/NFAT pathway further increases GAP-43 transcription in response to neurotrophin. Cultured E16 rat cortical neurons were transfected with GAP-43-Luc and treated with or without 10 ng/ml BDNF, 1 μm CsA, and 250 ng/ml FK506 for 16 h. All assays were performed in triplicate. The relative light unit was calculated as the ratio of firefly luciferase/Renilla luciferase signal. Signals were then normalized to reporter only transfection. Student's t test was performed to compare significant difference to reporter only transfections. p value: *, < 0.05, **, < 0.01. Error bars represent S.E.
Transfection
Either experiments were performed using Lipofectamine 2000 (Invitrogen) for PC-12 and rat cortical neurons 5 days in vitro (DIV 5), or electroporation with the rat neuron nucleofector kit (Amaxa Biosystems) was used according to the provided protocol. Briefly, for Lipofectamine experiments, PC-12 cells were seeded into 24-well plates in Dulbecco's modified medium, 1 day prior to transfection, or rat cortical neurons were seeded 5 days before transfection. Lipofectamine reagent to total DNA ratios were according to standard recommendation, with each well receiving 500 ng of the IL-2-(NFAT)-Luc reporter or 500 ng of the GAP-43-Luc reporters and 25 ng of pRL-TK-Renilla-luciferase (Promega) and 500 ng of pEGFP-NFAT-3 expression construct or empty pcDNA3.1(+) for a total of 1,025 ng of total DNA. After a 24 h-incubation, PC-12 cells were stimulated for another 24 h with 100 ng/ml of NGF (BD Biosciences). In rat neuron experiments, the following amounts of pEGFP-NFAT-3 DNA were used: 0, 125, 250, and 350 ng. For electroporation experiments, the standard Amaxa Biosystems rat protocol was followed. Briefly, five million neurons were used for each cuvette, with 2–4 μg of total DNA. Neurons were plated in 24-well plates at a density of 0.4 million cells/well and allowed to attach for 1 h, and then media were changed with and without 10 ng/ml BDNF (R&D Systems), 1 μm CsA, 250 ng/ml FK506 (LC Laboratories), or control DMSO vehicle and incubated for a total of 17 h. For both protocols, cells were harvested and lysed with 100 μl of passive lysis buffer, and luciferase activities were determined using the Dual-Luciferase kit (Promega).
Immunocytochemistry
Primary rat or mouse cortical neurons were grown on poly-d-lysine-coated coverslips and fixed with 4% paraformaldehyde, 4% sucrose. Neurons were blocked in 10% albumin and 0.2% Triton X-100 and incubated overnight at 4 °C with the appropriate primary antibodies. After several rinses in PBS, the coverslips were incubated with the appropriate secondary antibodies and Hoechst 33258 (Molecular Probes/Invitrogen) and washed in PBS before mounting on slides with Fluorsave (Calbiochem). Immunofluorescence was detected using a Zeiss microscope (Axiovert 200, Zeiss).
Immunohistochemistry
Mice were deeply anesthetized and perfused with 0.9% saline solution followed by 4% formaldehyde in 0.1 m phosphate buffer. Whole brain was removed, put in 4% formaldehyde again for 24 h, and then in 30% sucrose for an additional 24 h. The entire brain was frozen in 2-methylbutane on dry ice and stored at −80 °C. Frozen brains fixed with Jung tissue-freezing medium (Leica Microsystems) were cut into 12-μm-thick coronal sections and mounted on slides. Each slide was heated at 50 °C for 30 min, incubated for 30 min in 4% sucrose/PBS at room temperature, treated for 15 min with ice-cold 100% methanol, and washed three times with PBS for 10 min each time. Next, slides were blocked with 10% normal goat serum in 0.2% Triton X-100/PBS for 1 h at room temperature and probed with primary antibody in 1% normal goat serum/PBS overnight at 4 °C. On the next day, slides were washed three times with PBS, probed with secondary antibody in 1% normal goat serum/PBS for 1 h at room temperature, or blocked again and probed with another primary antibody overnight at 4 °C. Slides were finally washed three times in PBS, incubated with Hoechst for 45 s, washed one time for 5 min in PBS, and cleaned with Xylol. Samples were treated with fluorescent mounting medium (Dako), the coverslip was added, and samples were examined under a fluorescence microscope (Axiovert, Zeiss).
Immunoblotting
Mouse brains and cultured differentiated neurons were lysed in a solution containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1% Nonidet P-40, 0.1% SDS, and Complete protease inhibitor mixture (Roche Applied Science). Samples were further dissociated through a Dounce glass homogenizer. Extracts were left on ice for 30 min, and cell debris was removed by centrifugation. The protein concentration was determined by standard protein assay (Bio-Rad). Lysate (30–50 μg) was separated by SDS-PAGE and transferred to nitrocellulose. Detection was by HRP-chemiluminescence reagents (Thermo Fisher Scientific).
For Fig. 3C, quantitation of GAP-43 protein levels was performed by densitometry (FluorChem 8900) of three immunoblots for both GAP-43 and GAPDH. The resulting GAP-43 changes are expressed as a normalized ratio of GAP-43 to GAPDH levels.
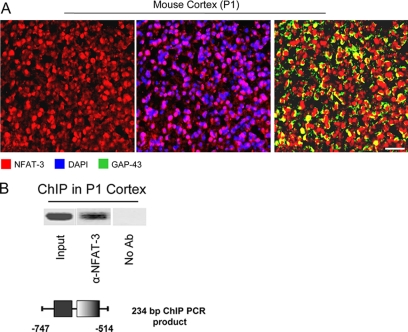
FIGURE 3.
NFAT-3 is nuclear and occupies the GAP-43 promoter in the developing mouse cortex. A, NFAT-3 is localized in the nucleus of cortical neurons. Fixed mouse P1 brain was probed for immunohistochemistry with α-NFAT-3, α-GAP-43, and DAPI nuclear staining. Scale bar: 30 μm. B, NFAT-3 occupies the GAP-43 promoter in the mouse brain. ChIP was performed for NFAT-3 using the same antibody as in Fig. 2, from fixed P1 mouse brain, and recovered DNA was measured by semiquantitative PCR. GAP-43-specific primers spanning the putative NFAT site were used. The resulting PCR amplicon containing the NFAT site of the GAP-43 promoter is depicted. No Ab, no antibody.
Antibodies
Immunoblotting
For immunoblotting, the following antibodies were used: α-NFAT-3 (Santa Cruz Biotechnology, sc-1153), α-GAP-43 (Chemicon/Millipore, AB5220), α-β-actin (Sigma, A5441) and α-GAPDH (Cell Signaling, 2118), α- rabbit-HRP (Thermo Fisher Scientific, 31460), α-mouse-HRP (Thermo Fisher Scientific, 31430).
Chromatin Immunoprecipitation Assays (ChIPs) Antibody
For the ChIP assays, the following antibody was used: α-NFAT-3 (Santa Cruz Biotechnology, sc-1153 X).
Immunocytochemistry and Immunohistochemistry
For immunocytochemistry and immunohistochemistry, the following antibodies were used: α-NFAT-3 (Affinity BioReagents, PA1-021), α-GAP-43 (Chemicon/Millipore, AB5220), α-rabbit Alexa Fluor 568 (Invitrogen, A11011), and α-mouse Alexa Fluor 488 (Invitrogen, A11001).
ChIP
ChIP assays were performed using the chromatin immunoprecipitation assay kit according to the protocol provided (Upstate Biotechnology/Millipore). Briefly, for experiments done with primary neurons and PC-12 cells, at least 1 × 106 cells were used for each assay and cross-linked with 1% formaldehyde for 10 min. For the experiment with whole brain, the brain was removed and fixed in 4% formaldehyde/PBS overnight and then minced, passed through a glass homogenizer, and sonicated until the DNA fragments were between 300 and 1,000 bp in length. Semiquantitative PCR was performed on immunoprecipitated DNA with the following GAP-43 ChIP primers spanning the putative NFAT site: 5′-GCAGCTGTAACTTGTGTGCA-3′ (sense), and 5′-TAAACACCTCCAATCTGGACC-3′ (antisense).
Real Time Reverse Transcription-PCR Analysis
Total RNA was extracted from wild-type and NFAT-3 null mice brain at E10, E13, postnatal day 1 (P1), P3, P7, and adult developmental stage using TRIzol reagent (Invitrogen), and cDNA was synthesized from 1 μg of RNA using random hexamers from the SuperScript II reverse transcriptase kit (Invitrogen). The subsequent cDNA was used in a real time QT-PCR using SYBR GreenER (Invitrogen) on an ABI Prism 7000 sequence detection system. Standard curve reactions were run with each primer set with identical cDNA to allow comparison between β-actin normalized values for each primer pair. The sequences of the primers used were: GAP-43 (sense), 5′-AAGCTACCACTGATAACTC-3′, GAP-43 (antisense), 5′-CTTCTTTACCCTCATCCTGTCG-3′; CAP-23 (sense), 5′-GGCGGCAGCGCTCCAACTCG-3′, CAP-23 (antisense), 5′-CCGCCTGGGGTTCGCTCTCC-3′; SCG-10 (sense), 5′-AGACTCCTCTCTCGCTCTCTCCGC-3′, SCG-10 (antisense), 5′-AGCCTCTTGAGACTTTCTTCGCTCCTC-3′; L1cam (sense), 5′-ATGCTGCGGTACGTGTGGCCTCTC-3′, L1cam (antisense), 5′-CCACTTGGGGGCACCCTCGG-3′; NFAT1 (sense), 5′-TGGCCCGCCACATCTACCCT-3′, NFAT1 (antisense), 5′-TGGTAGAAGGCGTGCGGCTT-3′; NFAT3 (sense), 5′-CTTTACCCAGCCAGTATGAG-3′, NFAT3 (antisense), 5′-CTGTAGCCTAGGAGCTTGAC-3′; β-actin (sense), 5′-ACAGCTTCACCACCACAGCTGA-3′, β-actin (antisense), 5′-GAGGTCTTTACGGATGTCAACGTC-3′.
Normalized expression was calculated as
RESULTS
Overexpressed NFAT-3 Represses GAP-43 Transcription
During our characterization of GAP-43 transcriptional regulation by p53 (18), we performed an in silico promoter analysis with an ad hoc software that utilizes bioinformatics matrixes to predict transcription binding sites (MatInspector, Genomatix). A search of the DNA sequence upstream of the translational start site of the GAP-43 gene, in addition to revealing a p53 binding site, also identified an NFAT binding site within the first 1,200 bp region upstream of the coding sequence (Fig. 1A). Because NFAT has been shown to be important in axon outgrowth during neuronal development (25) and GAP-43 is a well characterized gene whose expression is also actively regulated during axon outgrowth and neural maturation (6–8), we asked whether NFAT might be an important transcriptional regulator of GAP-43 expression.
To address the question of whether NFAT directly regulates GAP-43 transcription, we cloned a 550-bp region of the GAP-43 promoter that contained the putative NFAT site and the recently characterized p53 site into a luciferase reporter plasmid (18) (Fig. 1A). Next, using transient transfection assays in neuron-like PC-12 cells, we asked whether this region of the GAP-43 promoter is sufficient to respond to neurotrophin stimulation, and more importantly, to NFAT transcriptional activity. PC-12 cells are neuron-like cells that express GAP-43 in response to neurotrophin activation, and they are therefore a useful cell culture system to initially investigate the NFAT-dependent transcriptional regulation of GAP-43.
First, we transfected an artificial reporter containing three copies of the NFAT/AP1 composite site from the IL-2 promoter to confirm that NFAT transcriptional complexes are active and respond to neurotrophin stimulation in PC-12 cells. Then, the same experiment was performed by transfecting cells with the GAP-43 luciferase reporter. Not surprisingly, both the IL-2-(NFAT) and the GAP-43-(NFAT) reporters responded to NGF stimulation with an increase in luciferase signal (Fig. 1B). Next, we tested whether overexpression of NFAT-3 could drive transcription from these reporters. Overexpressed NFAT-3 in PC-12 cells increased IL-2-(NFAT) reporter signal 3.5-fold, and the addition of NGF further increased signal to 6.5-fold, but surprisingly, overexpressed NFAT-3 repressed basal GAP-43 reporter signal as well as NGF-mediated activation (Fig. 1B). Finally, having observed a possible role for NFAT-3 as a transcriptional repressor in PC-12 cells, we asked whether NFAT-3 might also repress the neurotrophin activation of the GAP-43 expression in cultured cortical neurons. We transfected the GAP-43 reporter into cultured cortical neurons and observed again that overexpression of NFAT-3 repressed reporter signal to similar levels as seen in PC-12 cells (Fig. 1C). Furthermore, although neurotrophin stimulation with BDNF modestly increased GAP-43 signal, inhibition of calcineurin-dependent NFAT activation with CsA/FK506 increased reporter signal further than that seen with only neurotrophin stimulation (Fig. 1D). Transfection experiments with NFAT-3 in both PC-12 cells and primary neurons did not show an increase in cell death when compared with control as analyzed by cell counting and nuclear morphological analysis by Hoechst staining (not shown). Overall, these data suggest that NFAT-3 might be a direct transcriptional repressor of GAP-43 expression.
NFAT-3 Is Nuclear and Occupies the GAP-43 Promoter
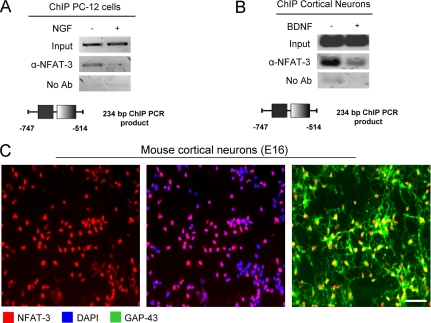
To demonstrate that NFAT-3 occupies the GAP-43 promoter region containing the putative NFAT binding site (Fig. 1A), we used ChIP and semiquantitative PCR to show promoter occupancy. We observed NFAT-3 at the GAP-43 promoter, and this occupancy was reduced upon NGF treatment in PC-12 cells (Fig. 2A). Next, the same experiment was performed in primary cultured neurons to confirm promoter occupancy in neurons. We again observed that NFAT-3 occupied the GAP-43 promoter and that BDNF, analogously to NGF in PC-12 cells, strongly reduced the capacity of NFAT-3 to occupy this promoter (Fig. 2B). Concurrently, we also examined NFAT-3 localization in cultured cortical neurons prior to ChIP and found that NFAT-3 is localized to the nucleus, which is a necessary condition for its transcriptional repression (Fig. 2C). As a final test to verify the presence of NFAT-3 in the nucleus and occupancy of the GAP-43 promoter in the mouse brain, we performed immunohistochemistry and ChIP assays from P1 mouse brains. Immunohistochemistry of P1 mouse brain revealed that NFAT-3 is indeed localized to the nucleus of cortical neurons (Fig. 3A), and a ChIP assay from fixed brain at the same developmental stage showed that NFAT-3 occupied the GAP-43 promoter in vivo (Fig. 3B). These molecular studies indicate that NFAT-3 occupies the GAP-43 promoter and that neurotrophin stimulation reduces promoter occupancy.
FIGURE 2.
NFAT-3 is nuclear and occupies the GAP-43 promoter in PC-12 cells, cultured neurons, and mouse brain. A, NFAT-3 occupies the GAP-43 promoter in PC-12 cells. ChIP on PC-12 cells for endogenous NFAT-3 was performed using α-NFAT-3 antibody. Cells were plated and stimulated with 100 ng/ml NGF or control for 24 h and then cross-linked and harvested. The amount of DNA recovered was measured by semiquantitative PCR. GAP-43-specific primers spanning the putative NFAT site were used. The resulting PCR amplicon containing the NFAT site of the GAP-43 promoter is depicted. No Ab, no antibody. B, NFAT-3 occupies the GAP-43 promoter in cultured cortical neurons. ChIP was performed for NFAT-3 on cultured E16 rat cortical neurons, which were treated with 10 ng/ml BDNF 1 h after plating for 5 days and harvested, and recovered DNA was measured by semiquantitative PCR. The resulting PCR amplicon containing the NFAT site of the GAP-43 promoter is depicted. C, immunocytochemistry of cultured cortical neurons shows nuclear localization for NFAT-3. Cultured E16 cortical neurons were fixed with 4% formaldehyde and probed with α-NFAT-3, α-GAP-43 antibodies, and DAPI nuclear staining. Scale bar: 20 μm.
NFAT-3 Represses GAP-43 Expression in the Developing Mouse Brain
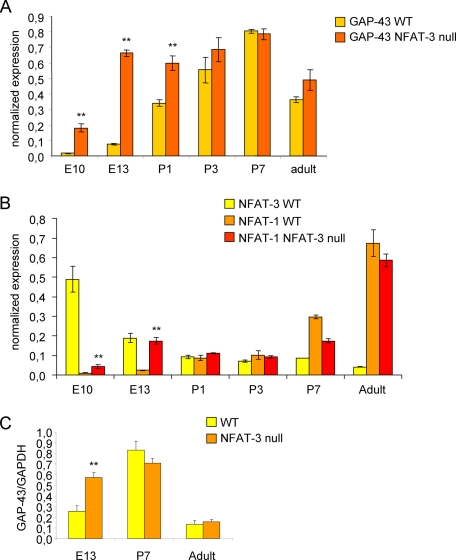
Next, we took advantage of NFAT-3 null mice to examine GAP-43 expression in both wild-type and null mice at various developmental stages from embryonic, postnatal, and adult brain. Because NFAT-3 has been shown to be expressed in the mouse brain, we hypothesized that ablation of NFAT-3 would result in derepression of GAP-43 expression, especially at those development stages where NFAT has been show to be transcriptionally active (25, 30, 42). We collected mouse brain at different developmental stages from embryonic day 10 (E10) to adult. Total RNA was extracted, and quantitative PCR was performed to examine GAP-43 expression levels. We found that mRNA levels of GAP-43 were higher at E10, E13, and P1 brains of NFAT-3 null mice. However, this difference is progressively reduced, mainly due to the postnatal increase in GAP-43 levels in wild-type brains (Fig. 4A). Significantly, this difference in GAP-43 mRNA levels between wild-type and NFAT-3 null brains during the E10–P1 developmental window corresponded to the highest expression levels of NFAT-3 mRNA in wild-type brain, which is greatly reduced by P1 (Fig. 4B). Therefore, the wild-type expression profile of NFAT-3 suggests that NFAT-3 plays an active role in repressing GAP-43 expression in the mouse brain during the E10–P1 developmental window as its down-regulation correlated with a reciprocal up-regulation of GAP-43 expression (compare Fig. 4A with 4B). Interestingly, the expression of NFAT-1 mRNA from wild-type brains followed an exact opposite profile when compared with NFAT-3; NFAT-1 mRNA progressively increased from E10 to adulthood (Fig. 4B). On the other hand, NFAT-1 expression in NFAT-3 null brains is enhanced at E10 and E13, which corresponded to the time when NFAT-3 expression peaked in wild-type brains (Fig. 4B). NFAT-4 mRNA was also measured, but its expression level was very low and did not change between wild-type and NFAT-3 null brains (data not shown).
FIGURE 4.
NFAT-3 represses GAP-43 expression during embryonic mouse brain development. A, GAP-43 is higher in NFAT-3 null brains during embryonic development. Quantitative PCR was performed for GAP-43 mRNA levels in NFAT-3 null and wild-type (WT) mice from E10, E13, P1, P3, P7, and adult brains. Total RNA was reverse-transcribed, and target mRNA was quantified. GAP-43 levels are normalized to β-actin mRNA levels. (Under “Experimental Procedures,” see Equations 1 and 2 for the formula to determine β-actin normalized expression levels.) B, NFAT-3 decreases significantly from embryonic to postnatal stages, whereas NFAT-1 increases. Quantitative PCR was performed for NFAT-3 and NFAT-1 mRNA levels in both NFAT-3 null and wild-type mice. (n = 3, triplicate). Student's t test was performed to compare significant difference between wild-type and null samples at each time point, p value: **, <0.01. Error bars represent S.D. C, GAP-43 protein is higher in E13 NFAT-3 null than wild-type brain. Immunoblot analysis was performed for GAP-43 protein from NFAT-3 null and wild-type mice brains. Lysates made from E13, P7, and adult brains was recovered and analyzed by immunoblot analysis with α-GAP-43 and α-GAPDH antibodies. The intensity of the α-GAP-43 immunoblot signal was quantified and normalized to α-GAPDH signal to calculate the -fold difference of GAP-43 protein levels between NFAT-3 null and wild-type brains. (n = 3, triplicate). Student's t test p value: **, <0.01. Error bars represent S.D.
We also examined the protein levels of GAP-43 at selected developmental stages, and we confirmed that the expression of GAP-43 protein was low at E13, peaked at P7, and was reduced in the adult (Fig. 4C). The difference in GAP-43 mRNA levels between null and wild-type mice was also evident at the protein level, where GAP-43 protein was 2-fold higher in the null mice at E13 and was not significantly different at P7 and in adult brains (Fig. 4C).
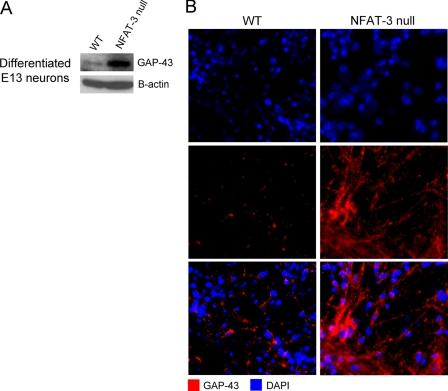
Finally, to directly demonstrate whether NFAT-3 represses GAP-43 expression during neuronal differentiation, we compared protein levels of GAP-43 by immunoblotting and immunocytochemistry of cultured differentiated neurons from wild-type and NFAT-3 null mice. As the difference in the expression of GAP-43 is maximal at E13 in the developing mouse brain between wild-type and NFAT-3 null mice, cells from E13 embryonic brains were cultured and differentiated and then lysed for immunoblotting or fixed for immunocytochemistry at DIV 7. We observed that GAP-43 protein levels from both lysate and fixed cells were higher in null mice when compared with wild-type levels (Fig. 5, A and B). The neuronal network also appeared more developed in NFAT-3 null when compared with wild-type cultures, suggesting a negative role of NFAT-3 in neuronal maturation. These results lead us to ask whether NFAT-3 represses the transcription of other genes functionally related to GAP-43 and involved in neuronal outgrowth and maturation.
FIGURE 5.
NFAT-3 represses GAP-43 expression during in vitro neuronal differentiation. A, enhanced GAP-43 protein expression in differentiating NFAT-3 null cultured cells following neuronal differentiation. Immunoblot analysis was performed on lysates of cultured and differentiated cells taken from E13 NFAT-3 null and wild-type (WT) brain and probed with α-GAP-43 and α-β-actin antibodies. B, GAP-43 is up-regulated in differentiating neurons at DIV 7 from NFAT-3 null brain. Immunocytochemistry of differentiating neurons from wild-type and NFAT-3 null mice was performed with α-GAP-43 antibody and DAPI nuclear staining. Scale bar: 10 μm.
An in silico promoter analysis identified three additional putative gene targets of NFAT-3: CAP-23, L1cam, and SCG-10 (supplemental Fig. 1A). Quantitative PCR was performed using RNA from the brains of wild-type and NFAT-3 null mice. We found that CAP-23 was significantly higher in NFAT-3 null brains between E10 and P1 (a profile analogous to GAP-43). Similarly, L1cam and SCG-10 were also higher at these embryonic stages in NFAT-3 null brains, but unlike GAP-43 and CAP-23, these differences persisted into early postnatal stages. The differential expression levels of all three genes did not appear to be maintained into adulthood (supplemental Fig. 1, B–D).
Taken together, these data strongly support an unexpected role for NFAT-3 as a transcriptional repressor of GAP-43 and suggest the potential of NFAT-3 to regulate the expression of other genes involved in neuronal outgrowth and maturation.
DISCUSSION
Others have observed that NFAT is required for proper neural development and functions downstream of neurotrophin signaling (25). In addition, NFAT has been shown to be expressed in the brain and spinal cord and to be involved in the regulation of presynaptic differentiation (21, 23, 27, 43). Analogously, GAP-43 expression promotes neuronal outgrowth and differentiation and is up-regulated upon neurotrophin stimulation (44–47). Recently, a putative NFAT binding site was revealed by in silico analysis during our characterization of GAP-43 regulation by p53.
Therefore, our initial hypothesis predicted that NFAT-3 would be a transcriptional activator of GAP-43 expression. However, to our surprise, we found that overexpressed NFAT-3 repressed GAP-43 reporter activity in transfection experiments in both PC-12 cells and primary cortical neurons (Fig. 1, B and C). One caveat of these types of experiments would be that the repression we observed upon overexpression of NFAT-3 was from a “squelching” of transcription machinery in general. However, we found that luciferase signal from the IL-2-(NFAT) reporter was not repressed at the same concentration of NFAT-3 DNA as that used with the GAP-43 reporter, and in fact, it is enhanced by NGF stimulation. We therefore believed that the overall repression seen in the NFAT-3 overexpression experiment with the GAP-43 reporters was not due to a squelching effect of general transcription machinery or coactivators by the overexpression of NFAT-3. We also observed that inhibition of NFAT-3 activity with CsA and FK506 further enhanced neurotrophin-activated GAP-43 transcription in primary neurons (Fig. 1D). Along with the finding that NFAT-3 was nuclear and occupied the GAP-43 promoter in primary neurons (Fig. 2, B and C) and in vivo in the mouse brain (Fig. 3, A and B), we propose the current hypothesis that NFAT-3 occupies a site adjacent to the recently described p53 site (18) within the GAP-43 promoter, prior to neurotrophins signaling, to repress GAP-43 transcription.
Interestingly, a recent study on splice variants of NFAT family members demonstrated that NFAT-3 is modestly expressed in the adult mouse brain, and more importantly, that the expression profile for NFAT-3 during brain development inversely matches the expression profile of GAP-43 (48). They found that NFAT-3 expression level peaks at E13 in wild-type brains and decreases through embryonic to postnatal development, which is in agreement with our findings. Their findings are also consistent with a model whereby overall levels of NFAT-3 function to repress GAP-43 expression as the down-regulation of NFAT-3 protein levels would result in the up-regulation of GAP-43.
Characterization of NFAT1, -3, and -4 triple null mice showed developmental abnormalities resulting in dramatic defects in axon outgrowth mainly in the root ganglia system (25). However, the triple null mice have little or no defects in neuronal differentiation or survival. Interestingly, the double null NFAT-3/4 mice showed disorganized vasculature patterns, and the authors concluded that calcineurin/NFAT-3/4 signaling during E8 was required to repress aberrant growth of vessels. Although two NFAT-3 null mice have been separately generated, subtle NFAT-3-dependent dysregulation of axon outgrowth and guidance during brain development has not been investigated (49, 50).
Our findings of increased GAP-43 expression in the single NFAT-3 null mice during the E10–P1 window support a role for NFAT-3 in repressing abnormal axon outgrowth and guidance during the latter part of embryonic brain development. Here, we note that the incorrect temporal expression of key axon outgrowth target genes, such as GAP-43, might result in subtle physiological axon outgrowth dysfunctions that are only revealed when the correct developmental stage is examined.
Our examination of cultured and differentiated neuronal cells from E13, a developmental stage where we believe NFAT-3 functions to repress GAP-43 expression, is also in agreement with our working model. We observed enhanced protein levels of GAP-43 in these cells in NFAT-3 null when compared with wild-type neurons (Fig. 5, A and B). In addition, we provide results that support the role of NFAT-3 as a general repressor of the neuronal outgrowth program. We also observed higher levels of CAP-23, SCG-10, and L1cam mRNA from NFAT-3 null when compared with wild-type brains (supplemental Fig. 1, B–D). We now believe that overall levels of NFAT-3 might function to repress genes involved in neuronal outgrowth and maturation during embryonic development.
In fact, both our in vitro and our in vivo data are all consistent with a model whereby NFAT-3 regulates neuronal outgrowth gene expression by repressing activation from the proximal promoter region during specific windows of embryonic brain development. Thus, NFAT-3 might represent a general negative transcriptional regulator of the neuronal outgrowth program.
In perspective, the current findings suggest that a more extensive investigation of NFAT-3 transcriptional targets during brain development is warranted. In addition, as developmentally regulated, transcription-dependent outgrowth pathways in neurons may be recapitulated following axonal injury in the adult (51), it is possible that NFAT-3 plays a role in axonal regeneration after axotomy by controlling the gene expression of neuronal outgrowth genes. Experiments are undergoing to test this hypothesis.
Supplementary Material
Acknowledgment
We thank Dr. Jeffery D. Molkentin for providing the NFAT-3 null mice.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 NS052640 (to S. D. I.). This work was also supported by grants from the Hertie Foundation; the Fortune Grant, University of Tuebingen; and Deutsche Forschungsgemeinschaft Grant DI 1497/1-1 (to S. D. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- NFAT
- nuclear factor of activated T cells
- CsA
- cyclosporin A
- BDNF
- brain-derived neurotrophic factor
- NGF
- nerve growth factor
- PBS
- phosphate-buffered saline
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- EGFP
- enhanced green fluorescent protein
- ChIP
- chromatin immunoprecipitation
- IL-2
- interleukin-2
- HRP
- horseradish peroxidase
- DMSO
- dimethyl sulfoxide
- DAPI
- 4′,6-diamidino-2-phenylindole
- E
- embryonic day
- P
- postnatal day
- DIV
- days in vitro.
REFERENCES
- 1.Chen L. (1999) Curr. Opin. Struct. Biol. 9, 48–55 [DOI] [PubMed] [Google Scholar]
- 2.Reményi A., Schöler H. R., Wilmanns M. (2004) Nat. Struct. Mol. Biol. 11, 812–815 [DOI] [PubMed] [Google Scholar]
- 3.Ogata K., Sato K., Tahirov T. H. (2003) Curr. Opin. Struct. Biol. 13, 40–48 [DOI] [PubMed] [Google Scholar]
- 4.Ma Q. (2006) J. Physiol. 575, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ooi L., Wood I. C. (2008) Biochem. J. 414, 327–341 [DOI] [PubMed] [Google Scholar]
- 6.Jacobson R. D., Virág I., Skene J. H. (1986) J. Neurosci. 6, 1843–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De la Monte S. M., Federoff H. J., Ng S. C., Grabczyk E., Fishman M. C. (1989) Brain Res. Dev. Brain Res. 46, 161–168 [DOI] [PubMed] [Google Scholar]
- 8.Mahalik T. J., Carrier A., Owens G. P., Clayton G. (1992) Brain Res. Dev. Brain Res. 67, 75–83 [DOI] [PubMed] [Google Scholar]
- 9.Benowitz L. I., Perrone-Bizzozero N. I., Neve R. L., Rodriguez W. (1990) Prog. Brain Res. 86, 309–320 [DOI] [PubMed] [Google Scholar]
- 10.Benowitz L. I., Routtenberg A. (1997) Trends Neurosci. 20, 84–91 [DOI] [PubMed] [Google Scholar]
- 11.Van der Zee C. E., Nielander H. B., Vos J. P., Lopes da Silva S., Verhaagen J., Oestreicher A. B., Schrama L. H., Schotman P., Gispen W. H. (1989) J. Neurosci. 9, 3505–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Namgung U., Routtenberg A. (2000) Eur. J. Neurosci. 12, 3124–3136 [DOI] [PubMed] [Google Scholar]
- 13.Nedivi E., Basi G. S., Akey I. V., Skene J. H. (1992) J. Neurosci. 12, 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhard E., Skene J. H. (1992) Perspect. Dev. Neurobiol. 1, 29–37 [PubMed] [Google Scholar]
- 15.Reinhard E., Nedivi E., Wegner J., Skene J. H., Westerfield M. (1994) Development 120, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 16.Starr R. G., Lu B., Federoff H. J. (1994) Brain Res. 638, 211–220 [DOI] [PubMed] [Google Scholar]
- 17.Di Giovanni S., Knights C. D., Rao M., Yakovlev A., Beers J., Catania J., Avantaggiati M. L., Faden A. I. (2006) EMBO J. 25, 4084–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedeschi A., Nguyen T., Puttagunta R., Gaub P., Di Giovanni S. (2009) Cell Death Differ. 16, 543–554 [DOI] [PubMed] [Google Scholar]
- 19.Wilkins B. J., Dai Y. S., Bueno O. F., Parsons S. A., Xu J., Plank D. M., Jones F., Kimball T. R., Molkentin J. D. (2004) Circ. Res. 94, 110–118 [DOI] [PubMed] [Google Scholar]
- 20.Plyte S., Boncristiano M., Fattori E., Galvagni F., Paccani S. R., Majolini M. B., Oliviero S., Ciliberto G., Telford J. L., Baldari C. T. (2001) J. Biol. Chem. 276, 14350–14358 [DOI] [PubMed] [Google Scholar]
- 21.Bradley K. C., Groth R. D., Mermelstein P. G. (2005) J. Neurosci. Res. 82, 762–770 [DOI] [PubMed] [Google Scholar]
- 22.Seybold V. S., Coicou L. G., Groth R. D., Mermelstein P. G. (2006) J. Neurochem. 97, 397–407 [DOI] [PubMed] [Google Scholar]
- 23.Groth R. D., Coicou L. G., Mermelstein P. G., Seybold V. S. (2007) J. Neurochem. 102, 1162–1174 [DOI] [PubMed] [Google Scholar]
- 24.Groth R. D., Weick J. P., Bradley K. C., Luoma J. I., Aravamudan B., Klug J. R., Thomas M. J., Mermelstein P. G. (2008) Eur. J. Neurosci. 27, 31–42 [DOI] [PubMed] [Google Scholar]
- 25.Graef I. A., Wang F., Charron F., Chen L., Neilson J., Tessier-Lavigne M., Crabtree G. R. (2003) Cell 113, 657–670 [DOI] [PubMed] [Google Scholar]
- 26.Graef I. A., Mermelstein P. G., Stankunas K., Neilson J. R., Deisseroth K., Tsien R. W., Crabtree G. R. (1999) Nature 401, 703–708 [DOI] [PubMed] [Google Scholar]
- 27.Groth R. D., Mermelstein P. G. (2003) J. Neurosci. 23, 8125–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao A., Luo C., Hogan P. G. (1997) Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 29.Macian F. (2005) Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 30.Graef I. A., Chen F., Crabtree G. R. (2001) Curr. Opin. Genet. Dev. 11, 505–512 [DOI] [PubMed] [Google Scholar]
- 31.Jain J., Burgeon E., Badalian T. M., Hogan P. G., Rao A. (1995) J. Biol. Chem. 270, 4138–4145 [PubMed] [Google Scholar]
- 32.Graef I. A., Gastier J. M., Francke U., Crabtree G. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogan P. G., Chen L., Nardone J., Rao A. (2003) Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 34.Jain J., McCaffrey P. G., Miner Z., Kerppola T. K., Lambert J. N., Verdine G. L., Curran T., Rao A. (1993) Nature 365, 352–355 [DOI] [PubMed] [Google Scholar]
- 35.Clipstone N. A., Crabtree G. R. (1992) Nature 357, 695–697 [DOI] [PubMed] [Google Scholar]
- 36.Okamura H., Garcia-Rodriguez C., Martinson H., Qin J., Virshup D. M., Rao A. (2004) Mol. Cell Biol. 24, 4184–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beals C. R., Clipstone N. A., Ho S. N., Crabtree G. R. (1997) Genes Dev. 11, 824–834 [DOI] [PubMed] [Google Scholar]
- 38.Ho S., Clipstone N., Timmermann L., Northrop J., Graef I., Fiorentino D., Nourse J., Crabtree G. R. (1996) Clin. Immunol. Immunopathol. 80, S40–45 [DOI] [PubMed] [Google Scholar]
- 39.Nguyen T., Di Giovanni S. (2008) Int. J. Dev. Neurosci. 26, 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H., Peisley A., Graef I. A., Crabtree G. R. (2007) Trends Cell Biol. 17, 251–260 [DOI] [PubMed] [Google Scholar]
- 41.Im S. H., Rao A. (2004) Mol. Cells 18, 1–9 [PubMed] [Google Scholar]
- 42.Horsley V., Pavlath G. K. (2002) J. Cell Biol. 156, 771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida T., Mishina M. (2005) J. Neurosci. 25, 3067–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gianola S., Rossi F. (2004) Eur. J. Neurosci. 19, 819–830 [DOI] [PubMed] [Google Scholar]
- 45.Aigner L., Arber S., Kapfhammer J. P., Laux T., Schneider C., Botteri F., Brenner H. R., Caroni P. (1995) Cell 83, 269–278 [DOI] [PubMed] [Google Scholar]
- 46.Aigner L., Caroni P. (1993) J. Cell Biol. 123, 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laux T., Fukami K., Thelen M., Golub T., Frey D., Caroni P. (2000) J. Cell Biol. 149, 1455–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vihma H., Pruunsild P., Timmusk T. (2008) Genomics 92, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graef I. A., Chen F., Chen L., Kuo A., Crabtree G. R. (2001) Cell 105, 863–875 [DOI] [PubMed] [Google Scholar]
- 50.Wilkins B. J., De Windt L. J., Bueno O. F., Braz J. C., Glascock B. J., Kimball T. F., Molkentin J. D. (2002) Mol. Cell Biol. 22, 7603–7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emery D. L., Royo N. C., Fischer I., Saatman K. E., McIntosh T. K. (2003) J. Neurotrauma. 20, 1271–1292 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.