Abstract
The molecular chaperone Hsp27 exists as a distribution of large oligomers that are disassembled by phosphorylation at Ser-15, -78, and -82. It is controversial whether the unphosphorylated Hsp27 or the widely used triple Ser-to-Asp phospho-mimic mutant is the more active molecular chaperone in vitro. This question was investigated here by correlating chaperone activity, as measured by the aggregation of reduced insulin or α-lactalbumin, with Hsp27 self-association as monitored by analytical ultracentrifugation. Furthermore, because the phospho-mimic is generally assumed to reproduce the phosphorylated molecule, the size and chaperone activity of phosphorylated Hsp27 were compared with that of the phospho-mimic. Hsp27 was triply phosphorylated by MAPKAP-2 kinase, and phosphorylation was tracked by urea-PAGE. An increasing degree of suppression of insulin or α-lactalbumin aggregation correlated with a decreasing Hsp27 self-association, which was the least for phosphorylated Hsp27 followed by the mimic followed by the unphosphorylated protein. It was also found that Hsp27 added to pre-aggregated insulin did not reverse aggregation but did inhibit these aggregates from assembling into even larger aggregates. This chaperone activity appears to be independent of Hsp27 phosphorylation. In conclusion, the most active chaperone of insulin and α-lactalbumin was the Hsp27 (elongated) dimer, the smallest Hsp27 subunit observed under physiological conditions. Next, the Hsp27 phospho-mimic is only a partial mimic of phosphorylated Hsp27, both in self-association and in chaperone function. Finally, the efficient inhibition of insulin aggregation by Hsp27 dimer led to the proposal of two models for this chaperone activity.
Oligomeric heat shock protein 27 (Hsp27)2 is a ubiquitous mammalian protein with a variety of functions in health and disease (1–8). These functions include ATP-independent chaperone activity in response to environmental stress, e.g. heat shock and oxidative stress, control of apoptosis, and regulation of actin cytoskeleton dynamics. Hsp27 is a member of the α-crystallin small heat shock protein family of which αB-crystallin is the archetype. These proteins are characterized by an α-crystallin domain of 80–90 residues consisting of roughly eight β-strands that form an intermolecular β-sheet interaction interface within a dimer, the basic building subunit of the oligomer (2, 4, 9–11).
Hsp27 is in equilibrium between high molecular weight oligomers and much lower molecular weight multimers. It has been reported that unphosphorylated Hsp27 includes predominantly a distribution of high molecular species ranging in size from 12-mer to 35-mer (12–19). Phosphorylation of Hsp27 at serines 15, 78, and 82 by the p38-activated MAPKAP-2 kinase (20–22) or the use of the triple Ser-to-Asp phospho-mimic results in a major shift in the equilibrium toward much smaller multimers (23) and in an alteration of its function (1, 3, 6, 7, 24, 25). The size distribution of the smaller species has been reported to be between monomer and tetramer (12–16, 18, 19).
Small heat shock proteins, including Hsp27, behave as ATP-independent molecular chaperones during cellular heat shock. They bind partially unfolded proteins and prevent their aggregation until the proteins can be refolded by larger ATP-dependent chaperones or are digested (7, 8, 26). This function includes the up-regulation and/or phosphorylation of Hsp27.
It is not entirely clear what the role of Hsp27 size and phosphorylation state plays in its heat shock function because there are conflicting results in the literature. Some in vitro studies concluded that the unphosphorylated oligomeric Hsp27 (or the murine isoform Hsp25) protects proteins against aggregation better than does the phosphorylation mimic (13, 19, 27), whereas others found no difference (16, 28, 29), and still other studies found that the mimic protects better than does the unphosphorylated wild type (27, 30, 31). In-cell studies found that phosphorylation of Hsp27 was essential for thermo-protection of actin filaments (32), and the Hsp27 phosphorylation mimic decreased inclusion body formation better than did unphosphorylated Hsp27 (33). This study was undertaken to investigate the molecular chaperone function of Hsp27 by correlating chaperone activity with Hsp27 size and by comparing fully phosphorylated Hsp27 with its phospho-mimic.
EXPERIMENTAL PROCEDURES
Expression and Purification of Hsp27
Hsp27 and its S15D/S78D/S82D phospho-mimic mutant were expressed and purified using the New England Biolabs IMPACT-CN (Intein-Mediated Purification with an Affinity Chitin-binding Tag) system. The details of this system are described in the manual from New England Biolabs. A brief description follows. The Hsp27 template DNA is cloned into the pTYB11 plasmid from New England Biolabs, using the SapI site and EcoRI cloning site. The SapI site places the Hsp27 template directly adjacent to the intein template on the plasmid. The Hsp27 is then expressed fused to the intein in BL21 cells induced with isopropyl 1-thio-β-d-galactopyranoside. The intein tag is a 454-amino acid residue sequence from the Saccharomyces cerevisiae VMA1 gene. It contains a chitin binding domain and is self-cleavable using the reducing agent dithiothreitol (DTT). Purification is done by running the cell lysate over a chitin bead column, binding the intein to the column, and then cleaving the intein from Hsp27 using DTT, thereby releasing the Hsp27 from the column. The pTYB11 plasmid allows the Hsp27 and intein to be directly adjacent, at the N terminus of Hsp27, so that after cleavage the Hsp27 does not contain any extra residues. The proteolytic inhibitor PMSF was included from the point of bacterial cell lysis throughout the remainder of the preparation and during storage of the protein. Without PMSF, Hsp27 became slowly digested to a product that ran slightly faster than full-length Hsp27 on SDS-PAGE. Hsp27 concentration was determined from the absorbance at 280 nm, after subtracting the density at 320 mm, using an extinction coefficient of A280 nm0.1% = 1.65.
Disulfide Cross-linking of Hsp27-3D
Hsp27 has a single Cys at position 137 that is in the dimer interface (9, 10) and as such can be disulfide cross-linked (34–37). Hsp27-3D was disulfide cross-linked by air oxidation by dialyzing versus a buffer in the absence of metal ion chelators, i.e. in 2 mm Mops, 0.01% NaN3, 50 μm PMSF, pH 7.5, for 4–7 days at 4 °C. This is essentially the method reported (35, 36).
Phosphorylation and Urea-PAGE of Hsp27
Hsp27 was phosphorylated in vitro by incubation with MAPKAP kinase-2 (Upstate Biotechnology, Inc.) (38). In the reaction mixture the concentration of Hsp27 was 4 μm and that of the kinase was 1.7 ng/ml. The reaction mixture was incubated at 30 °C for 5 h and then at room temperature for an additional 18 h. Aliquots were removed periodically and run on 10% (polyacrylamide) urea-PAGE according to the literature (39). In the preparation of large batches of phosphorylated protein, the kinase/Hsp27 ratio was reduced, to conserve the kinase, and the incubation time was increased for complete phosphorylation.
Analytical Ultracentrifugation and Data Analysis
Sedimentation velocity experiments were conducted in an Optima XL-I ultracentrifuge (Beckman, Palo Alto, CA), using either an An60 Ti four-hole rotor or an An50 Ti eight-hole rotor. Data were acquired with the interference optics system using sapphire cell windows. To improve the signal-to-noise ratio, the cell assembly was enhanced by using double-sector, meniscus-matching, 12-mm, aluminum-filled Epon centerpieces and interference slit window holders on the top window (Biomolecular Interaction Technologies Center, Durham, NH). Samples were subjected to overnight dialysis in the appropriate buffer and then spun at 6,000 rpm for 5 min to match the menisci. After this step, the rotor was removed, shaken, and placed back in the chamber to allow for the temperature to equilibrate for at least 1 h. Velocity experiments were conducted at 50,000 rpm at 20 or 37 °C. Sedimentation velocity data were analyzed by using the program SEDANAL (40) to test for interacting models. A curve was deemed acceptable only if the standard deviation was less than or equal to the previously measured optical noise of the system (1 × 10−2 fringes). The program SEDFIT (41) was used to calculate the least squares sedimentation coefficient distribution (ls-g(s)) and the weight average sedimentation coefficient from this distribution. The program SEDNTERP (42) was used to estimate hydration and to convert the sedimentation coefficients to values at the standard conditions of 20 °C in water, s20,w in Svedberg units.
Molecular Chaperone Activity
Bovine insulin (Sigma) or bovine calcium-depleted α-lactalbumin (Sigma) aggregation was initiated by chemical reduction with DTT and was followed by light scattering in a Varian Eclipse spectrofluorometer using an excitation and emission wavelength of 465 nm. Temperature was controlled with a Varian Peltier device. Activity for insulin was measured at 37 °C in 0.1 m NaCl, 20 mm Mops, pH 7.5 (43). Activity for α-lactalbumin was measured at 37 °C in 0.125 m NaCl, 20 mm Mes, pH 6.0 (27). Measurements were made in the absence and presence of Hsp27 at various ratios of insulin or α-lactalbumin to Hsp27. All components of the mixture, except DTT, were combined in fluorescence cells and incubated in the spectrofluorometer at 37 °C for about 30 min, whereupon the DTT was added, and the light scattering followed as a function of time.
RESULTS AND DISCUSSION
Urea-PAGE of Phosphorylated Hsp27
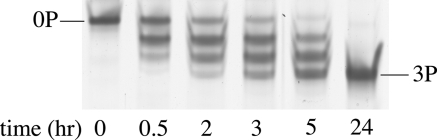
Because urea-PAGE is used to detect the phosphorylated states of proteins (39, 44–47), we tried the same technique to identify the phosphorylated states of Hsp27. Fig. 1 is urea-PAGE of the time course of the in vitro MAPKAP-2 phosphorylation of Hsp27, which is reported to take place at Ser-15, Ser-78, and Ser-82 (20). The single band of the unphosphorylated species develops with time into four equally spaced bands and finally becomes only the fastest moving band of the four (3P). On SDS-PAGE (data not shown), samples at these time points ran as a single band with the same migration. These results are consistent with the four bands representing the unphosphorylated, singly, doubly, and triply phosphosphorylated (3P) species in order of increasing gel migration. The equal separation of the bands attests to the addition of one equally charged phosphate group per band. The single SDS-PAGE band for all stages of phosphorylation indicates that the faster migrating urea-PAGE bands are not because of proteolysis. Phosphorylated Hsp27 such that it migrated as the single 3P band on urea-PAGE was used as phosphorylated Hsp27 in subsequent experiments. The phosphorylation of Hsp27 is usually followed by isoelectric focusing gel electrophoresis (22).
FIGURE 1.
Urea-PAGE of Hsp27 phosphorylation by MAPKAP-2 kinase as a function of time. 0P indicates the band representing unphosphorylated Hsp27, and 3P indicates the band representing triply phosphorylated Hsp27.
Hsp27 Size by Analytical Ultracentrifugation Sedimentation Velocity
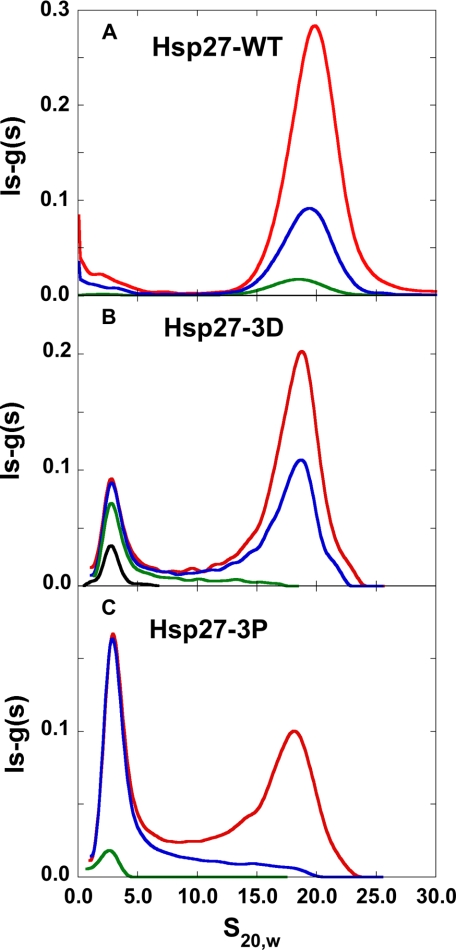
For unphosphorylated Hsp27 (Hsp27-wt), data acquired at 37 °C in physiological buffer and corrected to s20,w resulted in a sedimentation coefficient value close to 20 S (Fig. 2A). To determine whether the 20 S material represented a limiting size, peak s20,w values in Fig. 2A were extrapolated to infinite concentration by plotting 1/s20,w versus 1/c. The extrapolated value was 20 S to within the uncertainty of the measurements. This value is consistent with a spherical 22-mer (including 0.3 g of water/g of protein) and represents the minimum size (because of the assumption of spherical shape) of the largest oligomer, i.e. at infinite concentration, observable under these conditions. Because the sedimentation value of Hsp27-wt at the highest concentration studied (Fig. 2A) is close to the infinite concentration value, then at this concentration Hsp27-wt is an oligomeric distribution centered around a minimum size of a 22-mer. This size is consistent with a previous analytical ultracentrifugation study (19).
FIGURE 2.
Analytical ultracentrifugation sedimentation velocity ls-g(s) versus s20,w curves computed with SEDFIT. A, Hsp27-wt at 0.66 mg/ml (red), 0.22 mg/ml (blue), and 0.03 mg/ml (green). B, Hsp27-3D at 0.53 mg/ml (red), 0.32 mg/ml (blue), 0.09 mg/ml (green), and 0.03 mg/ml (black). C, Hsp27-3P at 0.47 mg/ml (red), 0.22 mg/ml (blue), and 0.02 mg/ml (green) at 37 °C in 120 mm NaCl, 25 mm Tris, 2 mm MgCl2, 0.1 mm EGTA, 0.01% NaN3, 50 μm PMSF, 5 mm DTT, pH 7.5.
On a priori grounds we would have expected the higher s20,w peak to be narrower than the lower s20,w value peak (Fig. 2, B and C) if the self-association were a simple two species equilibrium. Contrary to expectations, the higher s20,w peak is broader than the lower s20,w value peak, making it likely that there is a distribution of large oligomers of different sizes and/or shapes. This polydispersity is consistent with cryo-electron microscopy (17) and gel filtration studies (12–16).
Phosphorylation of Hsp27 (Hsp27-3P) results in the appearance of a much smaller species at an s20,w value of about 2.5 S in addition to the larger oligomer with the proportion of the smaller species increasing with decreasing concentration (Fig. 2C). The size distribution of the S15D/S78D/S82D phospho-mimic mutant (Hsp27-3D) is between Hsp27-wt and Hsp27-3P (Fig. 2B). The species present are in dynamic equilibrium as evidenced by the shift from larger to smaller species upon dilution. It is also clear that there are few intermediate species present between the high molecular weight oligomer distribution and the much smaller dissociated species, especially for Hsp27-wt and Hsp27-3D.
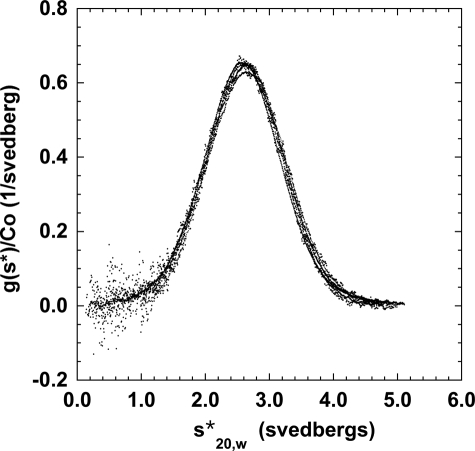
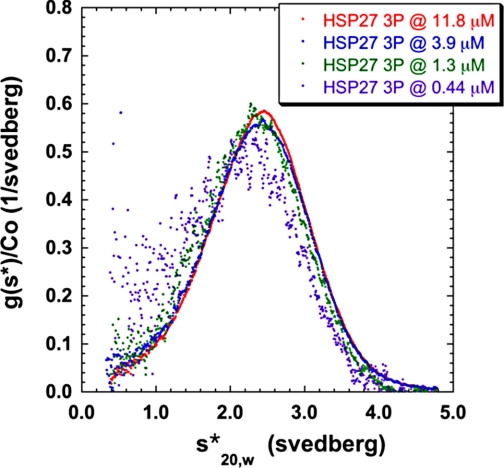
To help characterize the size of the small phosphorylated species, we attempted to produce a solution of Hsp27 that was monodisperse. After surveying a number of buffer conditions, we found that Hsp27-3D formed a monodisperse sedimentation profile with a sedimentation coefficient (s20,w) of about 2.6 S in the absence of added salt at 20 °C (Fig. 3). At this low salt condition (<1 mm ionic strength), the boundaries formed by Hsp27-3D did not shift with concentration (Fig. 3), and they fit to a single species of a molecular mass of 40,000 Da using SEDANAL. Because the molecular mass of Hsp27 is 23 kDa, this species is consistent with a tight dimer. Strong support for this assignment comes from the sedimentation velocity of intermolecularly disulfide cross-linked Hsp27-3D. Cross-linked Hsp27-3D sediments in this low salt buffer with an s20,w value of close to 2.6 S (Fig. 3), providing further proof that the former is a dimer. Phosphorylated Hsp27 under these conditions is mostly dimer but, unlike the phospho-mimic, shows some indication of dissociation because its profile is shifted to somewhat lower s values upon dilution (Fig. 4).
FIGURE 3.
Analytical ultracentrifugation sedimentation velocity of Hsp27-3D and its disulfide cross-linked product at 20 °C in the absence of added salt in 2 mm Tris, 0.1 mm EDTA, 0.01% NaN3, 50 μm PMSF (5 mm DTT in the uncross-linked sample only), pH 7.5. The concentration-normalized sedimentation distribution function (y axis) is plotted versus the apparent sedimentation coefficient (x axis). Hsp27-3D curves are at 23, 5.6, 2.7, and 1.1 μm; cross-linked Hsp27-3D curves are at 10.5, 3.9, and 1.2 μm. All curves are plotted as closed circles because they all very closely overlap. Any differences in overlap are random and are not systematic.
FIGURE 4.
Analytical ultracentrifugation sedimentation velocity of fully phosphorylated Hsp27 (Hsp27-3P) at 20 °C in the absence of salt in 2 mm Tris, 0. 1 mm EDTA, 0.01% NaN3, 50 μm PMSF, 5 mm DTT, pH 7.5. The concentration-normalized sedimentation distribution function (y axis) is plotted versus the apparent sedimentation coefficient (x axis).
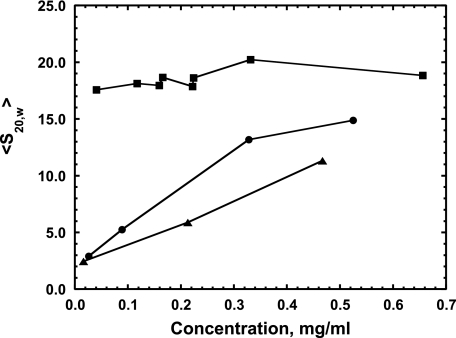
To determine the s value for the small Hsp27 species under more physiological conditions and to compare the size of the different forms of Hsp27, we plotted the weight average of the sedimentation coefficient (〈s20,w〉) as a function of Hsp27 concentration under such conditions (Fig. 5). 〈s20,w〉 was calculated by integrating the ls-g(s) curves (see Fig. 2) as a function of concentration for Hsp27-wt, -3D, and -3P. For unphosphorylated Hsp27, 〈s20,w〉 changes very little over the entire concentration range, indicating no appreciable dissociation of the large oligomers even at very low concentrations. For the phospho-mimic, 〈s20,w〉 is considerably less than that of Hsp27-wt at the high concentration end and then slowly decreases further with dilution. For phosphorylated Hsp27, 〈s20,w〉 is smaller yet than that of the phospho-mimic, and both reach an 〈s20,w〉 value of 2.5 S at the lowest concentrations studied. This is the value for the dimer that we have determined above, demonstrating that the dimer is the smallest species present for phosphorylated Hsp27 and its phospho-mimic under physiological conditions. The average size of phosphorylated Hsp27 is smaller than that of the phospho-mimic at all concentrations studied above about 0.03 mg/ml (Fig. 5).
FIGURE 5.
Weight average sedimentation coefficient 〈s20,w〉 versus concentration of Hsp27-wt (square), Hsp27-3D (circle), Hsp27-3P (triangle) sedimented at 37 °C in 120 mm NaCl, 25 mm Tris, 2 mm MgCl2, 0. 2 mm EGTA, 0.01% NaN3, 50 μm PMSF, 5 mm DTT, pH 7.5.
From the value of 2.5 S, the dimer axial ratio was estimated (42) to be 11:1 using a hydration of 0.4 g of water/g of protein calculated from the amino acid composition. This indicates that the dimer sediments as an elongated molecule because an Hsp27 dimer of two spherical structures would have a much smaller axial ratio of 2:1.
The fact that 〈s20,w〉 increases as a function of increasing Hsp27-3D concentration (Fig. 5) indicates a dynamic equilibrium between the dimer and oligomer species. In contrast, a previous analytical ultracentrifugation study (19) found that, under similar buffer conditions, 〈s20,w〉 for Hsp27-3D remained constant at about 2.6 S from 0 to 0.6 mg/ml, indicating that the dimer does not self-associate to form oligomer. The reason for this difference is not apparent. They also concluded, from modeling, that this value of about 2.6 S was a weight average value from a monomer ⇌ dimer equilibrium mixture, and we determined that the 2.5–2.6 S species was a dimer without any significant monomer present.
Molecular Chaperone Function by DTT-induced Aggregation of Insulin and α-Lactalbumin
The molecular chaperone activity of small heat shock proteins is often evaluated by DTT reduction of insulin (43). Reduction breaks the disulfide bonds holding together insulin chain A and chain B whereupon chain B aggregates (48).
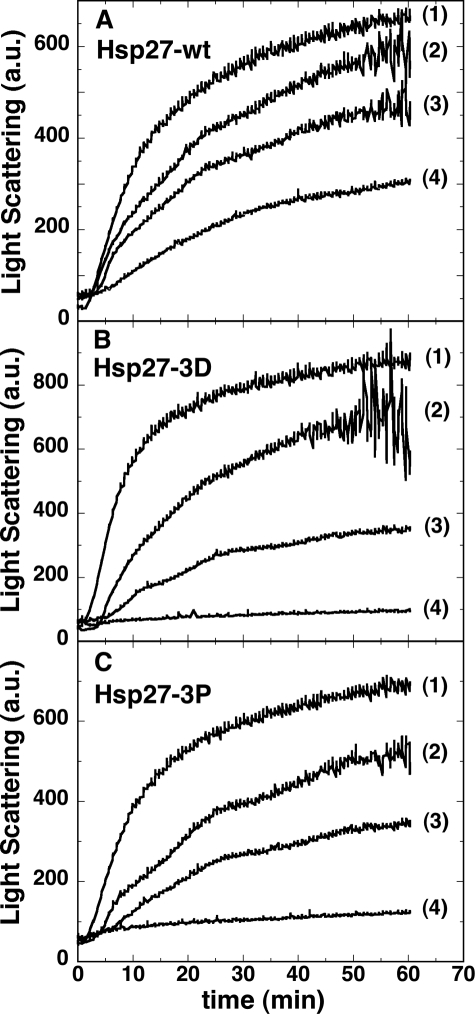
The molecular chaperone activity of Hsp27-3P was compared with Hsp27-wt and Hsp27-3D during the DTT-induced aggregation of insulin. Experiments in Fig. 6 were performed at an insulin concentration of 0.25 mg/ml (43.6 μm) and in the absence and presence of Hsp27 at insulin/Hsp27 monomer molar ratios of 164:1, 82:1, and 41:1. It is clear that all three forms of Hsp27 protect against aggregation with Hsp27-3D and Hsp27-3P protecting roughly equally but much better than Hsp27-wt. In fact, at an insulin/Hsp27 monomer molar ratio of 41:1, insulin plus DTT showed no increase in turbidity in the presence of Hsp27-3D and Hsp27-3P, although there was a substantial increase in the presence of the same amount of Hsp27-wt. In all cases there appears to be a lag before the onset of turbidity (Fig. 6), consistent with a slow nucleation phase followed by a rapid aggregation phase.
FIGURE 6.
Light scattering (arbitrary units) versus time of insulin at 43.6 μm (0.25 mg/ml) after addition of 20 mm DTT in the absence (trace 1) or presence of Hsp27 at insulin/Hsp27 monomer molar ratio of 164:1 (trace 2), 82:1 (trace 3), or 41:1 (trace 4) for Hsp27-wt (A), Hsp27-3D (B), or Hsp273P (C). At the longer times the light scattering was, in some cases, quite noisy because of heterogeneity of the insulin precipitate, which often formed large clumps.
At the Hsp27 concentrations used in Fig. 6 (≤0.025 mg/ml), it is clear from the centrifugation experiments (Fig. 5) that Hsp27-3D and Hsp27-3P are completely dissociated to dimers, whereas the Hsp27-wt is assembled in oligomeric structures. This indicates that the dimeric form of Hsp27 is the most active chaperone species for insulin, thus explaining the equal protection afforded by Hsp27-3P and Hsp27-3D at these low concentrations.
To test this hypothesis further, insulin aggregation experiments were performed at a higher Hsp27 concentration (about 0.17 mg/ml) where there is a relative increase in oligomeric species at the expense of dimer for both Hsp27-3P and Hsp27-3D with Hsp27-3P containing a higher proportion of dimer and less oligomer than Hsp27-3D (Fig. 5). The effectiveness of this higher Hsp27 concentration in inhibiting insulin aggregation was compared for Hsp27-wt, Hsp27-3D, and Hsp27-3P at an insulin/Hsp27 monomer molar ratio of 14.4:1 (Fig. 7). The relative turbidity inhibition by Hsp27 inversely correlates with the Hsp27 size, i.e. the smallest, Hsp27-3P, inhibits better than the larger Hsp27-3D, which inhibits better than the largest, Hsp27-wt. Thus the experiments at both relatively low (Fig. 6) and high (Fig. 7) concentrations support the hypothesis that the smaller the Hsp27 species the better is its molecular chaperone activity, with respect to insulin aggregation. This conclusion is consistent with the model suggesting that for small heat shock proteins the oligomer is a storage state for the more chaperone-competent smaller subunits (31). That Hsp27-3P is less effective in suppressing insulin aggregation at these higher concentrations is most likely because of the increased relative amount of Hsp27-3P oligomer compared with dimer. The experiment in Fig. 7 not only illustrates the role of size in the Hsp27 chaperone function, it also clearly demonstrates that the Hsp27-3D mutant only partially mimics the chaperone activity of phosphorylated Hsp27. We cannot entirely rule out that, in addition to its effect on Hsp27 size, phosphorylation plays some other specific role in the Hsp27 chaperone function.
FIGURE 7.
Light scattering (arbitrary units) versus time of insulin at 106.8 μm (0.6125 mg/ml) after addition of 20 mm DTT in the absence (trace 1) or presence of Hsp27-wt (trace 2), Hsp27-3D (trace 3), or Hsp27-3P (trace 4) at an insulin/Hsp27 monomer molar ratio of 14.4:1. In the absence of Hsp27 (trace 1), after about 16 min, the light scattering signal becomes very noisy and starts to drop precipitously and thus the rest of the trace is not shown. This phenomenon is because of large clumps of aggregates forming and settling below the light beam.
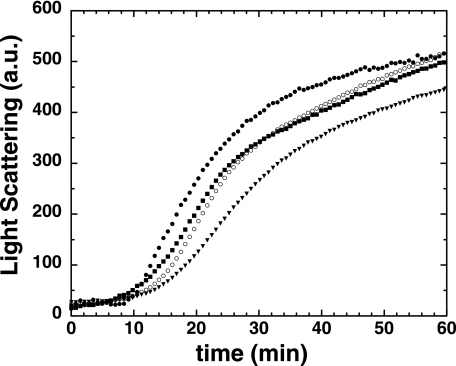
α-Lactalbumin similarly aggregates upon reduction of its intramolecular disulfide bonds with DTT (49). Fig. 8 demonstrates that Hsp27-3P limits α-lactalbumin aggregation better than does Hsp27-3D, which is slightly better than Hsp27-wt. These findings show again, with a protein other than insulin, that phosphorylated Hsp27 is a more effective molecular chaperone than is the unphosphorylated protein or the phospho-mimic.
FIGURE 8.
Light scattering (arbitrary units) versus time of α-lactalbumin at 14.3 μm (0.2 mg/ml) after addition of 20 mm DTT in the absence (closed circles) or presence of 1.4 μm (0.03 mg/ml) Hsp27-wt (closed squares), Hsp27-3D (open circles), or Hsp27-3P (closed triangles). α-Lactalbumin/Hsp27 monomer molar ratio = 10.2:1.
Phosphorylated Hsp27 was highly effective in inhibiting the aggregation of insulin at relatively low insulin and Hsp27 concentrations (Fig. 6) where Hsp27-3P was exclusively dimer. At a ratio of 1 Hsp27 monomer per 41 insulin molecules, the aggregation was completely inhibited. It would appear unlikely that about 40 individual chains of insulin are simultaneously bound to each Hsp27 molecule, considering that this is equivalent to an insulin/Hsp27-3P mass ratio of about 6:1 (assuming Hsp27-3P binds only insulin B chains). However, two hypothetical models might account for this potent activity. One mechanism is based on the postulate that, in general, protein aggregation passes through a slow nucleation step before rapid aggregation (50, 51). Insulin aggregation proceeding via insulin nucleation is consistent with the observed lag phase before aggregation (Figs. 6 and 7). Thus we propose that Hsp27-3P sequesters multimeric insulin nuclei that precede aggregation, thereby associating with many unfolded insulin molecules without binding to each individually. That the Hsp27 dimer is an elongated molecule further contributes to maximize insulin binding.
Another possibility, which may act together with or instead of the nucleation model, is that Hsp27-3P could change the conformation of bound unfolded insulin. This changed state of unfolded insulin would be aggregation-incompetent and remain as such for some time after dissociation from Hsp27 before it reverted back to the aggregation-competent structure. At the present time there are insufficient data to provide further support for either of these models.
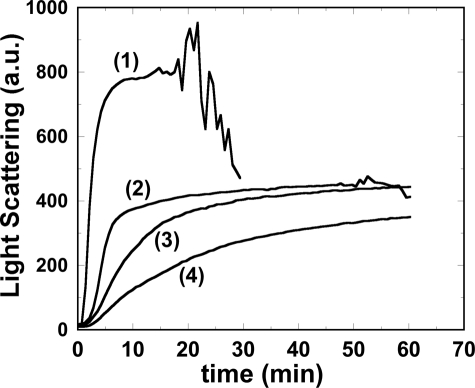
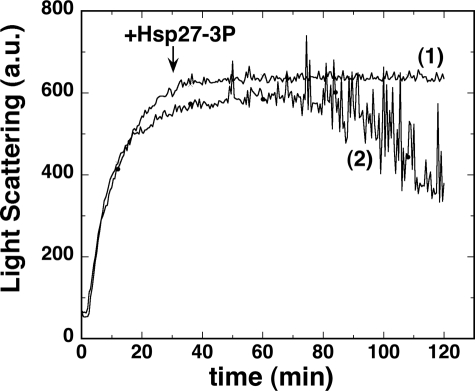
To know if Hsp27 can reverse protein aggregation, we added Hsp27-3P to DTT-aggregated insulin at an insulin/Hsp27-3P monomer molar ratio of 41:1, a level that fully protected insulin from aggregating (Fig. 6C). The Hsp27-3P was added just as the aggregation reached peak value (Fig. 9). The light scattering remained unchanged, indicating that Hsp27-3P cannot reverse insulin aggregation. However, for the insulin sample without added Hsp27-3P or with added buffer lacking Hsp27-3P, the light scattering trace eventually became extremely noisy, and the signal dropped dramatically (Fig. 9). A visual inspection of the samples after the run revealed that the Hsp27-3P-added sample was homogeneously cloudy, and the unaltered sample contained large aggregation clumps that had settled below the fluorometer light beam, thus explaining the erratic light scattering intensity. Hsp27-wt, at the same level, was able to similarly block large aggregate formation (data not shown).
FIGURE 9.
Light scattering (arbitrary units) versus time of insulin at 43.6 μm (0.25 mg/ml) after addition of 20 mm DTT. Trace 1, at 30 min Hsp27-3P was added at an insulin/Hsp27-3P monomer molar ratio of 41:1. Trace 2, no further addition.
Thus it appears that insulin aggregation occurs in two phases, first is the formation of smaller aggregates that then combine, in a second phase, to form much larger aggregates. Furthermore, Hsp27 was able to block the second phase from happening, in an apparently phosphorylation-independent manner. Does this two-phase aggregation phenomenon have an in vivo relevance? Does the second phase correspond, in some way, to inclusion body formation, and does Hsp27 play some protective role in this process? It has been reported that transfection of Hsp27 into cells does decrease inclusion body formation (33).
Our finding that Hsp27-3D and -3P protect against insulin aggregation better than Hsp27-wt is in contrast to the literature (13, 19). The reason for this discrepancy is not clear. As pointed out in the Introduction, there are several disagreements in the literature over whether unphosphorylated Hsp27 (or Hsp25) protects against protein aggregation better than does the phospho-mimic. There are several possible explanations for these differences as follows: some of these disagreements involved different target proteins; some studies used Hsp27, whereas others used Hsp25; and one study used tissue-prepared Hsp25, whereas all others used bacterially expressed Hsp27 (or Hsp25); and several studies employed Hsp27 expressed with additional residues at the N or C terminus. It has been reported that extra His residues at the N terminus alter the size distribution of Hsp27 (18), which could then alter the molecular chaperone activity. However, there are no clear trends between these possibilities and the different reported molecular chaperone activities. The resolution of these discrepancies awaits further investigation.
Conclusions
The following conclusions have been made. 1) Urea-PAGE is an effective technique for assaying the level of phosphorylation of Hsp27. 2) Although the Hsp27-3D mutant is widely used and assumed to functionally reproduce the phosphorylated molecule, the mutant is only a partial and qualitative mimic for phosphorylated Hsp27, i.e. the size of the phosphorylated protein is smaller than that of the mimic, and it protects against protein aggregation better than does the mimic. 3) The dimer is the minimum size of phosphorylated Hsp27 under physiological conditions and is an elongated molecule. 4) Unphosphorylated Hsp27 oligomer is a polydisperse distribution of species centered around a minimum 22-mer size. 5) There is a low population of species between the Hsp27 dimer and the large oligomeric species, indicating that intermediate sized multimers are energetically unfavorable. 6) Increasing molecular chaperone activity of Hsp27, with respect to insulin and α-lactalbumin aggregation, correlates with a decreasing size of Hsp27, with the Hsp27 dimer being the most active species. 7) Two possible models were proposed to describe the mechanism of the molecular chaperone activity of phosphorylated Hsp27 on insulin. 8) Although Hsp27 cannot reverse insulin aggregation, it does prevent smaller aggregates from combining to form larger aggregates.
Acknowledgments
We are grateful to Jennifer Horne for initiating Hsp27 expression and Dr. Zenon Grabarek for helpful discussions and a critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL66219 and AR41637. This work was also supported by an American Heart Association post-doctoral fellowship (to A. M.) and a Boston Biomedical Research Institute pilot grant.
- Hsp27
- heat shock protein 27
- Hsp27-wt
- wild type, unphosphorylated, Hsp27
- Hsp27-3P
- Hsp27 triply phosphorylated by MAPKAP-2 kinase
- Hsp27-3D
- Hsp27 S15D/S78D/S82D phospho-mimic mutant
- DTT
- dithiothreitol
- Mops
- 3-(N-morpholino)propanesulfonic acid
- Mes
- 2-(N-morpholino)ethanesulfonic acid
- PMSF
- phenylmethylsulfonyl fluoride.
REFERENCES
- 1.Welsh M. J., Gaestel M. (1998) Ann. N.Y. Acad. Sci. 851, 28–35 [DOI] [PubMed] [Google Scholar]
- 2.Van Montfort R., Slingsby C., Vierling E. (2001) Adv. Protein Chem. 59, 105–156 [DOI] [PubMed] [Google Scholar]
- 3.Mounier N., Arrigo A. P. (2002) Cell Stress Chaperones 7, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gusev N. B., Bogatcheva N. V., Marston S. B. (2002) Biochemistry 67, 511–519 [DOI] [PubMed] [Google Scholar]
- 5.Clark J. I., Muchowski P. J. (2000) Curr. Opin. Struct. Biol. 10, 52–59 [DOI] [PubMed] [Google Scholar]
- 6.Sun Y., MacRae T. H. (2005) FEBS J. 272, 2613–2627 [DOI] [PubMed] [Google Scholar]
- 7.Arrigo A. P., Simon S., Gibert B., Kretz-Remy C., Nivon M., Czekalla A., Guillet D., Moulin M., Diaz-Latoud C., Vicart P. (2007) FEBS Lett. 581, 3665–3674 [DOI] [PubMed] [Google Scholar]
- 8.Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 9.Mchaourab H. S., Berengian A. R., Koteiche H. A. (1997) Biochemistry 36, 14627–14634 [DOI] [PubMed] [Google Scholar]
- 10.Berengian A. R., Parfenova M., Mchaourab H. S. (1999) J. Biol. Chem. 274, 6305–6314 [DOI] [PubMed] [Google Scholar]
- 11.Van Montfort R. L., Bateman O. A., Lubsen N. H., Slingsby C. (2003) Protein Sci. 12, 2606–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavoie J. N., Lambert H., Hickey E., Weber L. A., Landry J. (1995) Mol. Cell. Biol. 15, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogalla T., Ehrnsperger M., Preville X., Kotlyarov A., Lutsch G., Ducasse C., Paul C., Wieske M., Arrigo A. P., Buchner J., Gaestel M. (1999) J. Biol. Chem. 274, 18947–18956 [DOI] [PubMed] [Google Scholar]
- 14.Lambert H., Charette S. J., Bernier A. F., Guimond A., Landry J. (1999) J. Biol. Chem. 274, 9378–9385 [DOI] [PubMed] [Google Scholar]
- 15.Chernik I. S., Panasenko O. O., Li Y., Marston S. B., Gusev N. B. (2004) Biochem. Biophys. Res. Commun. 324, 1199–1203 [DOI] [PubMed] [Google Scholar]
- 16.Thériault J. R., Lambert H., Chávez-Zobel A. T., Charest G., Lavigne P., Landry J. (2004) J. Biol. Chem. 279, 23463–23471 [DOI] [PubMed] [Google Scholar]
- 17.Haley D. A., Bova M. P., Huang Q. L., Mchaourab H. S., Stewart P. L. (2000) J. Mol. Biol. 298, 261–272 [DOI] [PubMed] [Google Scholar]
- 18.Lelj-Garolla B., Mauk A. G. (2005) J. Mol. Biol. 345, 631–642 [DOI] [PubMed] [Google Scholar]
- 19.Lelj-Garolla B., Mauk A. G. (2006) J. Biol. Chem. 281, 8169–8174 [DOI] [PubMed] [Google Scholar]
- 20.Stokoe D., Engel K., Campbell D. G., Cohen P., Gaestel M. (1992) FEBS Lett. 313, 307–313 [DOI] [PubMed] [Google Scholar]
- 21.Gaestel M., Schröder W., Benndorf R., Lippmann C., Buchner K., Hucho F., Erdmann V. A., Bielka H. (1991) J. Biol. Chem. 266, 14721–14724 [PubMed] [Google Scholar]
- 22.Landry J., Lambert H., Zhou M., Lavoie J. N., Hickey E., Weber L. A., Anderson C. W. (1992) J. Biol. Chem. 267, 794–803 [PubMed] [Google Scholar]
- 23.Kato K., Hasegawa K., Goto S., Inaguma Y. (1994) J. Biol. Chem. 269, 11274–11278 [PubMed] [Google Scholar]
- 24.Dillmann W. H. (1999) Ann. N.Y. Acad. Sci. 874, 66–68 [DOI] [PubMed] [Google Scholar]
- 25.MacRae T. H. (2000) Cell. Mol. Life Sci. 57, 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrnsperger M., Gräber S., Gaestel M., Buchner J. (1997) EMBO J. 16, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panasenko O. O., Seit Nebi A., Bukach O. V., Marston S. B., Gusev N. B. (2002) Biochim. Biophys. Acta 1601, 64–74 [DOI] [PubMed] [Google Scholar]
- 28.Knauf U., Jakob U., Engel K., Buchner J., Gaestel M. (1994) EMBO J. 13, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pivovarova A. V., Mikhailova V. V., Chernik I. S., Chebotareva N. A., Levitsky D. I., Gusev N. B. (2005) Biochem. Biophys. Res. Commun. 331, 1548–1553 [DOI] [PubMed] [Google Scholar]
- 30.Markov D. I., Pivovarova A. V., Chernik I. S., Gusev N. B., Levitsky D. I. (2008) FEBS Lett. 582, 1407–1412 [DOI] [PubMed] [Google Scholar]
- 31.Shashidharamurthy R., Koteiche H. A., Dong J., McHaourab H. S. (2005) J. Biol. Chem. 280, 5281–5289 [DOI] [PubMed] [Google Scholar]
- 32.Geum D., Son G. H., Kim K. (2002) J. Biol. Chem. 277, 19913–19921 [DOI] [PubMed] [Google Scholar]
- 33.Chiou Y. W., Hwu W. L., Lee Y. M. (2008) Biochim. Biophys. Acta 1782, 169–179 [DOI] [PubMed] [Google Scholar]
- 34.Miron T., Wilchek M., Geiger B. (1988) Eur. J. Biochem. 178, 543–553 [DOI] [PubMed] [Google Scholar]
- 35.Zavialov A., Benndorf R., Ehrnsperger M., Zav'yalov V., Dudich I., Buchner J., Gaestel M. (1998) Int. J. Biol. Macromol. 22, 163–173 [DOI] [PubMed] [Google Scholar]
- 36.Zavialov A. V., Gaestel M., Korpela T., Zav'yalov V. P. (1998) Biochim. Biophys. Acta 1388, 123–132 [DOI] [PubMed] [Google Scholar]
- 37.Diaz-Latoud C., Buache E., Javouhey E., Arrigo A. P. (2005) Antioxid. Redox. Signal. 7, 436–445 [DOI] [PubMed] [Google Scholar]
- 38.Larsen J. K., Yamboliev I. A., Weber L. A., Gerthoffer W. T. (1997) Am. J. Physiol. 273, L930–940 [DOI] [PubMed] [Google Scholar]
- 39.Perrie W. T., Perry S. V. (1970) Biochem. J. 119, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stafford W. F., Sherwood P. J. (2004) Biophys. Chem. 108, 231–243 [DOI] [PubMed] [Google Scholar]
- 41.Schuck P. (2003) Anal. Biochem. 320, 104–124 [DOI] [PubMed] [Google Scholar]
- 42.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Sciences (Harding S. E., Rowe A. J., Horton J. C. eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 43.Farahbakhsh Z. T., Huang Q. L., Ding L. L., Altenbach C., Steinhoff H. J., Horwitz J., Hubbell W. L. (1995) Biochemistry 34, 509–516 [DOI] [PubMed] [Google Scholar]
- 44.Pal B. K., McAllister R. M., Gardner M. B., Roy-Burman P. (1975) J. Virol. 16, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agrawal H. C., O'Connell K., Randle C. L., Agrawal D. (1982) Biochem. J. 201, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harisanova N. T., Ralchev K. H. (1986) Mol. Biol. Rep. 11, 199–203 [DOI] [PubMed] [Google Scholar]
- 47.Hayley M., Chevaldina T., Mudalige W. A., Jackman D. M., Dobbin A. D., Heeley D. H. (2008) J. Muscle Res. Cell Motil. 29, 101–107 [DOI] [PubMed] [Google Scholar]
- 48.Sanger F. (1949) Biochem. J. 44, 126–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuwajima K. (1996) FASEB J. 10, 102–109 [DOI] [PubMed] [Google Scholar]
- 50.Roberts C. J. (2007) Biotechnol. Bioeng. 98, 927–938 [DOI] [PubMed] [Google Scholar]
- 51.Morris A. M., Watzky M. A., Finke R. G. (2009) Biochim. Biophys. Acta 1794, 375–397 [DOI] [PubMed] [Google Scholar]